Abstract
Tospoviruses have the ability to infect plants and their insect vectors. Tomato spotted wilt virus (TSWV), the type species in the Tospovirus genus, infects its most important insect vector, Frankliniella occidentalis, the western flower thrips (WFT). However, no detrimental effects on the life cycle or cytopathological changes have been reported in the WFT after TSWV infection, and relatively few viral particles can be observed even several days after infection. We hypothesized that TSWV infection triggers an immune response in the WFT. Using subtractive cDNA libraries to probe WFT DNA macroarrays, we found that the WFT's immune system is activated by TSWV infection. The activated genes included (i) those encoding antimicrobial peptides, such as defensin and cecropin; (ii) genes involved in pathogen recognition, such as those encoding lectins; (iii) those encoding receptors that activate the innate immune response, such as Toll-3; and (iv) those encoding members of signal transduction pathways activated by Toll-like receptors, such as JNK kinase. Transcriptional upregulation of these genes after TSWV infection was confirmed by Northern analysis, and the kinetics of the immune response was measured over time. Several of the detected genes were activated at the same time that viral replication was first detected by reverse transcription-PCR. To our knowledge, this is the first report of the activation of an insect vector immune response by a plant virus. The results may lead to a better understanding of insects' immune responses against viruses and may help in the future development of novel control strategies against plant viruses, as well as human and animal viruses transmitted by insect vectors.
Tospoviruses infect hundreds of plant species, including the most important agricultural crops, commonly cause devastating damage, and represent one of the most economically important groups of plant pathogens worldwide (42). Tomato spotted wilt virus (TSWV) is the type species of the Tospovirus genus (12) and belongs to the Bunyaviridae family along with the Bunyavirus, Hantavirus, Nairovirus, and Phlebovirus genera (15). The family Bunyaviridae is made up mostly of animal viruses transmitted by arthropods, such as mosquitoes and ticks (15). Several bunyaviruses have been shown to infect and replicate in their insect vectors (6, 15, 19, 29). Tospoviruses, the only plant-infecting members of the family Bunyaviridae, also infect their insect vectors, including Frankliniella occidentalis Pergande (Thysanoptera: Thripidae), the western flower thrips (WFT). The WFT is a diminutive insect that constitutes a serious pest on its own and is considered the most important tospovirus vector given its global distribution and capacity to transmit most tospoviruses (48, 49). No detrimental effects on the life cycle or cytopathological changes have been found in the WFT after TSWV infection, and a relatively low virus titer is found in the insect body, with the exception of the salivary glands, even several days after infection (49, 52).
TSWV has four structural proteins (L, N, G1, and G2) and two nonstructural proteins (NSm and NSs). Both nonstructural proteins are detected in infected insect cells or tissues and have served as markers of virus infection in the WFT (30, 42, 47). TSWV infection in the WFT occurs after feeding on infected plants during the larval stages (49). Only WFT larvae are susceptible to TSWV infection and subsequent acquisition (uptake and ability to transmit the virus). The virus seems to invade the insect body via interaction of the viral glycoprotein(s), possibly G1, with a 50-kDa cellular receptor in the WFT midgut (32, 49). WFT adults are resistant to TSWV infection and consequently are refractory to virus acquisition owing to an unknown midgut barrier (35, 49).
Shortly after feeding on infected plants, nonstructural viral proteins (NSm and NSs) can be detected in the WFT, first in midgut epithelial cells and then in muscle cells surrounding the alimentary canal, followed by the putative ligaments joining the midgut to the salivary gland and, finally, the insect salivary glands (35, 36, 49). After infection of the salivary glands, TSWV can be inoculated into new host plants during WFT feeding by egestion of insect saliva into the plant tissue, completing the transmission cycle. TSWV infection is therefore a key determinant in successful acquisition and vectoring by the WFT (49). The TSWV-WFT relationship is defined as circulative-propagative, meaning that the virus circulates in several of the vector organs and replicates in its tissues (49).
It has long been known that insects lack an adaptive immune system, and therefore their innate immune system is solely responsible for their remarkable resistance to microbial infections (2, 21, 24). Activation of the insect innate system, like that of its mammalian counterpart, involves the recognition of microbial components by several types of cell surface receptors, including Toll-like receptors, which trigger the expression of an array of antimicrobial peptides (synthesized in the fat body), as well as initiate a variety of proteolytic cascades (2, 21).
The Drosophila response to bacterial and fungal infections is relatively well understood at the molecular level (21, 24). However, the immune response of insects to viral infections is poorly understood. It has been reported that cells of Helicoverpa zea larvae infected with Autographa californica polyhedrovirus are encapsulated by hemocytes and subsequently cleared, demonstrating that the insect cellular immune response is effective against viral pathogens (51). Only two virus-induced molecules, pherokine-2 and -3, were identified in Drosophila after intrathoracic injection of the Drosophila C virus (39). A lipase showing antiviral activity against Bombyx mori polyhedrovirus was purified from the digestive juice of B. mori larvae and detected into the insect midgut, the site of virus entry (37). In addition, it has been shown that Flock house virus is both an initiator and a target of RNA silencing in Drosophila host cells (27) and that several members of the baculovirus family of insect viruses are able to induce apoptosis during infection, resulting in limited viral replication (10).
We hypothesized that TSWV triggers an immune response in WFT tissues. To test this hypothesis, we constructed subtractive cDNA libraries from infected versus noninfected WFT larvae and used them to probe WFT DNA macroarrays. The data presented here indicate that TSWV elicits an immune response in the WFT and that this immune response involves, for example, the transcriptional upregulation of antimicrobial proteins (such as defensins) and other immune system-related proteins (such as Toll, Toll-like receptors, lectins, and complement-like protein). These findings may lead to a better understanding of insects' immune responses against viruses and, one hopes, a better understanding of virus-vector interactions and may help in the future development of novel antiviral strategies for insect-transmitted plant and animal viruses.
MATERIALS AND METHODS
Virus isolates and insect populations.
The TSWV BR-01 Brazilian original isolate was maintained in Datura stramonium plants under greenhouse conditions. A WFT population (from Vargem Bonita, Brasília-DF, Brazil), provided by T. Nagata (Universidade Católica, Brasília-DF, Brazil), was maintained in a growth chamber (Percival E-30B) at 26°C with a 16-h light-8-h dark photoperiod inside plastic cups and fed with fresh green bean pods. Bean pods containing first-instar larvae (about 12 h old) were transferred to healthy or TSWV-infected D. stramonium plants, kept in a separate growth chamber, for the construction of subtracted libraries. TSWV infection of WFT larvae was obtained via feeding on TSWV-infected D. stramonium plants. The time of virus exposure was calculated in terms of the amount of time that the WFT larvae were left to feed on D. stramonium plants. A Tobacco mosaic virus (TMV) isolate collected at Vargem Bonita in Nicotiana tabacum leaves was diagnosed by enzyme-linked immunosorbent assay and also used to infect D. stramonium plants. TMV does not infect the WFT and was used as an additional negative control.
Subtractive cDNA libraries.
Subtractive cDNA libraries were prepared as described previously (7, 8, 25, 28, 41), with some modifications. Total RNA was purified with a Total RNA Purification kit (Invitrogen) by using 200 mg of WFT larvae at the first and second instars (12 and 96 h old). Samples from the first instar were used as driver RNA (noninfected), and samples from the second instar were used as tester RNA (TSWV infected or from the WFT fed on TMV-infected plants). The time delay (from 12 to 96 h) was determined empirically, since this is the first study of the WFT immune system, and was used in an attempt to ensure the detection of eventual changes in gene expression. In addition, 96-h-old larvae are morphologically distinct from 12-h-old larvae (second instar versus first instar), which facilitates their visual separation.
Samples of WFT poly(A)+ RNA were obtained with oligo(dT)25-Dynabeads (Dynal). Independent driver RNA samples were obtained from (i) non-TSWV-infected WFT fed on healthy D. stramonium, (ii) non-TSWV-infected WFT fed on TMV-infected D. stramonium, and (iii) TSWV-infected WFT fed on TSWV-infected D. stramonium (infections were checked by reverse transcription [RT]-PCR; data not shown). Dynabead complexes were washed twice in RT buffer (50 mM Tris-HCl [pH 8], 75 mM KCl, 3 mM MgCl2). First-strand cDNA synthesis [with poly(A)+ RNA from noninfected WFT] was performed directly on the magnetic beads with oligo(dT)25-Dynabeads as primers and the cDNA Synthesis System Plus kit (Amersham). After first-strand cDNA synthesis, poly(A)+ RNA was removed by washing samples (three times) with EDTA at 95°C for 3 min each time. First-strand cDNA-Dynabead complexes were captured with a magnet and resuspended in hybridization buffer (120 mM NaH2PO4 [pH 6.8], 820 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS]). Poly(A)+ RNA (tester) isolated from 96-h-old TSWV-infected WFT larvae was then added (or RNA from WFT fed on TMV-infected plants or from WFT fed on healthy plants). The mixture was denatured for 3 min at 95°C and incubated at 65°C for 24 h in a rotary hybridization oven (Affymetrix). The poly(A)+ RNA-cDNA-Dynabead complexes were captured by magnetic separation and separated from the supernatant containing nonhybridized poly(A)+ RNA. The supernatants were subjected to a second hybridization reaction with the recycled cDNA-Dynabeads. Magnetic beads were recycled by two washes in buffer A (0.15 M LiCl, 0.01 M Tris-HCl [pH 8], 1 mM EDTA). This hybridization step was repeated three more times. The poly(A)+ RNA population present in the last supernatant was captured with fresh oligo(dT)25-Dynabeads and eluted three times in water at 55°C. First- and second-strand cDNA syntheses, with the final poly(A)+ RNA samples, were carried out with the cDNA Synthesis System Plus kit (Amersham).
In this way, four main subtracted libraries were obtained. Library 1 (noninfected; negative control) consisted of tester RNA from 96-h-old noninfected WFT added to cDNA derived from driver RNA from 12-h-old noninfected WFT (fed on healthy D. stramonium). Library 2 (noninfected but exposed to TMV; negative control) consisted of tester RNA from 96-h-old noninfected WFT added to cDNA derived from driver RNA from 12-h-old noninfected WFT (fed on TMV-infected D. stramonium). Library 3 (TSWV infected) consisted of tester RNA from 96-h-old TSWV-infected WFT added to cDNA from driver RNA from 12-h-old noninfected WFT (fed on TSWV-infected D. stramonium). Library 4 (double negative control) consisted of an additional negative control library was also used in preliminary experiments and was made with both tester and driver RNAs collected from 12-h-old WFT larvae.
DNA macroarrays.
WFT DNA macroarrays were prepared with plates containing 72 by 96 wells and positively charged nylon membranes (Amersham) measuring 7 by 11 cm with Q-Bot (Genetix) by following the manufacturer's instructions. In each membrane, 6,912 WFT PCR products were spotted in the central area of the nylon membranes. Ten membranes were used in three independent experiments, with a total of 69,120 clones screened in each experiment. To manufacture the DNA macroarrays, two independent WFT cDNA lambda libraries were combined: (i) a larval WFT cDNA lambda library obtained previously with the Lambda-ZAP II kit (Stratagene) as described before (31) and (ii) a newly obtained adult WFT cDNA lambda library (Lambda-ZAP II kit). The resultant combined lambda library was used to obtain a plasmid library with the ExAssist interference-resistant helper phage kit (Stratagene) by mass excision in accordance with the manufacturer's instructions. The plasmid library was used to transform Escherichia coli strain SOLR (Stratagene), which was plated in master plates (20 by 100 mm). Bacterial cultures were produced in plates with 72 by 96 wells, from the master plates, with the Q-Bot colony-picking function. PCR products were obtained in plates consisting of 72 by 96 wells by robotically dipping bacterial cultures in 25 μl of PCR solution (20 pmol each of the T3 and T7 primers, 4 mM MgCl2, 1.5 mM deoxynucleoside triphosphates, 1.25 μl of dimethyl sulfoxide, 0.1 U of Taq polymerase [Invitrogen]) with PCR System 9700 (Applied Biosystems). The following cycling parameters were used: 95°C for 5 min; 30 cycles of 95°C for 1 min, 55°C for 2 min, and 68°C for 4 min; and 68°C for 10 min. Twelve microliters of the PCR products obtained was then spotted onto each nylon membrane by using the Q-Bot grinding function. Membranes were incubated in denaturation buffer (0.5 N NaOH, 1.5 M NaCl) for 2 h, neutralized for 30 min in 0.5 M Tris-Cl (pH 7.5), rinsed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 2 h, UV cross-linked for 15 min at 120,000 μJ/cm2 (Stratalinker; Stratagene), and hybridized with [α-32P]rCTP-labeled riboprobes prepared from the subtractive libraries obtained. On each membrane, a WFT α-actin PCR product, from a clone obtained with α-actin primers designed from Drosophila sequences, was spotted for normalization (preliminary experiments showed no α-actin activation; data not shown). Each plate (72 by 96 wells) received a letter code (AA to AZ), and individual wells received four-digit numbers.
Hybridization.
Riboprobes were prepared from the four subtractive libraries obtained and were synthesized by random priming with T7 polymerase (Riboprobe System; Promega) in accordance with the manufacturer's instructions. The riboprobes obtained showed an activity of 1 × 107 to 6 × 107 cpm/μg of DNA. Membranes were prehybridized in 4× SSC-10× Denhardt's solution-50% formamide-0.1% SDS-50 mM Na2HPO4 (pH 7.2)-1 mM EDTA-100 μg of sheared herring sperm DNA (Invitrogen) per ml at 42°C for 2 h. Riboprobe solution was added at 5 × 106 cpm/ml, and the mixture was incubated overnight at 42°C. Membranes were washed twice at 65°C for 30 min each time in 2× SSC-0.1% SDS. Four different riboprobes were hybridized to each group of 10 membranes in each experiment, for a total of 40 membranes per experiment. Riboprobes were made from libraries 1, 2, 3, and 4 (described above).
Image acquisition and analysis.
After hybridization, membranes were exposed to a Storm 860 PhosphoImager screen (Amersham Pharmacia) for up to 12 h and scanned with Storm 860 software. Data were obtained as a list of integrated intensity values and array positions for each spot and saved in a spreadsheet (Microsoft Excel) to which the intensity values of each clone was linked. The intensity of the signal was first corrected by the average signals of three blank spots on the membrane where no PCR samples were applied. The corrected hybridization signals were normalized with an α-actin PCR product spotted on each corner of each membrane as described previously (25). The arrays were not used for absolute quantitative analysis, the relative hybridization levels were used as an indicator of differential expression. Only those clones that generated hybridization levels twofold or higher when probed with library 3 compared to negative control library 1 in all three independent experiments were selected for sequencing.
Sequence analysis.
Sequencing reactions were performed with the T7 and T3 universal primers, with plasmids purified from the bacterial culture plates, after clone selection. Sequences were identified by a BLASTn search at the National Center for Biotechnology Information homepage (http://www.ncbi.nlm.nih.gov). Genes were considered homologues when queries showed more than 60% nucleotide identity to known genes.
Northern analysis.
Five micrograms of each RNA sample collected from WFT fed on TSWV-infected D. stramonium plants for 0, 2, 4, 6, 12, 24, 96, or 240 h (time of virus exposure) was loaded into a 0.8% agarose formaldehyde gel, transferred overnight to a nylon membrane (Amersham), baked at 75°C for 1 h, and hybridized overnight at 65°C, after 3 h of preincubation in hybridization buffer, as described above. Riboprobes were synthesized from selected clones with the Riboprobe System (Promega), as described above, with 50 mCi of [α-32P]rCTP (3,000 Ci/mM). Membranes were washed three times as described above and analyzed with a Storm 860 PhosphorImager (Amersham Pharmacia).
RT-PCR.
Viral infections of both plants and insects were assayed by RT-PCR, as described before (30), with primers specific to the viral complementary M (medium) RNA containing the NSm sequence (5′ GCCATTGTTCTCCGGAAGATTCAG 3′ and 5′ CAACCCTAGTTAATTTCGTTGGTG 3′). For the WFT, about 100 dissected guts were used as the RNA source and RNA was extracted with a QIAshredder and RNeasy (Qiagen). Plant RNA samples were extracted with RNeasy (Qiagen). After feeding on TSWV-infected plants for the indicated time, WFT larvae were starved for 24 h prior to RT-PCR to avoid the detection of virus particles simply carried in WFT guts.
RESULTS
The WFT immune system is activated by TSWV.
Analysis of WFT DNA macroarrays hybridized to riboprobes made from WFT subtracted cDNA libraries showed that numerous genes homologous to genes previously identified with reported or putative immune system-related functions were consistently activated in three independent experiments (Table 1). A total of 51 genes consistently yielded a twofold or greater change in intensity in all of the experiments performed with TSWV-infected subtracted libraries. BLAST analysis showed that 27 of those genes are homologous to genes with known or putative immune functions (Table 1).
TABLE 1.
WFT immune system-related genes activated after TSWV infection
| Function,a WFT clone | Homologous function | Accession no. | FCb | % Identityc |
|---|---|---|---|---|
| Recognition/phagocytosis | ||||
| AA-0027 | Aedes aegypti type C lectin | AF466606 | 2.1 | 66 |
| AA-5991 | Drosophila macroglobulin, complement related | AE003619 | 2.2 | 72 |
| AG-0230 | Anopheles gambiae lectin | Z69982 | 2.7 | 69 |
| AH-0393 | Anopheles gambiae complement-like TEP1 | AF291654 | 2.5 | 67 |
| AJ-5617 | Drosophila lectin galC1 | NM057852 | 2.7 | 68 |
| Signal transduction | ||||
| AA-0013 | Drosophila MyD88 | NM136635 | 2.5 | 62 |
| AD-2420 | Drosophila Toll-3 | AF247765 | 3.2 | 61 |
| AF-6197 | Drosophila Toll like | S76155 | 2.4 | 69 |
| Toll pathway | ||||
| AJ-4782 | Aedes aegypti Relish | AF498105 | 2.7 | 75 |
| AJ-5388 | Drosophila 18w | NG000232 | 2.7 | 65 |
| AE-6899 | Drosophila Cactus | NM165152 | 2.1 | 61 |
| AD-0089 | Drosophila 18 wheeler | AC005286 | 3.2 | 69 |
| AI-3008 | Drosophila Spatzle | U05850 | 2.2 | 76 |
| AB-4991 | Drosophila Cactus kinase Ikkα | AF128403 | 2.1 | 71 |
| JNK pathway | ||||
| AD-1293 | Drosophila Bsk | NM164901 | 2.2 | 61 |
| AJ-4019 | Aedes albopictus Jnk | AF515780 | 2.4 | 68 |
| Antimicrobial | ||||
| AA-3277 | Aedes albopictus cecropin B | AF145803 | 2.3 | 74 |
| AC-0769 | Anopheles gambiae defensin | X93562 | 2.8 | 67 |
| AJ-4909 | Aedes aegypti defensin A | S82860 | 2.3 | 66 |
| AJ-5003 | Drosophila lysozyme C | NM080130 | 3.1 | 72 |
| AF-4571 | Aedes albopictus defensin D | AY005473 | 2.2 | 61 |
| Protease | ||||
| AI-2766 | Aedes aegypti trypsin | U28367 | 2.9 | 65 |
| AJ-5761 | Drosophila gamma Try | U04853 | 3.2 | 69 |
| Dipeptidyl-peptidase, AD-3384 | Drosophila dipep-peptidase IV | AE003533 | 2.7 | 77 |
| Storage protein, AD-4515 | Drosophila larval serum ptn1β | U63556 | 2.5 | 67 |
| Cytokine, AB-1952 | Drosophila activin | AE003582 | 2.9 | 79 |
| Immunoglobulin G superfamily, AI-6001 | Hyalophora cecropia hemolin | M63398 | 3.1 | 72 |
Putative or reported immune system-related function (references cited in the text). The genes encoding the proteins shown were consistently activated in all three independent experiments performed and showed homology to a known gene with a reported or putative immune-related function, with an average fold change of ≥2 in TSWV-infected versus noninfected WFT subtracted libraries.
FC, average fold change in signal intensity as calculated by Storm 860 software from three independent experiments.
Percent identity as calculated by BLASTn software.
When negative control subtracted libraries (noninfected WFT and WFT fed on TMV-infected plants) were used to make the riboprobes, only about 30 genes were detected (data not shown). These 30 genes did not show homology to genes with reported immune functions. These genes included those encoding transcription factors (11 genes) and structural proteins (7 genes); those involved in DNA repair (2 genes) and replication (1 gene), protein synthesis (8 genes), and metabolism (7 genes); those encoding kinases (4 genes), neuropeptides (3 genes), and insect hormones (4 genes); and those that showed no significant homology to genes previously described in the GenBank database (24 genes; data not shown). These genes probably represent the WFT gene pool upregulated during larval development since they were detected when RNA from a first-instar sample (driver) was extracted from the RNA pool of the second-instar sample (tester), resulting in an mRNA population that represents those genes whose expression is higher in second-instar WFT than in first-instar WFT.
RNA from WFT fed on TMV-infected plants was used to make an additional negative control subtracted library (library 2) because TMV does not infect the WFT; however, whether the presence of noninfecting viral particles would trigger a nonspecific immune response was unknown. This library also did not result in the detection of genes with known immune functions, and the same genes described above were detected (with putative developmental functions) (data not shown).
In addition, when RNA samples from the first instar were extracted from RNA samples also from the first instar (library 4, described above, as an additional negative control), no signals were detected, indicating that the subtraction method was efficient (data not shown).
Table 1 shows that WFT genes activated after TSWV infection included homologues of genes with reported or putative immune functions. These genes can be divided into the following categories: (i) those encoding proteins putatively involved in recognition and phagocytosis of insect-invading microorganisms, such as lectins (13, 20, 24, 45), Jnk kinase (33), and complement-like proteins (13, 24); (ii) those encoding Toll-like surface receptors (23), responsible for the activation of signal transduction pathways that trigger the innate immune response, such as Toll-3, which has been reported to participate in double-stranded RNA recognition in mammalian cells (1), and MyD88, which is required in the immune response against fungi and bacteria (44); (iii) those encoding proteins that participate in the subsequent Toll signaling cascade, such as Relish, 18w, Ikkα, Spatzle, and Cactus (23, 26, 38); (iv) those encoding antimicrobial peptides, such as defensins A and D, cecropin B, and lysozyme C, a family whose members have been reported in immune responses against viruses, bacteria, and fungi (21); (v) those encoding cytokines that have potential roles in activating humoral responses, such as activin (9, 24, 40); (vi) those encoding members of the immunoglobulin superfamily of proteins, such as hemolin (43); (vii) those encoding storage proteins, such as larval serum protein 1β, a protein similar to arylphorins and arylphorin receptors, whose specific immune function is unknown (5), and (viii) those encoding proteases, whose general role in insects' innate response, as well as many other functions, has also been reported (9, 21, 24, 51).
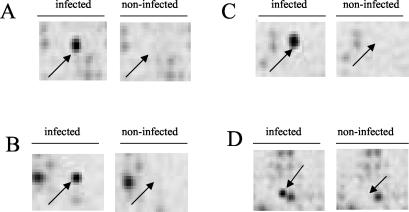
Figure 1 shows DNA macroarray hybridization results in selected areas of nylon membranes with some of the selected genes (those encoding defensin D, hemolin, Ikk kinase, and complement-like TEP1).
FIG. 1.
Selected areas of WFT DNA macroarrays after hybridization with riboprobes derived from noninfected versus TSWV-infected subtracted cDNA libraries. Selected regions of nylon membranes show the detection of defensin D (A), hemolin (B), Ikk kinase (C), and complement-like TEP-1 (D) (arrows).
Northern analyses confirm activation of immune system-related genes in the WFT after TSWV infection.
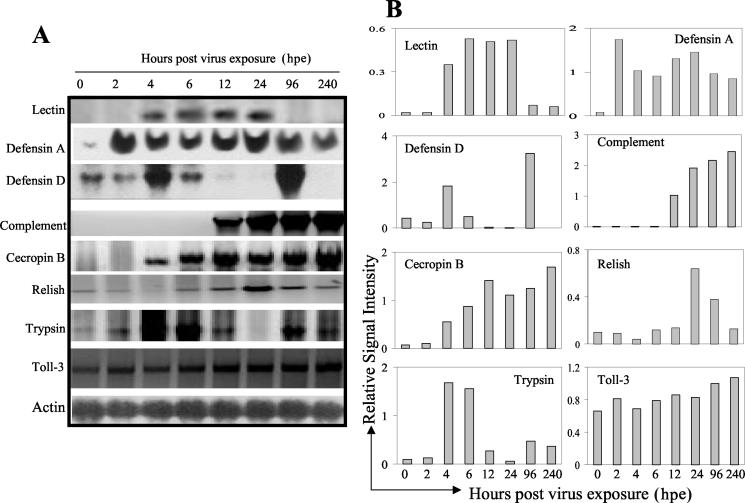
Figure 2 shows a Northern blotting analysis of a series of selected genes, detected previously by DNA macroarray hybridization (Fig. 1), in a time course experiment (0, 2, 4, 6, 12, 24, 96, and 240 h postexposure [hpe] to virus). Samples from 0 to 24 hpe represent larvae at the first-instar stage, samples at 96 hpe represent larvae at the second-instar stage, and samples at 240 hpe represent WFT adults (Fig. 2A and B). The lectin homologue showed activation at 4 hpe, with a peak at 12 hpe and subsequent downregulation after that point. The defensin A homologue showed upregulation at 2 hpe, followed by a slight decrease, a subsequent increase in expression at 12 hpe, and a decrease at the second instar (96 hpe). The defensin D homologue showed upregulation at 4 hpe, followed by a decrease and peak at the second instar (96 hpe). The macroglobulin complement-related homologue showed increasing activation starting at 12 hpe. The cecropin B homologue showed increasing activation starting at 4 hpe. The Relish homologue showed a very subtle activation, with a relatively small peak at 24 hpe. The trypsin homologue showed a peak at 6 hpe, a decrease, and a new peak at the second instar (96 hpe), and the Toll-3 homologue also showed a very slight increase in activation, with a peak only at the adult stage (240 hpe) (Fig. 2A and B). The α-actin probe was used as a negative and loading control (Fig. 2A), and the α-actin signal was used to normalize the data showed in Fig. 2B.
FIG. 2.
(A) Northern analysis of activated immune-related WFT genes after TSWV infection. Riboprobes are indicated at the left. Samples were collected from 0 to 240 hpe, meaning time feeding on TSWV-infected plants. Samples from 0 to 24 hpe represent insects at the first-instar larval stage, those from 96 hpe represent the second-instar larval stage, and those from 240 hpe represent the adult stage. Actin was used as a negative control for activation and as a loading control. (B) Graphic representation of the data showed in panel A. Relative signal intensity is the signal intensity of each gene in the Northern blot normalized to that of the corresponding actin signal, measured with a Storm 860 PhosphoImager.
RT-PCR analysis indicates that TSWV infection coincided with the activation of at least four of the detected genes.
Figure 3 shows that TSWV viral complementary M RNA, containing the NSm sequence, was detected by RT-PCR in dissected WFT guts at 4 hpe, suggesting that TSWV replication in and infection of WFT gut tissues took place at least 4 h after feeding on TSWV-infected plants. Four of the detected genes shown in Fig. 2 (those for lectin, defensin D, cecropin B, and trypsin) showed activation at 4 hpe (Fig. 2A and B), coinciding with the time of initial TSWV detection by RT-PCR (Fig. 3). The WFT midgut is the primary site of TSWV infection (49).
FIG. 3.
RT-PCR of dissected WFT guts with primers specific to the TSWV complementary M RNA. Samples were collected from 0 to 240 hpe, meaning time feeding on TSWV-infected plants.
DISCUSSION
We investigated the activation of immune system-related genes in F. occidentalis, the WFT, after infection with TSWV. TSWV is an important plant virus worldwide, and the WFT is its most important insect vector. Several investigators have reported TSWV infection and replication in the WFT (49). The data presented here (Table 1 and Fig. 1 and 2) indicate that TSWV infection triggers an immune response in the WFT, which may be responsible, for example, for the low virus titer found in the insect body, and in the hemolymph in particular (49). It is possible that reported differences in vector competence, observed in different WFT populations and different thrips species (36, 49), resulted, at least in part, from differentiated immune responses to TSWV infection. Other hypotheses that arose from this study are as follows: (i) the resistance of WFT adults to TSWV (49) could be the result of a potent immune response at that stage; (ii) the resistance of mosquitoes to bunyavirus superinfection (6) could be the result of immune system activation in mosquitoes; and (iii) the immune response of the WFT may vary in different organs.
Of the 51 genes activated after TSWV infection (Table 1), 24 were homologous to genes with no reported immune function, such as encoding DNA repair-related proteins, transcription factors, cuticle and structural proteins, DNA replication factors, and neuropeptides and insect hormones, and some showed no significant homology to genes previously described in the GenBank database (data not shown). The detection of the transcriptional upregulation of these genes probably represents WFT larval development from the first instar to the second instar, since the same genes, and similar genes in the same categories, were also detected when the negative control riboprobes were used (data not shown), and similar genes have also been detected in mosquito larval-development studies (25). The detection of these genes was possible because the driver RNA sample was collected from 12-h-old WFT larvae and the tester RNA sample was collected from 96-h-old larvae. Furthermore, the simple presence of noninfecting virus particles in the insect gut, in this case TMV, was not enough to trigger any immune response (data not shown).
Other studies, involving genomic-scale approaches to evaluate immune responses against viral infections, have used mostly cDNA microarrays (11, 16, 18, 34, 46, 54). The method used here, which combined subtracted cDNA libraries and DNA macroarrays, is simple, practical, and less expensive than microarrays. However, it is labor intensive and less sensitive in terms of signal intensity, the number of signals detected, and of course the number of genes that can be analyzed at once, which explains the relatively low number of genes detected in this study (around 50) compared to those detected with microarrays (often hundreds). In addition, because of the library construction strategy used here, detection of downregulated genes was not possible. However, the method could be particularly applicable to other insect-transmitted plant viruses that also infect their vectors, such as rhabdoviruses (22), and to human and animal mosquito-transmitted bunyaviruses (6, 15).
The Northern analysis shown in Fig. 2A and B corroborated the use of subtractive libraries with DNA macroarrays and illustrates a time frame similar to those previously reported for fungal and bacterial infections in Drosophila (13, 17). It has been found in Drosophila that defensins, Toll pathway genes, and complement genes can show sustained activation even at 96 h postinfection (13). Here, most genes showed higher expression even at the adult stage (Fig. 2A, 240 hpe).
TSWV replication in and infection of WFT guts, at least with respect to the presence of viral complementary M RNA containing the NSm sequence, was detected by RT-PCR at 4 h after feeding on TSWV-infected plants or at 4 hpe (Fig. 3). TSWV infection after only 3 h of feeding on infected plants has been reported previously (35). Also, it has been shown that the WFT midgut is the primary site of TSWV infection (47, 48, 49). Since WFT individuals were starved for 24 h before RNA sampling, the results indicate the detection of TSWV RNA inside the WFT gut tissues, not the detection of viral particles simply carried in the lumen of the WFT gut.
Figures 2 and 3 show a correlation between the detection of viral cRNA, which indicates virus replication and infection, and the activation of four of the selected defense genes, i.e., those that encode lectin, defensin D, cecropin B, and trypsin (Fig. 2B and 3). However, the activation of another four of the selected genes did not show a correlation with the detection of viral cRNA (those that encode complement, Relish, Toll-3, and defensin A). Apparent early defensin A activation may be related to the recognition of virus particles and/or products before any viral replication event takes place. Apparent late activation of complement, Relish, and Toll-3 may be related to insect immune system regulation and/or may be unnecessary before viral products reach a certain level in WFT tissues.
Defensins have been reported in antiviral immune responses in mammalian cells against adenovirus (4), rhinovirus (14), and retrovirus (50, 53). We are currently investigating if the defensins detected in this study have any anti-TSWV activity.
Another plant pathogen, the bacterium Erwinia carotovora, has been shown to activate the immune system of Drosophila melanogaster, its eventual insect vector (3). To our knowledge, this is the first report of the activation of an insect vector immune system by a virus and may help in the future development of novel antiviral strategies. An example is the expression of insect antiviral proteins, such as defensins, in transgenic plants.
Acknowledgments
We thank J. Figueiredo, M. Oliveira, and C. Castro (Universidade de Brasília) for technical assistance; T. Nagata (Universidade Católica, Brasília-DF, Brazil) for kindly providing a WFT population; Genaro R. Paiva (CENARGEN/EMBRAPA, Brasilia-DF, Brazil), Thomas German (University of Wisconsin—Madison), and Diane Ullman (University of California at Davis) for invaluable discussions; and D. Dickey for critical reading of the manuscript.
This research was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPQ, Centro Brasileiro-Argentino de Biotecnologia-CBAB and by the Decanato de Pesquisa e Pós-Graduação-DPP, Universidade de Brasília (R.B.M.).
REFERENCES
- 1.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 2.Barillas-Mury, C., B. Wizel, and Y. S. Han. 2000. Mosquito immune responses and malaria transmission: lessons from insect model systems and implications for vertebrate innate immunity and vaccine development. Insect Biochem. Mol. Biol. 30:429-442. [DOI] [PubMed] [Google Scholar]
- 3.Basset, A., R. S. Khush, A. Braun, L. Gardan, F. Boccard, J. A. Hoffmann, and B. Lemaitre. 2000. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. USA 97:3376-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastian, A., and H. Schafer. 2001. Human alpha-defensin 1 (HNP-1) inhibits adenoviral infection in vitro. Regul. Pept. 101:157-161. [DOI] [PubMed] [Google Scholar]
- 5.Beresford, P. J., J. M. Basinski-Gray, J. K. C. Chiu, J. S. Chadwick, and W. P. Aston. 1997. Characterization of hemolytic and cytotoxic gallysins: a relationship with arylphorins. Dev. Comp. Immunol. 21:253-266. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, D. H. L., and B. J. Beaty. 1986. Interference—immunity of mosquitoes to bunyavirus superinfection. Symp. Zool. Soc. Lond. 56:95-115. [Google Scholar]
- 7.Bonaldo, M. F., G. Lennon, and M. Bento Soares. 1996. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 6:791-806. [DOI] [PubMed] [Google Scholar]
- 8.Cecchini, E., P. J. Dominy, C. Geri, K. Kaiser, J. Sentry, and J. J. Milner. 1993. Identification of genes up-regulated in dedifferentiating Nicotiana glauca pith tissue, using an improved method for constructing a subtractive cDNA library. Nucleic Acids Res. 21:5742-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christophides, G. K., E. Zdobnov, C. Barillas-Mury, E. Birney, S. Blandin, C. Blass, P. T. Brey, F. H. Collins, A. Danielli, G. Dimopoulos, C. Hetru, N. Hoa, J. A. Hoffmann, S. M. Kanzok, I. Letunic, E. A. Levashina, T. G. Loukeris, G. Lycett, S. Meister, K. Michel, H. M. Muller, M. A. Osta, S. M. Paskewitz, J. M. Reichhart, A. Rzhetsky, L. Troxler, K. D. Vernick, D. Vlachou, J. Volz, C. von Mering, J. N. Xu, L. B. Zheng, P. Bork, and F. C. Kafatos. 2002. Immunity-related genes and gene families in Anopheles gambiae. Science 298:159-165. [DOI] [PubMed] [Google Scholar]
- 10.Clark, T. E., and R. J. Clem. 2003. Insect defenses against virus infection: the role of apoptosis. Int. Rev. Immunol. 5-6:401-424. [DOI] [PubMed] [Google Scholar]
- 11.Cuadras, M. A., D. A. Feigelstock, S. An, and H. B. Greenberg. 2002. Gene expression pattern in Caco-2 cells following rotavirus infection. J. Virol. 76:4467-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Avila, A. C., P. de Haan, R. Kormelink, R. O. Resende, R. W. Goldbach, and D. Peters. 1993. Classification of tospoviruses based on phylogeny of nucleoprotein gene sequences. J. Gen. Virol. 74:153-157. [DOI] [PubMed] [Google Scholar]
- 13.De Gregorio, E., P. T. Spellman, G. M. Rubin, and B. Lemaitre. 2001. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA 98:12590-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duits, L. A., P. H. Nibbering, E. van Strijen, J. B. Vos, S. P. Mannesse-Lazeroms, M. A. van Sterkenburg, and P. S. Hiemstra. 2003. Rhinovirus increases human beta-defensin-2 and -3 mRNA expression in cultured bronchial epithelial cells. FEMS Immunol. Med. Microbiol. 38:59-64. [DOI] [PubMed] [Google Scholar]
- 15.Elliot, R. M. 1996. The Bunyaviridae. Plenum Press, New York, N.Y.
- 16.Geiss, G. K., R. E. Bumgarner, M. C. An, M. B. Agy, A. B. van't Wout, E. Hammersmark, V. S. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2000. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarray. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 17.Gillespie, J. P., A. M. Bailey, B. Cobb, and A. Vilcinskas. 2000. Fungi as elicitors of insect immune responses. Arch. Insect Biochem. Physiol. 44:49-68. [DOI] [PubMed] [Google Scholar]
- 18.Glynne, R. J., and S. R. Watson. 2001. The immune system and gene expression microarrays. J. Pathol. 195:20-30. [DOI] [PubMed] [Google Scholar]
- 19.Hacker, J. K., and J. L. Hardy. 1997. Adsorptive endocytosis of California encephalitis virus into mosquito and mammalian cells: a role for G1. Virology 235:40-47. [DOI] [PubMed] [Google Scholar]
- 20.Hartshorn, K. L., U. Holmskov, S. Hansen, P. Zhang, J. Meshi, T. Mogues, M. R. White, and E. C. Crouch. 2002. Distinctive anti-influenza properties of recombinant collectin 43. Biochem. J. 366:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, Jr., and R. A. B. Ezekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 284:1313-1318. [DOI] [PubMed] [Google Scholar]
- 22.Hogenhout, S. A., M. G. Redinbaugh, and E.-D. Ammar. 2003. Plant and animal rhabdovirus host range: a bug's view. Trends Microbiol. 11:264-271. [DOI] [PubMed] [Google Scholar]
- 23.Imler, J. L., and J. A. Hoffmann. 2002. Toll receptors in Drosophila: a family of molecules regulating development and immunity. Curr. Top. Microbiol. Immunol. 270:63-79. [DOI] [PubMed] [Google Scholar]
- 24.Irving, P., L. Troxler, T. S. Heuer, M. Belvin, C. Kopczynski, J. M. Reichhart, J. A. Hoffmann, and C. Hetru. 2001. A genome-wide analysis of immune responses in Drosophila. Proc. Natl. Acad. Sci. USA 98:15119-15124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krebs, K. C., K. L. Brzoza, and Q. Lan. 2002. Use of subtracted libraries and macroarray to isolate developmentally specific genes from mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 32:1757-1767. [DOI] [PubMed] [Google Scholar]
- 26.Lemaitre, B., E. Nicolas, L. Michaut, J. M. Reichhart, and J. A. Hoffmann. 1996. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86:973-983. [DOI] [PubMed] [Google Scholar]
- 27.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 28.Lombardi, M. P., M. J. B. van den Hoff, J. M. Ruijter, M. Luijerink, A. A. Buffing, M. W. Markman, A. F. M. Moorman, and R. H. L. Deprez. 2003. Expression analysis of subtractively enriched libraries (EASEL): a widely applicable approach to the identification of differentially expressed genes. J. Biochem. Biophys. Methods 57:17-33. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig, G. V., B. A. Israel, B. M. Christensen, T. M. Yuill, and K. T. Schultz. 1991. Role of La Crosse virus glycoproteins in attachment of virus to host cells. Virology 181:564-571. [DOI] [PubMed] [Google Scholar]
- 30.Mason, G., P. Roggero, and L. Tavella. 2003. Detection of Tomato spotted wilt virus in its vector Frankliniella occidentalis by reverse transcription-polymerase chain reaction. J. Virol. Methods 109:69-73. [DOI] [PubMed] [Google Scholar]
- 31.Medeiros, R. B., L. Rasochova, and T. L. German. 2000. Simplified, rapid method for identification and cloning of putative viral receptors or virus-binding proteins via the far-Western screening of a cDNA expression library with purified virus particles. J. Virol. Methods 86:155-166. [DOI] [PubMed] [Google Scholar]
- 32.Medeiros, R. B., D. E. Ullman, J. L. Sherwood, and T. L. German. 2000. Immunoprecipitation of a 50 kDa protein: a candidate receptor component for tomato spotted wilt tospovirus in its thrips vector, Frankliniella occidentalis. Virus Res. 67:109-118. [DOI] [PubMed] [Google Scholar]
- 33.Mizutani, T., M. Kobayashi, Y. Eshita, K. Shirato, T. Kimura, Y. Ako, H. Miyoshi, T. Takasaki, I. Kurane, H. Kariwa, T. Umemura, and I. Takashima. 2003. Involvement of the Jnk-like protein of the Aedes albopictus mosquito cell line, C6/36, in phagocytosis, endocytosis and infection of West Nile virus. Insect Mol. Biol. 12:491-499. [DOI] [PubMed] [Google Scholar]
- 34.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagata, T., A. K. Inoue-Nagata, H. M. Smid, R. Goldbach, and D. Peters. 1999. Tissue tropism related to vector competence of Frankliniella occidentalis for Tomato spotted wilt tospovirus. J. Gen. Virol. 80:507-515. [DOI] [PubMed] [Google Scholar]
- 36.Nagata, T., A. K. Inoue-Nagata, J. van Lent, R. Goldbach, and D. Peters. 2002. Factors determining vector competence and specificity for transmission of Tomato spotted wilt virus. J. Gen. Virol. 83:663-671. [DOI] [PubMed] [Google Scholar]
- 37.Ponnuvel, K. M., H. Nakazawa, S. Furukawa, A. Asaoka, J. Ishibashi, H. Tanaka, and M. Yamakawa. 2003. A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus. J. Virol. 77:10725-10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutschmann, S., A. C. Jung, R. Zhou, N. Silverman, J. A. Hoffmann, and D. Ferrandon. 2000. Role of Drosophila IKK gamma in a Toll-independent antibacterial immune response. Nat. Immunol. 1:342-347. [DOI] [PubMed] [Google Scholar]
- 39.Sabatier, L., E. Jouanguy, C. Dostert, D. Zachary, J.-L. Dimarcq, P. Bulet, and J.-L. Imler. 2003. Pherokine-2 and -3, two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur. J. Biochem. 270:3398-3407. [DOI] [PubMed] [Google Scholar]
- 40.Schneider, D., and M. Shahabuddin. 2000. Malaria parasite development in a Drosophila model. Science 288:2376-2379. [DOI] [PubMed] [Google Scholar]
- 41.Sharma, P., A. Lonneborg, and P. Stougaard. 1993. PCR-based construction of subtractive cDNA library using magnetic beads. BioTechniques 15:610-611. [PubMed] [Google Scholar]
- 42.Sherwood, J. L., T. L. German, A. E. Whitfield, J. W. Moyer, and D. E. Ullman. 2001. Tospoviruses, p. 1034-1040. In O. C. Maloy and T. D. Murray (ed.), Encyclopedia of plant pathology. John Wiley & Sons, Inc., New York, N.Y.
- 43.Sun, S. C., I. Lindstrom, H. G. Boman, I. Faye, and O. Schmidt. 1990. Hemolin: an insect-immune protein belonging to the immunoglobulin superfamily. Science 250:1729-1732. [DOI] [PubMed] [Google Scholar]
- 44.Tauszig-Delamasure, S., H. Bilak, M. Capovilla, J. A. Hoffmann, and J. L. Imler. 2002. Drosophila MyD88 is required for the response to fungal and gram-positive bacterial infections. Nat. Immunol. 3:91-97. [DOI] [PubMed] [Google Scholar]
- 45.Thielens, N. M., P. Tacnet-Delorme, and G. J. Arlaud. 2002. Interaction of C1q and mannan-binding lectin with viruses. Immunobiology 205:563-574. [DOI] [PubMed] [Google Scholar]
- 46.Tian, B., Y. Zhang, B. A. Luxon, R. P. Garofalo, A. Casola, M. Sinha, and A. R. Brasier. 2002. Identification of NF-κB-dependent gene networks in respiratory syncytial virus-infected cells. J. Virol. 76:6800-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ullman, D. E., T. L. German, J. L. Sherwood, D. M. Wescot, and F. A. Cantone. 1993. Tospovirus replication in insect vector cells: immunocytochemical evidence that the nonstructural protein encoded by the S RNA of tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathology 83:456-463. [Google Scholar]
- 48.Ullman, D. E., J. L. Sherwood, and T. L. German. 1997. Thrips as vectors of plant pathogens, p. 539-565. In Lewis, T. (ed.), Thrips as crop pests. CAB-International, Wallingford, United Kingdom.
- 49.Ullman, D. E., R. B. Medeiros, L. R. Campbell, A. E. Whitfield, J. L. Sherwood, and T. L. German. 2002. Thrips as vectors of tospoviruses. Adv. Bot. Res. 36:113-140. [Google Scholar]
- 50.Wang, W., A. M. Cole, T. Hong, A. J. Waring, and R. I. Lehrer. 2003. Retrocyclin, an antiretroviral theta-defensin, is a lectin. J. Immunol. 170:4708-4716. [DOI] [PubMed] [Google Scholar]
- 51.Washburn, J. O., B. A. Kirkpatrick, and L. E. Volkman. 1996. Insect protection against viruses. Nature 383:767-769. [Google Scholar]
- 52.Wijkamp, I., R. Goldbach, and D. Peters. 1999. 1996. Propagation of Tomato spotted wilt virus in Frankliniella occidentalis does neither result in pathological effects nor in transovarial passage of the virus. Entomol. Exp. Appl. 81:285-292. [Google Scholar]
- 53.Zhang, L., W. Yu, T. He, J. Yu, R. E. Caffrey, E. A. Dalmasso, S. Fu, T. Pham, J. Mei, J. J. Ho, W. Zhang, P. Lopez, and D. D. Ho. 2002. Contribution of human α-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science 298:995-1000. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., B. A. Luxon, A. Casola, R. P. Garofalo, M. Jamaluddin, and A. R. Brasier. 2001. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J. Virol. 75:9044-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]