Abstract
The biological significance of the presence of a long cytoplasmic domain in the envelope (Env) transmembrane protein gp41 of human immunodeficiency virus type 1 (HIV-1) is still not fully understood. Here we examined the effects of cytoplasmic tail elongation on virus replication and characterized the role of the C-terminal cytoplasmic tail in interactions with the Gag protein. Extensions with six and nine His residues but not with fewer than six His residues were found to severely inhibit virus replication through decreased Env electrophoretic mobility and reduced Env incorporation compared to the wild-type virus. These two mutants also exhibited distinct N glycosylation and reduced cell surface expression. An extension of six other residues had no deleterious effect on infectivity, even though some mutants showed reduced Env incorporation into the virus and/or decreased cell surface expression. We further show that these elongated cytoplasmic tails in a format of the glutathione S-transferase fusion protein still interacted effectively with the Gag protein. In addition, the immediate C terminus of the cytoplasmic tail was not directly involved in interactions with Gag, but the region containing the last 13 to 43 residues from the C terminus was critical for Env-Gag interactions. Taken together, our results demonstrate that HIV-1 Env can tolerate extension at its C terminus to a certain degree without loss of virus infectivity and Env-Gag interactions. However, extended elongation in the cytoplasmic tail may impair virus infectivity, Env cell surface expression, and Env incorporation into the virus.
Primate lentiviruses such as human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (SIV) all have unusually long cytoplasmic domains, more than 150 amino acids, in their viral envelope (Env) transmembrane (TM) protein, gp41, compared to the 20 to 40 amino acids for most other retroviruses. The cytoplasmic domain of HIV-1 has been shown to provide several important functions, including virus replication, infectivity, transmission, and cytopathogenicity in the virus life cycle. Mutations in this domain may enhance Env fusion activity, but viral infectivity is greatly reduced in infected peripheral blood mononuclear cells, macrophages, and most T-cell lines (11, 13, 15, 49). The three highly conserved amphipathic α-helical segments, designated lentiviral lytic peptide 1 (LLP)-1, LLP-2, and LLP-3, located in the C terminus of the cytoplasmic tail are thought to be associated with the inner surfaces of viral and cellular membranes (33, 45). We previously showed that the multimerization potential and membrane binding ability of the cytoplasmic tail may play a crucial role in virus replication (3, 5, 7, 28) and that the N-terminal segment of LLP-1 contains a structural determinant critical for modulating Env stability (27).
In HIV-2 and SIV, the cytoplasmic tail of TM proteins also modulates virus replication and plays a crucial role in viral pathogenesis. Truncations of the cytoplasmic tail enhance infectivity, fusogenicity, and Env incorporation into virions and expand the host range (23, 35, 41, 50). Although passage of SIVmac in human T-cell lines selects for variants with premature stop codons that truncate the cytoplasmic tail (25), truncated cytoplasmic tails rapidly revert to full-length tails during replication in infected macaques (21, 25, 31). These results indicate a selective advantage to SIV replication conferred by the full-length cytoplasmic tail in vivo.
Another pivotal role played by the cytoplasmic tail is its specific association with the N-terminal region of the trimeric matrix (MA) protein (10, 29, 30, 40). This cytoplasmic tail-Gag interaction promotes recruitment of trimeric Env complexes into viral buds at the plasma membrane, from which mature virions are released into the medium. Deletions in the cytoplasmic tail and alterations to the MA protein impair Env incorporation into nascent virions (10, 11, 48, 49). Moreover, substitution of Glu for Leu at residue 12 or 30 in the MA region was found to block Env incorporation (14). However, the blocking of Env incorporation can in turn be overcome by deletions in the long cytoplasmic tail of Env (13, 14, 17, 32). These results support the notion that the long cytoplasmic tail of the trimeric Env has to fit the cage hole present in the lattice-like MA trimeric structure during virus assembly (20, 38). Nevertheless, whether the reduced Env incorporation observed with these mutants correlates with impaired Env-Gag interaction in cells was not addressed in those studies.
The precise sequences or residues located in the cytoplasmic tail that directly interact with the MA or that modulate Env-Gag interaction remain controversial (11, 13, 15, 49). A large central domain of the cytoplasmic tail, spanning residues 742 to 835, was implicated in interactions with Gag, as judged by the ability of the peptides to inhibit the immunocapture of Pr55Gag particles (22). Mutation and revertant studies have suggested that residues within or adjacent to the LLP-2 motif participate in the interaction with MA (13, 14, 36). On the other hand, analysis of a series of short in-frame deletions with the SIV TM protein cytoplasmic tail showed that deletions in the one-third C-terminal segment impair Env assembly into the virus (2). An in vitro direct protein-binding assay was employed to map the C-terminal 67-residue stretch of the cytoplasmic tail, including the entire LLP-1 sequence and the last few amino acids in the C-terminal portion of LLP-2, as the region interacting with MA; deletions at either the N or C terminus of this region significantly reduced cytoplasmic tail-MA associations (8). These studies indicated that the C-terminal cytoplasmic tail has a critical role in Env-Gag interactions.
Retention of a long cytoplasmic tail sequence in the TM proteins of HIV and SIV suggests that the extensive cytoplasmic tail may be essential as a key function in virus replication which has not yet been fully identified. Whether or not the presence of an intact, full-length cytoplasmic tail in the Env proteins of HIV and SIV is a prerequisite for Env functions and whether these viruses can tolerate a cytoplasmic tail longer than its native sequence are key issues for understanding the role of the cytoplasmic tail during the evolution of these primate immunodeficiency viruses.
Mutations, deletions, and truncations are often used to address the functions of viral membrane Env proteins of interest. However, elongation at the C terminus of Env proteins by addition of repeated single or dipeptide residues turns out to be a useful alternative for elucidating the role of cytoplasmic tail in virus replication. For instance, extension of the cytoplasmic tail of influenza virus hemagglutinin (HA) with one to six residues of His was found not to affect HA Env biogenesis; however, the syncytium-forming ability of HA was significantly reduced, depending on the number of His residues added (39). Similar results were also obtained when different dipeptide sequences were added to the C terminus of HA (39). Moreover, employing this cytoplasmic tail elongation approach allows us to look into the long-standing question of whether the C-terminal cytoplasmic tail is critical for Env-Gag interactions in cells.
Using HIV-1 in the present study as a model system, we examined the effects on virus replication of extension of the gp41 cytoplasmic tail at the C terminus by adding different numbers of His residues and other amino acid residues. Our results demonstrate that HIV-1 Env can tolerate extension at its C terminus to a certain degree without loss of virus infectivity and Env incorporation into the virus. The results also indicate that the immediate C-terminal portion of the cytoplasmic tail is not critical for Env-Gag interactions. However, extended elongation of the cytoplasmic tail may impair virus infectivity and Env intracellular transport and incorporation into the virus. Our study also suggests that HIV-1 may be constrained so as to preserve an appropriate length of its cytoplasmic domains during virus evolution.
MATERIALS AND METHODS
Cells and hybridomas.
293T, HeLa, and HeLa-CD4-LTR-β-gal cells were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum. CEM-SS, SupT1, and H938 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. Hybridoma 902 secretes a mouse monoclonal antibody (MAb) specific for the gp120 V3 region of the LAV and IIIB strains of HIV-1. Chessie 8 produces an MAb specific for gp160 and maps to amino acid residues 727 to 732 of HIV-1LAI. Hybridoma 183 (clone H12-5C) is a mouse MAb reactive with the HIV-1 p24. These hybridomas were maintained in RPMI 1640 containing 10% fetal bovine serum and injected intraperitoneally into BALB/c mice to produce ascites fluids.
Oligonucleotide-directed mutagenesis and construction of plasmids.
For the construction of cytoplasmic tail elongation mutants, an NruI site was first introduced in between the C-terminal Leu-856 codon and the stop codon of the env gene of the HXB2RU3 provirus (44) by oligonucleotide-directed, site-specific mutagenesis with PCR overlap extension as previously described (27). Oligonucleotides 5′-TTGGAAAGGATTTTGCTATCGCGATAAGATGGGTGGCAAGTGGTC-3′ (sense) and 5′-CACTTGCCACCCATCTTATCGCGATAGCAAAATCCTTTCCAAGCCCTG-3′ (antisense) (sequences in italic mark the NruI recognition site) were used as the internal paired primers to generate the NruI site. The amplified mutated BamHI-XhoI fragment was first ligated into pGEM-7Zf (Promega) to yield pGEM-BX(NruI). Amino acid residues as shown in Fig. 1 were added to the coding sequence of the C terminus of the cytoplasmic tail by PCR with the 8423f oligonucleotide as the sense primer and each of the oligonucleotides listed below as the antisense primer: His mutant, 5′-CGATTAGTGTAGCAAAATCCTTTCCAAGCCCTG-3′; 2His mutant, 5′-CGATTAGTGGTGTAGCAAAATCCTTTCCAAGCCCTG-3′; 3His mutant, 5′-CGATTAGTGGTGGTGTAGCAAAATCCTTTCCAAGCCCTG-3′; 6His mutant, 5′-CGATTAGTGGTGGTGGTGGTGGTGTAGCAAAATCCTTTCCAAGCCCTG-3′; 9Hismutant, 5′-CGATTAGTGGTGGTGGTGGTGGTGGTGGTGGTGTAGCAAAATCCTTTCCAAGCCCTG-3′; 3(Leu-Ile) mutant, 5′-CGATTAGATCAGGATCAGGATCAGTAGCAAAATCCTTTCCAAGCCCTG-3′; 3(Thr-Ser) mutant, 5′-CGATTAGGAGGTGGAGGTGGAGGTTAGCAAAATCCTTTCCAAGCCCTG-3′; 6Gly mutant, 5′-CGATTAGCCGCCGCCGCCGCCGCCTAGCAAAATCCTTTCCAAGCCCTG-3′; 6Gln mutant, CGATTCTGCTGCTGCTGCTGCTGTAGCAAAATCCTTTCCAAGCCCTG-3′; 6Pro mutant, CGATTATGGTGGTGGTGGTGGTGGTAGCAAAATCCTTTCCAAGCCCTG-3′; and 6Trp mutant, CGATTACCACCACCACCACCACCATAGCAAAATCCTTTCCAAGCCCTG-3′. These sequences encoded a stop codon immediately after the residues added and contained half of the NruI site for cloning. Italicized nucleotides mark the antisense sequences of the amino acids added. These amplified mutated BamHI-NruI fragments were cloned in the BamHI and NruI sites of pGEM-BX(NruI). The isolated mutated BamHI-XhoI fragments were then used to replace the homologous sequence in HXB2RU3 to generate various mutant proviruses.
FIG. 1.
Construction of HIV-1 cytoplasmic tail elongation mutants. The amino acid sequence in the single-letter code from residues 828 to 856 of Env of strain HXB2 is shown at the top. Extensions with different numbers of His residues (A) and residues other than His (B and C) at the C terminus of this domain were constructed as described in Materials and Methods. WT, wild type.
The KpnI-XhoI fragments isolated from His mutant proviruses were cloned in the corresponding sites in pSVE7puro to yield various HIV-1 long-terminal-repeat (LTR)-driven env expression plasmids. For generation of pEBG clones encoding glutathione S-transferase (GST)-cytoplasmic subdomain fusion proteins, a series of cytoplasmic tail truncation mutant pSVE7puro plasmids (5) were used as templates in PCR as previously described (28). For construction of pEBG constructs that encoded GST fusion proteins containing elongated cytoplasmic domains, 706fSpeI (28) and oligonucleotide 5′-CCCGGTACCGCTCCCACCCCATCTGCTGC-3′ were used as the sense and antisense primers, respectively, and various mutant HXB2RU3 proviruses were used as templates to generate the DNA fragments that were subsequently cloned into the pEBG vector. pHIVgptGag/30LE was constructed by substituting the NarI-SpeI fragment isolated from pNL4-3/30LE for the corresponding fragment in pHIVgptGag.
Plasmid DNA transfections.
To generate virus stocks for the infection studies and to examine viral protein expression, subconfluent 293T cells grown in 100-mm petri dishes were transfected with 20 μg of HXB2RU3 proviruses with the calcium phosphate coprecipitation method as previously described (3, 6). Subconfluent HeLa cells grown in 100-mm petri dishes were transfected with 8 μg of proviruses by the Lipofectamine Plus transfection method (Invitrogen). For preparation of vesicular stomatitis virus (VSV) glycoprotein G-pseudotyped virus stocks, 293T cells were cotransfected with 7.5 μg each of proviruses and pHCMV-VSV G, a human cytomegalovirus (CMV) promoter-directed VSV G protein expression plasmid. For Env cellular localization, HeLa cells were grown on coverslips placed in 60-mm petri dishes and cotransfected with 4 μg of pSVE7puro plasmids along with 1 μg of pIIIextat, an HIV-1 LTR-driven Tat expression plasmid. To examine the interaction between Gag and gp41 cytoplasmic subdomains in the format of a GST fusion protein, 293T cells were cotransfected with 10 μg each of pCMVgag and pEBG-cytoplasmic subdomain fusion plasmids. Alternatively, cells were cotransfected with 18 μg of pHIVgptGag or pHIVgptGag/30LE in the presence of 3 μg of pEBG or pEBG/706-856.
Virus infection studies.
Cell-free viruses containing 2 × 104 cpm of reverse transcriptase activity were used to challenge 106 CEM-SS or SupT1 cells. For VSV G-pseudotyped virus infection, 106 SupT1 cells were challenged with VSV G pseudotypes containing 2 × 106 cpm of reverse transcriptase activity. For assessment of single-cycle virus replication, viruses containing 2 × 106 cpm of reverse transcriptase activity were used to challenge 106 H938 cells. Infected cells were washed with phosphate-buffered saline on the second day and then maintained at 37°C for an additional 2 days before harvest for the Western blot analysis or chloramphenicol transferase (CAT) assay. For the env trans-complementation assay, Env-pseudotyped HXBCATΔBgl reporter viruses containing 2 × 105 cpm of reverse transcriptase activity were used to challenge subconfluent HeLa-CD4-LTR-β-gal cells grown in 60-mm dishes or 106 SupT1 cells. All infected cells were washed on the second day and then maintained at 37°C for an additional 2 days before harvest for the protein expression analysis or CAT assay.
Protein and enzymatic analyses.
Cell and virus lysates obtained from transfection or VSV G pseudotype infection were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting with murine Mabs 902, Chessie 8, and 183 as previously described (27). Virus production was measured by reverse transcriptase activity as previously described (4). CAT activity was assessed as previously described (3).
Syncytium formation assay.
To quantitate the syncytium-forming ability of His mutants, 293T cells were transfected with 4 μg each of the wild-type and mutant pSVE7puro plasmids together with 1 μg of pIIIextat. One day after transfection, 106 H938 cells were added to transfected cells. Two days after coculture, cell lysates were prepared, and CAT activity was measured.
Confocal microscopy and flow cytometry analyses.
For Env cellular localization, transfected HeLa cells were fixed with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100. After blocking, the slides were successively incubated with MAbs 902, Chessie 8, and fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G, and then analyzed by confocal microscopy as previously described (7). For total Env expression, VSV G-pseudotyped viruses containing 7 × 105 cpm of reverse transcriptase activity were used to infect CEM-SS cells. Infected cells were fixed, permeabilized, and incubated with MAb 902 and fluorescein isothiocyanate-conjugated second antibody. For cell surface Env expression, after blocking, infected cells were incubated with MAb 902, fixed, and then incubated with fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G. The immunostained cells were quantitated by fluorescence-activated cell sorting (FACS) analysis with a FACSCalibur (Becton Dickinson Immunocytometry Systems).
Isolation of the HIV-1 core structure.
Immature cores were isolated from cell-free, concentrated viruses according to a previously described procedure (47) except that 0.45 ml of 15% sucrose containing 1% Triton X-100 was overlaid on the top of the 30 to 70% linear density sucrose gradient. After fractionation, samples in each fraction were diluted and centrifuged at 100,000 × g at 4°C for 30 min to pellet the viruses. Viral proteins were then analyzed by Western immunoblotting.
Metabolic labeling and cell surface biotinylation.
Metabolic labeling with [35S]methionine and chase with excess unlabeled methionine were performed as previously described (27). For cell surface biotinylation, transfected cells were labeled with [35S]methionine for 3 h and then biotinylated with 0.25 mg of membrane impermeable sulfo-N-hydroxysuccinimide-biotin per ml on ice for 30 min as previously described (37). Cell lysates were immunoprecipitated with pooled anti-HIV-1 antisera preadsorbed onto protein A-Sepharose 4B beads. Equal volumes of the isolated immune complexes were directly resolved by SDS-PAGE or precipitated with 80 μl of 50% of neutravidin-agarose (Pierce) prior to SDS-PAGE.
N-linked glycosylation analyses.
For deoxymannojirimycin (DMM) treatment, HeLa cells transfected with the wild-type or 6His mutant provirus were grown in the presence or absence of 1 mM DMM (Sigma) for 48 h prior to harvesting for Western blot analysis. For endo-β-N-acetylglucosaminidase H (endo H) digestion, lysates of [35S]methionine-labeled HeLa cells transfected with proviruses were incubated with MAb 902-coated protein G-Sepharose, and the precipitated antigens were digested or not with 1 mU of endo H (Roche Molecular Biochemicals) at 37°C for 12 h as previously described (42). For glycopeptidase (PNGase) F digestion, the MAb 902-isolated antigens were digested in the presence or absence of 0.5 U of PNGase F (Sigma) at 37°C for 1.5 h. For neuraminidase treatment, the isolated Env antigens were digested or not with 0.05 U of neuraminidase (Sigma) at 37°C for 30 min.
RESULTS
Construction and infectivity assay of His mutant viruses.
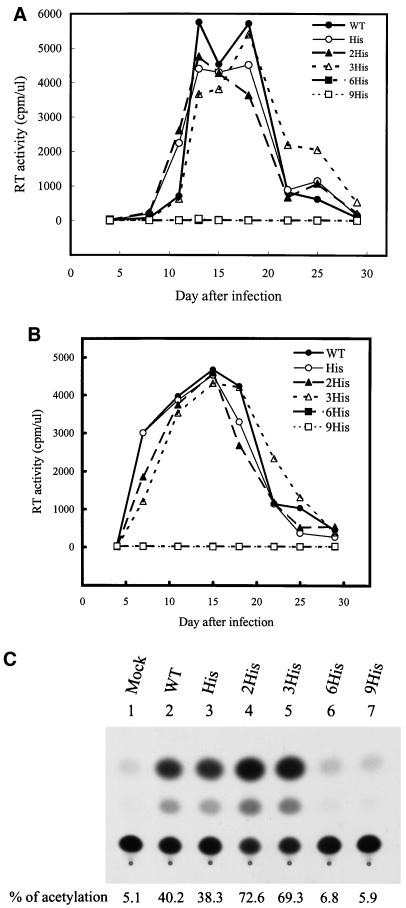
To understand the requirement for the native length of the gp41 cytoplasmic domain in HIV-1 replication, we first examined the effects of elongation of the cytoplasmic tail on virus infection by adding one, two, three, six, or nine His residues to the C terminus of this domain (Fig. 1A). His residues were previously used to address the effect of elongation of the cytoplasmic domain on the membrane fusion ability of influenza virus HA (39). Viruses were generated from 293T cells transfected with the wild-type or mutant proviruses by a standard calcium phosphate coprecipitation method. All of the wild-type, His, 2His, and 3His mutants replicated productively, as measured by reverse transcriptase activity, in CEM-SS cells, whereas the 6His and 9His mutants did not replicate even 29 days subsequent to virus challenge (Fig. 2A). The 6His and 9His mutants also did not establish productive replication in SupT1 cells, another CD4+ T-cell line (Fig. 2B). The infectivity of viruses was also titrated on H938, a cat-harboring reporter cell line (12). This cell line allows detection of productive infection of viral particles by the ability of Tat to transactivate HIV-1 LTR-linked cat gene expression in target cells upon virus infection. The 6His and 9His mutants showed no significant CAT activities compared to the wild-type and other His mutant viruses (Fig. 2C).
FIG. 2.
Replication kinetics of His mutants in CD4+ T cells. Cell-free wild-type (WT) and mutant viruses were used to challenge 106 CEM-SS cells (A) or SupT1 cells (B). Virus production as measured by reverse transcriptase (RT) activity was monitored after infection. Wild-type and mutant viruses were also used to infect 106 H938 cells, and CAT activity was monitored (C).
Exhibition by 6His and 9His mutants of decreased electrophoretic mobility and reduced Env incorporation into viruses.
To understand the defects of the 6His and 9His mutants, HeLa cells were transfected with wild-type and mutant proviruses, and cell and virus lysates were analyzed by SDS-PAGE followed by Western blotting with MAbs 902, Chessie 8, and 183 to detect gp120, gp41, and capsid protein p24, respectively. All of the wild-type and mutant viruses produced comparable amounts of intracellular and virion-associated Gag precursor Pr55 and its cleavage products, p41 and p24 (Fig. 3A). Also, the wild-type and all mutant viruses showed comparable amounts of intracellular gp160 and gp41 (Fig. 3A, lanes 2 to 7). Intracellular gp41 detected in the 6His and 9His mutants migrated more heterogeneously, with the predominant species migrating more slowly than those detected in the wild-type and other His mutant viruses (Fig. 3A, lanes 2 to 7). Intracellular gp120 of the 6His and 9His mutants, marked gp120*, also migrated more slowly than that of the wild-type and other mutant viruses (Fig. 3A, lanes 2 to 7). gp120 and gp41 were not or were only slightly detected in the 6His and 9His mutant virions but were clearly detected in the wild-type and other His mutant virions (Fig. 3A, lanes 9 to 14). The 6His and 9His mutants also exhibited these altered phenotypes in COS-1 and 293T cells (data not shown).
FIG. 3.
Viral protein expression of His mutants. (A) HeLa cells were transfected with the wild-type (WT) or mutant proviruses, and equal volumes of cell and virion lysates were analyzed by Western blotting with MAbs 183 and 902 (top panel). After stripping off of the membrane-bound antibodies, the same blot was reprobed with MAb Chessie 8 (bottom panel). (B) SupT1 cells were infected by wild-type and His mutant VSV G pseudotypes, and cell and virion lysates were analyzed by Western blotting with MAbs 902, Chessie 8, and 183, respectively. In each figure, the intracellular gp120 band observed in the 6His and 9His mutants is marked by an asterisk.
Next, cell-free wild-type and mutant viruses pseudotyped with VSV glycoprotein G after reverse transcriptase normalization were used to infect SupT1 cells. Again, none of the His mutants affected either Gag precursor synthesis, cleavage, virus assembly or budding, or release into the culture medium (Fig. 3B) or altered Env precursor synthesis and processing (Fig. 3B, lanes 1 to 6). However, the gp120 band detected in the 6His and 9His mutants exhibited decreased electrophoretic mobility (Fig. 3B, lanes 1 to 6). The intracellular and virion-associated gp41 species produced by these two mutants migrated more slowly than those produced by the wild-type and other mutant virions (Fig. 3B). These two mutants also displayed a striking reduction in the Env incorporation phenotype (Fig. 3B, lanes 7 to 12). Moreover, the 6His and 9His mutants exhibited decreased intracellular gp120 and gp41 electrophoretic mobility and reduced Env incorporation in CEM-SS cells (data not shown).
Specificity of amino acid extension at the C terminus of the cytoplasmic tail on virus infectivity.
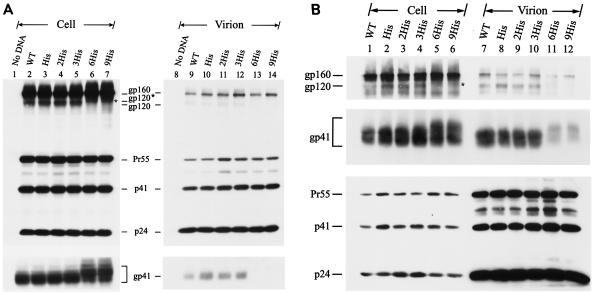
Mutant viruses encoding Env proteins in which the C terminus of the Env was separately extended with three repeats of Leu-Ile and Thr-Ser dipeptides and with six Gly residues, termed the 3(Leu-Ile), 3(Thr-Ser), and 6Gly mutants, respectively, were examined (Fig. 1B). All three mutants established productive replication in CEM-SS (Fig. 4A), and infected H938 cells as effectively as the wild-type and other mutant viruses (data not shown). Moreover, all three mutants showed normal Env precursor synthesis and processing (Fig. 4B, lanes 2 to 6). Although the 6Gly mutant exhibited inhibition of Env incorporation (Fig. 4B, lanes 8 to 12), the virion-associated gp120 and gp41 levels detected in this mutant were still higher than those detected in the 6His mutant virion (Fig. 4B, compare lane 12 to lane 9).
FIG. 4.
Replication and expression of cytoplasmic tail elongation mutants. (A and B) Analyses of the 3(Leu-Ile), 3(Thr-Ser), and 6Gly mutants. Cell-free wild-type (WT) and mutant viruses were used to challenge CEM-SS cells, and reverse transcriptase (RT) activity was monitored after infection (A). HeLa cells were transfected with the wild-type and mutant proviruses, and cell and virion lysates were analyzed by Western blotting with MAbs 902, Chessie 8, and 183 (B). (C and D) Analyses of the 6Gln, 6Pro, and 6Trp mutants. Replication of wild-type and mutant viruses in CEM-SS cells was assessed as described for panel A (C). Viral protein expression in HeLa cells was examined as described for panel B (D).
Next, mutant viruses bearing an extension with six residues of Gln, Pro, and Trp at the C terminus of the cytoplasmic tail were examined (Fig. 1C). The 6Pro mutant replicated with slightly delayed kinetics in CEM-SS cells and produced slightly lower CAT activity upon challenge to H938 cells compared to the wild-type and other two mutant viruses (Fig. 4C and data not shown). Consistent with these results, only the 6Pro mutant showed reduced gp120 and gp41 incorporation into the virus, even though Env precursor synthesis and processing in this mutant were normal (Fig. 4D).
Cellular localization of the 6His and 9His mutants.
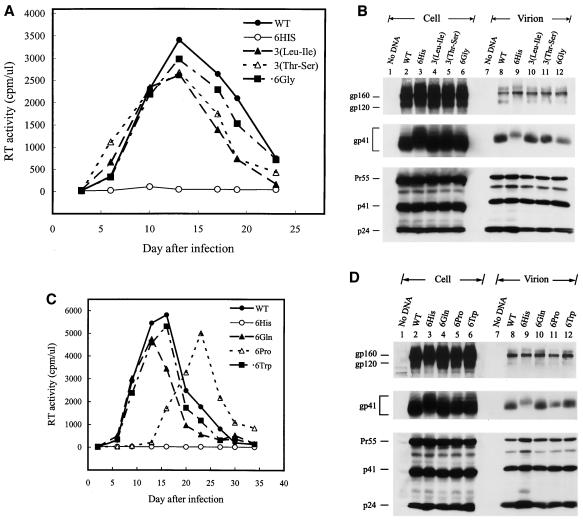
We then sought the molecular basis for the altered Env phenotypes of the 6His and 9His mutants. Confocal microscopy showed that the 6His and 9His mutant Env proteins, as in the case of the wild-type and 3His mutant Env, were perinuclearly stained, with speckles of fluorescence evident in the cytoplasm as well as peripherally on the plasma membrane (Fig. 5A). Cell surface biotinylation with sulfo-N-hydroxysuccinimide-biotin, a membrane-nonpermeating reagent previously used elsewhere (37), showed that gp120 could be detected at the plasma membrane of cells expressing the wild-type and 3His Env but not on the surface of cells expressing the 6His and 9His mutant Envs (Fig. 5B). When wild-type Env-expressing cells were treated with brefeldin A, which prevents the transport of glycoproteins out of the endoplasmic reticulum, prior to biotinylation, gp120 was not detected on the plasma membrane (data not shown), indicating that intracellular Env is not biotinylated by this reagent.
FIG. 5.
Analyses of His mutants. (A) Cellular localization of His mutants. HeLa cells cotransfected with pIIIextat and the wild-type (WT) and His mutant pSVE7puro plasmids were analyzed by confocal microscopy as described in Materials and Methods. (B) Cell surface biotinylation of the 6His mutant. HeLa cells transfected with pHXBCATΔBgl (3), marked Env−, which do not express a functional env gene, and wild-type or mutant proviruses as indicated were metabolically labeled and surface biotinylated, and cell lysates were precipitated with anti-HIV-1 preadsorbed to protein A-Sepharose. Equal amounts of the isolated Env proteins were subjected to SDS-PAGE (lanes 1 to 5) or precipitated with neutravidin-agarose prior to SDS-PAGE (lanes 6 to 10). (C) env trans-complementation assay of the His mutants. Cell-free, env-defective HIV-1 cat reporter viruses generated in the presence of wild-type or mutant Env coexpression were used to challenge HeLa-CD4-LTR-β-gal (top panel) and SupT1 (bottom panel) cells, and CAT activity was assessed. (D) Quantitation of the syncytium-forming ability of the mutant proteins. 293T cells were cotransfected with pIIIextat and the ΔKS, wild-type, and mutant pSVE7puro plasmids. One million H938 cells were added to transfected cells 1 day after transfection, and cell lysates were assayed for CAT activity after 2 days of coculture.
Functional characterization of the 6His and 9His mutant Env proteins.
To confirm the impaired infectivity and reduced Env incorporation phenotypes of the 6His and 9His mutants, the infectivity of these two mutants was assessed by a single-round virus replication assay with an env-defective, HIV-1 cat reporter virus, HXBCATΔBgl (19). Entry of defective viruses produced from cells coexpressing the 6His or 9His mutant Env into HeLa-CD4-LTR-β-gal cells (24) (Fig. 5C, top panel) and SupT1 cells (Fig. 5C, bottom panel) was severely impaired compared to that by the defective viruses pseudotyped with the wild-type or 3His mutant Env.
To examine whether the 6His and 9His mutant proteins expressed on the cell surface were still functional in mediating syncytium formation, 293T cells cotransfected with pIIIextat and each of the wild-type and mutant pSVE7puro plasmids were cocultured with HeLa-CD4-LTR-β-gal. Cells were then stained in situ for β-galactosidase activity. The 6His and 9His mutants as well as the wild-type and 3His Env were able to induce syncytium formation (data not shown). To quantitate the syncytium-forming ability of these two mutants, 293T cells coexpressing Tat along with each of the Env proteins were cocultured with H938 cells, and Tat-mediated cat gene expression in target cells was quantitated. The transcellular activity of Tat expressed in cells cotransfected with pIIIextat and the ΔKS env plasmid showed a low level of CAT expression in H938 target cells (Fig. 5D, lane 2). The two mutant 6His and 9His proteins expressed on the cell surface showed a lower CAT activity than the wild-type and 3His mutant proteins (Fig. 5D), which is in accordance with the reduced Env expression of these two mutants (Fig. 5B).
Distinct N-glycosylation patterns of the 6His mutant.
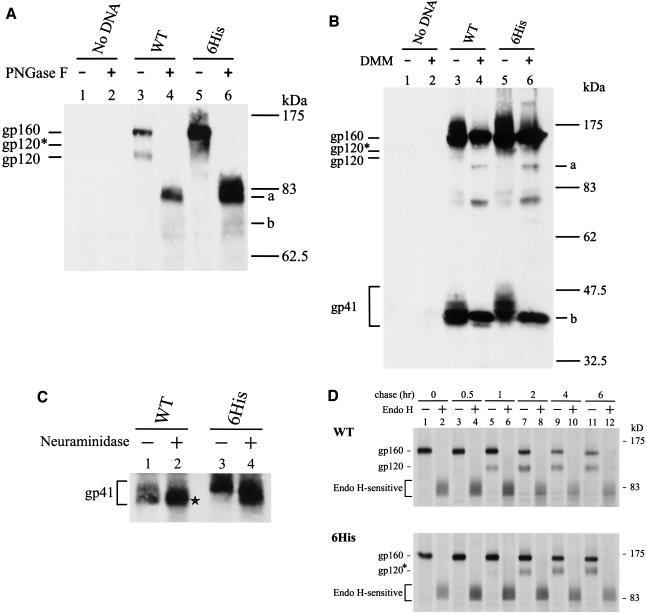
Since the 6His and 9His mutants exhibited the same altered Env phenotypes, further analyses were only performed on the 6His mutant. The MAb 902-precipitated wild-type and 6His mutant proteins were treated or not with PNGase F, which removes all N-glycans from proteins. After PNGase F treatment, the gp160 and gp120 bands of the wild-type and 6His mutant Envs migrated equivalently to the same positions in SDS-PAGE, as marked by bands a and b, respectively, in Fig. 6A. These species represented fully unglycosylated forms of gp160 and gp120, respectively. This observation indicated that the decreased mobility of the 6His mutant cannot be accounted for by the increased molecular mass of the six His residues added to the Env backbone.
FIG. 6.
Analyses of N-linked glycosylation of the 6His mutant. (A) PNGase F analysis. Equal volumes of MAb 902-isolated Env proteins from HeLa cells transfected with the wild-type (WT) or 6His mutant provirus were treated or not with PNGase F prior to Western blotting with MAb 902. Bands a and b represent the gp160 and gp120 forms without N-glycans, respectively. (B) DMM treatment. Cells transfected with the wild-type or mutant provirus were grown in the presence or absence of 1 mM DMM for 48 h, and cell lysates were analyzed by Western blotting with MAb 902. Bands a and b represent the high-mannose forms of gp120 and gp41, respectively. (C) Neuraminidase digestion. Equal volumes of the isolated wild-type and 6His mutant Env proteins were treated or not with neuraminidase prior to Western blotting with MAb Chessie 8. (D) Pulse-chase and endo H digestion. HeLa cells transfected with the wild-type or 6His mutant provirus were metabolically labeled and chased for different times. Equal volumes of MAb 902-isolated Env proteins were treated or not with endo H prior to Western blotting with MAb 902.
HIV-1 Env is highly glycosylated, and these N-linked carbohydrates are differentially processed in the Golgi apparatus, resulting in a mixture of complex, hybrid, and high-mannose carbohydrate structures (16, 34). HeLa cells transfected with the wild-type and 6His proviruses were grown in the presence or absence of 1 mM DMM, which inhibits mannosidase I in the Golgi apparatus, generating proteins containing only high-mannose N-glycans lacking sialic acid (43). As previously noted (18), the N-glycans of the wild-type gp120 were only partially converted to the complex type, and DMM blocked this process, resulting in higher electrophoretic mobility, as marked by band a in Fig. 6B (compared to lanes 3 and 4). The more slowly migrating gp120 of the 6His mutant, marked gp120* in Fig. 6B (compared to lanes 3 and 5), migrated equivalently to wild-type gp120 when both were made in the presence of DMM (Fig. 6B, lanes 4 and 6), indicating that gp120 of the 6His mutant contains more complex N-glycans than does wild-type Env. The wild-type gp41 made in the presence of DMM treatment, marked band b, migrated equivalently to the faster-migrating form of the wild-type gp41 made in the absence of DMM (Fig. 6B, lanes 3 and 4). The 6His mutant gp41 made in the presence of DMM also migrated equivalently to the faster-migrating form of the wild-type gp41 (Fig. 6B, band b in lanes 4 and 6). These results indicated that the more slowly migrating form of the wild-type gp41 and the vast majority of the 6His mutant gp41 contained complex N-glycans and that the faster-migrating form of the wild-type gp41 contained high-mannose N-glycans.
The predominant, faster-migrating species of the wild-type gp41 did not change mobility after treatment with neuraminidase (Fig. 6C, lanes 1 and 2), which removes sialic acid residues located on terminal branches of complex N-glycans. In contrast, the minor, slowly migrating form of the wild-type gp41 (Fig. 6C, lane 1) increased mobility upon neuraminidase treatment (Fig. 6C, lane 2, marked by an asterisk). Also, neuraminidase digestion increased the mobility of the 6His mutant gp41 (Fig. 6C, lanes 3 and 4). This study confirmed that most of the N-glycans on the wild-type gp41 do not contain sialic acids and that most of the N-glycans on the slower-migrating form of the wild-type and on the 6His mutant gp41 are of the complex type.
A pulse-chase coupled to digestion with endo H, which removes only high-mannose and hybrid N-glycans but not the complex carbohydrate structures from proteins, showed that most of the wild-type and 6His mutant precursors were converted to species with an apparent molecular mass of about 83 kDa (Fig. 6D). This observation indicated that most of the uncleaved wild-type and 6His mutant precursors are enriched with high-mannose N-glycans. The gp120 bands of the wild-type and 6His mutant were clearly detected 1 and 2 h, respectively, after the chase, implying that the 6His mutant is transported to the trans-Golgi network, where precursor processing is believed to occur, with slower kinetics than wild-type Env.
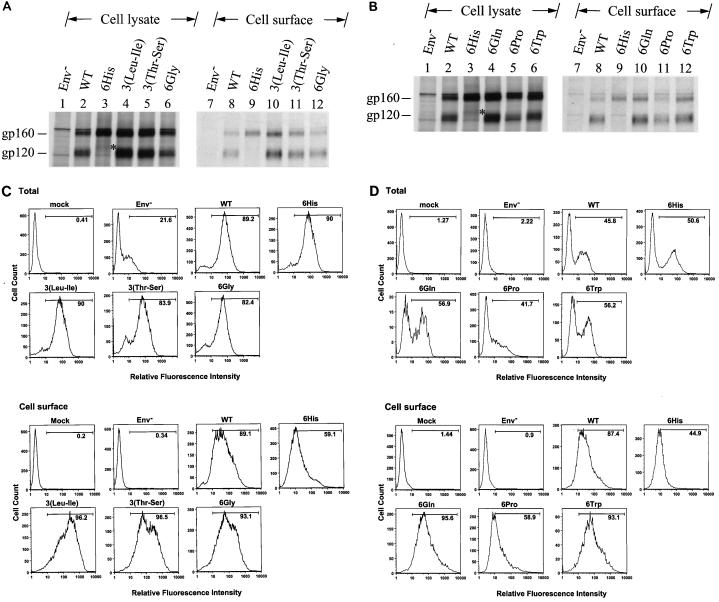
Cell surface expression of mutants other than His mutants.
The cell surface Env expressions of the 3(Leu-Ile), 3(Thr-Ser), and 6Gly mutants in HeLa cells was then compared with that of the wild type by surface biotinylation. All of these mutants showed levels of the intracellular gp160 precursor and cell surface gp120 comparable to those produced by the wild-type virus (Fig. 7A). Extension with six Gln and six Trp residues affected neither Env precursor synthesis nor cell surface Env expression (Fig. 7B). Although the 6Pro mutant produced an amount of gp160 comparable to that produced by the wild type, this mutant exhibited reduced gp120 expression on the cell surface (Fig. 7B).
FIG. 7.
Cell surface expression of mutants containing six extra residues at the C terminus of the cytoplasmic tail. (A and B) Surface biotinylation of the 6His, 3(Leu-Ile), 3(Thr-Ser), and 6Gly mutants and of the 6His, 6Gln, 6Pro, and 6Trp mutants, respectively. HeLa cells transfected with the wild-type (WT) or mutant proviruses as indicated were metabolically labeled and surface biotinylated. Equal volumes of anti-HIV-1-isolated Env proteins were resolved by SDS-PAGE without (lanes 1 to 6) or with (lanes 7 to 12) prior precipitation with neutravidin-agarose. (C and D) FACS analyses of the 6His, 3(Leu-Ile), 3(Thr-Ser), and 6Gly mutants and of the 6His, 6Gln, 6Pro, and 6Trp mutants, respectively. VSV G-pseudotyped wild-type and mutant viruses as indicated were used to challenge CEM-SS cells, and total and cell surface Env expression was analyzed by FACS. Mock infection and infection with pseudotyped virus produced from pHXBCATΔBgl cotransfection, labeled Env−, were used as negative controls.
The cell surface Env expressions of the 3(Leu-Ile), 3(Thr-Ser), and 6Gly mutants in CEM-SS were then compared with those of the wild type and 6His mutant by FACS. The wild type and 6His and all other mutant viruses showed similar levels of total Env in cells (Fig. 7C). Except for the 6His mutant, which showed a reduced surface Env level, all mutants produced a comparable or even slightly larger amount of Env than the wild type on the cell surface (Fig. 7C). Although the 6Pro mutant produced an amount of total Env comparable to that produced by the wild type, this mutant exhibited reduced Env cell surface expression (Fig. 7D). On the other hand, the 6Gln and 6Trp mutants produced a higher level of total Env than the wild type, which paralleled the increase in Env cell surface expression of these two mutants (Fig. 7D).
Interaction with Gag of the gp41 cytoplasmic tail in the format of a GST fusion protein.
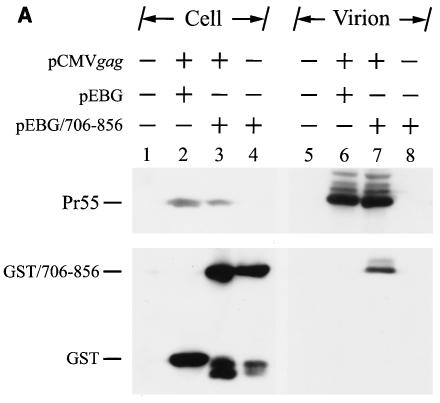
Next, we attempted to examine whether, in the absence of gp120, the ectodomain, and the TM region of the Env, elongation at the C terminus of the cytoplasmic tail affected cytoplasmic tail-Gag interaction. First, we examined whether the cytoplasmic tail could interact with Gag by examining the incorporation of the GST-cytoplasmic tail fusion protein into virions. 293T cells were cotransfected with pCMVgag in the presence of pEBG or pEBG/706-856. pCMVgag directs the synthesis of the Gag precursor driven by a CMV promoter. pEBG/706-856 encodes a mammalian elongation factor 1α promoter-driven cytoplasmic tail fragment fused to the C terminus of GST (28). GST/706-856 but not GST itself was incorporated into the Gag virus-like particles (Fig. 8A, lanes 7 and 6, respectively). GST/706-856 itself was not detected in the pelleted material when culture supernatant of cells expressing GST/706-856 alone was ultracentrifuged (Fig. 8A, lane 8).
FIG. 8.
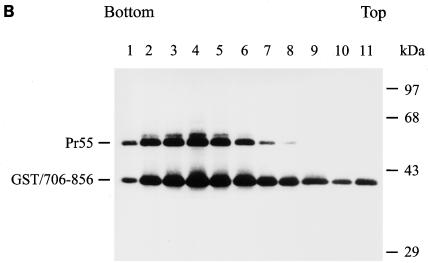
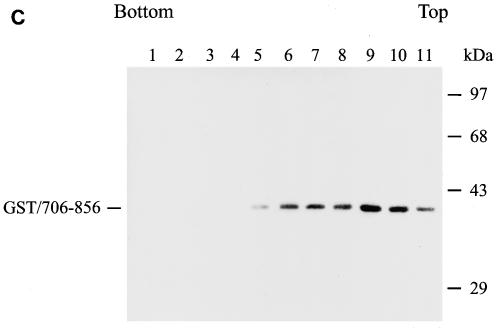
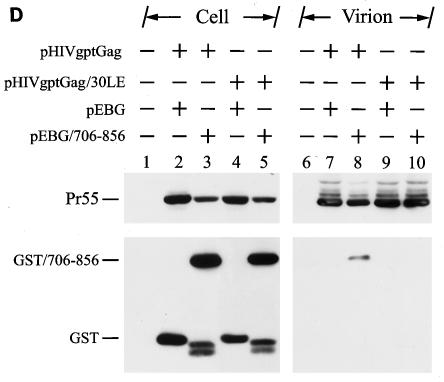
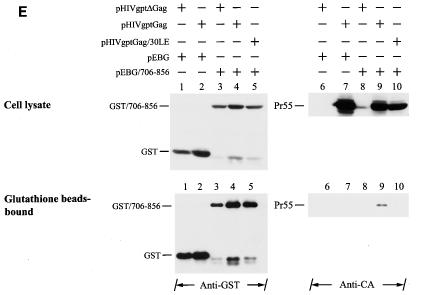
Examination of Gag-cytoplasmic tail interactions. (A) Incorporation of a cytoplasmic tail fusion protein into Gag particles. 293T cells were cotransfected with pCMVgag together with pEBG or pEBG/706-856 as indicated. Cell and virion lysates were analyzed by Western blotting with MAb 183 and a rabbit anti-GST antibody, respectively. Transfection with pEBG/706-856 in the presence of pCDNA3 was also performed in parallel (lanes 4 and 8). (B) Isolation of the immature HIV-1 core structure from Gag and GST/706-856 coexpression. Cell-free, concentrated viral particles obtained from Gag and GST/706-856 coexpression were sedimented through a linear 30 to 70% sucrose gradient containing a layer of 1% Triton X-100. Isolated core particles in each fraction were analyzed by Western blotting with MAbs 183 and Chessie 8. (C). Sedimentation analysis of GST/706-856. 293T cells expressing GST/706-856 were lysed with phosphate-buffered saline containing 1% Triton X-100, and the lysates were sedimented through a sucrose gradient lacking detergent. After fractionation, proteins in each fraction was concentrated by 10% cold trichloroacetic acid precipitation and then analyzed by Western blotting with Chessie 8. (D) Lack of GST/706-856 incorporation into the 30LE mutant virus-like Gag particles. Cells were cotransfected with wild-type or 30LE mutant pHIVgptGag in the presence of pEBG or pEBG/706-856. Cell and virus lysates were analyzed with MAb 183 and rabbit anti-GST, respectively, by Western blotting. (E) Lack of interaction of GST/706-856 with the 30LE mutant Gag. 293T cells were cotransfected with pHIVgptΔGag or with wild-type or 30LE mutant pHIVgptGag in the presence of pEBG or pEBG/706-856. Transfected cells were lysed with Tris-buffered saline containing 10 mM 3-[(cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). One fraction of the cell lysates was subjected directly to Western blotting with rabbit anti-GST and MAb 183 (top panels). Another fraction was first incubated with glutathione-Sepharose 4B, and the bound proteins were analyzed by Western blotting with anti-GST and MAb 183 (bottom panel).
To show that virion-associated GST/706-856 is indeed stably associated with the Gag protein, cell-free, virus-like particles collected from the culture supernatants of 293T cells coexpressing Gag and GST/706-856 were ultracentrifuged through a 30 to 70% linear sucrose gradient containing a layer of 1% Triton X-100. This method was used to determine the detergent-resistant interaction between gp41 and the isolated immature Gag core (47). The vast majority of the GST/706-856 fusion protein cosedimented with Pr55 in fractions 2 to 5, which corresponded to the isolated immature core (Fig. 8B). When cell lysate containing GST/706-856 alone was analyzed, this fusion protein sedimented to top fractions of the gradient (Fig. 8C).
To prove the fidelity of this method in detecting Gag-Env interactions, incorporation of GST/706-856 into the wild-type and 30LE virus-like Gag particles was examined. The 30LE mutant Gag encodes a Leu-to-Glu mutation at amino acid residue 30 of the MA that impairs Env assembly into the virus, presumably due to the lack of Gag-Env interactions (14). 293T cells were cotransfected with pEBG/706-856 together with the wild-type or 30LE mutant Gag plasmid pHIVgptGag. pHIVgptGag contains a functional gag gene but lacks the pol gene in the backbone of pHIVgpt, which encodes a simian virus 40 promoter-driven gpt drug resistance gene in the env position. GST/706-856 was incorporated into the wild-type but not the 30LE mutant Gag particles (Fig. 8D).
To confirm that GST/706-856 does not interact with the 30LE mutant Gag, 293T cells were cotransfected with pEBG or pEBG/706-856 together with pHIVgptΔGag, in which most of the gag coding sequence was deleted, or with the wild-type or 30LE pHIVgptGag plasmid (Fig. 8E, top panel). Gag was not bound to glutathione-Sepharose 4B beads when coexpressed with GST (Fig. 8E, bottom panel, lane 7). The wild-type Gag but not the 30LE mutant Gag bound to the glutathione beads (Fig. 8E, bottom panel, lanes 9 and 10, respectively). These results collectively indicated that GST/706-856 is specifically associated with wild-type Gag but not the 30LE mutant Gag, which allows the GST-cytoplasmic tail fusion protein to be incorporated into wild-type virus-like Gag particles.
Determination of interactions of elongated and truncated cytoplasmic tails with Gag.
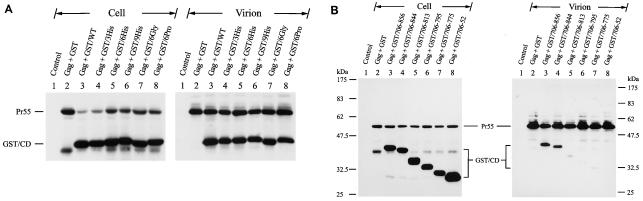
Since the 6His, 9His, 6Gly, and 6Pro mutants exhibited reduced Env incorporation, the interactions of these elongated cytoplasmic tails along with those of the wild-type and 3His mutant with Gag were examined. Comparable amounts of the wild-type and cytoplasmic tail elongation fusion proteins were detected in the virions (Fig. 9A).
FIG. 9.
Effects of alterations to the C-terminal cytoplasmic tail on interactions with Gag. (A) Virion incorporation of GST fusion proteins containing elongated cytoplasmic tails. 293T cells were cotransfected with pCMVgag and each of the pEBG and pEBG chimeras that encoded elongated cytoplasmic tails as indicated, and cell and virion lysates were analyzed by Western blotting with MAbs 183 and Chessie 8. (B) Incorporation of GST-cytoplasmic subdomain fusion proteins into Gag particles. Cell and virion lysates obtained from 293T cells coexpressing Gag and each of the GST fusion proteins that contained cytoplasmic subdomain fragments as indicated were analyzed by Western blotting with MAbs 183 and Chessie 8. Migration of GST-cytoplasmic tail fusion proteins is marked by GST/CD.
To further understand the involvement of the C-terminal cytoplasmic tail in cytoplasmic tail-Gag interactions, incorporation of GST fusion proteins containing cytoplasmic tails with a series of truncations from the C terminus of this domain was analyzed. Among these cytoplasmic subdomain fusion proteins, only GST/706-856 and GST/706-844 were effectively incorporated into the virus (Fig. 9B).
DISCUSSION
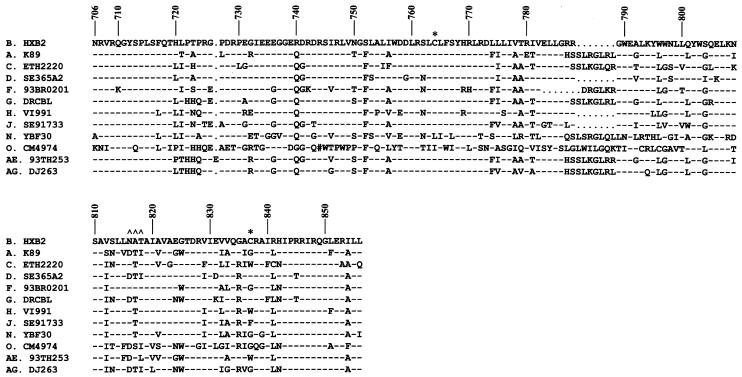
Previous mutation, deletion, and truncation studies have suggested that retention of an intact, native cytoplasmic tail is crucial for virus replication. Interestingly, the lengths of the cytoplasmic tails of different HIV-1 clades may differ (Fig. 10) (26). Although HIV-1 appears to be able to bear internal insertions into its cytoplasmic tail, it is not known whether HIV-1 can tolerate extension of the Env cytoplasmic tail at its C terminus without compromise of its infectivity.
FIG. 10.
Sequence comparison of the cytoplasmic tails of different HIV-1 serotypes. The amino acid sequences in the single-letter code of the entire cytoplasmic tails of representative isolates from different HIV-1 clades are aligned. The numbers at the top indicate the amino acid position in Env of the HXB2 strain. Dashes and dots indicate that the residue at that position is identical to that of or absent from the HXB2 strain, respectively. The two asterisks depict the Cys palmitoylation sites, and ∧∧∧ denotes the putative N-glycosylation site in the cytoplasmic tail of HXB2 Env. The # symbol in clade O CM4974 indicates a frameshift or that the codon in that position contains an N or illegal character.
In the present study, we examined the effect of extension of the gp41 cytoplasmic tail at the C terminus on virus replication. We first assessed the effects of extension with different numbers of His residues, which contain an imidazole structure, at the C terminus of this domain (Fig. 1A). Extension with six and nine residues but not with fewer than six residues remarkably reduced virus infectivity (Fig. 2), which was accompanied by decreased Env electrophoretic mobility and reduced Env incorporation into the virus by these two mutants (Fig. 3 and data not shown). To understand the specificity of the amino acids added to the C terminus of Env on virus replication, mutant viruses bearing extensions with residues other than His were examined. Extension with a stretch of three repeats of Leu-Ile and Thr-Ser and with six Gly residues, all of which are smaller than His, did not affect virus infectivity (Fig. 4A and data not shown). Trp, which contains an aromatic structure, and Gln, which contains an amide group, are bigger and larger than Pro, an α-helix breaker. Nevertheless, the two mutants 6Trp and 6Gln were sill infectious, whereas the 6Pro mutant virus showed delayed infectivity (Fig. 4C and data not shown). Although the 6Gly and 6Pro mutants showed inhibited Env assembly, these two mutants still produced a higher level of virion-associated Env than did the 6His mutant (Fig. 4B and Fig. 4D). Comparisons of the replication potentials and the levels of Env incorporation of these two mutants to those of the 6His mutant suggest that a threshold amount of virion-associated Env is required for virus infectivity (Fig. 4). The reduced Env incorporation of the 6Pro mutant (Fig. 4D) reflects the reduced Env cell surface expression of this mutant (Fig. 7B and Fig. 7D). Our study also indicates that the size and molecular weight of the residues extended are not the major factor determining impairment of virus infectivity and reduction of Env incorporation by cytoplasmic tail elongation.
The reduced Env electrophoretic mobility of the two mutants 6His and 9His was not due to the larger sizes of the amino acids added (Fig. 6A) but rather to the distinct N-glycosylation pattern of these two mutants (Fig. 6B and Fig. 6C). Most of the wild-type and 6His mutant Env precursors remained endo H sensitive (Fig. 6D), which is consistent with previous findings that the vast majority of the Env synthesized is retained in the endoplasmic reticulum or in a cis-Golgi compartment in an endo H-sensitive state (1, 9, 46). The reduced cell surface Env expression of these two mutants (Fig. 5B) was due to delayed kinetics of transport of these two mutants to the trans-Golgi network, where precursor processing is believed to take place (Fig. 6D). Nevertheless, once having arrived at the cell surface, these two mutants still retained the ability to cause syncytium formation with CD4+ T cells, although the degree of syncytium formation of these two mutants was correlated to their reduced cell surface expression (Fig. 5D). To our knowledge, these specific and unique phenotypes of the 6His and 9His mutants have not been reported previously for any other Env mutants.
Previous studies revealed that the cytoplasmic tail of HIV and SIV is not required for Env transport to the cell surface (11, 13, 37, 41, 49). The differential N-glycosylation patterns and transport kinetics of the 6His and 9His mutants suggest that elongation with six and nine His residues may trigger a conformational change in Env which interferes with recognition of the mutant Env by the cellular transport machinery, thereby slowing down Env targeting to the cell surface. Alternatively, the 6His and 9His mutants may interact with endoplasmic reticulum- or Golgi-resident cellular proteins which recognize the unique conformational changes induced by the addition of six and nine His residues, resulting in delayed Env transport to the plasma membrane. The increasingly severe effect of extension with six residues of Gly, Pro, and His on reduction of Env cell surface expression also indicates that extension of the cytoplasmic tail of HIV-1 Env at the C terminus can alter Env intracellular trafficking to the cell surface.
Given the results of virus infectivity and Env incorporation of the mutants analyzed here, we reasoned that there is flexibility in the length and structural modifications to the C terminus of the cytoplasmic tail which still allow functional interaction with Gag to take place. To dissect the role of the cytoplasmic tail from other regions within the Env in Env-Gag interactions, we examined incorporation into the virus of the cytoplasmic tail fused to the C terminus of GST. This assay detects the authentic Gag-Env interaction in cells (Fig. 8). Moreover, cytoplasmic tail elongation and truncation studies showed that the immediate C terminus of the cytoplasmic tail does not interact directly with MA but that the C-terminal 13 to 43 residues are critical for the Gag-cytoplasmic tail interaction (Fig. 9A and B, respectively). Based on the hypothesized trimeric MA-Env-interacting cage hole model, although extensions with six residues including His may render conformational changes to the C terminus of the cytoplasmic tail, it is likely that these elongated cytoplasmic tails are still able to pass through the trimeric MA cage hole without much steric hindrance. This allows the MA binding sites in the cytoplasmic tail to interact with the MA globular head. Alternatively, the C-terminal cytoplasmic tail may somehow project outward away from the trimeric MA cage hole during Env-Gag interaction.
Although the 6His mutant Env was expressed on the cell surface, albeit at a reduced level compared to wild-type Env (Fig. 5B and 7), this mutant Env was relatively ineffective at being incorporated into the virus compared to wild-type Env (Fig. 3). In contrast, GST fusion proteins containing six or nine His residues extending the C-terminal cytoplasmic tail still effectively interacted with Gag (Fig. 9A). Moreover, cell surface expression of the 6Gly mutant Env (Fig. 7A and Fig. 7C) and interaction of the GST-6Gly fusion protein with Gag (Fig. 9A) were normal, but Env incorporation into this mutant virus was reduced (Fig. 4B). It appears that Env incorporation into the virus may require more stringent conditions than GST-cytoplasmic tail incorporation, presumably due to the presence of gp120 and the ectodomain and TM region of gp41. These results collectively indicate that the cytoplasmic tail-Gag interaction itself is not sufficient for determining effective Env incorporation. Other factors, such as cellular proteins, may modulate the Env incorporation process as well. However, GST/706-856 incorporation into the virus might only be determined by specific interactions of the cytoplasmic tail with Gag.
Because the three-dimensional structure of the HIV-1 gp41 cytoplasmic domain is not available, precise residues or sequences in this domain that interact with MA cannot be established. In view of the controversial results in regions or sequences crucial for Env-Gag interactions, our study has implications for understanding the molecular basis underlying Gag-cytoplasmic tail interactions in cells and the spatial orientation of the C terminus of the cytoplasmic tail relative to other parts of the domain. The present study may also provide insight into why HIV-1 may have evolved to adopt a specific mechanism to retain a proper length of Env cytoplasmic tail for replication.
Acknowledgments
H938 was obtained from Barbara K. Felber and George N. Pavlakis through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We are grateful to the following individuals for providing reagents: pHXB2RU3 from Tun-Kou Lee; pNL4-3/30LE from Eric O. Freed; pCMVgag, pHIVgptΔGag, and pHIVgptGag from Chien-Tien Wang; pooled anti-HIV-1 antisera from Mao-Yuan Chen; and rabbit anti-GST antibody from Young-Sun Lin.
This work was supported by grants from Academia Sinica (AS-92-TP-A04) and from the National Health Research Institutes (NHRI-EX91-9136SN), Taipei, Taiwan, Republic of China.
REFERENCES
- 1.Berman, P. W., W. M. Nunes, and O. K. Haffar. 1988. Expression of membrane-associated and secreted variants of gp160 of human immunodeficiency virus type 1 in vitro and in continuous cell lines. J. Virol. 62:3135-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Celma, C. C., J. M. Manrique, J. L. Affranchino, E. Hunter, and S. A. Gonzalez. 2001. Domains in the simian immunodeficiency virus gp41 cytoplasmic tail required for envelope incorporation into particles. Virology 283:253-261. [DOI] [PubMed] [Google Scholar]
- 3.Chen, S. S.-L., A. A. Ferrante, and E. F. Terwilliger. 1996. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology 226:260-268. [DOI] [PubMed] [Google Scholar]
- 4.Chen, S. S.-L., C.-N. Lee, W.-R. Lee, K. McIntosh, and T.-H. Lee. 1993. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane glycoprotein. J. Virol. 67:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, S. S.-L., S.-F. Lee, C.-K. Chuang, and V. S. Raj. 1999. trans-Dominant interference with human immunodeficiency virus type 1 replication and transmission in CD4+ cells by an envelope double mutant. J. Virol. 73:8290-8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, S. S.-L., S.-F. Lee, H.-J. Hao, and C.-K. Chuang. 1998. Mutations in the leucine zipper-like heptad repeat sequence of human immunodeficiency virus type 1 gp41 dominantly interfere with wild-type virus infectivity. J. Virol. 72:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. S.-L., S.-F. Lee, and C.-T. Wang. 2001. Cellular membrane-binding ability of the C-terminal cytoplasmic domain of human immunodeficiency virus type 1 envelope transmembrane protein gp41. J. Virol. 75:9925-9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosson, P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 15:5783-5788. [PMC free article] [PubMed] [Google Scholar]
- 9.Crise, B., L. Buonocore, and J. K. Rose. 1990. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J. Virol. 64:5585-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman, T., F. Mannano, W. A. Haseltine, and H. G. Göttlinger. 1994. Role of matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 68:1689-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubay, J. W., S. J. Robers, B. H. Haln, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felber, B. K., and G. N. Pavlakis. 1988. A quantitative bioassay for HIV-1 based on trans-activation. Science 239:184-187. [DOI] [PubMed] [Google Scholar]
- 13.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed, E. O., and M. A. Martin. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabuzda, D. H., A. Lever, E. Terwilliger, and J. Sodroski. 1992. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 66:3306-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geyer, H., C. Holschbach, G. Hunsmann, and J. Schneider. 1988. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J. Biol. Chem. 263:11760-11767. [PubMed] [Google Scholar]
- 17.Gonzalez, S. A., A. Burny, and J. L. Affranchino. 1996. Identification of domains in the simian immunodeficiency virus matrix protein essential for assembly and envelope glycoprotein incorporation. J. Virol. 70:6384-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruters, R. A., J. J. Neefjes, M. Tersmette, R. E. de Goede, A. Tulp, H. G. Huisman, F. Miedema, and H. L. Ploegh. 1987. Interference with HIV-induced syncytium formation and viral infectivity by inhibitors of trimming glucosidase. Nature (London) 330:74-77. [DOI] [PubMed] [Google Scholar]
- 19.Helseth, E., M. Kowalski, D. Gabuzda, U. Olshevsky, W. Haseltine, and J. Sodroski. 1990. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J. Virol. 64:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, C. P., D. W. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch, V. M., P. Edmondson, M. Murphey-Corb, B. Arbeille, P. R. Johnson, and J. I. Mullins. 1989. SIV adaptation to human cells. Nature (London) 341:573-574. [DOI] [PubMed] [Google Scholar]
- 22.Hourioux, C., D. Brand, P. Y. Sizaret, F. Lemiale, S. Lebigot, F. Barin, and P. Roingeard. 2000. Identification of the glycoprotein 41(TM) cytoplasmic tail domains of human immunodeficiency virus type 1 that interact with Pr55Gag particles. AIDS Res. Hum. Retrovir. 16:1141-1147. [DOI] [PubMed] [Google Scholar]
- 23.Johnston, P. B., J. W. Dubay, and E. Hunter. 1993. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J. Virol. 67:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodama, T., D. P. Wooley, Y. M. Naidu, H. W. Kestler 3rd, M. D. Daniel, Y. Li, and R. C. Desrosiers. 1989. Significance of premature stop codons in env of simian immunodeficiency virus. J. Virol. 63:4709-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korber, B., C. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. Mellors, and J. Sodroski. 1998. Amino acid alignment, p. IIA-1-IIA-77. In B. Korber, C. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS 1998: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 27.Lee, S.-F., C.-Y. Ko, C.-T. Wang, and S. S.-L. Chen. 2002. Effect of point mutations in the N terminus of the lentivirus lytic peptide-1 sequence of human immunodeficiency virus type 1 transmembrane glycoprotein gp41 on Env stability. J. Biol. Chem. 277:15363-15375. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S.-F., C.-T. Wang, J. Y.-P. Liang, S.-L. Hong, C.-C. Huang, and S. S.-L. Chen. 2000. Multimerization potential of the cytoplasmic domain of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J. Biol. Chem. 275:15809-15819. [DOI] [PubMed] [Google Scholar]
- 29.Lodge, R., H. G. Götlinger, D. Gabuzda, E. A. Cohen, and G. Lemay. 1994. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J. Virol. 68:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lodge, R., J.-P. Lalonde, G. Lemay, and E. A. Cohen. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 16:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luciw, P. A., K. E. Shaw, B. L. Shacklett, and M. L. Marthas. 1998. Importance of the intracytoplasmic domain of the simian immunodeficiency virus (SIV) envelope glycoprotein for pathogenesis. Virology 252:9-16. [DOI] [PubMed] [Google Scholar]
- 32.Mannano, F., E. Kondo, J. Sodroski, A. Bukovsky, and H. G. Göttlinger. 1995. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J. Virol. 69:3824-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, M. A., and R. C. Montelaro. 1992. Amphipathic helical segments of HIV-1 transmembrane (TM) proteins and their potential role in viral cytopathicity, p. 351-364. In R. C. Aloia (ed.), Advances in membrane fluidity, vol. 6. A. R. Liss, New York, N.Y. [Google Scholar]
- 34.Mizuochi, T., T. J. Matthews, M. Kato, J. Hamako, K. Titani, J. Solomon, and T. Feizi. 1990. Diversity of oligosaccharide structures on the envelope glycoprotein gp 120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. Presence of complex-type oligosaccharides with bisecting N-acetylglucosamine residues. J. Biol. Chem. 265:8519-8524. [PubMed] [Google Scholar]
- 35.Mulligan, M. J., G. V. Yamshchikov, G. D. Ritter, Jr., F. Gao, M. J. Jin, C. D. Nail, C. P. Spies, B. H. Hahn, and R. W. Compans. 1992. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 66:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and α-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nermut, M. V., D. J. Hockley, J. B. M. Jowett, I. M. Jones, M. Garrequ, and D. Thomas. 1994. Fullerene-like organization of HIV gag-protein shell in virus-like particles produced by recombinant baculovirus. Virology 198:288-296. [DOI] [PubMed] [Google Scholar]
- 39.Ohuchi, M., C. Fischer, R. Ohuchi, A. Herwig, and H. D. Klenk. 1998. Elongation of the cytoplasmic tail interferes with the fusion activity of influenza virus hemagglutinin. J. Virol. 72:3554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owens, R. J., J. W. Dubay, E. Hunter, and R. W. Compans. 1991. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc. Natl. Acad. Sci. USA 88:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritter, G. D., Jr., M. J. Mulligan, S. L. Lydy, and R. W. Compans. 1993. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology 197:255-264. [DOI] [PubMed] [Google Scholar]
- 42.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sears, P., and C. H. Wong. 1998. Enzyme action in glycoprotein synthesis. Cell. Mol. Life Sci. 54:223-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trujillo, J. R., W. K. Wang, T.-H. Lee, and M. Essex. 1996. Identification of the envelope V3 loop as a determinant of a CD4-negative neuronal cell tropism for HIV-1. Virology 217:613-617. [DOI] [PubMed] [Google Scholar]
- 45.Venable, R. M., R. W. Pastor, B. R. Brooks, and F. W. Carson. 1989. Theoretically determined three-dimensional structures for amphipathic segments of HIV-1 gp41 envelope protein. AIDS Res. Hum. Retrovir. 5:7-22. [DOI] [PubMed] [Google Scholar]
- 46.Willey, R. L., J. S. Bonifacino, B. J. Potts, M. A. Martin, and R. D. Klausner. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA 85:9580-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of gp41 with Pr55Gag in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, X., X. Yuan, Z. Matsuda, T.-H. Lee, and M. Essex. 1992. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J. Virol. 66:4966-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, X., X. Yuan, M. F. McLane, T.-H. Lee, and M. Essex. 1993. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J. Virol. 67:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zingler, K., and D. R. Littman. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J. Virol. 67:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]