Abstract
Studying signal transduction in skin-resident T cells (sr-T cells) can be limited by the small size of clinical biopsies. Here, we isolated sr-T cells from clinical samples and analyzed signaling protein complexes by multiplex immunoprecipitation detected by flow cytometry (mIP-FCM). In samples from two independent donors, antigenic stimulation induced signaling proteins to join shared complexes that were observed in 7 pair-wise combinations among 5 proteins. This demonstrates that sr-T cells isolated from small clinical samples provide sufficient material for mIP-FCM-based analysis of signaling-induced protein complexes. We propose that this strategy may be useful for gaining improved mechanistic insight of sr-T cell signal transduction associated with dermatological disease.
Background
Skin-resident (sr) T cells are vital for immunological surveillance, but can be pathogenic in dermatologic diseases such as alopecia areata, psoriasis, and allergic contact dermatitis, [1, 2]. Differential activity of signaling pathways may play a causal role in determining these healthy vs disease outcomes. However, protein complexes important for signal transduction in sr-T cells are difficult to study, largely due to limited cell numbers that can be obtained from normal or diseased skin biopsies.
Clark et al. [3] described a method for isolating sr-T cells by culturing skin biopsies on a porous substrate for three weeks. During this time, fibroblasts exit the skin, attach to the substrate, release chemokines, and attract sr-T cells that migrate from the skin sample into the media. This procedure for sr-T cell isolation has subsequently been referred to as the “crawl-out” method [4]. While this represents a significant advance in the ability to isolate intact cutaneous T lymphocytes, the number of T cells obtained can still be limiting (~1–5 × 105 cells per 4-mm punch biopsy) for standard analyses of protein complexes, such as those involving immunoprecipitation and/or Western blotting.
Multiplex immunoprecipitation detected by flow cytometry (mIP-FCM) is a novel biochemical technique that measures the extent to which proteins bind each other and are expressed in shared complexes [5]. Because low cell numbers have been shown to be sufficient for IP-FCM analysis [6–8], we hypothesized that the multiplex approach could be applied to examine signaling-induced protein complexes from sr-T cells that were isolated from small clinical samples.
Question Addressed
Can the crawl-out method yield sufficient primary human sr-T cells for protein complex analysis using mIP-FCM?
Experimental Design
Identification of antibodies, and description of the methods used for T cell harvesting and stimulation are described in supplementary online materials.
Multiplex Immunoprecipitation Detected by Flow Cytometry (mIP-FCM)
The technical details and protocols of singleplex IP-FCM [6–8] and multiplex IP-FCM [5] have been previously published. Briefly, after incubation on ice for 20 min, lysates were centrifuged at 21,000 × G for 10 min in order to discard nuclei. Multiplex immunoprecipitation was performed by adding to each post-nuclear lysate a mix of five specific antibody-conjugated Luminex beads. Following overnight incubation at 4C, beads were washed on a magnetic plate washer (Biorad) and incubated with PE-conjugated probe antibodies, or with biotinylated probe antibodies followed by streptavidin-PE. After washing, beads were resuspended in IP-FCM buffer (150 mM NaCl, 50 mM Tris-HCl, 1% bovine serum albumin (Fraction-V, Sigma-Aldrich)) and analyzed on a Bioplex 200 multiplex bead reader.
Statistical Analysis
Duplicate mIP-FCM samples were acquired for all data associated with each donor. The median fluorescence intensity (MFI) of corresponding duplicate data was averaged, and pair-wise protein complex data was expressed as the ratio of stimulated/unstimulated to indicate fold-change. To statistically analyze data from both donors together, two-tailed Student’s t-test was used to determine whether significant changes in protein complexes occurred in stimulated vs unstimulated conditions. Results are reported at both p < 0.05 level, and the Bonferroni-adjusted level p < 0.005 (based on 10 pair-wise comparisons).
Results
The crawl-out method of cutaneous T cell isolation yielded on average 1.7 ± 0.2 × 105 cells from the Cytomatrix supports (mean ± SEM). Cellular flow cytometry showed that greater than 90% of isolated cells were CD3+ (data not shown), with approximately 71% of T cells being CD4+ and 22% CD8+ (illustrated for one donor, Fig 1A). Furthermore, approximately 90% of T cells were clearly CLA+CCR4+ (Fig 1B). These data are similar to previous reports using the crawl-out method [3, 4], indicating that isolation of primary human sr-T cells was achieved as expected.
Figure 1. The crawl-out method yields sr-T cells suitable for mIP-FCM.
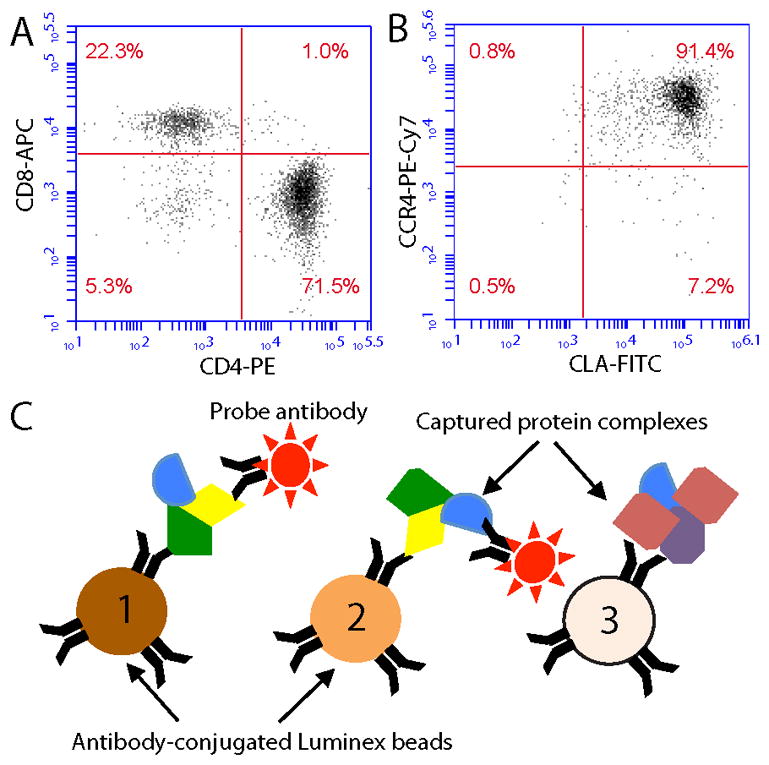
A) Representative FACS analysis of CD4 and CD8 expression on skin-resident (sr) CD3+ T cells isolated via the crawl-out method. B) FACS analysis of CLA and CCR4 surface expression on these CD3+ sr-T cells. C) Schematic diagram of multiplex IP-FCM (mIP-FCM). Each of several different Luminex bead color-classes is coupled to a unique immunoprecipitating Ab. Beads are incubated with lysates so that immunoprecipitated proteins and their co-associated binding partners are captured on the beads. Probe Abs then detect co-associated proteins on the beads.
Multiplex IP-FCM captures target proteins on Luminex microspheres, and then detects co-associated proteins using fluorophore-conjugated probe antibodies (Fig 1C). This allows flow cytometry-based semi-quantitative analysis of proteins bound to each other in shared complexes. We performed mIP-FCM on lysates of freshly-harvested sr-T cells that were subjected to either unstimulated negative-control conditions, or 5 minutes of anti-CD3 + anti-CD28 antigenic stimulus. Analysis focused on 5 members of the proximal T cell antigen receptor (TCR)/CD3 signaling pathway: TCR, Linker of activated T cells (LAT), Zeta-associated protein 70 (ZAP70), Phospholipase C gamma (PLCg), and Phosphoinositide 3-kinase (PI3K). These proteins were chosen because they are known to inducibly associate in shared complexes and mediate signal transduction upon antigenic stimulation (Fig 2A) [9]. Thy1 glycoprotein was also analyzed, its lack of association in these signaling complexes serving as a specificity control for protein complex formation. The results from two individual donors analyzed by mIP-FCM are summarized in Fig 2B, with data expressed as fold-change in stimulated vs. unstimulated conditions. The donors and experiments were each independent, with samples acquired and analyses performed on different days. We observed that protein complexes from both donors displayed similar induction in response to stimulation (Donor 1, Donor 2, Fig. 2B). For unified analysis of data from both donors, their corresponding fold-changes were averaged, and raw values of stimulated vs. unstimulated conditions were compared by Student’s t-test (Overall, Fig. 2B). We observed 7 protein pairs whose quantities in shared complexes increased in response to antigenic stimulation by ≥ 1.4-fold on average (Overall, Fig. 2B). Defining significance as p < 0.05, all seven increases in signaling complexes were significant (red + orange, Fig. 2B), while using a more stringent Bonferroni-adjusted cut-off p < 0.005, five of the increases remained significant (red, Fig. 2B). Overall, these results show that mIP-FCM can be applied to measure changes in protein complexes using sr-T cells obtained via the crawl-out method from small clinical skin samples.
Figure 2. Protein complex analysis from primary human sr-T cells using mIP-FCM.
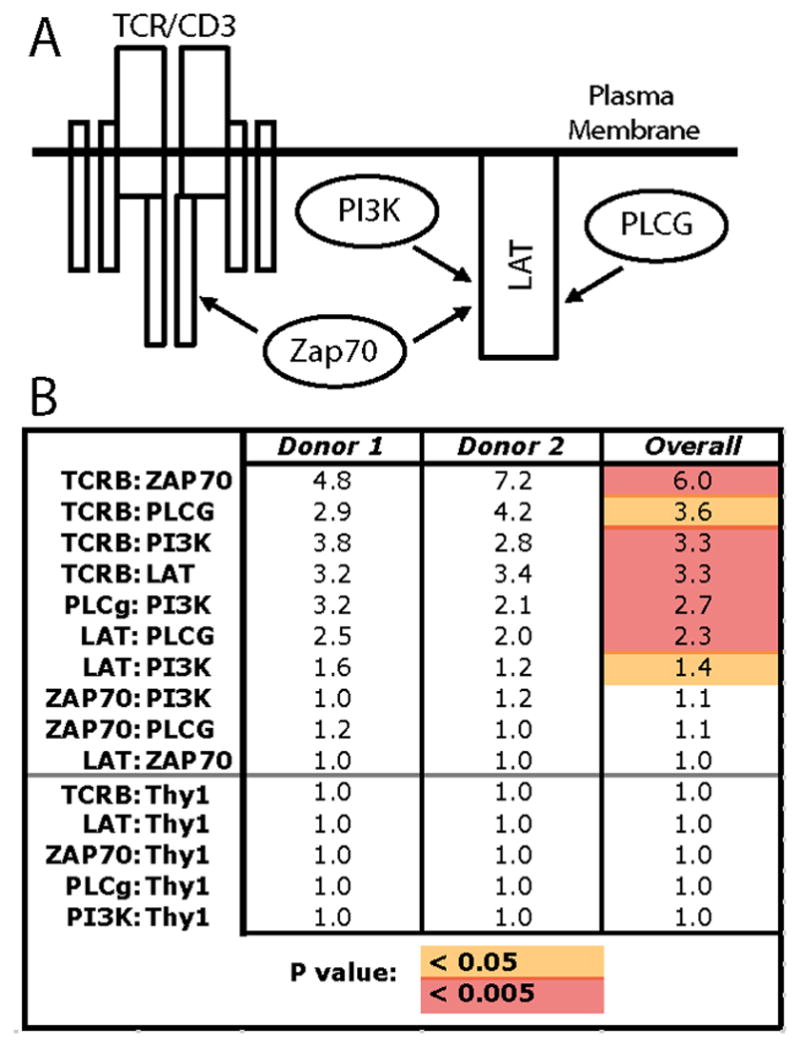
A) Target proteins are known to form shared complexes that mediate signal transduction upon T cell antigenic stimulation. B) Skin-resident T cells from 2 separate individuals were isolated via the crawl-out method, and then either left unstimulated, or stimulated with anti-CD3 + anti-CD28 for 5 minutes. Following lysis, mIP-FCM was was performed. Numbers represent the fold-change of protein pairs observed in shared complexes in stimulated vs unstimulated conditions. For unified analysis of data from both donors, their corresponding fold-changes were averaged, and raw values of stimulated vs. unstimulated conditions were compared by Student’s t-test (Overall; orange, p < 0.05; red, p < 0.005).
Conclusions
In this study, we stimulated sr-T cells isolated from human skin and used mIP-FCM to detect increases in signaling protein complexes. This type of analysis would not be feasible using traditional methods such as IP-Western blotting, because of the small amount of biological material available. Our data suggest that mIP-FCM could be a useful tool for analyzing protein complexes from small clinical biopsies, enabling improved study of the molecular machinery of T cell signaling in T cell-mediated dermatologic disease.
Supplementary Material
Acknowledgments
We thank Rachael Clark, Thomas Kupper, and Jessica Teague (Brigham and Women’s Hospital, Harvard University) for generously sharing protocols for the crawl-method. This work was funded by National Institutes of Health grant 1R01GM103841-01A1, a generous philanthropic gift for research directed toward Alopecia Areata by Mr. Bernard Fineman (AGS), and Mayo Foundation (AGS, DG).
Footnotes
Author Contributions
SEPS, SCN, MRP, DG, and AGS proposed and designed experiments, and/or interpreted data; SEPS and TRD performed experiments; SEPS and AGS wrote the manuscript.
Conflict of Interest
The authors declare no conflicts of interest.
References
- 1.Clark RA, Chong B, Mirchandani N, et al. The vast majority of CLA+ T cells are resident in normal skin. J Immunol. 2006;176(7):4431–9. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- 2.Kupper TS, Fuhlbrigge RC. Immune surveillance in the skin: mechanisms and clinical consequences. Nat Rev Immunol. 2004;4(3):211–22. doi: 10.1038/nri1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark RA, Chong BF, Mirchandani N, et al. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J Invest Dermatol. 2006;126(5):1059–70. doi: 10.1038/sj.jid.5700199. [DOI] [PubMed] [Google Scholar]
- 4.Poot J, Thewissen M, Booi D, et al. Characterization of skin-resident T cells using a crawl-out method and flow cytometry. Exp Dermatol. 2013;22(8):554–5. doi: 10.1111/exd.12191. [DOI] [PubMed] [Google Scholar]
- 5.Bida AT, Gil D, Schrum AG. Multiplex IP-FCM (immunoprecipitation-flow cytometry): Principles and guidelines for assessing physiologic protein-protein interactions in multiprotein complexes. Methods. 2011;56(2):154–60. doi: 10.1016/j.ymeth.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrum AG, Gil D, Dopfer EP, et al. High-sensitivity detection and quantitative analysis of native protein-protein interactions and multiprotein complexes by flow cytometry. Sci STKE. 2007;2007(389):pl2. doi: 10.1126/stke.3892007pl2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis TR, Schrum AG. IP-FCM: immunoprecipitation detected by flow cytometry. J Vis Exp. 2010:46. doi: 10.3791/2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SE, Bida AT, Davis TR, et al. IP-FCM Measures Physiologic Protein-Protein Interactions Modulated by Signal Transduction and Small-Molecule Drug Inhibition. PLoS One. 2012;7(9):e45722. doi: 10.1371/journal.pone.0045722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


