Abstract
Hexanucleotide repeat expansions in chromosome 9 open reading frame 72 (C9orf72) have recently been linked to frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS), and may be the most common genetic cause of both neurodegenerative diseases. Genetic variants at TMEM106B influence risk for the most common neuropathological subtype of FTLD, characterized by inclusions of TAR DNA binding protein of 43kDa (FTLD-TDP). Previous reports have shown that TMEM106B is a genetic modifier of FTLD-TDP caused by progranulin (GRN) mutations, with the major (risk) allele of rs1990622 associating with earlier age at onset of disease. Here we report that rs1990622 genotype affects age at death in a single-site discovery cohort of FTLD patients with C9orf72 expansions (n=14), with the major allele correlated with later age at death (p=0.024). We replicate this modifier effect in a 30-site international neuropathological cohort of FTLD-TDP patients with C9orf72 expansions (n=75), again finding that the major allele associates with later age at death (p=0.016), as well as later age at onset (p=0.019). In contrast, TMEM106B genotype does not affect age at onset or death in 241 FTLD-TDP cases negative for GRN mutations or C9orf72 expansions. Thus, TMEM106B is a genetic modifier of FTLD with C9orf72 expansions. Intriguingly, the genotype that confers increased risk for developing FTLD-TDP (major, or T, allele of rs1990622) is associated with later age at onset and death in C9orf72 expansion carriers, providing an example of sign epistasis in human neurodegenerative disease.
Keywords: TMEM106B, C9orf72, frontotemporal dementia, frontotemporal lobar degeneration, amyotrophic lateral sclerosis, genetic modifier
INTRODUCTION
Frontotemporal lobar degeneration (FTLD) is the second most common dementia in individuals under 65 years of age [30]. The most common neuropathological subtype is frontotemporal lobar degeneration with TAR DNA-binding protein of 43kDa (TDP-43) inclusions (FTLD-TDP) [30]. We previously reported the minimally characterized gene, TMEM106B, as a risk factor for FTLD-TDP by genome-wide association study (GWAS) [38], and this association has been verified independently [12,39]. In our GWAS, three SNPs reached genome-wide significance for association with FTLD-TDP [38]; all are located within a 36kb haplotype block that contains TMEM106B and no other genes. The major alleles of all three SNPs are associated with increased risk of FTLD-TDP (p=1.08×10−11, odds ratio=1.64 for major allele of rs1990622, the top GWAS SNP) [38].
Several studies have begun to elucidate the role TMEM106B plays in FTLD-TDP. TMEM106B levels have been shown to be increased in FTLD-TDP brains [5,38], and risk-associated alleles resulting in amino acid variation in the TMEM106B protein have been reported to result in higher steady-state levels of TMEM106B through slower protein degradation [26]. In addition, the major allele of rs1990622 has been associated with reduced plasma progranulin (PGRN) levels in both healthy individuals and in individuals with FTLD-TDP caused by mutations in GRN, the gene encoding progranulin [9,12]. Mutations in GRN are a major cause of familial FTLD-TDP [14], and are thought to cause disease via haploinsufficiency of the progranulin protein [14,31]. Interestingly, among GRN mutation carriers with FTLD (GRN(+) FTLD), TMEM106B rs1990622 major alleles have been reported to associate with earlier age at disease onset [9]. Experiments in cell culture systems have also demonstrated that TMEM106B and PGRN co-localize in several cell types, including neurons, and that over-expression of TMEM106B alters intra- and extracellular levels of PGRN [3,5,26]. Therefore, increased expression of TMEM106B may confer risk for FTLD-TDP by altering PGRN levels.
While GRN mutations account for ~5% of clinical FTLD cases [14], and other rarer, monogenic causes of FTLD are known (including mutations in MAPT, CHMP2B and VCP) [17,33,41], a substantial proportion of familial cases were until recently of unknown cause. This changed in late 2011 when two groups reported that hexanucleotide repeat expansions in the C9orf72 gene are perhaps the most common cause of familial FTLD, familial amyotrophic lateral sclerosis (ALS), and familial FTLD with motor neuron disease (FTLD-MND) [11,28]. Although these mutations display an autosomal dominant mode of inheritance, 3–6% of apparently sporadic cases of FTLD and ALS harbor C9orf72 expansions as well, which may be explained by genetic anticipation, de novo mutation, or incomplete penetrance [11,28].
The function(s) of C9orf72 and its role in disease are currently areas of ongoing research [10], with evidence for both loss-of-function [8,11,15,28] and gain-of-toxic-function [1,13,25] mechanisms. At a neuropathological level, C9orf72 expansion positive FTLD (C9orf72(+) FTLD) and ALS (C9orf72(+) ALS) cases exhibit TDP-43 pathology reminiscent of GRN(+) FTLD, as well as mutation-negative ALS and FTLD, although C9orf72(+) FTLD and ALS cases show unique pathological features as well [2,34,35].
Here, we assess whether TMEM106B risk genotypes exert a genetic modifier effect in C9orf72(+) FTLD and ALS, GRN(+) FTLD, and FTLD cases without either mutation. We also investigate whether these genotypes are associated with disease status in C9orf72(+) FTLD and with plasma progranulin levels in C9orf72(+) expansion carriers.
METHODS
Patient cohorts
FTLD and ALS cases with C9orf72 expansions of greater than 30 hexanucleotide repeats were identified from among cases in the Integrated Neurodegenerative Disease Database at the University of Pennsylvania (UPenn) to form a discovery cohort [37,44]. Patients were initially seen at the UPenn Frontotemporal Degeneration Center (FTDC), Amyotrophic Lateral Sclerosis Center (ALSC), or Alzheimer’s Disease Center (ADC); all were collected with Institutional Review Board Approval. In addition to having a C9orf72 expansion, the criteria for selection of FTLD cases was a pathological diagnosis of FTLD-TDP (n=10) or a clinical diagnosis of FTLD or FTLD-MND (n=19), according to published criteria [16,22–24,27,36]. C9orf72(+) ALS cases (n=55) all met El Escorial-revised criteria [4]. Twenty of the 55 ALS cases had autopsy confirmation of ALS pathology. For both FTLD and ALS cases, only probands were selected. In situations where patients exhibited both dementia and motor neuron disease (MND), cases were assigned to FTLD-MND if the initial presentation was cognitive and to ALS if the initial presentation was MND. All C9orf72(+) FTLD and C9orf72(+) ALS cases meeting these criteria were included without bias for familial-vs.-apparently-sporadic patterns of inheritance, and without prior knowledge of TMEM106B genotype.
The C9orf72(+) FTLD discovery cohort is 93.5% white (6.5% unknown ethnicity) and 54.8% male. The C9orf72(+) ALS cohort is 87.2% white, 5.6% black, 3.5% Latino, and 3.7% unknown ethnicity with 59.8% males. Age at onset and age at death were collected, but both were not available on all subjects (e.g. no age at death for living subjects, and sometimes no known age at onset for autopsy cases), therefore the numbers of cases from each cohort vary depending on the data needed for analysis. For the discovery cohort, age at onset was defined as the age at initial complaint, based on review of medical records.
The previously published and publicly available FTLD-TDP GWAS from the International Collaboration for Frontotemporal Lobar Degeneration was used as a replication cohort [38]. As previously described [38], all cases of this postmortem cohort were self-described as White, of European ancestry. In addition, samples were screened by principle components analysis of genomewide genotyping data, and at >200 ancestry informative markers, to reduce effects of population stratification. Only those cases with >90% inferred CEU (based on HapMap CEU population of Utah residents with ancestry from Northern and Western Europe) ancestry were included in the original GWAS [38], from which all cases of the current replication cohort are derived.
A subset of the FTLD-TDP cases were known from the original study to have a pathogenic GRN mutation (n=116) and are used here as a comparison group [7,38]. The majority of cases lacking a GRN or VCP mutation (n=321) were screened for C9orf72 expansions either by the contributing site or by UPenn, using published methods [11,28]. 80 FTLD-TDP cases with C9orf72 expansions were identified from 30 clinical sites that agreed to collaborate on this project (see Acknowledgement section for a full listing of clinical sites). Of the 80 cases, 5 UPenn cases overlapped with the UPenn discovery cohort and were removed, leaving 75 C9orf72 expansion cases for analysis in the replication cohort. In addition, 241 cases were formally tested for (and found negative for) C9orf72 expansions, and these were used as the mutation-negative FTLD-TDP cohort. We note that there were additional C9orf72(+) FTLD-TDP cases in the GWAS, but only those cases from sites agreeing to collaborate on this study (constituting >80% of the total FTLD-TDP GWAS C9orf72(+) cases) are included here.
For the replication cohort, age at onset and age at death were provided by the contributing clinical site.
Genotyping
DNA from UPenn cases, extracted from blood or brain samples as previously described [38], was tested for rs1990622 genotype using one of two methods: TaqMan chemistry-based allelic discrimination assays as previously described [5,38], or a custom Sequenom MassArray genotyping panel that includes PCR and extension primers for rs1990622. PCR and extension primer sequences for the Sequenom panel are available on request. Both genotyping methods were compared and found to be concordant (data not shown) [37].
Plasma progranulin measurement
Plasma samples were collected from UPenn ALS and FTLD discovery cohort patients, aliquotted, and stored at −80°C as previously described [6]. Progranulin levels were measured using a commercially available sandwich ELISA (Human progranulin ELISA kit, AdipoGen), according to manufacturer instructions.
Statistical analyses
Linear regression analyses evaluating the association of TMEM106B genotype with age at death or age at disease onset were performed in R, with or without covariates as described in the text. Two-tailed p-values are reported for the discovery cohort, and one-tailed p-values are reported for the FTLD-TDP GWAS replication cohort, since the expected directionality was known. For the combined dataset, survival analyses (Kaplan-Meier method) were also performed in Prism, and two-tailed p-values from the log-rank test for trend are reported.
Where indicated, codominant, major-allele-dominant, and minor-allele dominant models of genetic effect were investigated.
In addition, we tested for association between TMEM106B genotype and disease for genetically-defined subsets of FTLD (C9orf72(+) FTLD, GRN(+) FTLD, or individuals without C9orf72 expansions or GRN mutations). Chi-square statistics were calculated for rs1990622 using the FTLD-TDP GWAS cases and controls [38].
For plasma progranulin analyses, Kruskal-Wallis tests were used to compare plasma progranulin measures among carriers of different TMEM106B genotypes under a codominant model, and Mann-Whitney tests were used to compare different TMEM106B genotypes under major-allele-dominant and minor-allele dominant models. In addition, multivariate linear regressions predicting plasma progranulin levels from TMEM106B genotype were used to adjust for sex, age, duration of disease, or clinical manifestation as described in the text.
R-scripts for analyses are available upon request.
RESULTS
TMEM106B genotype at rs1990622 influences age at death in a discovery cohort of C9orf72(+) FTLD
TMEM106B genotype has been shown to demonstrate a genetic modifier effect in FTLD-TDP caused by autosomal dominant mutations in the progranulin gene (GRN) [9]. We therefore asked whether genetic variation at TMEM106B influences age at death or age at onset in C9orf72(+) FTLD or ALS disease cases. We assumed a codominant model for these initial analyses.
In C9orf72(+) FTLD (n=14), age at death was significantly correlated with TMEM106B genotype at rs1990622, the SNP previously found in our GWAS to associate most strongly with FTLD-TDP risk (p=0.024, Table 1). Adjusting for sex and presence/absence of co-existing MND did not affect this association. Moreover, the direction of association was surprising; specifically, the major allele of rs1990622 (C) was associated with later age at death in C9orf72(+) FTLD. In our GWAS, the major allele of rs1990622 was found to be associated with increased risk for the development of FTLD.
Table 1. TMEM106B genotype affects age at death in C9orf72 expansion carriers with FTLD or FTLD-TDP in a discovery cohort.
Linear regressions were used to evaluate the effect of TMEM106B genotype at rs1990622 on the age at death or age at onset in C9orf72 expansion carriers from a discovery cohort. In individuals who presented with clinical FTLD or FTLD-TDP, rs1990622 genotype was significantly associated with age at death in both univariate models and models adjusting for age and presence/absence of motor neuron disease (MND). In individuals who presented with ALS, rs1990622 genotype was not significantly associated with age at death, with a trend towards association with age at onset. Asterisks denote significance.
| Disease | Outcome | Predictors | Beta (rs1990622, each major allele) | R2 for model | P-value (rs1990622) |
|---|---|---|---|---|---|
| FTLD and FTLD-TDP | Age at Death (n=14) | rs1990622 | +6.278 | 0.303 | 0.024 * |
| rs1990622, Sex, MND | +5.297 | 0.393 | 0.049 * | ||
| Age at Onset (n=26) | rs1990622 | n.s. | |||
| rs1990622, Sex, MND | n.s. | ||||
| ALS | Age at Death (n=39) | rs1990622 | n.s. | ||
| rs1990622, Sex, FTD | n.s. | ||||
| Age at Onset (n=47) | rs1990622 | −4.264 | 0.044 | 0.085 n.s. | |
| rs1990622, Sex, FTD | −4.900 | 0.075 | 0.048 * |
In contrast, rs1990622 genotype did not affect age at death in C9orf72(+) ALS (n=39, Table 1). In this discovery cohort, rs1990622 genotype did not affect age at onset for C9orf72 expansion carriers who presented with either ALS (n=47) or FTLD (n=26). However, a statistically significant association emerged when we performed a multivariate analysis controlling for gender and presence of FTD in the clinical ALS cases, with the major allele associating with earlier age at onset (n=47, Table 1).
TMEM106B genotype at rs1990622 influences age at onset and age at death in a replication cohort of C9orf72(+) FTLD
We sought to replicate the genetic modifier effect of TMEM106B in C9orf72(+) FTLD in an independent cohort of patients. Since the majority of cases from our GWAS had been screened for the presence of C9orf72 expansions, these cases provided an ideal replication cohort to evaluate the effect of TMEM106B rs1990622 genotype on age at death in C9orf72(+) FTLD for three key reasons. First, since the FTLD-TDP GWAS predated the discovery of C9orf72 expansions as a cause of FTLD, this large, international cohort was unbiased in enrollment with respect to C9orf72 status. Second, all cases were neuropathologically confirmed to have FTLD-TDP, ensuring neuropathological homogeneity. Third, because all cases had undergone genome-wide genotyping and filtering for effects from population stratification, we could be certain that effects from cryptic familial relationships or population stratification would be minimal.
As shown in Table 2, rs1990622 genotype was again correlated with age at death in this cohort (n=75), in both univariate analyses (p=0.016) and linear regression models adjusting for sex and the presence or absence of MND (p=0.019). Moreover, in this larger replication cohort, rs1990622 genotype was also correlated with age at onset (n=68 with age at onset data, p=0.019 for univariate analyses and p=0.032 for multivariate analyses adjusting for sex and presence or absence of MND). Consistent with the results from our discovery cohort, the major allele (T) of rs1990622 was associated with later age at death, as well as later age at onset. Indeed, patients showed later disease onset and later death by more than three years for each additional major allele at rs1990622 carried.
Table 2. TMEM106B genotype affects age at death and age at onset in C9orf72 expansion carriers in a multi-site FTLD-TDP replication cohort.
Linear regressions were used to evaluate the effect of TMEM106B genotype at rs1990622 on the age at death or age at onset in C9orf72(+) FTLD from a multi-site replication cohort of FTLD-TDP cases. rs1990622 genotype was significantly associated with both age at death and age at onset, in both univariate models and models adjusting for age and presence/absence of motor neuron disease (MND). Asterisks denote significance.
| Disease | Outcome | Predictors | Beta (rs1990622, each major allele) | R2 for model | P-value (rs1990622) |
|---|---|---|---|---|---|
| FTLD-TDP | Age at Death (n=75) | rs1990622 | +3.342 | 0.048 | 0.016 * |
| rs1990622, Sex, MND | +3.413 | 0.032 | 0.019 * | ||
| Age at Onset (n=68) | rs1990622 | +3.473 | 0.049 | 0.019 * | |
| rs1990622, Sex, MND | +3.198 | 0.057 | 0.032 * |
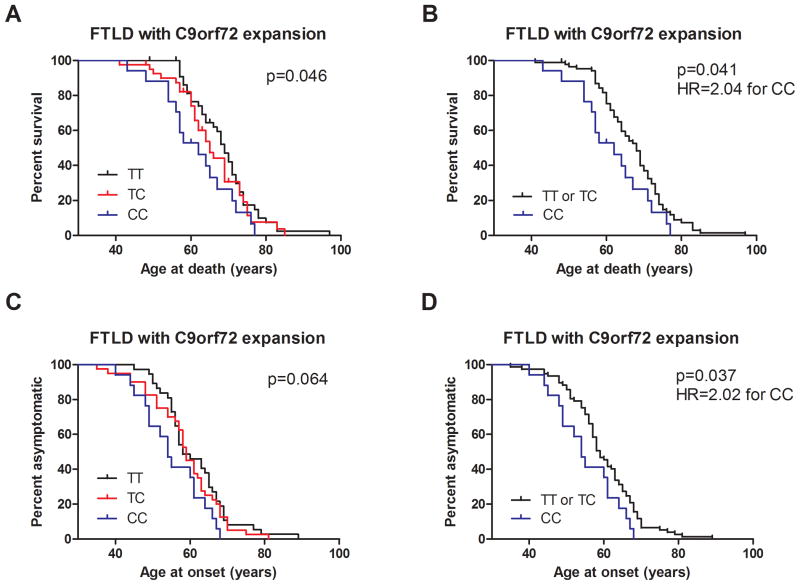
We further examined this genetic modifier effect using Kaplan-Meier survival analyses performed on the combined cohort (discovery plus replication, n=89 for age at death analysis, n=94 for age at onset analysis) of C9orf72(+) FTLD cases. As shown in Fig. 1, TMEM106B genotypes at rs1990622 were significantly associated with age at death (Fig. 1A, p=0.046, log rank test for trend), with a trend towards association for age at onset (Fig. 1C, p=0.064) in this combined cohort. In addition, we observed that the curve separation between rs1990622 minor allele homozygotes (CC) and heterozygotes (TC) was greater than the separation between heterozygotes (TC) and major allele homozygotes (TT). We therefore re-analyzed our data under a major-allele dominant model for rs1990622 and observed a stronger effect of TMEM106B genotype on age at death (p=0.041, log rank test for trend) and age at onset (p=0.037, log rank test for trend) in C9orf72(+) FTLD. Indeed, at any given age, minor allele (C) homozygotes at rs1990622 had more than twice the risk of manifesting disease (Fig. 1D, HR 2.022, 95% CI 1.042–3.925), and more than twice the risk of death (Fig. 1B, HR 2.039, 95% CI 1.031–4.033), compared to other genotypes.
Fig. 1. TMEM106B genotype influences age at death and age at onset in C9orf72(+) FTLD.
All survival analyses were performed in 104 total C9orf72(+) FTLD cases, from the combined discovery and replication cohorts. Of these 104 total cases, 89 had available age-at-death data, and 94 had age-at-onset data.
A) Age at death was significantly associated with TMEM106B genotype at rs1990622, the top SNP associated with FTLD-TDP in our prior GWAS. Log rank test for trend two-tailed p=0.046, assuming a codominant model.
B) Under a major-allele-dominant model, TMEM106B rs1990622 genotype was even more significantly associated with age at death, with more than twice the risk of death at any given age for CC carriers compared to carriers of one or more T alleles (two-tailed p=0.041, HR=2.039, 95% CI 1.031–4.033).
C) Age at onset showed a trend towards association with TMEM106B genotype at rs1990622. Log rank test for trend two-tailed p=0.064, assuming a codominant model.
D) Under a major-allele-dominant model, TMEM106B rs1990622 genotype showed a significant association with age at disease onset, with more than twice the risk of disease onset at any given age for CC carriers compared to carriers of one or more T alleles (two-tailed p=0.037, HR=2.022, 95% CI 1.042–3.925)
TMEM106B genotype does not exert a genetic modifier effect in C9orf72 expansion negative FTLD-TDP cases
We next asked whether the TMEM106B genetic modifier effect observed for C9orf72(+) FTLD extended to FTLD-TDP cases without C9orf72 expansions, again using FTLD-TDP cases from the FTLD-TDP GWAS for which C9orf72 and/or GRN mutation status was known. We considered cases with and without GRN mutations separately.
As shown in Fig. 2A, TMEM106B rs1990622 genotype did not affect age at death in FTLD-TDP cases without C9orf72 expansions or GRN mutations (n=241). In the subset of GRN-related FTLD-TDP (n=116, Fig. 2B), only one rs1990622 CC individual had age at death information available, so we could only compare TT and TC individuals, who did not differ significantly in age at death. Similar results were obtained for age-at-onset analyses (data not shown).
Fig. 2. TMEM106B genotype does not affect age at death or age at onset for FTLD-TDP without C9orf72 expansions.
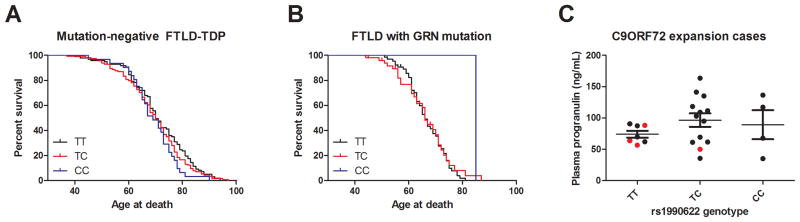
A) In 241 FTLD-TDP cases negative for GRN mutations or C9orf72 expansions, TMEM106B genotype at rs1990622 did not affect age at death.
B) In 116 FTLD-TDP cases with GRN mutations, we found no significant difference in age at death comparing TT and TC carriers at rs1990622. In this cohort, only one individual had the CC genotype, precluding our ability to evaluate the influence of this genotype.
C) Plasma progranulin levels were measured in a convenience subset of 24 C9orf72 expansion carriers by ELISA. Progranulin levels did not differ significantly by TMEM106B rs1990622 genotype, although the TT carriers exhibited significantly less variance in their progranulin levels. Black dots indicate individuals who presented with ALS, while red dots indicate individuals who presented with FTLD.
TMEM106B genotype is associated with FTLD-TDP in C9orf72 expansion carriers
The observed genetic modifier effect for TMEM106B in C9orf72(+) FTLD is surprising in its direction. Specifically, the rs1990622 major allele associated with increased risk of FTLD-TDP by GWAS is correlated with older age at onset and death among C9orf72(+) FTLD cases, implying a beneficial effect in this mutation subgroup. We therefore examined TMEM106B rs1990622 allele frequencies in 116 GRN(+) FTLD cases, 80 C9orf72(+) FTLD cases, and 241 FTLD-TDP cases in which mutations in GRN and expansions in C9orf72 had been excluded. As with the age-at-onset and age-at-death analyses, FTLD-TDP cases were from our prior FTLD-TDP GWAS, although numbers in each group are slightly higher because individuals with genotypes but lacking age-at-death or age-at-onset data could be included. As shown in Table 3, TMEM106B rs1990622 genotype was significantly associated with FTLD-TDP in all three subgroups, with the same direction of association in all three subgroups. In each case, the major allele of rs1990622 was enriched in disease.
Table 3. TMEM106B rs1990622 genotype is associated with FTLD-TDP in all genetic subgroups.
Chi-square tests were performed to evaluate for association between disease and rs1990622 genotype for FTLD-TDP subgroups defined by the presence of GRN mutations (GRN(+) FTLD-TDP), presence of C9orf72 expansions (C9orf72(+) FTLD-TDP), or the absence of both genetic mutations (FTLD-TDP (no mutation)). The major allele was significantly associated with disease in all three subgroups. Allele frequencies for normal controls provided here are from our previously published GWAS.
| Disease status | N | rs1990622 Major allele T | rs1990622 Minor allele C | p-value | Odds ratio | 95% CI |
|---|---|---|---|---|---|---|
| Normal | 2509 | 0.564 | 0.436 | - | ||
| GRN(+) FTLD-TDP | 116 | 0.776 | 0.224 | <0.0001 | 2.675 | 1.955–3.660 |
| C9orf72(+)FTLD-TDP | 80 | 0.669 | 0.331 | 0.008 | 1.560 | 1.117–2.179 |
| FTLD-TDP (no mutation) | 241 | 0.640 | 0.360 | 0.001 | 1.375 | 1.131–1.671 |
TMEM106B genotype is not associated with plasma progranulin levels in C9orf72 expansion carriers
TMEM106B genotype has been reported to influence plasma progranulin levels in healthy individuals and GRN+ FTLD, with the rs1990622 major allele associated with decreased progranulin expression. We evaluated whether this relationship was also true in C9orf72 expansion carriers. In a convenience subset of 24 C9orf72 expansion carriers (20 with C9orf72(+) ALS and 4 with C9orf72(+) FTLD) from the UPenn discovery cohort for whom we had plasma samples, we measured progranulin levels using an enzyme-linked immunosorbent assay (ELISA). As shown in Fig. 2C, there were no significant differences in plasma progranulin levels comparing C9orf72 expansion carriers with TT, TC, and CC genotypes at rs1990622. Adjusting for sex and age at plasma sampling or duration of disease did not affect this result. Additionally adjusting for clinical manifestation as FTLD or ALS did not affect this result.
DISCUSSION
In the current study, we find that TMEM106B is a genetic modifier for C9orf72(+) FTLD, demonstrating a significantly later age at death and age at onset for TMEM106B rs1990622 major allele (T) carriers. This effect appears to be specific to C9orf72(+) FTLD, since C9orf72(−)FTLD cases do not differ in age at death depending on rs1990622 genotype. In addition, rs1990622 major allele carriers are significantly enriched in C9orf72(+) FTLD, compared to neurologically normal controls. Finally, among C9orf72 expansion carriers, we do not see a clear effect of rs1990622 genotype on plasma progranulin levels.
We observe that TMEM106B genotypes exert a genetic modifier effect in C9orf72(+) FTLD. Examples of common risk variants acting as genetic modifiers in Mendelian subgroups of disease are increasingly being described. In the field of neurodegeneration, one well-known example is the age-at-onset modifying effect of Apolipoprotein E (APOE) isoform in PSEN2-related-Alzheimer’s Disease [43]. Moreover, in GRN+ FTLD, TMEM106B has been reported as a genetic modifier affecting both age-at-onset and circulating levels of progranulin [9,12].
What is more unusual in this case is the direction of the genetic modifier effect. Specifically, the TMEM106B allele that is associated with increased risk of developing FTLD-TDP [38] (and earlier age at onset in GRN+ FTLD [9]) appears to ameliorate the disease phenotype (associating with later age at death and onset) in C9orf72(+) FTLD. This effect may be an example of the general phenomenon of sign epistasis, in which a genetic variant is beneficial on some genetic backgrounds but deleterious in others. In this case, the genetic variant in question is TMEM106B genotype at rs1990622 (and linked SNPs), and the genetic backgrounds demonstrating opposing effects are (1) C9orf72(+) individuals -- where the major allele at rs1990622 and linked SNPs is protective in modulating the severity of FTLD manifestation, as demonstrated by older age at onset and age at death and (2) C9orf72(−) individuals -- where the major allele at rs1990622 and linked SNPs is harmful in conferring increased risk of developing FTLD.
Sign epistasis has its conceptual underpinnings in the evolutionary biology literature [42]. With the advent of modern experimental tools, sign epistasis has been demonstrated in lower organisms such as bacteria [32], with reports for this phenomenon in the realm of human genetics and human disease genetics as well [18,19]. In the few reported empirically-derived examples of sign epistasis, the two (or more) genetic loci involved converge mechanistically in, for example, antibiotic resistance pathways [29] or enzyme-substrate interactions [45]. Thus, the observed epistasis between TMEM106B and C9orf72 suggests that these two proteins may have convergent functions in the pathophysiology of FTLD-TDP. Intriguingly, TMEM106B has been linked to endosomal-lysosomal pathways [3,5,20,26]. The largely uncharacterized protein C9orf72 is structurally related to DENN protein family members [21]. DENN proteins function in the regulation of Rab GTPases, which in turn regulate the many membrane trafficking events needed for proper function of the endosomal-lysosomal pathway.
We note that TMEM106B rs1990622 genotypes differ in allelic frequencies between C9orf72(+) FTLD-TDP and normal controls; this situation in which a common variant shows allelic association with disease even in a monogenic, highly-penetrant subgroup of disease has been reported in GRN+ FTLD-TDP as well [12,38]. In the case of the GRN mutants, a potential explanation may lie in ascertainment bias, since TMEM106B risk variant carriers may manifest disease at an earlier age [9], making it more likely for them to be included in a cross-sectional sampling of diseased individuals. Alternately, the protective effect of the modifier locus (e.g. TMEM106B) may be significant enough to counter-act the disease-causing effects of the Mendelian genetic cause (e.g. GRN), such that carriers of protective variants never manifest clinically despite possessing a highly-penetrant genetic mutation. Such an argument cannot explain our current result, however, since the rs1990622 major allele (found by genome-wide association to be enriched in FTLD-TDP) appears to delay age at death and age at onset in C9orf72(+) FTLD cases. An alternate explanation may lie in the fact that C9orf72 expansions have a broad range of phenotypic expression, manifesting as ALS, FTLD, or a syndrome combining both motor neuron disease and dementia. We have previously shown that ALS patients who are major allele carriers at rs1990622 are more likely to demonstrate cognitive impairment [40]. Thus, it is possible that TMEM106B genotype modulates the phenotypic expression of C9orf72 expansions, with rs1990622 major allele carriers more likely to manifest clinically with dementia. Whether an effect of directing regional pathology towards cognitive regions rather than motor regions also underlies the apparently protective effect on age at death for TMEM106B rs1990622 major allele carriers with C9orf72 expansions remains to be seen.
It is notable that we were able to replicate the genetic modifier effect of TMEM106B genotype in C9orf72(+) FTLD in a 30-site, international cohort of subjects. Undoubtedly, site-to-site variation in methods of ascertaining age at onset would contribute to noise, and site-to-site variation in practice with respect to aggressiveness of clinical care with a fatal neurodegenerative disease would contribute to differences in age at death in such a dataset. The ability to see a significant genetic modifier effect of TMEM106B on C9orf72 in such a cohort, nonetheless, may have been helped by the fact that our replication cohort was homogeneous with respect to neuropathology (all FTLD-TDP), and genome-wide genotyping in these individuals allowed us to exclude important potential sources of noise, such as population stratification and cryptic familial relationships among individuals. In any case, the international, multi-site nature of our replication cohort increases our confidence that our findings are not due to artifact.
The current study has several limitations. First, while we did not see an age-at-death-modifying effect for TMEM106B in C9orf72 expansion-associated ALS, our sample size was small (n=39) and likely underpowered to adequately address this question. Thus, future studies examining this relationship in more C9orf72-expansion-related ALS cases would be a valuable addition to the data presented here. Second, we did not see a clear modifier effect of TMEM106B genotype in the GRN(+) FTLD-TDP cases in this study, as has been previously reported [9]. However, our study had only one rs1990622 minor allele homozygote in the GRN+ FTLD subgroup, precluding our ability to examine TMEM106B genotype effect in a major-allele-dominant model. Third, we were able to obtain plasma samples on 24 C9orf72 expansion carriers, in whom we measured progranulin levels. Plasma progranulin levels did not differ by TMEM106B genotype in this set of samples, which could reflect either insufficient sample size or a biologically-relevant finding. Should further studies in larger sample sizes corroborate our result, this would suggest that C9orf72 expansions may interrupt the means by which TMEM106B affects circulating progranulin levels. Finally, our study was a targeted evaluation of one locus (TMEM106B) for genetic modifier effect in C9orf72 expansion carriers, rather than a comprehensive screen for genetic modifiers in C9orf72(+) FTLD or ALS. It is entirely possible that other loci with epistatic effects exist and also play an important role in modulating the phenotype associated with C9orf72 expansions. In conclusion, we demonstrate here that TMEM106B is the first reported genetic modifier in C9orf72 expansion-related FTLD. Our findings suggest a previously unsuspected link between these two proteins in the pathophysiology of FTLD and open up new directions for the development of disease-modifying therapy
Acknowledgments
FUNDING
Contributing sites that provided C9orf72 genetic data included: Erasmus University, Rotterdam, The Netherlands; Indiana University, Indianapolis, Indiana; Banc de Teixits Neurologics-Biobanc-Hospital Clinic-IDIBAPS, Barcelona, Spain; Kings College, London, UK; UCL Institute of Neurology, Queen Square, London, UK; Ludwig-Maximilians University, Munich, Germany; University of New South Wales, Sydney, Australia; VIB, University of Antwerp, Antwerp, Belgium; Massachusetts General Hospital, Boston, Massachusetts; University of Sheffield, Sheffield, UK; Institut National de la Santé et de la Recherche Laboratoire de Neuropathologie, Paris, France.
Contributing sites with C9orf72(+) cases identified at UPenn included: Sydney Brain Bank, Australia; Boston University, Boston, Massachusetts; Duke University, Durham, North Carolina; Emory University, Atlanta; Georgia; Karolinska Institute, Stockholm, Sweden; Mt. Sinai School of Medicine, Bronx, New York; Oregon Health Sciences University, Portland, Oregon; University of Pittsburgh, Pittsburgh, Pennsylvania; Rush University, Chicago, Illinois; University of Texas Southwestern, Dallas, Texas; University of Toronto, Toronto, Canada; University of California (Davis, Irvine, San Diego campuses), California; University of Michigan, Ann Arbor, Michigan; University of Kuopio, Finland; University of Southern California, Los Angeles, California; Washington University, St. Louis, Missouri; University of Pennsylvania, Philadelphia, Pennsylvania.
Sources of support for this project include the NIH (AG033101, NS082265, P50 AG005133), The Neurological Tissue Bank of the Biobanc-HC-IDIBAPS, Hersenstichting project BG2010.02, Alzheimer Nederland/NIBC 056-13-018, Stichting Dioraphte projectnr 0802100, The National Institute for Health Research, SOPHIA, EuroMotor, National Health and Medical Research Council of Australia (NHMRC) (FTLD cases supported by NHMRC program grant 1037746), and Neuroscience Research Australia, University of New South Wales. The Antwerp site is in part funded by the MetLife Foundation, USA; the Interuniversity Attraction Poles program of the Belgian Science Policy Office (BELSPO), the Europe Initiative on Centers of Excellence in Neurodegeneration (CoEN) and the Methusalem program supported by the Flemish Government; the Foundation Alzheimer Research (SAO/FRA); the Medical Foundation Queen Elisabeth; the Research Foundation Flanders (FWO); the Agency for Innovation by Science and Technology Flanders (IWT), the University of Antwerp Research Fund, Belgium. The FWO provided a postdoctoral fellowship to J.v.d.Z and I.G. Alice Chen-Plotkin is also supported by the Burroughs Wellcome Fund Career Award for Medical Scientists, a Doris Duke Clinician Scientist Development Award, and the Benaroya Fund. Glenda Halliday holds a NHMRC Senior Principal Research Fellowship. Jonathan D. Rohrer and Martin Rosser are supported by the NIHR Queen Square Dementia Biomedical Research unit and work at the UCL Institute of Neurology Dementia Research Centre which is supported by Alzheimer’s Research UK, Brain Research Trust, and The Wolfson Foundation.
We thank Travis Unger and Beth McCarty Wood for technical assistance. We thank our patients and their families for their participation in this research.
INTERNATIONAL COLLABORATION FOR FRONTOTEMPORAL LOBAR DEGENERATION
The International Collaboration for Frontotemporal Lobar Degeneration consisted of clinical sites collaborating to collect cases for an FTLD-TDP genomewide association study (GWAS); this GWAS led to the discovery that common variants in TMEM106B are a genetic risk factor for FTLD-TDP [38]. Members of the Collaboration who contributed C9orf72(+)FTLD-TDP cases for the current study include Irina Alafuzoff, Anna Antonell, Nenad Bogdanovic, William Brooks, Nigel Cairns, Johnathan Cooper-Knock, Carl W. Cotman, Patrick Cras, Marc Cruts, Peter P. De Deyn, Charles DeCarli, Carol Dobson-Stone, Sebastiaan Engelborghs, Nick Fox, Douglas Galasko, Marla Gearing, Ilse Gijselinck, Jordan Grafman, Paivi Hartikainen, Kimmo J. Hatanpaa, J. Robin Highley, John Hodges, Christine Hulette, Paul G. Ince, Lee-Way Jin, Janine Kirby, Julia Kofler, Jillian Kril, John J. B. Kwok, Allan Levey, Andrew Lieberman, Albert Llado, Jean-Jacques Martin, Eliezer Masliah, Christopher J. McDermott, Catriona McLean, Ann C. McKee, Simon Mead, Carol A. Miller, Josh Miller, David Munoz, Jill Murrell, Henry Paulson, Olivier Piguet, Martin Rossor, Raquel Sanchez-Valle, Mary Sano, Julie Schneider, Lisa Silbert, Salvatore Spina, Julie van der Zee, Tim Van Langenhove, Jason Warren, Stephen B. Wharton, Charles L. White III, Randall Woltjer.
References
- 1.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boxer AL, Mackenzie IR, Boeve BF, Baker M, Seeley WW, Crook R, Feldman H, Hsiung GY, Rutherford N, Laluz V, Whitwell J, Foti D, McDade E, Molano J, Karydas A, Wojtas A, Goldman J, Mirsky J, Sengdy P, Dearmond S, Miller BL, Rademakers R. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatry. 2011;82:196–203. doi: 10.1136/jnnp.2009.204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady OA, Zheng Y, Murphy K, Huang M, Hu F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet. 2013;22:685–695. doi: 10.1093/hmg/dds475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks BR, Miller RG, Swash M, Munsat TL World Federation of Neurology Research Group on Motor Neuron Diseases . El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 5.Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, Busch JI, Akle S, Grossman M, Van Deerlin V, Trojanowski JQ, Lee VM. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci. 2012;32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen-Plotkin AS, Hu WT, Siderowf A, Weintraub D, Goldmann Gross R, Hurtig HI, Xie SX, Arnold SE, Grossman M, Clark CM, Shaw LM, McCluskey L, Elman L, Van Deerlin VM, Lee VM, Soares H, Trojanowski JQ. Plasma epidermal growth factor levels predict cognitive decline in Parkinson disease. Ann Neurol. 2011;69:655–663. doi: 10.1002/ana.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen-Plotkin AS, Martinez-Lage M, Sleiman PM, Hu W, Greene R, Wood EM, Bing S, Grossman M, Schellenberg GD, Hatanpaa KJ, Weiner MF, White CL, 3rd, Brooks WS, Halliday GM, Kril JJ, Gearing M, Beach TG, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Pickering-Brown SM, Snowden J, van Swieten JC, Heutink P, Seelaar H, Murrell JR, Ghetti B, Spina S, Grafman J, Kaye JA, Woltjer RL, Mesulam M, Bigio E, Llado A, Miller BL, Alzualde A, Moreno F, Rohrer JD, Mackenzie IR, Feldman HH, Hamilton RL, Cruts M, Engelborghs S, De Deyn PP, Van Broeckhoven C, Bird TD, Cairns NJ, Goate A, Frosch MP, Riederer PF, Bogdanovic N, Lee VM, Trojanowski JQ, Van Deerlin VM. Genetic and clinical features of progranulin-associated frontotemporal lobar degeneration. Arch Neurol. 2011;68:488–497. doi: 10.1001/archneurol.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, Kabashi E. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Ann Neurol. 2013 doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 9.Cruchaga C, Graff C, Chiang HH, Wang J, Hinrichs AL, Spiegel N, Bertelsen S, Mayo K, Norton JB, Morris JC, Goate A. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol. 2011;68:581–586. doi: 10.1001/archneurol.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruts M, Gijselinck I, Van Langenhove T, van der Zee J, Van Broeckhoven C. Current insights into the C9orf72 repeat expansion diseases of the FTLD/ALS spectrum. Trends Neurosci. 2013 doi: 10.1016/j.tins.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 11.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L, Crook J, Finger E, Hantanpaa KJ, Karydas AM, Sengdy P, Gonzalez J, Seeley WW, Johnson N, Beach TG, Mesulam M, Forloni G, Kertesz A, Knopman DS, Uitti R, White CL, 3rd, Caselli R, Lippa C, Bigio EH, Wszolek ZK, Binetti G, Mackenzie IR, Miller BL, Boeve BF, Younkin SG, Dickson DW, Petersen RC, Graff-Radford NR, Geschwind DH, Rademakers R. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76:467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, Crook R, Melquist S, Kuntz K, Petersen R, Josephs K, Pickering-Brown SM, Graff-Radford N, Uitti R, Dickson D, Wszolek Z, Gonzalez J, Beach TG, Bigio E, Johnson N, Weintraub S, Mesulam M, White CL, 3rd, Woodruff B, Caselli R, Hsiung GY, Feldman H, Knopman D, Hutton M, Rademakers R. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- 15.Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, Engelborghs S, Sieben A, De Jonghe P, Vandenberghe R, Santens P, De Bleecker J, Maes G, Baumer V, Dillen L, Joris G, Cuijt I, Corsmit E, Elinck E, Van Dongen J, Vermeulen S, Van den Broeck M, Vaerenberg C, Mattheijssens M, Peeters K, Robberecht W, Cras P, Martin JJ, De Deyn PP, Cruts M, Van Broeckhoven C. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 16.Gorno-Tempini ML, Hillis AE, Weintraub S, Kertesz A, Mendez M, Cappa SF, Ogar JM, Rohrer JD, Black S, Boeve BF, Manes F, Dronkers NF, Vandenberghe R, Rascovsky K, Patterson K, Miller BL, Knopman DS, Hodges JR, Mesulam MM, Grossman M. Classification of primary progressive aphasia and its variants. Neurology. 2011;76:1006–1014. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 18.Kern AD, Kondrashov FA. Mechanisms and convergence of compensatory evolution in mammalian mitochondrial tRNAs. Nat Genet. 2004;36:1207–1212. doi: 10.1038/ng1451. [DOI] [PubMed] [Google Scholar]
- 19.Kondrashov AS, Sunyaev S, Kondrashov FA. Dobzhansky-Muller incompatibilities in protein evolution. Proc Natl Acad Sci U S A. 2002;99:14878–14883. doi: 10.1073/pnas.232565499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang CM, Fellerer K, Schwenk BM, Kuhn PH, Kremmer E, Edbauer D, Capell A, Haass C. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem. 2012;287:19355–19365. doi: 10.1074/jbc.M112.365098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29:499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litvan I, Agid Y, Calne D, Campbell G, Dubois B, Duvoisin RC, Goetz CG, Golbe LI, Grafman J, Growdon JH, Hallett M, Jankovic J, Quinn NP, Tolosa E, Zee DS. Clinical research criteria for the diagnosis of progressive supranuclear palsy (Steele-Richardson-Olszewski syndrome): report of the NINDS-SPSP international workshop. Neurology. 1996;47:1–9. doi: 10.1212/wnl.47.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, Perry RH, Trojanowski JQ, Mann DM, Lee VM. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–113. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ Work Group on Frontotemporal Dementia and Pick’s Disease . Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch Neurol. 2001;58:1803–1809. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- 25.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson AM, Finch NA, Wojtas A, Baker MC, Perkerson RB, Castanedes-Casey M, Rousseau L, Benussi L, Binetti G, Ghidoni R, Hsiung GY, Mackenzie IR, Finger E, Boeve BF, Ertekin-Taner N, Graff-Radford NR, Dickson DW, Rademakers R. TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem. 2013 doi: 10.1111/jnc.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, van Swieten JC, Seelaar H, Dopper EG, Onyike CU, Hillis AE, Josephs KA, Boeve BF, Kertesz A, Seeley WW, Rankin KP, Johnson JK, Gorno-Tempini ML, Rosen H, Prioleau-Latham CE, Lee A, Kipps CM, Lillo P, Piguet O, Rohrer JD, Rossor MN, Warren JD, Fox NC, Galasko D, Salmon DP, Black SE, Mesulam M, Weintraub S, Dickerson BC, Diehl-Schmid J, Pasquier F, Deramecourt V, Lebert F, Pijnenburg Y, Chow TW, Manes F, Grafman J, Cappa SF, Freedman M, Grossman M, Miller BL. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ ITALSGEN Consortium. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schenk MF, Szendro IG, Salverda ML, Krug J, de Visser JA. Patterns of Epistasis between Beneficial Mutations in an Antibiotic Resistance Gene. Mol Biol Evol. 2013 doi: 10.1093/molbev/mst096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seelaar H, Rohrer JD, Pijnenburg YA, Fox NC, van Swieten JC. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: a review. J Neurol Neurosurg Psychiatry. 2011;82:476–486. doi: 10.1136/jnnp.2010.212225. [DOI] [PubMed] [Google Scholar]
- 31.Shankaran SS, Capell A, Hruscha AT, Fellerer K, Neumann M, Schmid B, Haass C. Missense mutations in the progranulin gene linked to frontotemporal lobar degeneration with ubiquitin-immunoreactive inclusions reduce progranulin production and secretion. J Biol Chem. 2008;283:1744–1753. doi: 10.1074/jbc.M705115200. [DOI] [PubMed] [Google Scholar]
- 32.Silva RF, Mendonca SC, Carvalho LM, Reis AM, Gordo I, Trindade S, Dionisio F. Pervasive sign epistasis between conjugative plasmids and drug-resistance chromosomal mutations. PLoS Genet. 2011;7:e1002181. doi: 10.1371/journal.pgen.1002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, Hodges JR, Spillantini MG, Thusgaard T, Brandner S, Brun A, Rossor MN, Gade A, Johannsen P, Sorensen SA, Gydesen S, Fisher EM, Collinge J. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 34.Snowden JS, Rollinson S, Thompson JC, Harris JM, Stopford CL, Richardson AM, Jones M, Gerhard A, Davidson YS, Robinson A, Gibbons L, Hu Q, DuPlessis D, Neary D, Mann DM, Pickering-Brown SM. Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain. 2012;135:693–708. doi: 10.1093/brain/awr355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart H, Rutherford NJ, Briemberg H, Krieger C, Cashman N, Fabros M, Baker M, Fok A, DeJesus-Hernandez M, Eisen A, Rademakers R, Mackenzie IR. Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathol. 2012;123:409–417. doi: 10.1007/s00401-011-0937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strong MJ, Grace GM, Freedman M, Lomen-Hoerth C, Woolley S, Goldstein LH, Murphy J, Shoesmith C, Rosenfeld J, Leigh PN, Bruijn L, Ince P, Figlewicz D. Consensus criteria for the diagnosis of frontotemporal cognitive and behavioural syndromes in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;10:131–146. doi: 10.1080/17482960802654364. [DOI] [PubMed] [Google Scholar]
- 37.Toledo JB, Van Deerlin VM, Lee EB, Suh E, Baek Y, Robinson JL, Xie SX, McBride J, Wood EM, Schuck T, Irwin DJ, Gross RG, Hurtig H, McCluskey L, Elman L, Karlawish J, Schellenberg G, Chen-Plotkin A, Wolk D, Grossman M, Arnold SE, Shaw LM, Lee VM, Trojanowski JQ. A platform for discovery: The University of Pennsylvania Integrated Neurodegenerative Disease Biobank. Alzheimers Dement. 2013 doi: 10.1016/j.jalz.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, Arnold SE, Mann DM, Pickering-Brown SM, Seelaar H, Heutink P, van Swieten JC, Murrell JR, Ghetti B, Spina S, Grafman J, Hodges J, Spillantini MG, Gilman S, Lieberman AP, Kaye JA, Woltjer RL, Bigio EH, Mesulam M, Al-Sarraj S, Troakes C, Rosenberg RN, White CL, 3rd, Ferrer I, Llado A, Neumann M, Kretzschmar HA, Hulette CM, Welsh-Bohmer KA, Miller BL, Alzualde A, Lopez de Munain A, McKee AC, Gearing M, Levey AI, Lah JJ, Hardy J, Rohrer JD, Lashley T, Mackenzie IR, Feldman HH, Hamilton RL, Dekosky ST, van der Zee J, Kumar-Singh S, Van Broeckhoven C, Mayeux R, Vonsattel JP, Troncoso JC, Kril JJ, Kwok JB, Halliday GM, Bird TD, Ince PG, Shaw PJ, Cairns NJ, Morris JC, McLean CA, DeCarli C, Ellis WG, Freeman SH, Frosch MP, Growdon JH, Perl DP, Sano M, Bennett DA, Schneider JA, Beach TG, Reiman EM, Woodruff BK, Cummings J, Vinters HV, Miller CA, Chui HC, Alafuzoff I, Hartikainen P, Seilhean D, Galasko D, Masliah E, Cotman CW, Tunon MT, Martinez MC, Munoz DG, Carroll SL, Marson D, Riederer PF, Bogdanovic N, Schellenberg GD, Hakonarson H, Trojanowski JQ, Lee VM. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Zee J, Van Langenhove T, Kleinberger G, Sleegers K, Engelborghs S, Vandenberghe R, Santens P, Van den Broeck M, Joris G, Brys J, Mattheijssens M, Peeters K, Cras P, De Deyn PP, Cruts M, Van Broeckhoven C. TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain. 2011;134:808–815. doi: 10.1093/brain/awr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vass R, Ashbridge E, Geser F, Hu WT, Grossman M, Clay-Falcone D, Elman L, McCluskey L, Lee VM, Van Deerlin VM, Trojanowski JQ, Chen-Plotkin AS. Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:373–380. doi: 10.1007/s00401-010-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 42.Weinreich DM, Watson RA, Chao L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- 43.Wijsman EM, Daw EW, Yu X, Steinbart EJ, Nochlin D, Bird TD, Schellenberg GD. APOE and other loci affect age-at-onset in Alzheimer’s disease families with PS2 mutation. Am J Med Genet B Neuropsychiatr Genet. 2005;132B:14–20. doi: 10.1002/ajmg.b.30087. [DOI] [PubMed] [Google Scholar]
- 44.Xie SX, Baek Y, Grossman M, Arnold SE, Karlawish J, Siderowf A, Hurtig H, Elman L, McCluskey L, Van Deerlin V, Lee VM, Trojanowski JQ. Building an integrated neurodegenerative disease database at an academic health center. Alzheimers Dement. 2011;7:e84–93. doi: 10.1016/j.jalz.2010.08.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Dourado DF, Fernandes PA, Ramos MJ, Mannervik B. Multidimensional epistasis and fitness landscapes in enzyme evolution. Biochem J. 2012;445:39–46. doi: 10.1042/BJ20120136. [DOI] [PubMed] [Google Scholar]