Abstract
The herpes simplex virus type 1 (HSV-1) UL20 protein is an important determinant for virion morphogenesis and virus-induced cell fusion. A precise deletion of the UL20 gene in the HSV-1 KOS strain was constructed without affecting the adjacent UL20.5 gene. The resultant KOS/UL20-null virus produced small plaques of 8 to 15 cells in Vero cells while it produced wild-type plaques on the complementing cell line G5. Electron microscopic examination of infected cells revealed that the KOS/UL20-null virions predominantly accumulated capsids in the cytoplasm while a small percentage of virions were found as enveloped virions within cytoplasmic vacuoles. Recently, it was shown that UL20 expression was necessary and sufficient for cell surface expression of gK (T. P. Foster, X. Alvarez, and K. G. Kousoulas, J. Virol. 77:499-510, 2003). Therefore, we investigated the effect of UL20 on virus-induced cell fusion caused by syncytial mutations in gB and gK by constructing recombinant viruses containing the gBsyn3 or gKsyn1 mutations in a UL20-null genetic background. Both recombinant viruses failed to cause virus-induced cell fusion in Vero cells while they readily caused fusion of UL20-null complementing G5 cells. Ultrastructural examination of UL20-null viruses carrying the gBsyn3 or gKsyn1 mutation revealed a similar distribution of virions as the KOS/UL20-null virus. However, cytoplasmic vacuoles contained aberrant virions having multiple capsids within a single envelope. These multicapsid virions may have been formed either by fusion of viral envelopes or by the concurrent reenvelopment of multiple capsids. These results suggest that the UL20 protein regulates membrane fusion phenomena involved in virion morphogenesis and virus-induced cell fusion.
Herpes simplex viruses (HSV) specify at least 11 virally encoded glycoproteins as well as several nonglycosylated membrane-associated proteins that are pivotal in membrane fusion processes during a productive viral infection, including virus-induced cell-to-cell fusion, pH-independent virus entry via fusion of the virion envelope with cellular membranes, morphogenesis and egress of infectious virion particles, and cell-to-cell spread. The prevailing model for morphogenesis and egress of infectious herpes virions into the extracellular space involves multiple membrane-associated events. These include primary envelopment by budding of capsids assembled in the nuclei through the inner nuclear leaflet, de-envelopment by fusion of viral envelopes with the outer nuclear leaflet, reenvelopment of cytoplasmic capsids into Golgi or trans-Golgi network-derived vesicles, and transport of enveloped viruses within cytoplasmic transport vesicles to extracellular spaces (reviewed in references 21, 23, and 28).
Spread of infectious virus occurs either by release of virions to extracellular spaces or through virus-induced cell-to-cell fusion. Mutations that cause extensive virus-induced cell fusion predominantly arise in four HSV genes, UL20 (4, 22), UL24 (19, 27), UL27 encoding glycoprotein B (gB) (6, 24), and UL53 coding for glycoprotein K (gK) (5, 7, 17, 25, 26). Of these four membrane-associated proteins, UL20 and gK are essential for the intracellular transport of virions to extracellular spaces (4, 12, 16, 18, 20). Recently, UL20 was shown to be required for Golgi-dependent glycosylation and cell surface expression of gK, suggesting a functional interdependence between gK and UL20 for virus egress and cell-to-cell fusion (9, 10).
The UL20 gene encodes a 222-amino-acid nonglycosylated transmembrane protein that is conserved by all alphaherpesviruses. Initial attempts to delete the HSV type 1 (HSV-1) UL20 gene were unsuccessful, indicating that the UL20 protein was essential for virus replication (22). However, subsequent experiments utilizing 143TK− cells indicated a cell type-dependent replication for the HSV-1(F) strain UL20-null virus (4). Electron microscopic examination of this virus on nonpermissive Vero cells revealed a block in virus transport to extracellular spaces caused by a profound entrapment of enveloped virions within perinuclear spaces (4). Similar to the HSV-1 UL20-null virus, pseudorabies virus (PRV) UL20-null virions exhibited a complete inhibition of virus transport to extracellular spaces. However, PRV enveloped virions accumulated within cytoplasmic vesicles, and cytoplasmic nonenveloped capsids were also observed (16).
Because UL20 expression was found to be necessary for intracellular transport and cell surface expression of gK in transient expression experiments (10), we investigated the role of the UL20 protein (UL20p) in virus-specified glycoprotein transport and virus-induced cell fusion. Herein, we show that in viral infections UL20p is required for gK transport to cell surfaces as well as virus-induced cell fusion caused by syncytial mutations in either gB or gK. Moreover, electron microscopic analysis indicated that UL20p plays crucial roles in cytoplasmic virion morphogenesis. Specifically, the results suggest that UL20p may function in the reenvelopment step or to prevent de-envelopment of virions contained within cytoplasmic vesicles.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Rockville, Md.). The Vero-based UL20 complementing cell line, G5, was a gift of P. Desai (Johns Hopkins Medical Center) and was maintained as previously described (8, 10, 12). The parental wild-type strain used in this study, HSV-1 KOS, was originally obtained from P. A. Schaffer (Harvard Medical School). The UL53-gK protein C epitope-tagged virus, gKD3protC, and the V5 epitope-containing viruses, gKV5DI and gKsyn20DIV5, were propagated in Vero cells as described previously (10, 15).
Plasmids.
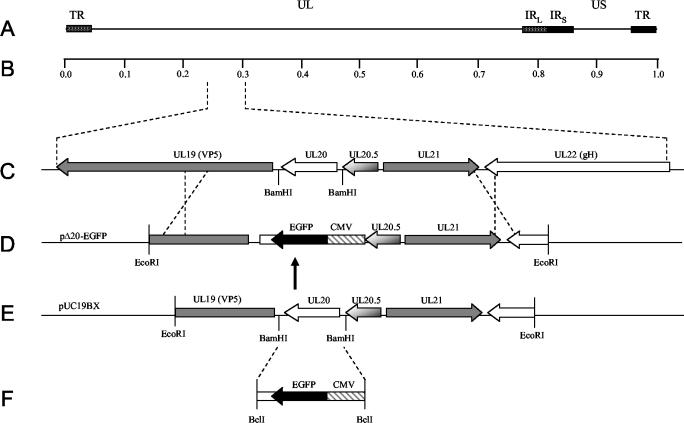
To generate a recombination plasmid that would delete the UL20 open reading frame (ORF) and preserve the UL20.5 and UL19 gene regulatory sequences, puc19 was digested with BamHI and the overhangs were filled in to inactivate the BamHI site, yielding plasmid puc19BX. Primers UL20flank5′EcoRI (5′-TTTCTTGAATTCCGGGTGGCCGGGGGCGGGGAC-3′) and UL20flank3′EcoRI (5′-CTTCTTGAATTCCCCATCACCCACAACGCCAGC-3′) were used to PCR amplify a 5,148-bp product from KOS viral DNA that spanned a region from UL19 to UL22. The PCR product was digested with EcoRI and ligated into EcoRI-digested puc19BX to generate p20F (Fig. 1D). Primers 5′CMVEGFP20.5BclI (5′-TTCTTCTTCTTCTGATCATGCTGACGTAAGTAGTTATTAATAGTAATCAATTACGGGG-3′)and 3′CMVEGFPUL19pro (5′-CCCTCTTGATGTTATGAGTTTGGACAAACCACAACTAGA-3′) were used to PCR amplify a cytomegalovirus (CMV)-enhanced green fluorescent protein (EGFP) gene cassette from pTF9200 (11, 14). Primers 5′UL19proBclI (5′-CCCGGGGACGTGTGATCACAGGCCTTATATACC-3′) and 3′UL19proEGFPpA (5′-GTCCAAACTCATAACATCAAGAGGGATCGTCTGCC-3′) were used to PCR amplify the UL19 promoter region. To delete the UL20 ORF without disrupting UL19 regulatory sequences, splice overlap extension PCR was used to fuse to the UL19 promoter region with the CMV-EGFP gene cassette (Fig. 1E). The resultant fusion product was digested with BclI and ligated into p20F that was BamHI restricted and ciprofloxacin treated. The resultant plasmid was designated pΔ20-EGFP.
FIG. 1.
Schematic of the construction of UL20-null viruses. (A) The top line represents the prototypic arrangement of the HSV-1 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. (B) Shown below is an expanded genomic region between map units 0.25 and 0.3 containing the UL19, UL20, UL20.5, UL21, and UL22 genes. (C) The recombination plasmid, pΔ20-EGFP, which contains UL20 flanking sequences for recombination with the viral genome, was generated by inserting a CMV-EGFP gene cassette (E) between the BamHI restriction sites flanking the UL20 gene (D).
Generation of UL20-null viruses.
Viruses that contained the CMV-EGFP gene cassette and, therefore, contained a UL20 gene deletion were generated by homologous recombination with plasmid pΔ20-EGFP and the respective parental virus. 143TK− cells were transfected with pΔ20-EGFP, and at 6 h posttransfection, they were infected at a multiplicity of infection (MOI) of 0.1 with either gKDIV5, gKD3prtC, gKsyn20DIV5, or tsB5 (gBsyn3) to generate UL20-null viruses that were designated Δ20DIV5, Δ20D3prtc, Δ20syn20DIV5, and Δ20gBsyn3, respectively. Recombinant viruses were plaque isolated on G5 cells by fluorescence microscopy and sequentially plaque purified at least five times. Insertion of the CMV-EGFP gene cassette and the concomitant deletion of the UL20 gene were confirmed by diagnostic PCR, DNA sequencing, and restriction endonuclease analysis.
One-step growth kinetics and plaque morphology.
Analysis of one-step growth kinetics of total infectious virus was as described previously (10, 13, 15). Briefly, each virus at an MOI of 5 was adsorbed to approximately 8 × 105 cells of the indicated cell line at 4°C for 1 h. Thereafter, prewarmed media was added, and virus was allowed to penetrate for 2 h at 37°C. Any remaining extracellular virus was inactivated by low-pH treatment (0.1 M glycine, pH 3.0). Cells and supernatants were harvested immediately thereafter (0 h) or after 4-, 8-, 12-, or 24-h incubations at 37°C. Virus titers were determined by endpoint titration of virus stocks on G5 cells. For plaque morphology, Vero or G5 cells were infected at an MOI of 0.001 with the indicated virus and visualized by immunohistochemistry at 36 h postinfection (hpi) by utilizing horseradish peroxidase-conjugated anti-HSV antibodies (DAKO) and Novared (VectorLabs) substrate development (10, 13).
Confocal microscopy.
Cell monolayers grown on coverslips in six-well plates were infected with the indicated virus at an MOI of 10. For cell surface biotinylation, prior to fixation, cells were washed with Tris-buffered saline (TBS)-Ca/Mg and incubated for 15 min at room temperature in EZ-Link Sulfo-NHS-LC biotin cell impermeable biotinylation reagent (Pierce Chemical), which reacts with primary amines on cell surface proteins. Cells were washed with TBS, fixed with electron microscopy (EM)-grade 3% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, Pa.) for 15 min, washed twice with phosphate-buffered saline-50 mM glycine, and permeabilized with 1.0% Triton X-100. Monolayers were subsequently blocked for 1 h at 4°C with 7% normal goat serum and 7% bovine serum albumin in TBS (TBS blocking buffer) before incubation for 5 h at 4°C with either anti-V5 (Invitrogen, Carlsbad, Calif.) for recognition of gK, anti-gD, or anti-gB (Rumbaugh-Goodwin Institute, Plantation, Fla.) diluted 1:500 in TBS blocking buffer. Cells were then washed extensively and incubated for 1 h with Alexa Fluor 594-conjugated anti-immunoglobulin G diluted 1:500 in TBS blocking buffer. After incubation, excess antibody was removed by washing five times with TBS. For cell surface labeling, biotinylated cells were reacted with 1:1,000-diluted Alexa Fluor 488-conjugated streptavidin for 20 min and visualized in the red channel. Cells were examined with a TCS SP2 laser scanning microscope (Leica Microsystems, Exton, Pa.) fitted with a 63× Leica objective (1.4 numerical aperture; Planachromatic). Individual optical sections in the z axis, averaged six times, were collected at 2× zoom in series in the different channels at 1,024- by 1,024-pixel resolution as described previously (10, 13). Images were compiled and rendered in Adobe Photoshop.
Cell surface immunohistochemistry.
Detection of cell surface and total glycoprotein distribution by immunohistochemistry was essentially as described previously (10, 13). Briefly, cell monolayers in six-well plates were infected with the indicated virus at an MOI of 5 and incubated at 37°C for 12 h. Infected monolayers were washed with TBS-Ca/Mg and subsequently incubated at room temperature for 1 h with the specified antibody diluted 1:500 in TBS-Ca/Mg supplemented with 5% bovine serum albumin and 3% normal goat serum. Cell monolayers were extensively washed with TBS-Ca/Mg, fixed with EM-grade 3% paraformaldehyde, permeabilized with 1% Triton X-100, and blocked for 1 h in TBS blocking buffer. For total glycoprotein detection, cells were fixed and permeabilized prior to incubation with the primary antibody. Immunohistochemistry was performed by using the Vector Laboratories Vectastain Elite ABC kit according to the manufacturer's directions and as described previously (10, 13).
EM.
Cell monolayers were infected with the indicated virus at an MOI of 5. All cells were prepared for transmission EM examination at 16 hpi. Infected cells were fixed in a mixture of 2% paraformaldehyde and 1.5% glutaraldehyde in 0.1 M NaCaC buffer, pH 7.3. Following osmication (1% OsO4) and dehydration in an ethanol series, the samples were embedded in Epon-Araldyte resin and polymerized at 70°C. Thin sections were made on an MTXL Ultratone (RMC Products), stained with 5% uranyl acetate and citrate nitrate acetate (CNA) lead, and observed with a Zeiss 10 transmission electron microscope as described previously (12, 13).
RESULTS
Construction and characterization of UL20-null viruses specifying syncytial mutations in gB or gK.
Previous investigations revealed that UL20p expression was required for gK Golgi-dependent processing and cell surface expression in transient expression experiments (9, 10). To verify this potential interdependence between gK and UL20 in the context of viral infections and the effect of UL20 on virus-induced cell fusion, recombinant viruses lacking the UL20 gene were constructed in the presence or absence of either the gBsyn3 or gKsyn1 mutation. Mutant viruses carrying either the gBsyn3 mutation (R-to-H amino acid change in the carboxyl terminus of gB) or the gKsyn1 mutation (A-to-V amino acid change in the amino terminus of gK) cause extensive virus-induced cell fusion and the formation of large syncytial plaques (6, 7). To facilitate homologous recombination with virus genomes and deletion of the UL20 gene without disrupting proximal genes or their gene regulatory sequences, a CMV-EGFP gene cassette was inserted in place of the UL20 ORF within a cloned DNA fragment, resulting in the EGFP gene cassette flanked by HSV-1 upstream and downstream sequences (Fig. 1). Previously, our laboratory showed that in-frame insertion of epitope tags did not adversely affect gK structure and function, whereas they allowed efficient detection of gK in transfected or infected cells (10, 15). Therefore, viruses gKD3prtc and gKDIV5 containing the prtC and V5 epitope tags (10, 15), respectively, were used to generate UL20-null recombinant viruses Δ20D3prtc and Δ20DIV5, respectively. A similar construction strategy was used to generate UL20-null viruses in the tsB5 (syn3) or KOS (gKsyn20DIV5) genetic backgrounds and were designated Δ20gBsyn3 and Δ20syn20DIV5, respectively. Recombinant viruses were isolated via fluorescence microscopy and plaque purified on G5 cells (8), which complemented the UL20-null defect. Individual virus isolates were confirmed by diagnostic PCR and DNA sequencing of targeted DNA fragments.
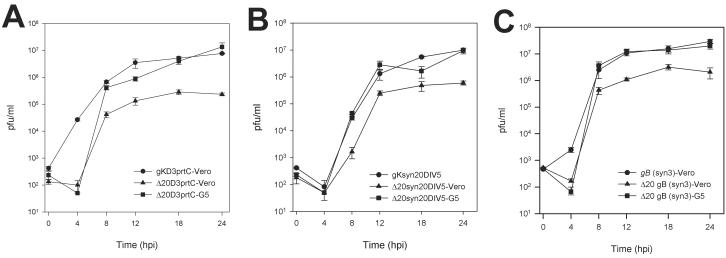
To assess the effect of the deletion of the UL20 gene on cell-to-cell spread and viral replication, the plaque morphology and kinetics of viral replication for Δ20D3prtc, Δ20DIV5, Δ20syn20DIV5, and Δ20gBsyn3 virus strains in Vero and G5 cells were examined. UL20-null viruses, Δ20DIV5, Δ20D3prtc, Δ20syn20DIV5, and Δ20gBsyn3, replicated significantly less efficiently than their corresponding parental viruses. However, replication of all four UL20-null viruses was restored to near wild-type levels in complementing G5 cells (Fig. 2). Although peak infectious titers produced by the nonsyncytial Δ20DIV5 and Δ20D3prtc viruses were reduced approximately 50-fold in Vero cells as opposed to G5 cells, both viruses exhibited similar viral replication kinetics (Fig. 2A and data not shown). By comparison, the syncytial viruses Δ20syn20DIV5 and Δ20gBsyn3 (Fig. 2B and C) replicated to approximately fivefold-greater titers than those observed for the nonsyncytial virus isolates (compare Fig. 2A to B and C).
FIG. 2.
Comparison of virus replication characteristics of KOS gKD3prtC-tagged viruses (A), syn20DIV5 viruses (B), and gBsyn3 viruses (C). One-step kinetics of infectious virus production were calculated for the respective parental viruses on Vero cells (•), UL20-null viruses on Vero cells (▴), and UL20-null viruses on G5 cells (▪).
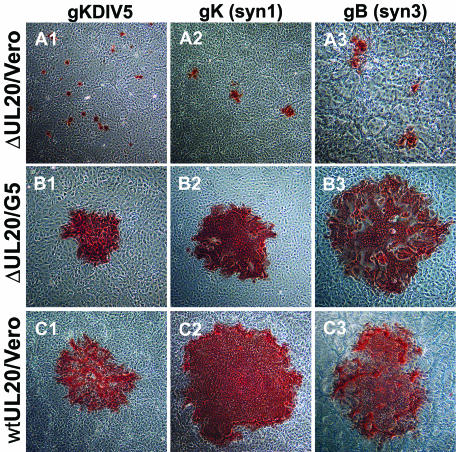
Deletion of the UL20 gene resulted in substantial changes in virus plaque morphology for both wild-type and syncytial mutant viruses (Fig. 3). Parental virus strains syn20DIV5 (gKsyn1) and tsB5 (gBsyn3), which contain syncytial mutations in gK and gB, respectively, produced large syncytial plaques in both Vero cells (Fig. 3C2 and C3, respectively) and G5 cells (data not shown). Similarly, gK-tagged KOS viruses, gKDIV5 and gKD3prtC, generated plaques of approximately wild-type size in Vero cells (Fig. 3C1 and data not shown, respectively). Viruses containing deletions in UL20 produced small nonsyncytial plaques on Vero cells containing approximately 8 to 30 cells even in a genetic context that contained syncytial mutations in gB and gK (Fig. 3A). In contrast, gB- or gK-mediated syncytia formation and virus spread were restored to nearly wild-type levels in UL20 complementing G5 cells (Fig. 3B). Both Δ20syn20DIV5 and Δ20gBsyn3 produced larger plaques than their nonsyncytial KOS counterparts on average (Fig. 3A2 and A3, respectively, compared to A1). Taken together, the growth curves and plaque morphologies induced by these viruses suggest that the presence of the gBsyn3 or gKsyn1 syncytial mutations may enable more-efficient infectious virus production and spread of UL20-null viruses. The data also indicate that UL20 is necessary for gB- and gK-mediated cell-to-cell fusion.
FIG. 3.
Plaque morphology of UL20-null viruses on Vero cells (A), UL20-null viruses on G5 cells (B), and UL20-null rescued viruses on Vero cells (C). Confluent cell monolayers were infected with Δ20gKDIV5 virus (A1 and B1), Δ20syn20DIV5 virus (A2 and B2), or Δ20gBsyn3 virus (A3 and B3) or the corresponding UL20-null rescued virus (C1, C2, and C3) at an MOI of 0.001, and viral plaques were visualized by immunohistochemistry at 36 hpi.
Characterization of cell surface expression of viral glycoproteins in Vero and G5 cells infected with UL20-null viruses.
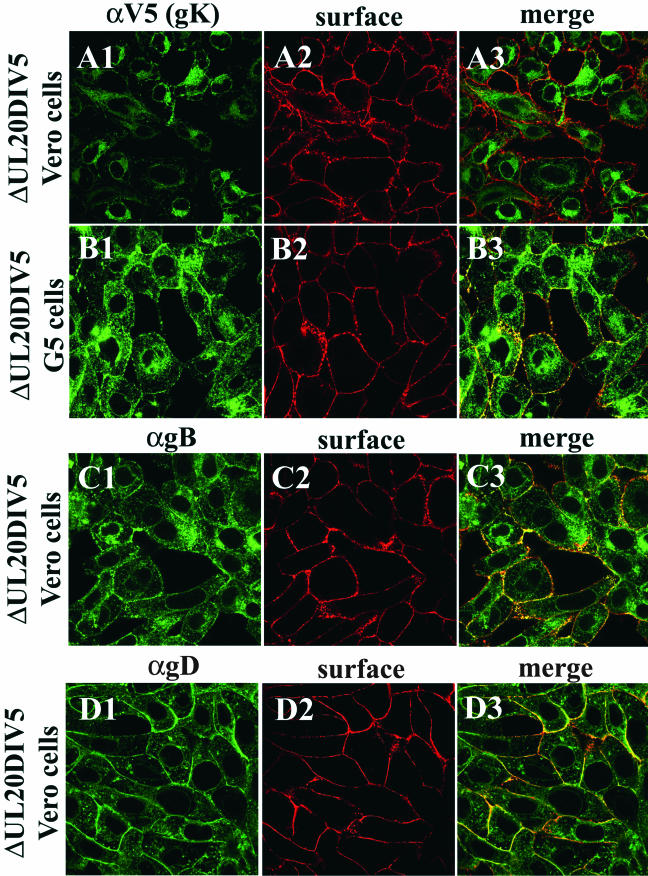
Recently, our laboratory showed that transport of gK to cell surfaces requires UL20 expression (10) and that inhibition of HSV-1 gK cell surface expression eliminated gK-mediated cell-to-cell fusion (10, 13). To determine if the observed lack of syncytial formation by the Δ20gBsyn3 and Δ20syn20DIV5 viruses was due to a defect in either gB or gK glycoprotein transport, cell surface localization of gB, gK, and gD was assessed by confocal microscopy and immunohistochemistry. Confocal microscopy revealed that HSV-1 gK was localized to the juxtanuclear region of the cell, reminiscent of endoplasmic reticulum localization, and failed to be transported to Vero cell surfaces infected with UL20-null viruses (Fig. 4A). In contrast, gK exhibited a broader cellular distribution in UL20-null infected G5 cells, and cell surface transport and localization of gK was restored (Fig. 4B). These results were in agreement with previously published observations that HSV-1 gK required UL20p for transport to cell surfaces in transient coexpression experiments (10). In contrast, gB and gD were transported to cell surfaces in the absence of UL20p (Fig. 4C and D), indicating that UL20p is required for gK but not gB or gD cell surface expression.
FIG. 4.
Confocal microscopic visualization of cell surface and intracellular distribution of gK (A and B), gB (C), and gD (D) in either Vero (A, C, and D) or G5 (B) cells infected with UL20-null Δ20gK DIV5 at an MOI of 10. Infected cell surfaces were labeled with biotin (red) under live conditions. Cells were fixed and processed for confocal microscopy and labeled with anti-gD (αgD, green) (D1 and D3), anti-gB (αgB, green) (C1 and C3), or anti-V5 (αV5) for gK (green) (A1, A3, B1, and B3). Superimpositions of red and green images for each group are shown (A3, B3, C3, and D3).
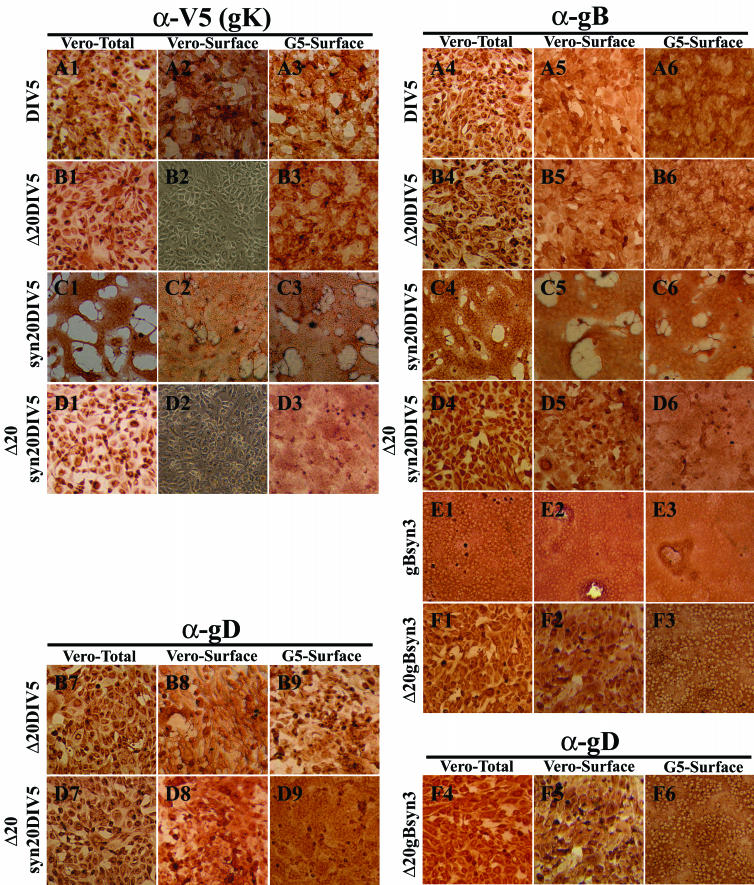
Similar results were observed by immunohistochemical detection of cell surface-expressed glycoproteins. Specifically, HSV-1 gK was not transported to infected Vero cell surfaces in the absence of the UL20 gene (Fig. 5B2 and D2). In comparison, gK was readily detected in G5 cells infected with the UL20-null viruses (compare Fig. 5B2 and D2 with B3 and D3, respectively). Furthermore, infection of G5 cells with UL20-null viruses containing the syn3 and syn1 mutations resulted in extensive cell-to-cell fusion (compare Fig. 5D2 and F2 with D3 and F3, respectively). In agreement with the confocal microscopy results, overall levels of cell surface-expressed gB and gD appeared to be similar in Vero (noncomplementing) and G5 (complementing) cells (Fig. 5). These data suggest that UL20 is required for virus-induced cell fusion caused by syncytial mutations in gK because gK cannot be expressed on cell surfaces in the absence of the UL20p. However, a similar argument cannot be made for gBsyn3-mediated cell fusion, since gB was efficiently expressed on cell surfaces in the absence of UL20p. Therefore, the role of UL20p on gB-mediated cell fusion was not readily apparent.
FIG. 5.
Cell surface immunohistochemical detection of gK (A1 to A3, B1 to B3, C1 to C3, and D1 to D3), gB (A4 to A6, B4 to B6, C4 to C6, D4 to D6, E1 to E3, and F1 to F3), or gD (B7 to B9, D7 to D9, and F4 to F6). Vero (1, 2, 4, 5, 7, and 8) or G5 (3, 6, and 9) cells were infected with DIV5 (A1 to A6), Δ20DIV5 (B1 to B9), syn20DIV5 (C1 to C6), Δ20syn20DIV5 (D1 to D9), gBsyn3 (E1 to E3), or Δ20gBsyn3 (F1 to F6) at an MOI of 10, and cells were immunohistochemically processed at 12 hpi under either live (2, 3, 5, 6, 8, and 9) or fixed and permeabilized (1, 4, and 7) conditions. α-V5, anti-V5; α-gB, anti-gB; α-gD, anti-gD.
UL20 is required for egress from infected cells.
The initial characterization of an HSV-1(F) UL20-null mutant revealed a cell type dependency for virus replication and egress (4). Electron microscopic examination of nonpermissive Vero cells infected with the HSV-1(F) UL20-null virus exhibited a drastic inhibition of intracellular virion transport. This was manifested by entrapment of enveloped virions within what appeared to resemble the perinuclear space and numerous unenveloped capsids in the adjacent cytoplasmic space (4). In contrast, PRV UL20-null enveloped virions accumulated within cytoplasmic vesicles, but similarly failed to egress from infected cells (16).
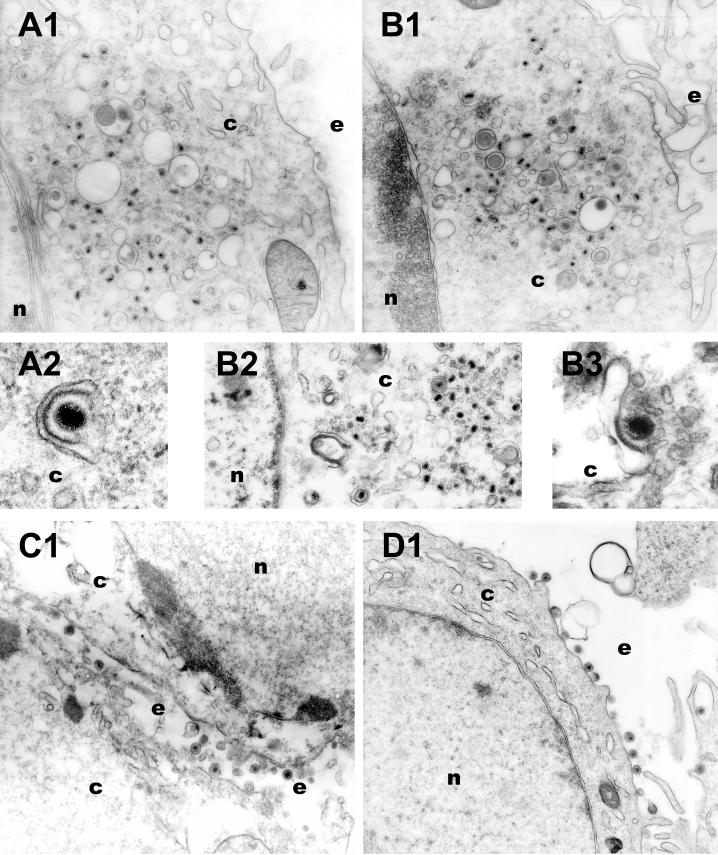
The HSV-1(F) UL20-null virus (R7225) was constructed by insertional inactivation of the UL20 gene and subsequent removal of the inserted thymidine kinase cassette (4). At the time, the presence of the adjacent UL20.5 gene was not known (30). DNA sequencing of the flanking sequences of the R7225 virus revealed that deletion of the TK cassette resulted in an in-frame fusion of the UL20.5 ORF with a gene segment coding for the carboxyl terminus of UL20 (data not shown). Therefore, we examined the effect of the UL20-specific deletion on virion morphogenesis and egress. The Δ20DIV5 and Δ20D3prtC viruses carry a deletion of the UL20 gene in the KOS genetic background. These viruses were precisely engineered to delete the UL20 gene without affecting the proximal UL20.5 gene (Fig. 1). Vero cells (Fig. 6A and B) or G5 cells (Fig. 6C and D) were infected with either Δ20DIV5 (Fig. 6A and C) or Δ20D3prtC (Fig. 6B and D) and examined by EM at 16 hpi. Consistent with the known egress defects of UL20-null viruses, there were no virions present on extracellular spaces of infected Vero cells (Fig. 6A1 and B1). In comparison, enveloped virions were readily observed on G5 cell surfaces (Fig. 6C1 and D1). In thin sections of infected Vero cells, capsids of the UL20-null viruses were observed within the cytoplasm, relatively few enveloped virions were found within cytoplasmic vesicles, and enveloped virions were not observed within the perinuclear space in the examined sections (Fig. 6A1, B1, and B2). In addition, numerous cytoplasmic capsids appeared localized proximal and in contact with cytoplasmic vesicles (Fig. 6A2 and B3). These ultrastructural morphologies are consistent with either virus failing to undergo secondary reenvelopment or, alternatively, enveloped virions within cytoplasmic vesicles undergoing de-envelopment.
FIG. 6.
Electron micrographs of Vero (A and B) and G5 (C and D) cells infected with either Δ20DIV5 (A and C) or Δ20D3prtC (B and D) virus. Confluent cell monolayers were infected at an MOI of 5, incubated at 37°C for 16 h, and prepared for transmission EM. Nuclear (n), cytoplasmic (c), and extracellular (e) spaces are marked.
Deletion of UL20 in a gB or gK syncytial genetic background results in multiple capsids within a single envelope.
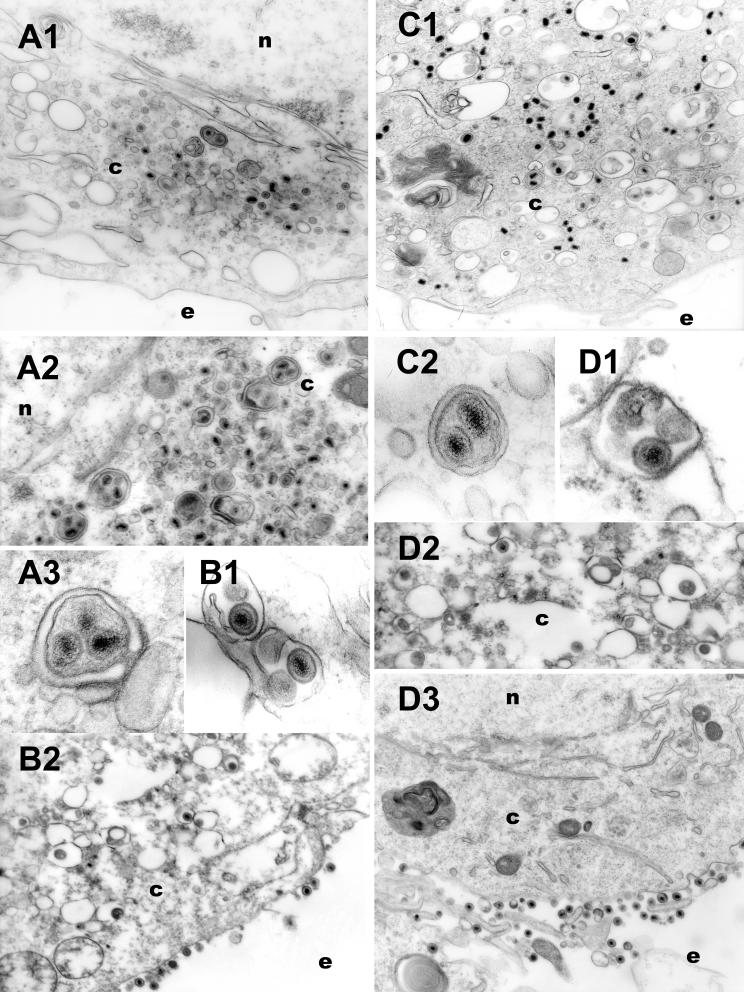
Recombinant viruses lacking the UL20 gene in the presence of gBsyn3 or gKsyn1 mutations seemed to replicate and spread more efficiently than their nonsyncytial counterparts. Therefore, the intracellular distribution of Δ20gBsyn3 and Δ20syn20DIV5 was examined via EM. Vero cells (Fig. 7A and C) or G5 cells (Fig. 7B and D) were infected with recombinant viruses Δ20syn20DIV5 (Fig. 7A and B) or Δ20gBsyn3 (Fig. 7C and D). As seen with the nonsyncytial UL20-null viruses, there were no enveloped virions visualized in extracellular spaces of Δ20gBsyn3- and Δ20syn20DIV5-infected Vero cells (Fig. 7A1 and C1) and unenveloped capsids accumulated primarily within the cytoplasm (Fig. 7A1, A2, and C1). However, multiple capsids with a single viral envelope were found within cytoplasmic vesicles (Fig. 7A2, A3, and C2). In contrast, infection of the complementing G5 cells produced fully enveloped virions containing single capsids within cytoplasmic vesicles (Fig. 7B1, D1, and D2). Furthermore, in G5 cells there were relatively few capsids in the cytoplasm (Fig. 7B2 and D2), and enveloped virions were readily visualized on extracellular surfaces (Fig. 7B2 and D3). These results suggest that the UL20p is essential for preservation of enveloped virion structure and may prevent either aberrant envelopment or fusion between enveloped virions.
FIG. 7.
Electron micrographs of Vero (A and C) and G5 (B and D) cells infected with either Δ20syn20DIV5 (A and B) or Δ20gBsyn3 (C and D) virus. Confluent cell monolayers were infected at an MOI of 5, incubated at 37°C for 16 h, and prepared for transmission EM. Nuclear (n), cytoplasmic (c), and extracellular (e) spaces are marked.
DISCUSSION
Our laboratory previously showed that the UL20 protein is required in transient expression assays for the Golgi-dependent glycosylation and cell surface expression of gK, implying a functional association between the UL20 protein and gK (10). Furthermore, the UL20 gene has been implicated in virion egress, virus-induced cell fusion, Golgi fragmentation, and Golgi-dependent glycosylation of HSV-1 viral glycoproteins (1, 3, 4, 16). In this report, we further clarify the role of the UL20 protein in virion egress and glycoprotein transport by producing and characterizing new UL20-null mutants with a precise deletion of the UL20 gene. Lack of UL20p resulted in accumulation of unenveloped capsids in the cytoplasm of infected cells, suggesting that the protein plays a role in either secondary envelopment or in preventing aberrant de-envelopment from cytoplasmic vesicles. Furthermore, lack of UL20p prevented virus-induced cell fusion caused by syncytial mutations in either gB or gK, suggesting that UL20p plays important regulatory roles in membrane fusion phenomena associated with virion morphogenesis and virus-induced cell fusion.
To investigate the role of the UL20 gene in glycoprotein transport, virion egress, and cell fusion, we generated a precise deletion of the UL20 gene without affecting the adjacent UL20.5 gene. The KOS/UL20-null virus exhibited similarities, as well as important differences to the previously constructed HSV-1(F) UL20-null virus (4). (i) Both UL20-null viruses produced small plaques on Vero cells containing on the average 8 to 30 cells at 48 hpi. (ii) All UL20-null virions failed to be transported to extracellular spaces, indicating that UL20 functions in intracellular virion transport. (iii) Although the HSV-1(F) R7225 UL20-null virions accumulated within perinuclear spaces (4), the KOS UL20-null virus accumulated capsids within the cytoplasm with a few enveloped virions found within intracytoplasmic vacuoles. (iv) All KOS UL20-null viruses, irrespective of the presence of either gB or gK syncytial mutations, failed to cause cell fusion. In contrast, the HSV-1(F) UL20-null virus produced syncytial plaques in non-Golgi fragmenting cell lines, such as 143TK− cells (4). (v) In cells infected with the UL20-null virus, glycoproteins gB and gD (Fig. 4 and 5) as well as gH and gC (data not shown) were efficiently transported to cell surfaces, whereas gK was unable to be transported to cell surfaces. In contrast, viral glycoproteins specified by the HSV-1(F) UL20-null virus were fully glycosylated (4) but lacked sialic acid residues and were present in reduced amounts on cell surfaces (3).
Role of UL20p in cytoplasmic virion morphogenesis.
The previously constructed HSV-1(F) UL20-null virus R7225 was generated by first inserting the thymidine kinase gene adjacent to the UL20 locus and subsequently deleting a portion of the UL20 gene (4). At the time of construction, it was not known that the UL20.5 gene was immediately upstream of the UL20 gene (30). We have recently determined that this construction resulted in an in-frame fusion of the carboxyl-terminal amino acids of UL20 with the amino terminus of UL20.5 (data not shown). Although the function of UL20.5 remains unknown, it has been shown to target specifically to nuclear membranes (30). Therefore, the retention of enveloped virions within the perinuclear spaces may be due to aberrant functions of the UL20-UL20.5 fusion protein at nuclear membranes. Alternatively, we considered that that the HSV-1(F) genetic background may have contributed to the observed ultrastructural phenotype exhibited by the UL20-null mutant virus R7225. However, construction of a mutant HSV-1(F) virus with a precise deletion of the UL20 gene exhibited plaque and ultrastructural phenotypes similar to those of the KOS UL20-null virus (data not shown).
The ultrastructural phenotype of the KOS UL20-null virus was similar to that of the PRV UL20-null virus in that both viruses accumulated enveloped virions in intracytoplasmic vacuoles. However, the HSV-1 (KOS) UL20-null virus seemed to produce more capsids in the cytoplasm than the PRV UL20-null virus (16). The static nature of the electron micrographs prevents any conclusive interpretation of the directionality of the morphogenetic events associated with the UL20-null defect. Therefore, accumulation of capsids in the cytoplasm of UL20-null infected Vero cells may be due either to an inability of these capsids to reenvelop following interaction with intracellular vacuolar membranes, or alternatively, cytoplasmic capsids may originate from the fusion of enveloped virions with cytoplasmic vacuoles, releasing capsids back into the cytoplasm. In support of this latter hypothesis, certain enveloped virions appeared to be in the process of de-enveloping from cytoplasmic vacuolar membranes. In this regard, the presence of UL20p may prevent aberrant fusion of virion envelopes with vacuolar membranes, implying that UL20p acts as an inhibitor of membrane fusion, presumably carried out by the viral fusogenic machinery associated with gB, gD, and gH/gL teins. Additional support for this hypothesis, is provided by the ultrastructural phenotype of UL20-null viruses specifying syncytial mutations in either gBsyn3 or gKsyn1. Ultrastructural examination of Vero cells infected with the UL20-null viruses carrying either the gBsyn3 or gKsyn1 mutations revealed the presence of virion envelopes containing multiple capsids within intracellular vacuoles. These virions may have formed either via aberrant reenvelopment of multiple capsids or by fusion of virion envelopes within cytoplasmic vacuoles. In either scenario, UL20 must play a crucial role in facilitating and maintaining a single envelope for each virion capsid. Recently, gK was shown to inhibit virus-free cell fusion produced by transient cotransfection of gB, gD gH, and gL (2). Therefore, since gK cannot be properly processed and transported in the absence of UL20p (9, 10), it is conceivable that lack of gK may be responsible for fusion among enveloped virions. Given the fact that UL20p and gK seem to be intimately associated with each other, it is also possible that a functional interaction between these two proteins may be required for prevention of aberrant fusion events.
The HSV-1 UL20 membrane protein has been associated with the integrity of the Golgi apparatus late in infection in a cell type-specific manner. Specifically, the HSV-1(F) UL20-null virus could replicate and egress from 143TK− cells but not from Vero, BHK, and Hep-2 cells (1, 3). These results indicated that 143TK− cells partially complemented the lack of UL20p in intracellular virion transport and egress from the infected cells. Lack of UL20p was also functionally associated with a concomitant alteration of late glycoprotein maturation. Specifically, in infections with the HSV-1(F) UL20-null virus, viral glycoproteins were processed in the Golgi but lacked sialic acid residues, which are added in the trans-Golgi network. Furthermore, these glycoproteins were not efficiently transported to infected cell surfaces (3). In contrast to these results, viral glycoproteins gB, gD, and gH, but not gK, were efficiently transported to cell surfaces of cells infected with the KOS UL20-null virus. These results are in agreement with those obtained with the PRV UL20-null virus, which was shown to affect only Golgi-mediated processing of gK and not any other viral glycoproteins (9, 16). Recently, we showed that overexpression of gK in the gK-transformed cell line gK9 (Vero) caused drastic inhibition of viral glycoprotein processing and transport to cell surfaces, as well as retention of enveloped virions within the perinuclear space, due to the collapse of the Golgi apparatus to the endoplasmic reticulum (13). Therefore, it is conceivable that nuclear retention of the UL20-UL20.5 fusion protein specified by the HSV-1(F) R7225 UL20-null virus may alter the structure and function of gK, which results in similar defects to those observed for gK9 cells.
Role of UL20p in virus-induced cell fusion.
Transient coexpression of gB, gD, gH, and gL is necessary and sufficient for the induction of cell-to-cell fusion (29). However, in the context of viral infections, UL20p expression is necessary for both gBsyn3- and gKsyn1-mediated cell fusion. The most plausible explanation for the inability of gKsyn1 to cause fusion in the absence of UL20 is that gKsyn1 does not get expressed on infected cell surfaces. In contrast, gB was expressed on infected cell surfaces in the absence of UL20p, suggesting that either UL20p and/or the absence of gK from cell surfaces affected the ability of gBsyn3 to cause virus-induced cell fusion. These results suggest a functional linkage and potential regulatory interactions among UL20p, gK, and gB for virus-induced cell fusion.
HSV-1 regulates membrane fusion events during different steps of its life cycle to achieve virus entry into cells, to spread from cell to cell, and to facilitate intracellular virion transport and morphogenesis. In principle, the virus-encoded membrane fusion machinery must be highly regulated as evidenced by the following: (i) wild-type viruses cause limited virus-induced cell fusion in contrast to syncytial strains, which cause extensive syncytial formation, (ii) enveloped virions do not fuse to intracellular membranes, causing aberrant de-envelopment, (iii) enveloped virions do not fuse to each other even when present in close proximity within intracellular vesicles, and (iv) intracellular compartments loaded with virus-encoded fusogenic machineries do not exhibit aberrant membrane fusion. In this regard, the highly hydrophobic, multimembrane-spanning proteins gK and UL20p may exert site-specific modulation of virus-induced membrane fusion phenomena mediated by fusogenic glycoproteins, such as gB, that are crucial for virion morphogenesis, intracellular virus transport, and egress.
Acknowledgments
We thank Olga Borkhsenious for expert technical assistance with EM.
This work was supported by the National Institute of Allergy and Infectious Diseases grant AI43000 to K.G.K. We acknowledge financial support by the LSU School of Veterinary Medicine to BIOMMED.
REFERENCES
- 1.Avitabile, E., G. S. Di, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., G. Lombardi, and G. Campadelli-Fiume. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J. Virol. 77:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avitabile, E., P. L. Ward, C. Di Lazzaro, M. R. Torrisi, B. Roizman, and G. Campadelli-Fiume. 1994. The herpes simplex virus UL20 protein compensates for the differential disruption of exocytosis of virions and viral membrane glycoproteins associated with fragmentation of the Golgi apparatus. J. Virol. 68:7397-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond, V. C., and S. Person. 1984. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology 132:368-376. [DOI] [PubMed] [Google Scholar]
- 6.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 7.Debroy, C., N. Pederson, and S. Person. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 145:36-48. [DOI] [PubMed] [Google Scholar]
- 8.Desai, P., N. A. DeLuca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietz, P., B. G. Klupp, W. Fuchs, B. Kollner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, T. P., X. Alvarez, and K. G. Kousoulas. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77:499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, T. P., V. N. Chouljenko, and K. G. Kousoulas. 1999. Functional characterization of the HveA homolog specified by African green monkey kidney cells with a herpes simplex virus expressing the green fluorescence protein. Virology 258:365-374. [DOI] [PubMed] [Google Scholar]
- 12.Foster, T. P., and K. G. Kousoulas. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 73:8457-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, T. P., G. V. Rybachuk, X. Alvarez, O. Borkhsenious, and K. G. Kousoulas. 2003. Overexpression of gK in gK-transformed cells collapses the Golgi apparatus into the endoplasmic reticulum inhibiting virion egress, glycoprotein transport, and virus-induced cell fusion. Virology 317:237-252. [DOI] [PubMed] [Google Scholar]
- 14.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 1998. Expression of the enhanced green fluorescent protein by herpes simplex virus type 1 (HSV-1) as an in vitro or in vivo marker for virus entry and replication. J. Virol. Methods 75:151-160. [DOI] [PubMed] [Google Scholar]
- 15.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 75:12431-12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchinson, L., K. Goldsmith, D. Snoddy, H. Ghosh, F. L. Graham, and D. C. Johnson. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchinson, L., and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 69:5401-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 20.Jayachandra, S., A. Baghian, and K. G. Kousoulas. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 71:5012-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacLean, C. A., S. Efstathiou, M. L. Elliott, F. E. Jamieson, and D. J. McGeoch. 1991. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72:897-906. [DOI] [PubMed] [Google Scholar]
- 23.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogue-Geile, K. L., G. T. Lee, S. K. Shapira, and P. G. Spear. 1984. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology 136:100-109. [DOI] [PubMed] [Google Scholar]
- 26.Ryechan, W. T., L. S. Morse, D. M. Knipe, and B. Roizman. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J. Virol. 29:677-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders, P. G., N. M. Wilkie, and A. J. Davison. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63:277-295. [DOI] [PubMed] [Google Scholar]
- 28.Tomishima, M. J., G. A. Smith, and L. W. Enquist. 2001. Sorting and transport of alpha herpesviruses in axons. Traffic 2:429-436. [DOI] [PubMed] [Google Scholar]
- 29.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward, P. L., B. Taddeo, N. S. Markovitz, and B. Roizman. 2000. Identification of a novel expressed open reading frame situated between genes U(L)20 and U(L)21 of the herpes simplex virus 1 genome. Virology 266:275-285. [DOI] [PubMed] [Google Scholar]