Abstract
Actinomycetes, a group of filamentous, Gram-positive bacteria, have long been a remarkable source of useful therapeutics. Recent genome sequencing and transcriptomic studies have shown that these bacteria, responsible for half of the clinically used antibiotics, also harbor a large reservoir of gene clusters, which have the potential to produce novel secreted small molecules. Yet, many of these clusters are not expressed under common culture conditions. One reason why these clusters have not been linked to a secreted small molecule lies in the way that actinomycetes have typically been studied: as pure cultures in nutrient-rich media that do not mimic the complex environments in which these bacteria evolved. New methods based on multispecies culture conditions provide an alternative approach to investigating the products of these gene clusters. We have recently implemented binary interspecies interaction assays to mine for new secondary metabolites and to study the underlying biology in inter-actinomycete interactions. Here we describe the detailed biological and chemical methods comprising these studies.
Keywords: secondary metabolites, interspecies interactions, amychelin, Streptomyces coelicolor, Amycolatopsis sp. AA4
1. Introduction
The majority of antibacterial compounds used clinically since the 1950s have been derived from actinomycetes, a group of filamentous, high G+C Gram-positive bacteria long known as prolific producers of secondary metabolites (Newman and Cragg, 2012, Newman et al., 2002, Berdy, 2005, Fischbach and Walsh, 2009). Recent genome sequencing efforts have brought this production capacity into focus by revealing dozens of easily identifiable secondary metabolite gene clusters per sequenced strain, indicating that actinomycetes contain vast reservoirs of potentially useful therapeutic agents (Bentley et al., 2002, Oliynyk et al., 2007, Copeland et al., 2009, Zhao et al., 2010, Nett et al., 2009). However, the bulk of these recognizable clusters appear to be inactive under the conditions in which actinomycetes have been examined. As a consequence, these clusters are not associated with cognate small-molecule products (Nett et al., 2009). Whether they represent gene clusters that are actively transcribed but not yet linked to a small molecule, or silent gene clusters that are inactive under the conditions examined, remains largely unknown. In some cases, transcriptomic or gene knockout studies have already allowed a distinction between these cluster types. The preponderance of biosynthetic gene clusters without known small-molecule products found in Saccharopolyspora erythraea and Streptomyces griseus, the erythromycin and streptomycin producers, respectively, is especially astonishing (Oliynyk et al., 2007, Ohnishi et al., 2008, Nett et al., 2009). These strains have been studied under a large number of conditions for several decades and >75% of their gene clusters still have no known small-molecule product (Nett et al., 2009). Clearly, alternative approaches are needed to explore metabolite production in actinomycetes.
The number of secondary metabolite gene clusters revealed by genome sequencing and the growing appreciation that the exchange of small molecules mediates inter- and intra-species communication suggest that bacterial interaction assays should be re-assessed as a potential conduit for the discovery of new small molecules (Fischbach and Krogan, 2010, Shank and Kolter, 2009, Straight and Kolter, 2009, Schmidt, 2008). Several features of actinomycete biology lend themselves well to interaction screens, as previously demonstrated by Ueda, Beppu, and coworkers (Ueda et al., 2000, Yamanaka et al., 2005). Actinomycete colonies often have striking morphologies that are visible to the naked eye, including pigmentation (by one or more compounds), complex colony structure (i.e. wrinkles or other features), aerial hyphae, and pigmented spores (Chater, 1998). Alterations in any of these visible phenotypes can be due to compounds secreted by a nearby colony. Thus, these phenotypic changes can serve as sensors for small molecules which might not be found in screens that only search for inhibition of bacterial growth. At the same time, several new lines of evidence suggest that co-culture may actually stimulate activation of silent gene clusters.
With these possibilities in mind, we began a series of interaction assays within a group of 20 sequenced actinomycetes to look for molecules whose effects could be recognized through altered morphology as well as molecules whose production might be stimulated through activation of transcription of otherwise silent gene clusters (Seyedsayamdost et al., 2011). Here, we describe detailed procedures for the methods we have recently used to gain insight into novel bioactive molecules that affect developmental changes within co-cultured actinomycetes. These screens are described on a scale that allows a single researcher to evaluate hundreds to thousands of interspecies interactions for new secondary metabolites.
2. Setting Up binary actinomycete interactions
Actinomycetes have traditionally been isolated from soil, which typically contains hundreds or thousands of different strains of these bacteria per gram (Curtis et al., 2002). With such staggering biological diversity, the soil environment has been vastly under-sampled with regards to its total actinomycete capacity (Lazzarini et al., 2000, Baltz, 2008). Nonetheless, the re-discovery rate of secondary metabolites produced by streptomycetes is relatively high (Watve et al., 2001, Baltz, 2008). This high rate in part reflects the inherent bias in most commonly used isolation techniques, which rely on two important features, (1) sporulation and (2) utilization of complex mixtures of starch and proteins for growth. Most isolation protocols employ heat and desiccation steps to select only for microbes capable of forming resistant spores, followed by growth on media where starch and/or proteins are the major substrates, thereby selecting for strains which produce extracellular amylases and proteases, a hallmark of actinomycetes. Efforts to maximize the diversity of cultured actinomycetes have included development of protocols for isolating strains outside the genus Streptomyces. These are the so-called ‘rare’ actinomycetes (Lazzarini et al., 2000), including those from the genera Actinoplanes, Saccharopolyspora, Amycolatopsis, and Streptosporangium. More recently, isolation of actinomycetes from more varied sources has also led to discovery of new compounds. Important new environmental sources include the ocean (Goodfellow and Fiedler, 2010), both from sediments (Jensen et al., 2005a, Jensen et al., 2005b) and bacterial symbionts of marine organisms (Jiang et al., 2007, Kurahashi et al., 2010), and insects, especially ants(Oh et al., 2009a), beetles (Scott et al., 2008, Oh et al., 2009b), and wasps (Oh et al., 2011, Poulsen et al., 2011). A number of culture conditions for isolating a variety of actinomycetes are listed in Table 1. As a starting point, we include here a basic protocol for the isolation of actinomycetes using starch-casein agar.
Table 1.
Culture media for isolation of various clades of actinomycetes.
| Reference | Target actinobacterial clade | Agar type | Selection criteria |
|---|---|---|---|
| (Wellington and Toth, 1994) | General actinomycete | Starch-casein | Starch and protein as primary growth substrates |
| Difco | General actinomycete | Proprietary | Proprietary, includes glycerol |
| (Hamaki et al., 2005) | Streptosporangium, Actinomadura | Soil extract | Growth on soil extract, small colonies picked under microscopic evaluation |
| (Hop et al., 2011) | Actinoplanes, Kineosporia, Cryptosporangia | Humic acid-vitamin agar, nalidixic acid, kabicidin | Rehydration-centrifugation to select for species with motile spores |
| (Hop et al., 2011) | Micromonospora, Nonomuraea, Streptomyces | Humic acid-vitamin agar, nalidixic acid, kabicidin | Sodium dodecyl sulfate-yeast extract dilution method |
| (Tan et al., 2006) | Amycolatopsis | SM1, SM2, SM3 | Combinations of C-sources and antibiotics: sorbitol, melezitose, neomycin, naladixic acid, novobiocin |
| (Kim et al., 2011) | Dactylosporangium | Streptomyces isolation agar | Gentamicin or oxytetracycline resistance |
2.1. Actinomycete isolation medium
- To one liter of distilled water add the following:
- 10.0 g soluble starch (Fisher #S-516)
- 0.30 g Casein Hydrolysate (Sigma #C-9386)
- 2.0 g KNO3
- 2.0 g NaCl
- 2.0 g K2HPO4
- 0.05 g MgSO4_7H2O
- 0.02 g CaCO3
- 0.01 g FeSO4_7H2O
- 15.0 g granulated agar
Boil the agar solution, allow to cool slightly and adjust to pH 7 with HCl or NaOH.
Autoclave at 121 °C for 15 min. After cooling to approximately 45 °C, add 5 mg cycloheximide in 1.0 mL methanol, mix thoroughly, and pour plates.
2.2. Desiccation of soil
Place 1–5 g of soil in an empty, sterile Petri dish.
Place dish at 50 °C for 48 hrs or until soil is completely dry.
Begin a dilution series by adding 0.1 g of soil to 0.9 mL sterile water. Mix by vortexing and continue dilutions to 10−6.
Plate 100 μL of the 10−4–10−6 dilutions on the starch-casein plates and allow to dry.
Incubate plates at 30 °C for 5 days or longer.
Re-streak colonies that exhibit actinomycete morphology (i.e. tough or brittle colony consistency, visible white aerial hyphae, etc.) for isolation.
Once pure cultures are obtained, proceed with making spore stocks for use in interaction screens.
2.3. Generation of spore stocks for long-term storage of actinomycetes
For most actinomycetes, it is possible to make frozen stocks of vegetative mycelium or spores. Vegetative mycelial stocks are more prone to loss of viability with repeated freeze-thaw cycles than spore stocks. In addition to remaining viable almost indefinitely if stored at −20 or −80 °C, spore stocks give more reliable colony forming unit counts than mycelial stocks, which tend to be composed of clumps of biomass.
Once a pure culture of an actinomycete has been obtained, a densely covered plate is desired to provide ample spore numbers for production of a highly concentrated spore stock. This can be achieved by taking an initial inoculum of spores from the pure culture plate and suspending it in 100–500 μL sterile water.
Vortex vigorously, and spread 100 μL of this solution onto the sporulation medium of choice. If robust development was observed on starch-casein (see above), then this medium may be used in step 1. Alternatively, ISP2 (Becton Dickinson #277010), and Oatmeal agar (Becton Dickinson #255210) are also common media that can promote robust actinomycete sporulation.
Incubate the plates at 30 °C until sporulation occurs, often marked by the production of powdery grey material after the formation of fuzzy white aerial hyphae. Some actinomycetes do not produce pigmented spores, and in these cases, the plates will remain white even though mature spores have formed. Microscopic examination of the eventual spore stock can be useful in verifying the presence of spores as opposed to only aerial hyphae.
Place 500 μL of sterile 20% glycerol into a cryovial.
Pipette 1 mL of sterile 20% glycerol onto the spore-covered plate surface. In most cases the glycerol solution will quickly form beads as a result of the hydrophobic surface layer covering spores and aerial hyphae.
Using a sterile cotton swab, gently rub the surface of the plate until the swab is completely covered with spores.
Transfer spores by twisting the swab against the interior wall of the cryovial until a dense spore solution is obtained. Repeat as necessary.
The concentration of the spore stock may be determined by serial dilution techniques followed by counting the numbers of the resulting colonies.
2.4. Calculation and set-up of interaction arrays
Once spore stocks of an actinomycete have been obtained and quantitated, the layout of the interaction array must be considered. The equation for calculating unique combinations from a larger set of items is:
| (1) |
where n = total number of items, and r = number of items in each combination. For example, if a total of eight actinomycetes will be assayed in pairs, then there will be:
| (2) |
It is recommended to conduct screens in duplicate so that any observed interaction phenotypes can be verified. The simplest way to set up an interaction array on this scale is to spot 1 μL drops of dense spore solutions 0.5 cm apart on an agar surface. Square 12-inch Petri dishes can accommodate many interactions on this scale when each row of interactions is spaced 1 cm apart, laterally. The procedure for setting up screens in an eight-by-eight format, where all interactions within a set of eight actinomycete strains are monitored, follows:
Make 1 mL of a 1:100 spore stock dilution in 20% sterile glycerol.
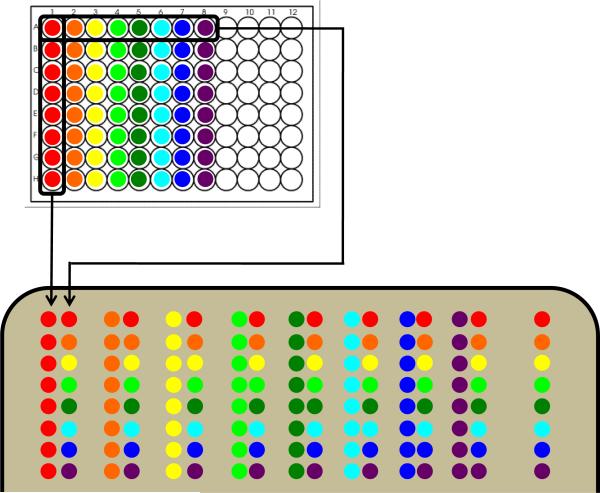
Array 100 μL aliquots of each spore stock into the first eight columns in a 96 well plate as shown in the top panel of Fig. 1.
Using an eight-lane multichannel pipettor, place 1 μL drops from the first column onto the agar surface.
Place 1-μL drops from the first row onto the agar surface in parallel (0.5 cm away) to the drops from the previous step.
Repeat in the pattern shown in the bottom panel of Fig. 1. This yields an array that includes each of the 28 interactions twice and each strain vs. itself once.
A row containing each of the strains alone is offset to the right to serve as a control, and to check for interactions that may be occurring vertically.
Incubate plates at 30 °C for 4–5 days or until developmental progress is visible in control colonies.
Figure 1.
Cartoon outline for generating screens within a group of eight bacterial strains. The strains of interest are arrayed in a 96-well plate as shown in the top panel. These are then transferred to an agar plate as shown in the bottom panel yielding an array that includes each of the 28 possible interactions twice and each strain vs. itself once. A column to the right includes each strain by itself for comparison.
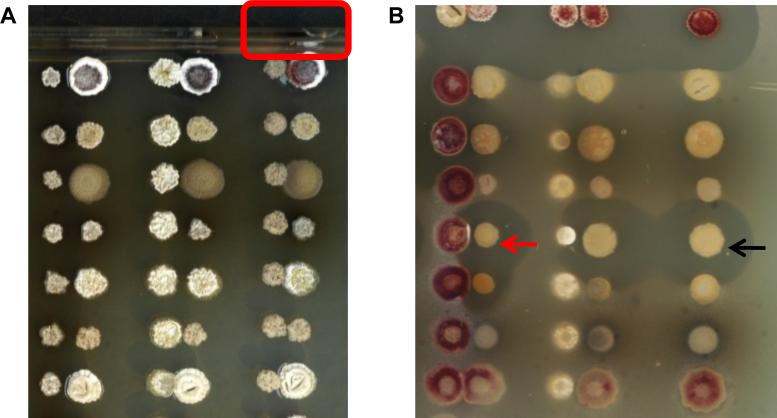
Interactions between actinomycetes can alter many phenotypes that are visible on a macro scale, including development (i.e. aerial hyphae formation and sporulation), pigmentation, and growth. A typical set of interactions is shown using R2YE agar (Fig. 2). An example of an altered developmental phenotype can be seen in Fig. 2A (red outline), where Amycolatopsis sp. AA4 (hereafter AA4) inhibited aerial hyphae formation on the adjacent colony of Streptomyces coelicolor M145 (John Innes Centre). This interaction was chosen for chemical analysis and prompted the discovery of a new Amycolatopsis siderophore, amychelin (Seyedsayamdost et al., 2011). The chemical and biological experiments which led to the isolation and characterization of amychelin are detailed in the following sections.
Figure 2.
Binary actinomycete interaction assays. (A) Shown are binary interactions of Streptomyces tendae (left column), Streptomyces albus (middle column), and Amycolatopsis sp. AA4 (right column) with eight actinomycete strains. The interaction between S. coelicolor and AA4, chosen for further study, is outlined at the top. (B) Binary actinomycete assays are overlaid with S. aureus to detect anti-Staphylococcal compounds that are produced as a function of co-culture. The black arrow points to a colony that does not generate an anti-Staphylococcal compound in isolation. In co-culture with another actinomycete, however, a halo in the overlaid S. aureus lawn is observed (red arrow), indicating production of an antibiotic.
Antibiotic production may also be stimulated or repressed by an interaction. This can be observed by overlaying the entire interaction plate with a layer of agar containing a test organism such as Staphylococcus aureus, as seen in Fig. 2B.
An overlay can be made by following these steps:
Grow an overnight culture of the test organism in LB (Becton Dickinson #244620) or other suitable broth (OD >1.5).
Autoclave a 1% agar solution and cool to ~45°C.
Add 1% inoculum (v/v) from the overnight culture and mix thoroughly.
Pour over interaction array and allow to cool.
Incubate overnight at 37 °C or appropriate temperature.
Check for altered zones of inhibition.
Alternatively, from step 3:
-
4
Pour into a 12-inch Petri plate at a thickness of ~2 mm, and allow to cool.
-
5
Transfer this agar layer onto a sheet of plastic wrap by overturning the plate and separating the agar layer with a metal spatula.
-
6
Place the interaction array plate face down over the top of the agar layer on the plastic wrap.
-
7
Invert the entire assembly, firmly pressing the agar layer containing the test organism onto the surface of the interaction array.
This method has the advantage of not displacing spores that may be present on some actinomycete colonies. Spores picked up in poured agar may germinate overnight, obscuring or further altering zones of inhibition.
3. Activity-guided fractionation for amychelin isolation
As described above, our interpretation of the phenotypes observed in the binary assays is that small molecules produced by one actinomycete affect developmental or other cellular processes in the neighboring actinomycete. To identify the molecule responsible for these changes, activity-guided fractionation may be carried out. In this method, various purification steps are carried out concomitant with a specific activity assay to identify the fraction containing the active ingredient. This procedure is carried out repeatedly until a pure compound is obtained. The assay may vary depending on the interaction but is critical in guiding the purification of small quantities of a compound present in a pool of many other, more abundant, compounds. To identify the small molecule(s) that inhibited development in S. coelicolor in the interaction shown in Fig. 2A (red outline), we carried out activity-guided fractionation using the phenotypic effect on S. coelicolor as a read-out.
3.1. Biological assays to guide purification of small molecules
To ensure that liquid AA4 cultures also produce the compound that inhibits development in S. coelicolor, prepare several 3-mL cultures of AA4 in R2YE medium (Seyedsayamdost et al., 2011) and incubate in a rotating drum incubator at 30 °C.
At various time points (3–8 days), isolate the supernatant by centrifugation (2 min, 15000 × g, room temperature), followed by filtration through a 0.2-μm Nalgene membrane.
Prepare a lawn of S. coelicolor by plating ~105 spores, prepared as described in Section 2.3, on solid R2YE agar medium followed by incubating the plate for 16 h at 30 °C.
Punch 1-cm wells into the agar using the round end of a sterile, plastic 1-mL pipette tip. Then mix 50–100 μL of the supernatant with 200–250 μL of sterile, melted 0.7% agar solution and add the combined 300 μL mixture into the agar well.
Monitor the S. coelicolor morphology over the next 24–48 h.
3.2. Large-scale fermentation of AA4 and activity-guided fractionation
Here, purification of the bioactive small molecule amychelin, which inhibits development in S. coelicolor (Fig. 2A), is described. The procedure may be used as a general template for activity-guided purification, though compound-specific modifications will likely become necessary at each step.
- For large-scale cultivation of AA4, prepare 2 L of a minimal medium consisting of 25 mM N-[tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid (TES), pH 7.2, 2.34 mM MgSO4, 0.5 mM NaH2PO4, 0.5 mM K2HPO4, 55.4 mM glucose, 85.5 mM NaCl, and 66.5 mM glycine. Add 1 mL of R2YE trace element solution to each liter of the minimal medium. The trace element solution includes the following components:
Trace elements solution components (1 L) Amount (mg) ZnCl2 40 FeCl3•6H2O 200 CuCl2•2H2O 10 MnCl2•4H2O 10 Na2B4O7•10H2O 10 (NH4)6Mo7O24•4H2O 10 Inoculate four 2.8-L baffled Fernbach flasks, each containing 0.5 L of the minimal medium described above, with 20 μL of a concentrated spore solution and incubate on a shaker/incubator for 8 days at 240 rpm and 30 °C.
After 8 days, isolate the supernatant by centrifugation (10000 × g, 15 min, room temperature) and further by filtration through a 0.2-μm Nalgene filter unit.
Extract the filtrate with an equal volume of ethyl acetate in a separatory funnel. Carry out the S. coelicolor agar assay described above on the aqueous and organic fractions. Continue with the fraction that contains the active component.
Analyze the active fraction, in this case the aqueous phase, by HPLC-MS using a Phenomenex Luna C18 analytical column (5 μm, 4.6 × 100 mm) operating at 0.7 mL/min with a gradient of 10% MeCN in H2O to 100% MeCN over 25 min. Water and MeCN each contain 0.1% formic acid.
Concentrate the aqueous phase to 0.35 L in vacuo and fractionate using solid-phase extraction: wash a 0.5 g C18 Sep-Pak cartridge (Waters) with 10 column volumes (CV, 20 mL) of methanol, followed by 10 CV of 10% MeOH in water. Pass the aqueous phase through the column and collect the flow-through.
Elute bound material using 10 CV of 20%, 40%, 60%, 80% MeOH in water, followed by MeOH. Carry out the assay described above on each of the fractions and continue purifying the fraction that contains the active component.
Fractionate the flow-through further by another solid-phase extraction step with a 0.5-g C8 Sep-Pak cartridge (Waters) using the same procedure as for the C18 cartridge. Carry out the S. coelicolor developmental assay on each of the fractions.
Complete the purification of the active component by HPLC using a Phenomenex Luna C18 column (5 μm, 9.4 × 250 mm) operating at 3 mL/min eluting with a gradient of 10% MeCN in H2O to 26% MeCN in H2O over 20 min. Water and MeCN contain 0.1% formic acid.
3.3. Generation and purification of Ga- or Fe-amychelin complexes
The interaction in Fig. 2A (red outline) is mediated by the novel siderophore, amychelin. Certain functional groups in siderophores, such as oxazolines, thiazolines, or cyclic hydroxamates, may not be stable under the conditions typically employed in small-molecule isolations. In such cases, a siderophore may be stabilized by formation of its metal complexes. The Ga-complex is often generated to facilitate structural elucidation by NMR (Mohn et al., 1994, Sharman et al., 1995). The ferric Fe-complex may also be generated and used in structural elucidation by X-ray crystallography (Zalkin et al., 1964, Van der Helm and Poling, 1976, Miller et al., 2006).
Transfer an 8-mL aliquot of the mixture from step 7 in Section 3.2, which contains ~1 mg/mL amychelin, into a 40-mL scintillation vial equipped with a magnetic stir bar, and begin to gently stir at room temperature.
Slowly add a 10-fold excess of solid GaBr3 (or FeCl3) over five minutes. [Alternatively, Ga may be added from a stock solution as described in detail, previously (Sharman et al., 1995)].
Continue to stir for an additional 10 minutes at room temperature.
Incubate the reaction at 4 °C overnight.
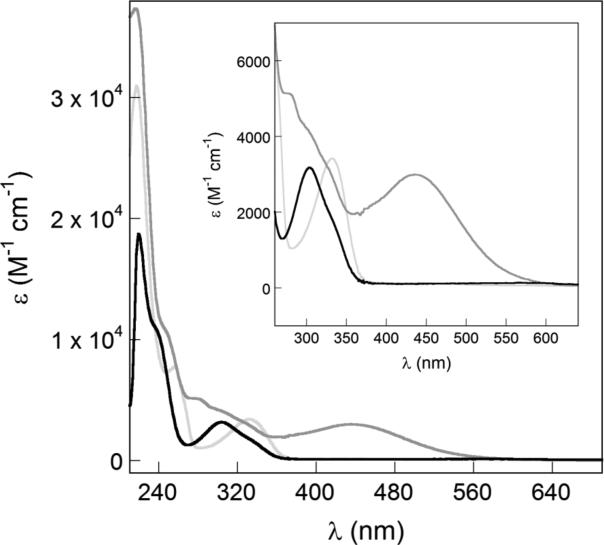
Addition of Fe leads to formation of a charge-transfer band (435 nm) which indicates Fe complexation (Fig. 3). A charge-transfer band is not formed with Ga because of the fully occupied d orbitals of the d10 GaIII center. A red-shift is observed in the absorbance spectrum of Ga-amychelin, likely due to deprotonation of the hydroxyl group of the phenyloxazoline moiety.
Purify Ga- or Fe-amychelin by HPLC using a Phenomenex Luna C18 column (5 μm, 21.2 × 250 mm) operating at 13 mL/min eluting with a gradient of 10% MeCN in H2O to 26% MeCN in H2O over 14 min. Water and MeCN contain 0.1% formic acid. Monitor the elution of Fe-amychelin at 435 nm; Ga-amychelin may be monitored using its 335-nm absorption band.
Figure 3.
UV-visible spectra of various forms of amychelin. Spectra for apo-amychelin (black trace), Ga-amychelin (light gray trace), and Fe-amychelin (dark gray trace). Ga- and Fe-amychelin were prepared as described in Section 3.3. The inset shows a magnified view of the charge transfer band for Fe-amychelin.
4. Determining the affinity of amychelin for Fe
The scarcity of free Fe in the environment and its requirement in actinomycete development sets up a fierce competition for its acquisition (Hider and Kong, 2010, Miethke and Marahiel, 2007, Challis and Hopwood, 2003). Our studies on the AA4–S. coelicolor interactions have pointed to an arms race in which bacteria evolve siderophores with ever-increasing Fe affinities in order to ensure access to Fe, and to prevent competitors from acquiring Fe. Studying the role of Fe in actinomycete development and competition requires determination of the affinity of siderophores for Fe. Two different constants, Kf and pFe, may be determined to quantitate the ability of a siderophore to bind Fe (Miethke and Marahiel, 2007, Harris et al., 1979b, Harris et al., 1979a, Abergel et al., 2008). Kf is the formation constant of the Fe-siderophore complex and it is generally reported as the fully deprotonated siderophore binding to Fe. Because these conditions are not physiologically relevant, determination of pFe may be preferred. pFe is defined as –log[FeIII], when, according to convention, Fe and siderophore are present at 1 μM and 10 μM, respectively. It is usually determined at pH 7.5 and does not require knowledge of the pKas of the chelating groups.
The assay is carried out in Hepes buffer consisting of 10 mM Hepes, 100 mM KCl, pH 7.5.
Generate and purify Fe-amychelin according to the procedure described above. Exchange Fe-amychelin into Hepes buffer to a stock concentration of 1 mM.
Determine the extinction coefficient, ε, of Fe-amychelin in Hepes buffer as a function of wavelength, λ, using the Beer-Lambert equation.
Carry out steps (2) and (3) for Fe-EDTA.
Make a stock solution of EDTA at 10–100 mM.
The assay can be carried out in a 96-well plate format or in a series of Eppendorf tubes. In a 96-well format, each well will contain 100 μL buffer, Fe-amychelin at a concentration of 10 or 100 μM and varied concentrations of EDTA ranging from 0.2-fold to 100-fold vs. Fe-amychelin, including a sample that contains only Fe-amychelin in Hepes buffer and no EDTA (control). The volume of each well is kept at 150 μL using Hepes buffer.
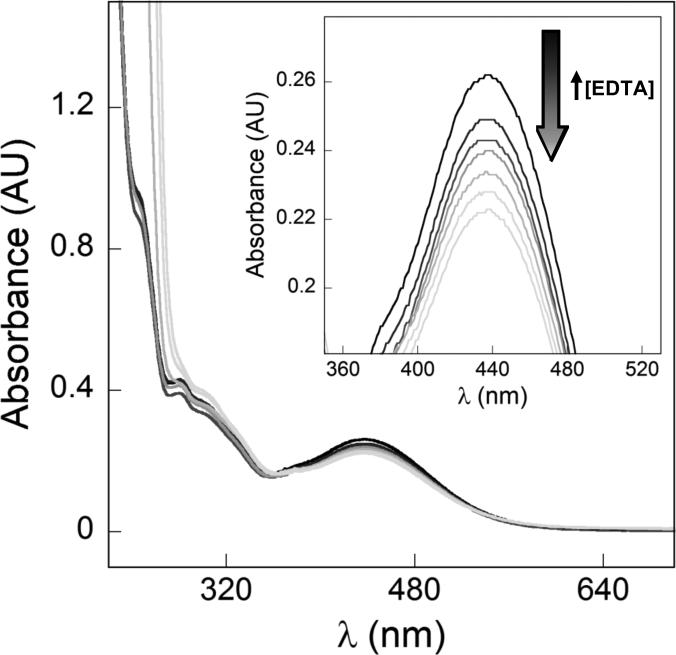
Cover the plate with a lid or a nylon cover to prevent evaporation. Allow the reactions to equilibrate at room temperature for at least 24 h. Determine the UV-vis spectrum of each well. Each spectrum is a composite of UV-vis traces for Fe-amychelin, Fe-EDTA, free amychelin and free EDTA (Fig. 4). Only Fe-amychelin has a charge-transfer band at 435 nm.
For each sample, subtract the Fe-amychelin control spectrum from the composite spectrum. This gives the concentration of Fe-amychelin using the Beer-Lambert equation. Compute the concentration of free amychelin by subtracting the concentration of Fe-amychelin from total amychelin equation (4).
Use the subtraction spectrum to compute the concentration of Fe-EDTA, and subsequently free EDTA using equation (5).
- Plot log {[apo-EDTA] / [apo-amychelin]} vs. log {[Fe-EDTA] / [Fe-amychelin]} and fit the data to equation (3), which has previously been derived, using non-linear regression analysis.
(3) (4) (5)
Figure 4.
EDTA competition assay to determine the pFe for amychelin. Fe-amychelin is incubated with various [EDTA]. Increasing EDTA concentrations result in disappearance of the Fe-amychelin charge transfer band, indicating conversion of Fe-amychelin into apo-amychelin and Fe-EDTA. The data can be deconvoluted as described in Section 4 to obtain a pFe for amychelin.
5. Identifying the amychelin biosynthetic gene cluster
The main advantage in using the Broad Institute actinomycete library (http://www.broadinstitute.org/annotation/genome/streptomyces_group) in the binary assays above is that the genome sequences of these strains have been determined, allowing us to connect the small molecules to their biosynthetic clusters. To link amychelin to its biosynthetic cluster, we first focused on the N-terminal hydroxybenzoyl group. The enzymes that incorporate this moiety into non-ribosomal peptide synthetase (NRPS)-derived natural products have been well-studied. Performing a BLAST alignment of the protein sequence of one such enzyme from the actinomycete Mycobacterium smegmatis against the predicted proteins from the genome of AA4 led us to a gene cluster which appeared to contain the necessary genes for generating a hydroxybenzoyl-containing siderophore. Bioinformatic analysis of the genes in the cluster and gene interruption studies confirmed that this cluster was responsible for amychelin biosynthesis, allowing us to propose a model for the biosynthesis of amychelin. This general procedure allows rudimentary identification of possibly interesting biosynthetic steps that may be studied further by enzymology or in in vivo studies. Because these studies may reveal new and unusual enzymatic transformations, such biosynthetic gene clusters are not only reservoirs of new small molecules but also reservoirs of new enzymatic reactions. In Table 2, we summarize a number of online bioinformatic tools that may be used for initial analyses of the genes within secondary metabolite biosynthetic gene clusters.
Table 2.
Bioinformatic tools for functional prediction of genes in biosynthetic gene clusters.
| Bioinformatic tool (website) | Brief tool description |
|---|---|
| ClustScan (http://bioserv.pbf.hr/cms/) | A database containing genetic and biochemical information on secondary metabolites derived from PKS, NRPS, and hybrid PKS/NRPS clusters. Allows identification and annotation of biosynthetic clusters in a genome or metagenome sequence. |
| BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) | Comparison of protein or DNA sequence to a database of available sequences. Allows identification of genes or proteins that most closely resemble the query sequence. |
| FASTA (http://www.ebi.ac.uk/Tools/sss/fasta/) | Homology search database, similar to BLAST. FASTA and BLAST are comparable for highly similar sequences, though BLAST is faster without significant loss of sensitivity. FASTA may be better for less similar sequences. |
| InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/) | InterPro contains a number of member databases, which provide amino acid sequence motifs for known protein families or domains. These identifiable features in known proteins are applied to a query sequence to provide a functional prediction. |
| PKS/NRPS Analysis (http://nrps.igs.umaryland.edu/nrps/) | Prediction of various NRPS and PKS domains (condensation/ketosynthase, adenylation/acyltransferase, etc.) within an NRPS or PKS protein sequence. Provides a predicted specificity for NRPS A-domains by comparison of the active site residues of known A-domains to those of a query sequence. |
| SBSPKS (www.nii.ac.in/sbspks.html) | Sequence and structure based analysis of PKSs. Query sequence is compared to a database of experimentally characterized PKSs with known reactions and protein structures. |
| Integrated Microbial Genomes (http://img.jgi.doe.gov/) | Platform for genome browsing and annotation including a number of useful and user-friendly tools for comparative analysis of genes, gene clusters, genomes, and protein function. |
6. Transcriptomic analysis using Nanostring technology
Gene expression at the level of transcription is among the most commonly measured experimental variables in modern biology. Methods for measuring transcriptomic output range from global, such as RNAseq and microarrays, down to single gene using qRTPCR. Recently, a new methodology, called Nanostring, has become available (Geiss et al., 2008). Nanostring allows for the quantification of transcripts of up to 800 genes in a single reaction, placing it as an intermediate technology (between global and single gene techniques) in terms of the number of genes per assay. This technology works by annealing fluorescently bar-coded reporter oligonucleotides to gene transcripts in the test sample, and subsequently capturing and counting the complex of gene transcript and the labeled oligonucleotides. The result is a direct digital count of the number of transcripts in a sample, with no bias introduced from reverse transcriptase amplification. Perhaps even more appealing is the fact that traditional RNA isolation is not required, adding to the relative ease and speed of obtaining transcriptomic data. Here we provide a general method for preparing actinomycete samples from colonies grown on solid medium for Nanostring analysis. More information regarding the method and data analysis can be found at the manufacturer's website: http://www.nanostring.com/.
Grow actinomycetes on solid medium until the desired time-point.
Scrape colony(s) from agar using a sterile razor blade, minimizing excess agar.
Transfer the colony onto the end of a clean ceramic pestle.
Pour a small amount of liquid nitrogen into a clean ceramic mortar.
Slowly submerge the tip of the pestle containing the colony into the liquid nitrogen, allowing the biomass to freeze.
Once frozen, grind the colony biomass thoroughly as the liquid nitrogen evaporates.
Add 150 μL Buffer RLT (lysis buffer available from Qiagen) to the pestle.
Grind until an even-looking biomass crude lysate is obtained.
Use a pipette to transfer as much of the lysate as possible to a sterile 1.4-mL tube. 20–50 μL is more than enough for Nanostring analysis.
Freeze the samples immediately by dipping the tube into liquid nitrogen, or by placing immediately at −80 °C. Store until use.
Add 1–2 μL of lysate to Nanostring reactions as indicated in the crude lysate manufacturer protocols.
Summary
This chapter provides a guide for creating efficient screens to examine the biology and chemistry of binary interactions among morphologically-complex actinomycetes. Such a screen led to the observation that Amycolatopsis sp. AA4 inhibited development of S. coelicolor aerial hyphae. The chemical characterization of this interaction, which led to the discovery of a siderophore, amychelin, provides an example of how such interactions can drive novel compound discovery. While the format suggested here is focused on actinomycetes, one can imagine how these regimes could be adapted for other fungi or bacteria, for example those of the human gut microbiota, or from other varied and complex environments. Bacteria of many phylogenetic backgrounds are capable of producing pigments or have complex colony morphologies that could be altered through interspecies interactions. Genetically tractable test organisms can also be engineered such that fluorescent protein expression driven from a promoter of interest can be assayed in the context of chemically-mediated interactions. Creative variations of these screens may thus open a door into the previously unexplored chemistry of interactions between microorganisms; a chemical realm potentially occupied by many useful and unique metabolites.
Acknowledgments
This work was supported by the National Institutes of Health (grant GM82137 to R.K., and grants AI057159 and GM086258 to J.C.). M.R.S. is supported by a NIH K99 Pathway to Independence Award (Grant 1K99 GM086799-01). M.F.T. is a NIH Postdoctoral Fellow (grant 5F32GM089044-02).
References
- Abergel RJ, Zawadzka AM, Raymond KN. Petrobactin-mediated iron transport in pathogenic bacteria: coordination chemistry of an unusual 3,4-catecholate/citrate siderophore. J. Am. Chem. Soc. 2008;130:2124–2125. doi: 10.1021/ja077202g. [DOI] [PubMed] [Google Scholar]
- Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 2008;8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Berdy J. Bioactive Microbial Metabolites. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Challis GL, Hopwood DA. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl. 2):14555–14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater KF. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- Copeland A, Lapidus A, Glavina Del Rio T, Nolan M, Lucas S, Chen F, Tice H, Cheng JF, Bruce D, Goodwin L, Pitluck S, Mikhailova N, et al. Complete genome sequence of Catenulispora acidiphila type strain (ID 139908). Stand. Genomic. Sci. 2009;1:119–125. doi: 10.4056/sigs.17259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Krogan NJ. The next frontier of systems biology: higher-order and interspecies interactions. Genome. Biol. 2010;11:208. doi: 10.1186/gb-2010-11-5-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Goodfellow M, Fiedler HP. A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek. 2010;98:119–142. doi: 10.1007/s10482-010-9460-2. [DOI] [PubMed] [Google Scholar]
- Hamaki T, Suzuki M, Fudou R, Jojima Y, Kajiura T, Tabuchi A, Sen K, Shibai H. Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J. Biosci. Bioeng. 2005;99:485–492. doi: 10.1263/jbb.99.485. [DOI] [PubMed] [Google Scholar]
- Harris WR, Carrano CJ, Raymond KN. Coordination chemistry of microbial iron transport compounds. 16. Isolation, characterization, and formation constants of ferric aerobactin. J. Am. Chem. Soc. 1979a;101:2722–2727. [Google Scholar]
- Harris WR, Carrano CJ, Raymond KN. Spectrophotometric determination of the proton-dependent stability constant of ferric enterobactin. J. Am. Chem. Soc. 1979b;101:2213–2214. [Google Scholar]
- Hider RC, Kong X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- Hop DV, Sakiyama Y, Binh CT, Otoguro M, Hang DT, Miyadoh S, Luong DT, Ando K. Taxonomic and ecological studies of actinomycetes from Vietnam: isolation and genus-level diversity. J. Antibiot. 2011;64:599–606. doi: 10.1038/ja.2011.40. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ. Microbiol. 2005a;7:1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Mincer TJ, Williams PG, Fenical W. Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek. 2005b;87:43–48. doi: 10.1007/s10482-004-6540-1. [DOI] [PubMed] [Google Scholar]
- Jiang S, Sun W, Chen M, Dai S, Zhang L, Liu Y, Lee KJ, Li X. Diversity of culturable actinobacteria isolated from marine sponge Haliclona sp. Antonie Van Leeuwenhoek. 2007;92:405–416. doi: 10.1007/s10482-007-9169-z. [DOI] [PubMed] [Google Scholar]
- Kim BY, Kshetrimayum JD, Goodfellow M. Detection, selective isolation and characterisation of Dactylosporangium strains from diverse environmental samples. Syst. Appl. Microbiol. 2011;34:606–616. doi: 10.1016/j.syapm.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Kurahashi M, Fukunaga Y, Sakiyama Y, Harayama S, Yokota A. Euzebya tangerina gen. nov., sp. nov., a deeply branching marine actinobacterium isolated from the sea cucumber Holothuria edulis, and proposal of Euzebyaceae fam. nov., Euzebyales ord. nov. and Nitriliruptoridae subclassis nov. Int. J. Syst. Evol. Microbiol. 2010;60:2314–2319. doi: 10.1099/ijs.0.016543-0. [DOI] [PubMed] [Google Scholar]
- Lazzarini A, Cavaletti L, Toppo G, Marinelli F. Rare genera of actinomycetes as potential producers of new antibiotics. Antonie Van Leeuwenhoek. 2000;78:399–405. [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Parkin S, Fetherston JD, Perry RD, Demoll E. Crystal structure of ferric-yersiniabactin, a virulence factor of Yersinia pestis. J. Inorg. Biochem. 2006;100:1495–1500. doi: 10.1016/j.jinorgbio.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Mohn G, Koehl P, Budzikiewicz H, Lefevre JF. Solution structure of pyoverdin GM-II. Biochemistry. 1994;33:2843–2851. doi: 10.1021/bi00176a014. [DOI] [PubMed] [Google Scholar]
- Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural Products As Sources of New Drugs over the 30 Years from 1981 to 2010. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM, Holbeck S, Sausville EA. Natural products and derivatives as leads to cell cycle pathway targets in cancer chemotherapy. Curr. Cancer. Drug Targets. 2002;2:279–308. doi: 10.2174/1568009023333791. [DOI] [PubMed] [Google Scholar]
- Oh DC, Poulsen M, Currie CR, Clardy J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 2009a;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DC, Poulsen M, Currie CR, Clardy J. Sceliphrolactam, a polyene macrocyclic lactam from a wasp-associated Streptomyces sp. Org. Lett. 2011;13:752–755. doi: 10.1021/ol102991d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DC, Scott JJ, Currie CR, Clardy J. Mycangimycin, a polyene peroxide from a mutualist Streptomyces sp. Org. Lett. 2009b;11:633–636. doi: 10.1021/ol802709x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 2008;190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- Poulsen M, Oh DC, Clardy J, Currie CR. Chemical analyses of wasp-associated streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS One. 2011;6:e16763. doi: 10.1371/journal.pone.0016763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EW. Trading molecules and tracking targets in symbiotic interactions. Nat. Chem. Biol. 2008;4:466–473. doi: 10.1038/nchembio.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR. Bacterial protection of beetle-fungus mutualism. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost MR, Traxler MF, Zheng SL, Kolter R, Clardy J. Structure and biosynthesis of amychelin, an unusual mixed-ligand siderophore from Amycolatopsis sp. AA4. J. Am. Chem. Soc. 2011;133:11434–11437. doi: 10.1021/ja203577e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharman GJ, Williams DH, Ewing DF, Ratledge C. Isolation, purification and structure of exochelin MS, the extracellular siderophore from Mycobacterium smegmatis. Biochem. J. 1995;305:187–196. doi: 10.1042/bj3050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight PD, Kolter R. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol. 2009;63:99–118. doi: 10.1146/annurev.micro.091208.073248. [DOI] [PubMed] [Google Scholar]
- Tan GY, Ward AC, Goodfellow M. Exploration of Amycolatopsis diversity in soil using genus-specific primers and novel selective media. Syst. Appl. Microbiol. 2006;29:557–569. doi: 10.1016/j.syapm.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Ueda K, Kawai S, Ogawa H, Kiyama A, Kubota T, Kawanobe H, Beppu T. Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. J. Antibiot. 2000;53:979–982. doi: 10.7164/antibiotics.53.979. [DOI] [PubMed] [Google Scholar]
- Van der Helm D, Poling M. The crystal structure of ferrioxamine E. J. Am. Chem. Soc. 1976;98:82–86. doi: 10.1021/ja00417a014. [DOI] [PubMed] [Google Scholar]
- Watve MG, Tickoo R, Jog MM, Bhole BD. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- Wellington EMH, Toth IK. Actinomycetes. In: Weaver RW, Angle S, Bottomley P, Bezdicek S, Smith S, Tabatabai A, Wollum A, editors. Methods of Soil Analysis. SSSA Book Series (No. 5) Soil Science Society of America; Madison, WI: 1994. pp. 269–290. [Google Scholar]
- Yamanaka K, Oikawa H, Ogawa HO, Hosono K, Shinmachi F, Takano H, Sakuda S, Beppu T, Ueda K. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology. 2005;151:2899–2905. doi: 10.1099/mic.0.28139-0. [DOI] [PubMed] [Google Scholar]
- Zalkin A, Forrester JD, Templeton DH. Crystal and Molecular Structure of Ferrichrome A. Science. 1964;146:261–263. doi: 10.1126/science.146.3641.261. [DOI] [PubMed] [Google Scholar]
- Zhao W, Zhong Y, Yuan H, Wang J, Zheng H, Wang Y, Cen X, Xu F, Bai J, Han X, Lu G, Zhu Y, et al. Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res. 2010;20:1096–1108. doi: 10.1038/cr.2010.87. [DOI] [PubMed] [Google Scholar]