Summary
Staphylococcus aureus virulence is regulated when secreted autoinducing peptides (AIPs) are recognized by a membrane-bound receptor histidine kinase (RHK), AgrC. Some AIPs are agonists of virulence gene expression, while others are antagonists. It is unclear how AIP binding regulates AgrC activity. Here, we reconstitute an AgrC family member, AgrC-I, using nanometer-scale lipid bilayer discs. We show that AgrC-I requires membranes rich in anionic lipids to function. The agonist, AIP-I, binds AgrC-I non-cooperatively in a 2:2 stoichiometry, while an antagonist ligand, AIP-II, functions as an inverse agonist of the kinase activity. We also demonstrate the kinase and sensor domains in AgrC are connected by a helical linker whose conformational state exercises rheostat-like control over the kinase activity. Binding of agonist or inverse-agonist peptides results in twisting of the linker in different directions. These two observations provide a view of the molecular motions triggered by ligand binding in an intact membrane-bound RHK.
Introduction
The staphylococcal agr locus encodes a quorum sensing (QS) system that controls the expression of virulence and other accessory genes (Novick and Geisinger, 2008; Thoendel et al., 2011). It consists of two oppositely oriented transcription units, of which one encodes four proteins, AgrBDCA, involved in producing or sensing of the autoinducer peptide, AIP, and the other encodes a regulatory RNA that is the effector of target gene regulation (Figure S1A). Importantly, as with peptide ligands in QS modalities in other Gram-positive (G+) bacteria, the AIP is proteolytically processed from a ribosome-translated precursor. Agr is universally conserved among the Staphylococci and its variation is thought to be the driving force behind speciation within the genus (Dufour et al., 2002). In the commensal pathogen Staphylococcus aureus, agr occurs in 4 allelic variants that differ in their ligand-receptor specificities, such that heterologous receptor-ligand interactions are in general inhibitory (Ji et al., 1997). Sensing of AIP is achieved through the AgrCA two-component system (TCS) in which AgrC is a membrane-bound receptor-histidine kinase (RHK) that undergoes autophosphorylation following cognate AIP binding and then transfers the phosphoryl group to a conserved response regulator (RR), AgrA. Phosphorylated AgrA binds to the region between the agr promoters, activating both, thus completing a strong positive feedback loop (Reyes et al., 2011). Considerable effort has gone into defining the structure-activity relationships within the AIPs and AgrC that govern agonism and antagonism (George et al., 2008; Lyon et al., 2002; Mayville et al., 1999; Tal-Gan et al., 2013). These studies have exclusively relied on cell-based assays involving coupled transcriptional readouts. Consequently, many questions remain unanswered related to the thermodynamics and kinetics of AgrC activation/inhibition by AIPs.
AgrC from S. aureus is a 430-residue, 6-span integral membrane protein that adopts a modular architecture shared among all RHK proteins, with a N-terminal sensor domain connected to a conserved C-terminal histidine kinase (HK) domain (Figure 1A). Notably, the protein apparently lacks any canonical transmission modules frequently found in other RHKs between the sensor and HK (Gao and Stock, 2009). Genetic complementation studies indicate that AgrC forms obligate dimers, and that auto-phosphorylation between the protomer subunits can occur in trans (Cisar et al., 2009). The AgrC HK domain (AgrC-IHK) possesses all enzymatic activities required for the TCS signaling, which are tightly regulated by AIP binding to the membrane-embedded sensor domain. However, we currently have no mechanistic information on how this regulation is achieved. Structural and functional studies on isolated HK domains have given rise to models of receptor activation involving rotation of helices that comprise the so-called dimerization and histidine phosphotransfer (DHp) subdomain, a helical hairpin region in the HK involved in the homodimerization of RHKs and containing the histidine phospho-acceptor residue (Albanesi et al., 2009; Casino et al., 2009; Marina et al., 2005). These helical movements are thought to modify packing of the DHp leading to a repositioning of the catalytic and ATP-binding (CA) subdomain such that phosphorylation of the His is, or is not, permitted on the basis of spatial proximity (Casino et al., 2010). While this ‘helix rotation’ model is attractive, there is a paucity of supporting biochemical data in the context of native, full-length RHK proteins, especially those with membrane-embedded sensors like AgrC. Studying such receptors in vitro entails biochemical reconstitution into a lipid bilayer environment, which is technically challenging. Although liposomal systems have been employed in the reconstitution of a few RHKs that recognize membrane-permeable autoinducers (Timmen et al., 2006; Wei et al., 2012), the closed topology of liposomes prevents any definitive characterization of receptors that transduce membrane-impermeable signals. In the present study, we have reconstituted the complete S. aureus AgrCA TCS module using nanometer-scale lipid bilayer discs, or nanodiscs. We use this system to comprehensively characterize the biochemistry of the TCS and uncover several novel properties, namely: AgrC lacks phosphatase activity, while AgrA is capable of relatively rapid auto-catalyzed dephosphorylation; agonist AIP binding shifts the AgrC autokinase equilibrium in favor of phosphorylation, while antagonist AIP binding can inhibit both AgrC phosphorylation and phosphoryl group transfer from AgrC to AgrA. Importantly, we show that binding of agonist versus antagonist AIP twists the helix linking the sensor and HK domains of AgrC-I in opposite directions, resulting in either activation or suppression of the autokinase activity of AgrC-I, respectively.
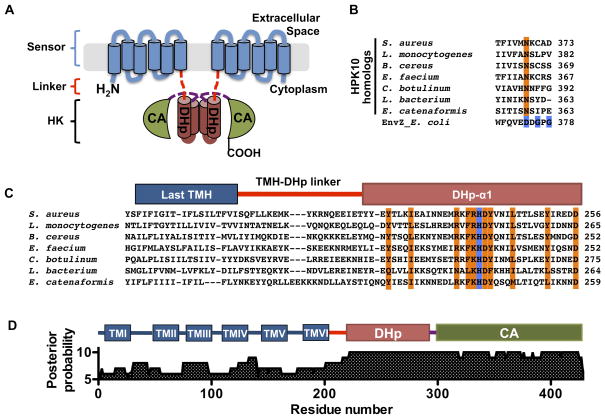
Figure 1. Domain architecture of AgrC-I, a member of the HPK10 subfamily, see also Figure S1 and Table S1.
(A) Domain architecture of AgrC-I. The protein is shown as a homodimer with the integral membrane sensor domain and cytoplasmic DHp and CA subdomains colored in blue, brown and green, respectively. TMH-DHp and DHp-CA linkers regions are depicted as red and purple dashed lines. (B) Alignment of the G1-box region of selected members of the HPK10 subfamily: conserved residues in canonical G1-box are highlighted in blue for EnvZ; ‘G1-box Asn’ is in orange. (C) HPK10 subfamily alignment from the last TMH to the first DHp helix: highly conserved residues and the phospho-acceptor histidine are highlighted in orange and blue, respectively. (D) Posterior probabilities of the 18-sequence sub-alignment according to the profile HMM: positions in the HMM are displayed with numbering corresponding to residues in AgrC-I; predicted domains of AgrC-I are shown above.
Results
AgrC-I is a member of a distinct RHK subfamily
We elected to focus on the TCS from a group 1 agr variant of S. aureus (Figure 1A and S1A). The cognate RHK, AgrC-I, falls within a rare RHK subfamily (originally referred to as the group 10 histidine protein kinase, or HPK10 subfamily) that differs from most other HKs by having a distinct sequence pattern, F[RK]HDYxN, around the histidine phosphoryl-acceptor residue (known as the H-box), and by the absence of a DxGxG motif conserved in the CA subdomain of most other subfamilies (known as the G1-box) (Grebe and Stock, 1999). As the number of sequenced bacterial genomes has grown dramatically since the original classification of HKs, we first attempted to identify more HPK10 homologs using a BLAST search of RHK sequences that match a slightly less restricted H-box pattern, [FILV][RK]H[DE][YF]. The search returned 330 non-redundant RHK sequences, 326 of which shared two additional distinct features (Table S1). Firstly, consistent with the initial classification, the sequence hits lacked a canonical G1-box: glycine is strongly under-represented at both of its conserved loci (< 5%) and, more surprisingly, the Asp residue is replaced by an invariant Asn, which we refer to as the ‘G1-box Asn’ (Figure 1B). Secondly, although covering 194 species from 22 taxonomic families, all hits come from low-GC G+ bacteria in line with 15 founding members of the HPK10 subfamily. Interestingly, the four sequence hits that did not conform to these two criteria contain both a canonical G1-box and originate from Gram-negative (G−) bacteria or archaea.
Among the 326 sequences, 318 were predicted to span the membrane at least once and 286 to span five times or more (Figure S1B and Table S1). Thus, the majority of the subfamily is probably involved in sensing extracellular or intra-membrane signals with a membrane-embedded sensor domain. To better delineate the domain boundaries in AgrC-I, we performed multiple sequence alignment for those 286 RHKs with five or more putative transmembrane helices (TMH), and analyzed a sub-alignment of 18 sequences, each representing a taxonomic family, using the hidden Markov model (HMM) generated from the overall alignment. These analyses revealed a less conserved, short linker between the well-aligned last TMH and the DHp subdomain in all sequences (Figure 1C, S1C and S1D). Specifically in AgrC-I, the conserved DHp subdomain that aligns to the HMM with high confidence begins at residue 222 rather than at residue 200 immediately after the last TMH (Figure 1D). We therefore refer to AgrC-I1-200 as the sensor domain and AgrC-I201-221 as the TMH-DHp linker region. As described previously, the DHp and CA subdomains are defined by residues AgrC-I222-290 and AgrC-I293-430, respectively, with a very short inter-subdomain linker between them (Thoendel et al., 2011).
Functional AgrC-I dimer embedded in nanodiscs
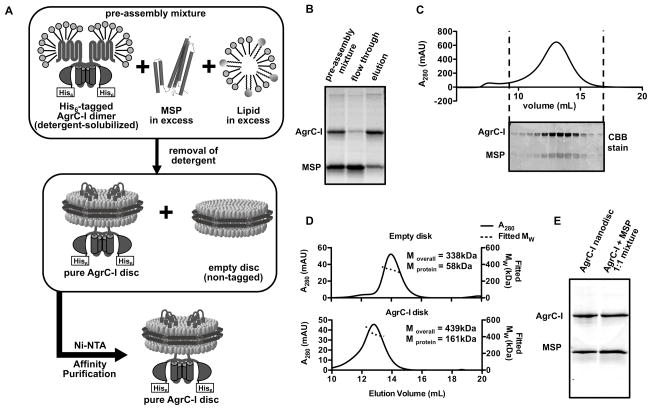
To reconstitute the AgrCA TCS in vitro, we overexpressed AgrC-I and AgrA-I in E. coli as His6-tagged recombinant proteins and purified them (Figure S2A and B). The detergent-solubilized AgrC-I did not have any autokinase activity regardless of the presence of AIP (data not shown). We therefore attempted to restore the activity by reconstituting the receptor into nanodiscs. Each nanodisc consists of a piece of disc-shaped lipid bilayer surrounded and stabilized by two copies of a membrane scaffold protein (MSP) (Denisov et al., 2004). To prepare nanodiscs containing one copy of the AgrC-I dimer per disc, purified AgrC-I was added to mixed micelles containing excess MSP and lipids (DMPC/DMPG, see below), lowering the chance of incorporating more than one copy of AgrC-I dimer in each nanodisc (Figure 2A). Detergent was then removed to drive self-assembly, and nanodiscs containing AgrC-I (AgrC-I discs) were separated from empty discs using Ni-NTA affinity purification (Figure 2A and B). Purified AgrC-I discs gave a single, symmetric peak on size-exclusion chromatography (SEC) from which the peak fractions were pooled for further studies (Figure 2C and S2C). The product has a narrow molecular weight distribution analyzed by SEC in tandem with multi-angle light scattering (SEC-MALS, Figure 2D). The expected stoichiometry of one copy of AgrC-I dimer per disc was also confirmed in the SEC-MALS experiment, as well as using SDS-PAGE employing ratiometric standards (Figure 2E).
Figure 2. Reconstitution of AgrC-I dimers into nanodiscs, see also Figure S2.
(A) Schematic showing the workflow used for reconstituting AgrC-I dimers into nanodiscs. (B) Purification of reconstituted AgrC-I discs by Ni-NTA affinity chromatography as analyzed by SDS-PAGE with Coomassie-brilliant blue (CBB) staining. (C) AgrC-I discs analyzed by SEC. Fractions collected between the dashed lines were further analyzed by SDS-PAGE with CBB staining (bottom). (D) SEC-MALS analysis of empty and purified AgrC-I discs. Moverall, average molecular weight (Mw) of the peak; Mprotein, Mw of proteins in each case; AgrC-I = 51 kDa, MSP = 29 kDa. (E) Stoichiometry of AgrC-I discs as analyzed by SDS-PAGE with CBB staining: AgrC-I discs (left lane) and an equimolar mixture of purified AgrC-I and MSP (right lane).
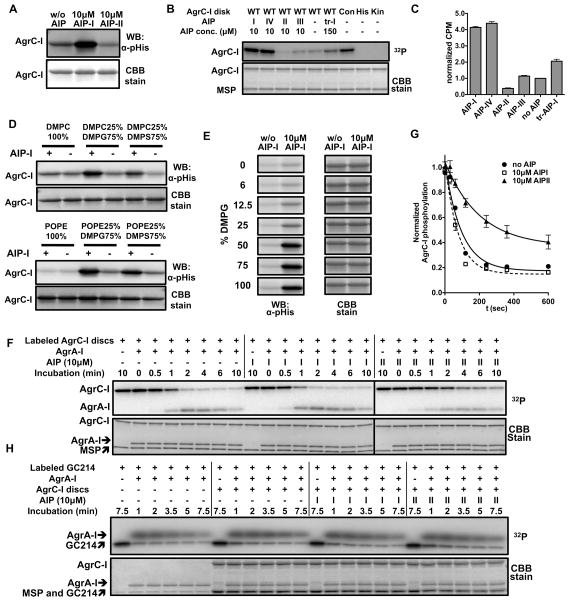
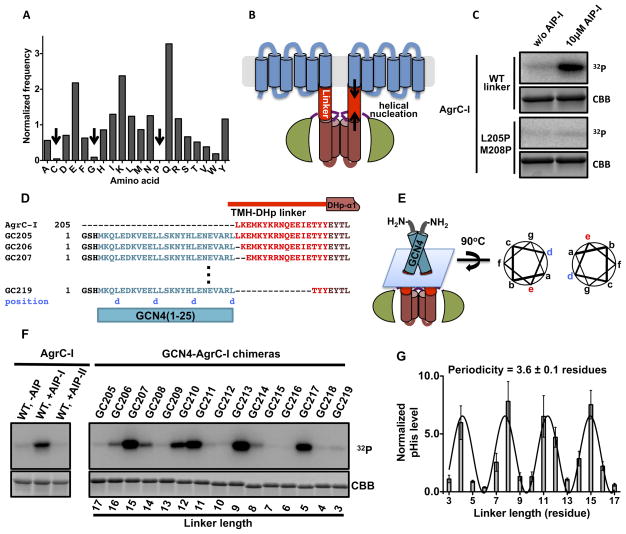
The autokinase activity of AgrC-I was investigated using two readouts: autoradiography/scintillation counting employing [γ-32P]-ATP and immunoblotting employing a newly developed, anti-phosphohistidine (pHis) antibody (Kee et al., 2013). Both readouts indicated that the nanodisc embedded AgrC-I dimer had a basal level of autokinase activity (Figure 3A and 3B). Native AIPs from all four S. aureus agr variants also behaved in line with what has been observed previously in cell-based studies (Lyon et al., 2002): AIP-I, the cognate AIP, strongly stimulated the autokinase activity of AgrC-I; AIP-II suppressed AgrC-I activity below the basal level and so acted as an inverse agonist, AIP-III behaved as a neutral antagonist and so did not effect the basal activity, and AIP-IV was an agonistic ligand with similar efficacy to AIP-I (Figure 3B, lanes 1–5 and Figure 3C). Interestingly, a truncated AIP analog, tr-AIP-I, behaved as a partial agonist of AgrC-I when added at saturating concentrations (Figures 3B, 3C and S3A). Importantly, autophosphorylated AgrC-I was stable to common buffer molecules, but was labile to hydroxylamine treatment, a signature of pHis-containing proteins (Figure S3B and S3C). The phosphorylation level also decreased upon incubation with ADP, which is indicative of the reverse autokinase reaction seen in other systems (Grimshaw et al., 1998) (Figure S3C). As expected, autokinase activity was abolished when we reconstituted AgrC-I carrying inactivating mutations in the DHp subdomain (H239Q) or CA subdomains (G394A/G396A) (Figure 3B, lanes 8 and 9). Conversely, incorporation of a constitutively active AgrC-I mutant, R238H, into nanodiscs led to significantly elevated levels of basal activity (Figure 3B, lane 7), providing further evidence that this in vitro reconstitution system replicates the cellular behavior of this RHK.
Figure 3. Biochemical activities associated with reconstituted AgrC-I, see also Figure S3.
(A) AgrC-I autokinase activity: AgrC-I discs were phosphorylated with 1 mM ATP at 37 °C for 3 minutes. AIP peptide was included as indicated. pHis levels in AgrC-I were analyzed by anti-pHis western blotting (upper). The blot was CBB stained thereafter as a loading control (lower). (B and C) Autokinase assays with AgrC-I variants or AIP analogs: nanodiscs containing AgrC-I wild-type (WT), constitutive mutant R238H (Con), DHp-inactive mutant H239Q (His) or CA-inactive mutant G394A/G396A (Kin) were phosphorylated with 20 μM [γ-32P] ATP at 37 °C for 40 minutes. WT-AgrC-I was treated with indicated AIP peptides; truncated AIP-I is referred to as tr-I or tr-AIP-I. pHis levels were analyzed by autoradiography in (B) or quantified using scintillation counting in (C). Bargraph shows CPM values normalized to the AIP-free reaction. Error bars = SD (n = 3). (D and E) Lipid dependence of AgrC-I autokinase activity: AgrC-I discs reconstituted with various lipids (D) or DMPC and DMPG at a series of ratios (E) were autophosphorylated in the presence or absence of excess AIP-I and analyzed by anti-pHis western-blotting as in A. (F) Phospho-relay from AgrC-I to AgrA-I: AgrA-I was incubated with AgrC-I discs labeled with [γ-32P]-ATP and indicated AIPs. Aliquots removed at various time-points were analyzed by SDS-PAGE followed by autoradiography. (G) Quantification of the autoradiograms in panel F showing decay of pHis levels in AgrC-I (normalized to t = 0) as a function of time. Error bars = range (n = 2). (H) Dephosphorylation of AgrA-I: AgrA-I (2 μM) was phosphorylated by incubation with the constitutively active chimeric protein GC214 (0.1 μM dimer – see text for details) labeled with a [32P]-phosphoryl group. This reaction was then supplemented with 2μM AgrC-I discs and AIPs as indicated.
The lipid mixture used for nanodisc reconstitution consisted of DMPC and DMPG at a molar ratio of 1:3; chosen to mimic the highly anionic membrane environment native to S. aureus (Beining et al., 1975). To investigate the influence of lipid composition on AgrC-I activity, we performed assays with receptors embedded in nanodiscs with differing lipid composition. Zwitterionic bilayers consisting of either DMPC or POPE eliminated the ability of AIP-I to activate the receptor (Figure 3D, lanes 1 and 2), despite the fact that AgrC-I interacted with the AIP activator in these discs, as shown by co-migration assays (Figure S3D). However, lipid environments enriched for anionic DMPG or DMPS largely restored the responsiveness of the receptor to AIP (Figure 3D, lanes 3 to 6). To further dissect the role of zwitterionic and anionic lipids in AgrC-I activation, we reconstituted the receptor into nanodiscs consisting of DMPC and/or DMPG at various ratios. The magnitude of AIP-I-dependent activation increased gradually as the DMPG content increased, reaching a maximal at a DMPG content of 75% (Figure 3E). These data suggest that a reasonably high negative-charge density, rather than the presence or absence of any specific lipid per se, is required for full AgrC-I activity.
Phospho-relay from AgrC-I to AgrA-I was reconstituted by adding the purified full-length RR to AgrC-I discs that had been pre-phosphorylated with [γ-32P]-ATP and then exchanged into an ATP-free buffer. Time courses revealed efficient phosphoryl group transfer between the RHK and RR, with a t1/2 of 66 seconds (Figure 3F, lanes 2–8 and Figure 3G). The inclusion of AIP-1 in the mix had little effect on the rate of transfer (Figure 3F, lanes 10–16 and Figure 3G, t1/2 = 52 sec), indicating that the agonist AIP acts solely at the level of AgrC phosphorylation. Surprisingly, we observed that the presence of the inhibitor AIP-II slowed down the rate of transfer (Figure 3F, lanes 18–24 and Figure 3G, t1/2 = 140 sec). Thus, this antagonist AIP can inhibit the TCS both at the level of HK autophosphorylation and phospho-relay to the RR.
Next we asked if AgrC possesses any phosphatase activity against phosphorylated AgrA, an activity common among RHKs (Gao and Stock, 2009). Since AgrA-I forms precipitate quickly following acetyl-phosphate treatment (data not shown), we employed an enzymatic approach to phosphorylate AgrA-I in situ using a soluble, highly active AgrC variant (GC214, vide infra) as the phospho-donor. We then monitored the decay of [32P]-phosphorylated AgrA-I in the presence or absence of AgrC-I disks. Surprisingly, we found no acceleration of AgrA-I dephosphorylation in the presence of AgrC-I disks at 20-fold excess to the GC214 phospho-donor (Figure 3H, compare lanes 1–6 to 7–12, and Figure S3E). Inclusion of either AIP-I or AIP-II along with AgrC-I discs also had no effect (Figure 3H, lanes 13–18, 19–24 and Figure S3E). Using the same strategy, we ruled out any detectable phosphatase activity in GC214 (data not shown). Consequently, the decrease in AgrA-I phosphorylation levels is due to the self-catalyzed dephosphorylation of the RR, with a t1/2 of 3.9 minutes (Figure S3F and Figure S3G). We therefore conclude that AIP signals do not regulate agr signaling at the level of RR dephosphorylation.
AgrC-I binds to AIP-I at 2:2 stoichiometry
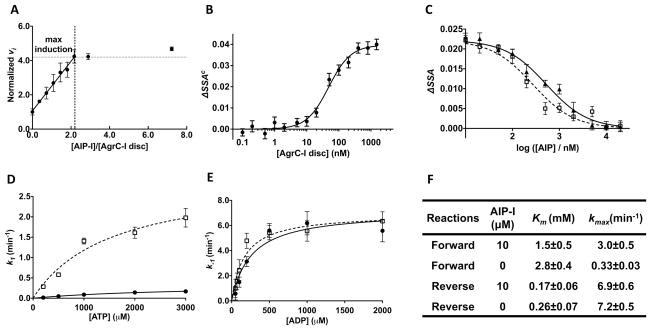
Next we turned to the binding interaction between AgrC-I and its AIP ligands. First we showed that the chemically labile thiolactone bond in AIP-I or AIP-II is stable upon incubation with AgrC-I discs (Figure S4A). To determine the stoichiometry of the AIP-AgrC interaction, we titrated AIP into AgrC-I nanodiscs fixed at a concentration of 1.25 μM, about two orders of magnitude higher than the EC50 value for AIP-I (Lyon et al., 2002). Assuming that the EC50 value is a reasonable estimate of the Kd value (vide infra), the AIP-I added to the system should bind to AgrC-I in a linear fashion until saturation of binding sites is achieved. The autokinase activity of AgrC-I was used as a measure of the level of AIP-I binding. As predicted, the initial autokinase velocity of the system first increased linearly and then reached a plateau after the AIP-I concentration reached ~2 equivalents with respect to AgrC-I nanodiscs (Figure 4A). These observations suggest that each AgrC-I dimer possesses two binding sites for AIP-I, and more intriguingly, that AIP-I binding at one site causes the same level of autokinase activation regardless of the occupancy of the other site – i.e., there is no evidence of cooperativity.
Figure 4. Interaction between AIPs and AgrC-I, see also Figure S4.
(A) Stoichiometry of the AgrC-I/AIP-I interaction. AgrC-I discs were incubated with 20 μM [γ-32P]-ATP along with AIP-I at different concentrations and in each case the initial autokinase velocity (vi) was determined by monitoring build-up of radio-labeled AgrC-I over time using autoradiography. Plot shows vi (normalized to the AIP-free reaction) versus [AIP-I]/[AgrC-I disc] ratio. Error bars = SD (n = 3). (B) Equilibrium binding of FAM-AIP-I to AgrC-I discs. Corrected steady-state anisotropy change (ΔSSAc, see Extended Experimental Procedures) of FAM-AIP-I fluorescence was plotted as a function of added AgrC-I discs and fit to a Hill equation. One representative titration of 4 is shown. Error bars (technical) = SEM (n = 6). (C) Equilibrium binding of native AIPs to AgrC-I discs based on competitive displacement of FAM-AIP-I from AgrC-I. ΔSSA of FAM-AIP-I is plotted as a function of added AIP-I (open squares and the dashed curve) or AIP-II (closed triangles and the solid curve). The data are fit to a competitive binding model (see Extended Experimental Procedures). One representative titration of 3 is shown. Error bars (technical) = SEM (n = 6). (D and E) Michaelis-Menten plots for the forward (D) and reverse (E) autokinase reaction of AgrC-I. The first-order kinetic constant for the each reaction is plotted as a function of substrate (ATP or ADP) in the presence (open square, dashed curve) or absence (closed circles, solid curve) of 10 μM AIP. Error bar = SD (n = 3). (F) Summery of kinetic parameters obtained from (D and E): kmax, the maximal first-order kinetic constant.
To measure the equilibrium binding constant of the AIP-AgrC interaction, we exploited a fluorescein-labeled AIP-I analog (FAM-AIP-I) that has AgrC-I agonist activity indistinguishable from that of the native AIP-I (Figure S4B and S4C). The fluorescence-anisotropy of FAM-AIP-I was measured as a function of added AgrC-I discs, yielding a Kd for FAM-AIP-I binding to AgrC-I of 122 ± 26nM (n=4), with a Hill coefficient of 0.97 ± 0.20 (Figure 4B). Importantly, we observed no change in anisotropy when FAM-AIP-I was titrated with nanodiscs containing an AgrC-IS109VS116I mutant that is defective in AIP-I-dependent activation (Geisinger et al., 2008) (Figure S4D and S4E). The lack of cooperativity in the interaction between AIP-I and AgrC-I correlates well with the independent, but additive, role of the two binding sites in the autokinase activation of AgrC-I. The Kd for native AIP-I and AIP-II were 63 ± 13nM (n=3) and 160 ± 20nM (n=3), respectively, obtained from competitive titrations of a pre-formed complex between FAM-AIP-I and AgrC-I (Figure 4C).
Phosphorylation of AgrC-I is reversible and energetically uphill
Atomic-resolution structures of CA subdomains in complex with adenosine nucleotides reveal a role for the conserved ‘G1 box’ residues in nucleotide binding: the amine group of the adenine ring hydrogen bonds to the Asp residue, while the negative charge on the Asp is stabilized by interactions with the backbone amides of the two glycine residues (Marina et al., 2005). As noted above, AgrC-I lacks these key residues suggesting that the interaction between CA subdomain and ATP might be altered. We determined the apparent Km values for the cofactor in the presence or absence of AIP-I using dot-blot-based kinetic assays (Figure S4F). The first-order kinetic constant (k1) for AgrC-I auto-phosphorylation was determined at a series of ATP concentrations and fit to the Michaelis-Menten kinetic model (Figure 4D). As expected, the presence of AIP-I stimulated receptor autophosphorylation, reflected in an ~8-fold increase in the maximum first-order rate constant, kmax, compared to the apo receptor (Figure 4F). The apparent Km was 2.8 mM for apo-AgrC-I and 1.5 mM for AIP-I-bound AgrC-I, both at least an order of magnitude larger than for RHKs with normal G1 boxes (Grimshaw et al., 1998; Kenney, 1997).
A similar kinetic characterization was performed for the reverse autokinase reaction, in which phosphorylated receptor transfers the phosphoryl group back to ADP (Grimshaw et al., 1998). AgrC-I discs were pre-phosphorylated and used as the substrate upon purification (Figure S4G). Kinetic constants for the reverse reaction were determined at a series of ADP concentrations in the presence or absence of AIP-I (Figure S4H and S4I). The reverse reaction was significantly more efficient: the Km for ADP was lower while the kmax was higher than the forward reaction (Figure 4E and 4F). This observation is consistent with the more favorable free energy of hydrolysis of phosphohistidine compared to ATP (−13 kcal/mol vs −7.3 kcal/mol). More surprisingly, and in stark contrast to the forward reaction, the presence of AIP-I had no significant effect on the efficiency of the reverse reaction. Thus, AIP-I binding to AgrC-I shifts the equilibrium in the favor of the forward reaction.
The TMH-DHp linker has strong helical propensity
We next turned to the question of how conformational changes in the sensor domain, as a result of AIP binding, might be transmitted to the AgrC-IHK. The RHK subfamily to which AgrC belongs has a short peptide sequence between the sensor and HK domains that we refer to as the TMH-DHp linker region. Analysis of the amino acid composition of this linker region across the entire subfamily revealed under-representation of three amino acids - cysteine, glycine and proline - compared to their average frequency in proteins (Mccaldon and Argos, 1988) (Figure 5A). Indeed, not a single proline residue was found in any linker sequence in the subfamily. The absence of glycine and proline, known alpha-helical breakers (Pace and Scholtz, 1998), led us to wonder if the linker region may be helical and perhaps propagate the helical segments that flank it, namely the last TMH of the sensor and the first helix of the DHp (DHp-α1) (Figure 5B). To test this idea, we synthesized two peptides corresponding to the TMH-DHp linker, one with the wild-type sequence and the other with two residues, Leu 205 and Met 208, mutated to proline (Figure S5A). Circular dichroism spectroscopy analysis revealed high helical propensity in the wild-type peptide, which is greatly attenuated in the case of the mutant (Figure S5B–D). The possibility of a contiguous helix connecting the sensor and DHp is attractive since it would provide a ready conduit for conformational changes in the sensor, resulting from AIP binding, being transmitted to the HK via the ‘helix rotation’ mechanism (Casino et al., 2010). Consistent with helical propensity in this region being important, the L205P/M208P mutations that diminished helicity of the linker peptide also abolished the AIPI-dependent kinase activation in full-length AgrC-I (Figure 5C).
Figure 5. Function of the TMH-DHp linker in signal transduction, see also Figure S5.
(A) Amino acid frequency in the TMH-DHp linkers within HPK10 sequences. Frequency of each amino acid is normalized to its overall composition in proteins. Under-represented amino acids are indicated with arrows. (B) Cartoon of full-length AgrC-I showing an α-helical TMH-DHp linker region. Helical nucleation that would stabilize the linker helix is highlighted with arrows. (C) Autokinase activation of AgrC-IL205P/M208P mutant: nanodiscs containing wild-type (WT) or mutant AgrC-I dimers were phosphorylated with [γ-32P]-ATP and analyzed as in Figure 3B. (D) Design of GCN4-AgrC-I chimera proteins. GCN41-25 (in cyan) was sequentially fused to residues in the TMH-DHp linker (in red). GCN4 residues registering at coiled-coil position ‘d’ are noted underneath the sequence in blue. (E) Cartoon of a GCN4-AgrC-I chimera with the interface between the GCN4 and AgrC-I sequences indicated by a blue plane. Right panel, coiled-coil helical wheel representation of the interface highlighting the location of the last residue of GCN4 (‘d’, in blue) and the junction residue (‘e’, in red). (F) Autokinase activities of GCN4-AgrC-I chimeras: dimeric chimeras and AgrC-I discs (with or without added AIP as indicated) were treated with [γ-32P]-ATP and analyzed as in Figure 3B. Linker length (in residues) is indicated underneath the loading control. (G) Quantification of the data in panel F. Plot shows normalized phosphorylation levels of the chimeras (normalized to that of the apo-AgrC-I discs) as a function of TMH-DHp linker length. Curve is a sinoidal function fit to the data. Error bars = SD (n = 3)
Twisting the helical register of the linker changes the activity of AgrC-IHK
We set out to investigate whether changes in the linker conformation correlate with AgrC-I autokinase activity. Our approach was to perturb the resting conformation of the linker region by fusion to a rigid body, the homodimeric GCN4 leucine zipper (O’Shea et al., 1991). We prepared 15 chimeric proteins, each comprising an N-terminal GCN4 coiled-coil sequence and a C-terminal AgrC-IHK separated by an intervening TMH-DHp linker segment of variable length. These protein chimeras were termed GC205 to GC219 after GCN4-AgrC-I and the position of the first AgrC-I residue fused to GCN4, to which we refer as the junction residue (Figure 5D). Since the C-terminal residue of the GCN4 portion registers at the ‘d’ position in the classic coiled-coil helical wheel, we inferred that the junction residue in each chimera should be moved to the ‘e’ position (Figure 5E). Therefore, the twisting force applied to TMH-DHp linker, as a result of this movement, is contingent on the resting register of the junction residue.
We compared the autokinase activities of these chimeras in the absence of any added AIP to that of full-length apo-AgrC-I, as well as the AIP-I and AIP-II bound states of the RHK. Strikingly, the phosphorylation levels of these GCN4-AgrC-I fusion proteins fluctuated periodically with the length of the linker (Figure 5F). The periodicity, obtained from fitting the data to a sine wave function, was 3.6, constituting one turn of an α-helix (Figure 5G). In addition, fine-tuning of the autokinase activity of AgrC-IHK could be realized through symmetrically twisting the linker helix to a wide range of angles. Chimeras with activity equivalent to that of AgrC-I in all three ligand states were found (Figure 5F), suggesting that the ligand-dependent differential activities of the full-length receptor might also be achieved through adjusting the linker conformation.
Binding of AIPs to the AgrC-I dimer induces rotation in the linker helix
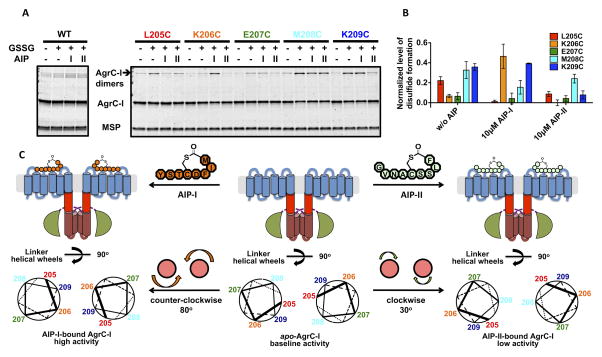
A cysteine crosslinking strategy was used to study the conformational changes in the linker region of full-length AgrC-I that accompany AIP binding. Based on the helical twisting model that emerged from the GCN4 chimera data, we hypothesized that, depending on the location of the cysteine and the ligand state of the receptor, inter-subunit disulfide bonds would be more or less likely to form under oxidizing conditions. The absence of native cysteine residues in the linker region facilitated this approach. Five AgrC-I mutants with cysteine replacements at residues 205-209 were expressed, purified and incorporated into nanodiscs. The mutants behaved similarly to wild-type AgrC-I in terms of their autokinase response to agonist and antagonist AIPs (Figure S6A). Non-reducing SDS-PAGE analysis of the mutants following oxidation with glutathione revealed a marked difference in the amount of covalent dimer formed depending on the mutant and the presence or absence of AIP (Figure 6A). The yield of covalent dimer from each sample was normalized for loading, and grouped according to three activity states (apo, AIP-I-bound or AIP-II-bound) as in Figure 6B. This revealed a periodicity within each state that, analogous to the GCN4 chimera data, could be interpreted by assigning parallel, juxtaposed helical structure to the linkers (Figure 6C). When analyzed in this way, the linker helices within the apo-AgrC-I dimer are oriented such that positions 205, 208 and 209 are juxtaposed and able to form crosslinks when cysteines are placed at these sites. Binding of the AIP-I activator leads to a ~80° counter-clockwise rotation of the two linker helices in the dimer such that residues 206 and 209 are now in optimal position to form crosslinks. Conversely, binding of the AIP-II inhibitor leads to a ~30° clockwise rotation of the helices from the resting state resulting in residue 208 being optimal for crosslinking.
Figure 6. Effect of AIP binding on conformation of the AgrC-I linker helices, see also Figure S6.
(A and B) Cysteine crosslinking of AgrC-I protomers bearing single cysteine point mutants in the TMH-DHp linker region: nanodiscs incorporated with wild-type or mutant AgrC-I dimers were mock-treated or incubated with 10 mM oxidized glutathione (GSSG) along with indicated AIP. (A) Formation of covalent dimers as analyzed by SDS-PAGE with CBB stain. (B) Quantification of the crosslinking results. Plot shows normalized levels of disulfide-linked dimer formation, grouped according to the three ligand states (see Experimental Procedures). Error bars = SD (n = 3). (C) Rotation of the TMH-DHp linker helix in AgrC-I induced by AIP binding. Top: Cartoons of AgrC-I dimer in the apo- (middle), AIP-I-bound (left) and AIP-II-bound (right) states. Bottom: The corresponding conformation of the linker helices is depicted with helical wheels showing the positions 205-209 according to the cysteine crosslinking results. Ligand binding and the consequent rotation of linker helices are highlighted between panels.
This interpretation of the crosslinking data was used to further analyze the differential activity associated with GCN4-chimeras. As noted above, the junction residue in each chimera can be assigned to the ‘e’ position of the coiled-coil helical wheel. Based on this logic, we predict that GC209, GC210 and GC205 should be conformational surrogates of the apo-, AIP-I- and AIP-II-bound forms of AgrC-I, respectively (Figure S6B to S6E). Remarkably, these chimeras accurately phenocopy the three activity states of AgrC-I: compared to GC209, GC210 had much higher autokinase activity while GC205 was not as active (Figure S6F). We therefore conclude that ligand binding regulates the autokinase activity of AgrC-I by inducing symmetric rotation in the helical TMH-DHp linker region.
Discussion
Implications of AgrC biochemical properties for QS in S. aureus
In this study, we have presented a comprehensive biochemical characterization of the AgrCA TCS. We found that AgrC-I dimers reconstituted into lipid nanodiscs possess a substantial level of basal activity. The cognate autoinducer non-cooperatively binds to AgrC-I in a 2:2 stoichiometry with nanomolar affinity, with individual binding events adding equally to stimulate the autokinase activity of the RHK. The pHis moiety on autophosphorylated AgrC-I is stable under physiological conditions, but rapidly transfers the phosphoryl group to AgrA-I. Remarkably, AgrC-I lacks any detectable phosphatase activity, leaving self-catalyzed AgrA-I dephosphorylation the only reaction that counteracts the phospho-relay cascade. These biochemical properties are likely to play essential roles in controlling the onset of virulence in S. aureus. At low cell density, for instance, the basal autokinase activity of AgrC maintains a low level of phosphorylated AgrA that, due to its differential affinity to the regulatory elements in the P2 and P3 promoter regions, enables baseline transcription of the P2 operon rather than P3 (Reyes et al., 2011). AIP, produced as a consequence, would accumulate to levels that significantly activate the AgrCA TCS during log-phase bacterial growth. The strong affinity of AIP for AgrC, the lack of phosphatase activity in AgrC, and the positive feedback of the autoinduction loop collectively leads to a burst of RNAIII production and thereby an abrupt switch into a high-density, virulent mode of life, usually within less than one hour during growth in rich media (Ziebandt et al., 2004). Finally, the t1/2 of phosphorylated AgrA is a mere 3.9 minutes at 37°C, shorter than most characterized transcription-factor RRs (Thomas et al., 2008). This enables agr-activated cells to rapidly turn off virulon expression and revert to vegetative growth if separated from the quorum.
AgrC-I is also distinguished from most other characterized RHKs by its dramatically lowered affinity for adenosine nucleotides, likely due to substitutions in the G1-box. We reason that this atypically low affinity might allow AgrC to sense cellular ATP levels, which vary significantly between optimal and stress conditions. Indeed, down regulation of accessory gene expression in times of nutrient or oxygen deprivation had been reported (Ziebandt et al., 2004). Since the cellular ATP concentration data is not available for S. aureus, the values for E. coli must be used as a reference (Tran and Unden, 1998). With abundant carbon source, E. coli contains approximately 3.5 mM cellular ATP, which decreases to sub-millimolar levels at stationary phase, or after switching to a weaker electron acceptor for respiration. Assuming similar variations in S. aureus, a drastic decrease in the autokinase activity would be expected for AgrC (apparent Km for AgrC-I ~2 mM) under nutrient-limiting or anaerobic conditions. Thus, we propose that inactivation of the virulon under stress conditions is at least partly a consequence of the acute sensitivity of AgrC to falling ATP levels, ultimately leading to a down-regulation of the agr response. This sensitivity may extend to processes mediated by other receptors of the HPK10 subfamily given the conserved nature of the altered G1-box.
The HPK10 subfamily and peptide-mediated quorum sensing
During the course of our reconstitution studies, a remarkable dependence on anionic bilayer environment was found for AgrC-I activation. The exact basis of this lipid-sensitivity in AgrC-I activation remains unclear. We did, however, rule out the possibility of global misfolding for AgrC-I embedded in 100% DMPC nanodiscs in that it forms dimers that bind to a fluorescent AIP-I analog, albeit without any resulting autokinase stimulation (Figure 3D and S3D). Given the highly basic amino acid sequence of the AgrC-I sensor domain (theoretical pI = 9.3), we speculate that anionic lipids enable competent conformational coupling between the AIP-binding site and AgrC-IHK by neutralizing excessive positive charges in the sensor domain. Intriguingly, the strongly anionic cell membrane found in S. aureus is shared among other species from the low-GC G+ phylum (Firmicutes), and distinguishes them from those representing major G− phyla (Gunstone et al., 1994). In addition, our bioinformatic analysis revealed a striking restriction of the HPK10 subfamily, to which AgrC belongs, to the phylum Firmicutes. All characterized RHKs from this subfamily are receptors of peptidic pheromones, and, perhaps not coincidentally, peptide-mediated QS is not known in any G− species. A plausible explanation for this taxonomic boundary is that HPK10 subfamily receptors are all evolved in, and presumably well adapted to, the anionic lipid environment. Such adaptation, if generally present, would create a considerable barrier to horizontal gene transfer of HPK10 homologs into G− species, which have PE-rich inner membranes. Biochemical studies into additional HPK10 homologs, perhaps using the nanodisc technology employed herein, might lend further support to the idea that peptide-responsive RHKs are restricted to G+ bacteria because of the lipid composition of their membranes.
Conformational coupling between the linker and the AgrC-IHK
Recent structural studies of fragments of diverse RHK proteins generally support the idea that the linkers connecting the HK and sensor domains adopt parallel helical conformations (Diensthuber et al., 2013; Marina et al., 2005). Despite these high-resolution snapshots, movement of the linker as a consequence of native signal inputs remains speculative in the context of full-length RHKs. Our studies on AgrC-I indicate that the TMH-DHp linker assumes a helical conformation whose structural properties are intimately tied to the activity state of the receptor. Cysteine crosslinking studies in the context of full-length AgrC-I revealed distinct crosslinking patterns for the resting, activated and inhibited states, indicating a substantial reorganization of the structure in the linker in response to binding of both activator and inhibitor AIPs (Figure 6). Interestingly, the first of these patterns (resting state) is not a simple weighted average of the latter two, which is inconsistent with a model wherein the TMH-DHp linkers are in equilibrium between ‘on’ and ‘off’ conformations with ligand binding altering the equilibrium position of the system (Figure S6G). Rather, this crosslinking data is more consistent with discrete linker conformations for AgrC in all three activity states (Figure 6C).
Our analysis of a series of GCN4-AgrC-I chimeras provided further insight on the signal-response behavior of AgrC-IHK. Assuming that the TMH-DHp linker adopts a parallel helical conformation, and that GCN4 consistently positions the junction residue at the ‘e’ register as illustrated in Figure 5E, then AgrC-IHK activity is seen to change gradually as a function of the twisting angle away from the resting state (Figure S5E and S5F). Intriguingly, this rheostat-like behavior extends beyond the range of twisting angles associated with AIP binding to AgrC-I as indicated by our crosslinking data. Our chimera approach has some interesting parallels with the work of Moffat and coworkers on FixL from Bradyrhizobium japonicum (Moglich et al., 2009). Similar to AgrC, the linker region in FixL connecting the sensor and HK domains is proposed to be helical. Chimeras between the FixL HK domain and a LOV light sensor domain from a heterologous protein (YtvA from B. subtilus) are active only when the FixL residue at the junction occupies two periodically equivalent positions (‘i’ and ‘i + 4’) in each haptad repeat of the putative linker coiled-coil. Maintaining the appropriate periodicity of the helical linker is therefore thought to be critical to the construction of functional RHKs via shuffling between sensor and HK domains, including such events as they occur during evolution (Capra and Laub, 2012). In stark contrast to the YtvA-FixL chimeras, our GCN4-AgrC-I constructs exhibited many intermediate activities between the two extremes: the system is remarkably plastic. Shuffling between the AgrC-IHK (or other HK domains with similar behaviors) and heterologous sensory modules could therefore result in new RHKs exhibiting variable degrees of basal kinase activity and possibly dynamic range, depending on the fusion junction and the structural rearrangement of the sensor between signaling states. Conceivably, such functional plasticity would be beneficial during RHK evolution by increasing the chance of generating competent chimeric receptors that could serve as starting points for further selection for an optimal working range for the new sensory function.
The DHp-α1 helix is kinked at a position slightly downstream of the phospho-acceptor histidine as a result of either a proline residue (Wang et al., 2013) or a sequence pattern of hydrophobic residues (Albanesi et al., 2009). This kink serves as a pivot point that allows the N-terminal half of DHp-α1 to bend or rotate relative to the rest of the DHp four-helix bundle. The hydrophobic pattern is also found in AgrC-I (residues F237, Y241 and L245), suggesting that its DHp-α1 region has similar behavior to its counterparts in other RHKs. Bioinformatics and structural studies on diverse HK-RR pairs pinpoint the C-terminal half of the DHp-α1 as the RR interaction epitope (Casino et al., 2009; Skerker et al., 2008). Due to the aforementioned plasticity in the DHp-α1 helix, the conformation of this region should be relatively insensitive to structural input from the sensor (propagated by the linker helix). Indeed, regulatory compounds of several TCSs have been shown to have neutral effect on the actual phospho-relay step (Timmen et al., 2006; Wei et al., 2012), which is also what we found for AgrC-I/AgrA-I in the presence of AIP-I. By contrast, the inverse agonist, AIP-II, clearly slows down phospho-relay to AgrA-I. While the structural basis of this effect remains unclear, we imagine that this effect must originate from either perturbed binding of the RR and/or a steric protection of the pHis.
In conclusion, we have succeeded in reconstituting a complete TCS involved in quorum sensing in a G+ bacterium using purified components, namely AIP, AgrC and AgrA. Key to this achievement was implementation of the nanodisc technology, which allowed biochemical access to both sides of the membrane-bound RHK. The ability to activate or inhibit the TCS in vitro, depending on the input AIP, was completely consistent with what is known about regulation of the agr system from in vivo studies, and importantly provided mechanistic insights previously obscured by the reliance of these cell-based studies on transcriptional readouts. Our studies raise several new questions about the system, including the precise location of the AIP binding site in the sensor domain, how occupancy of this site is communicated to the TMH-DHp linker and why anionic lipids are required for this transaction. The defined biochemical system described in this study provides the starting point by which to address these problems.
Experimental Procedures
Database search for identification of HPK10 homologs
Two rounds of database search were performed. In the first round, the sequence of AgrC-I205-430 was used as the query to search the reference sequence protein database (Refseq_protein) with Pattern Hit Initiated BLAST (PHI-BLAST) (Zhang et al., 1998). The initial pattern was [FILV][RK]H[DE][YF]. In order to preserve the initial pattern, no subsequent PSI-BLAST was performed. Hits with E-values below 0.05 were pooled and redundant hits were removed with a sequence identity cutoff of 80%. False (non-RHK) hits lacking an intact CA domain were detected and removed as described in Extended Experimental Procedures. Out of 183 sequences obtained in the first round, 12 distantly related to AgrC-I were chosen as queries for a second round of database search with similar strategies (see Table S1). Hit sequences obtained from both rounds were pooled and, upon removal of redundant and false hits, resulted in the ‘330 non-redundant sequences’ that appeared in the main-text.
Recombinant protein preparation and nanodisc formation
All AgrC-I variants, GCN4-AgrC-I chimeras and AgrA-I were expressed in E. coli and purified using standard protocols as detailed in Extended Experimental Procedures. For nanodisc assembly, MSPE3D1, dimeric AgrC-I and lipids were mixed at a molar ratio of 4:1:540 with dimyristoyl-phospholipids and 4:1:440 for palmitoyl-2-oleoyl-phospholipids (Ritchie et al., 2009). Upon detergent removal using SM-2 Adsorbent Bio-Beads (Bio-Rad), the post-assembly mixture was subjected to Ni-NTA affinity purification. Elution fractions containing AgrC-I-incorporated nanodiscs were resolved on Superose6 SEC in nanodisc buffer (20 mM HEPES-Na pH = 7.0, 100 mM NaCl, 1mM TCEP) from which peak fractions were pooled as product.
SEC-MALS analysis of nanodisc particle size and protein content
Nanodisc samples were resolved on a Superose6 SEC equilibrated with the nanodisc buffer. The eluate was directed to a DAWN HELEOS-II multi-angle light scattering instrument in tandem with OptiLab TrEX differential refractometer for real-time analysis. Fitting was accomplished using the “protein conjugate” method in the ASTRA 6.0 (Wyatt Technology) software package, which calculates both the averaged particle size and the protein content utilizing UV, MALS and dRI signals simultaneously. Input for refractive index increment, dn/dc, was 0.185 mL/g for protein and 0.135 mL/g for the modifier (treating lipids as the modifier). Input for extinction coefficient of protein was 0.898 for empty discs (corresponding to MSP) and 0.806 (an equimolar mixture of MSP and AgrC-I) for AgrC-I discs, both in the unit of mL/(mg·cm).
Biochemical assays
Biochemical reactions involving autokinase, phospho-relay and reverse autokinase activities of AgrC-I variants or GCN4-AgrC-I chimeras were setup with appropriate ligands, co-factors and/or AgrA-I as detailed in Extended Experimental Procedures. Time courses of AgrC-I or AgrA-I phosphorylation as analyzed by anti-pHis immunoblot, autoradiography or scintillation counting were quantified and fit to kinetic models of the reactions.
Fluorescent-anisotropy-based binding assays
FAM-AIP-I was synthesized as previously described (Lyon et al., 2002). Binding isotherms were generated by monitoring the change in steady-state fluorescence anisotropy of FAM-AIP-I as a function of added AgrC-I discs or, in the case of competition experiments, added AIP-I or AIP-II to preformed FAM-AIP-I/AgrC-I complexes. Datasets were fit to modified Hill equations as detailed in Extended Experimental Procedures.
Cysteine crosslinking of AgrC-I cysteine point mutants
Nanodiscs incorporated with WT or cysteine-mutant AgrC-I proteins were prepared at 1.0 μM in a buffer containing 20 mM HEPES pH 7.0, 100 mM NaCl and 0.2 mM TCEP. These nanodisc stocks were incubated with either AIP-I, AIP-II or vehicle (DMSO). The final concentration of each AIP was 10 μM. To initiate oxidation, 10 μL from each aliquot was mixed with 2 μL GSSG solution (60 mM, adjusted to pH=7.0). These mixtures were incubated at 37°C for 20min before mixing with thiol-free SDS sample buffer. A ‘non-oxidized’ control reaction was prepared by mixing AIP-free nanodiscs with SDS sample buffer and GSSG solution simultaneously to control for spurious oxidation post-denaturation. Oxidation reactions were subject to non-reducing SDS-PAGE. The gel was stained with Coomassie blue, and intensity of each AgrC dimer band was quantified and normalized to the MSP band in the same lane. Level of disulfide formation was calculated by subtracting normalized dimer band intensity of the non-oxidized discs from the oxidized bands for each AgrC-I mutant.
Supplementary Material
Highlights.
A quantitative study of the signal transduction in AgrC incorporated to nanodiscs
Rheostat-like activity control in response to twisting force on interdomain linkers
Linker rotation in opposite directions with agonist or inverse-agonist binding
AgrC’s low affinity to ATP: a probable reason of agr shutdown under energy stress
Acknowledgments
We would like to acknowledge the Sligar Lab (University of Illinois at Urbana-Champaign) for sharing the expression plasmid of the MSP, Jung-Min Kee and Rob Oslund (Princeton University) for assistance with purification of the anti-pHis antibody and Manuel Muller, Neel Shah (Princeton University) and Beat Fierz (Ecole polytechnique fédérale de Lausanne) for discussions. This work was supported by NIH grants AI042783 and GM095880.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albanesi D, Martin M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, de Mendoza D, Buschiazzo A. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. P Natl Acad Sci USA. 2009;106:16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beining PR, Huff E, Prescott B, Theodore TS. Characterization of Lipids of Mesosomal Vesicles and Plasma-Membranes from Staphylococcus-Aureus. J Bacteriol. 1975;121:137–143. doi: 10.1128/jb.121.1.137-143.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra EJ, Laub MT. Evolution of Two-Component Signal Transduction Systems. Annual Review of Microbiology. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. Structural Insight into Partner Specificity and Phosphoryl Transfer in Two-Component Signal Transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. The mechanism of signal transduction by two-component systems. Current opinion in structural biology. 2010;20:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Cisar EA, Geisinger E, Muir TW, Novick RP. Symmetric signalling within asymmetric dimers of the Staphylococcus aureus receptor histidine kinase AgrC. Mol Microbiol. 2009;74:44–57. doi: 10.1111/j.1365-2958.2009.06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- Diensthuber RP, Bommer M, Gleichmann T, Moglich A. Full-Length Structure of a Sensor Histidine Kinase Pinpoints Coaxial Coiled Coils as Signal Transducers and Modulators. Structure. 2013;21:1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Dufour P, Jarraud S, Vandenesch F, Greenland T, Novick RP, Bes M, Etienne J, Lina G. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol. 2002;184:1180–1186. doi: 10.1128/jb.184.4.1180-1186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological Insights from Structures of Two-Component Proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, George EA, Muir TW, Novick RP. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem. 2008;283:8930–8938. doi: 10.1074/jbc.M710227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EA, Novick RP, Muir TW. Cyclic peptide inhibitors of staphylococcal virulence prepared by Fmoc-based thiolactone peptide synthesis. J Am Chem Soc. 2008;130:4914–4924. doi: 10.1021/ja711126e. [DOI] [PubMed] [Google Scholar]
- Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- Grimshaw CE, Huang SM, Hanstein CG, Strauch MA, Burbulys D, Wang L, Hoch JA, Whiteley JM. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry-Us. 1998;37:1365–1375. doi: 10.1021/bi971917m. [DOI] [PubMed] [Google Scholar]
- Gunstone KD, Harwood JL, Padley FB. The Lipid Handbook. 2. London; New York: Chapman and Hall; 1994. p. 214. [Google Scholar]
- Ji GY, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- Kee JM, Oslund RC, Perlman DH, Muir TW. A pan-specific antibody for direct detection of protein histidine phosphorylation. Nat Chem Biol. 2013;9:416–U428. doi: 10.1038/nchembio.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney LJ. Kinase activity of EnvZ, an osmoregulatory signal transducing protein of Escherichia coli. Arch Biochem Biophys. 1997;346:303–311. doi: 10.1006/abbi.1997.0315. [DOI] [PubMed] [Google Scholar]
- Lyon GJ, Wright JS, Muir TW, Novick RP. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry-Us. 2002;41:10095–10104. doi: 10.1021/bi026049u. [DOI] [PubMed] [Google Scholar]
- Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. Embo J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayville P, Ji GY, Beavis R, Yang HM, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. P Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccaldon P, Argos P. Oligopeptide Biases in Protein Sequences and Their Use in Predicting Protein Coding Regions in Nucleotide-Sequences. Proteins. 1988;4:99–122. doi: 10.1002/prot.340040204. [DOI] [PubMed] [Google Scholar]
- Moglich A, Ayers RA, Moffat K. Design and Signaling Mechanism of Light-Regulated Histidine Kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum Sensing in Staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- O’Shea EK, Klemm JD, Kim PS, Alber T. X-Ray Structure of the Gcn4 Leucine Zipper, a 2-Stranded, Parallel Coiled Coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Reyes D, Andrey DO, Monod A, Kelley WL, Zhang GY, Cheung AL. Coordinated Regulation by AgrA, SarA, and SarR To Control agr Expression in Staphylococcus aureus. J Bacteriol. 2011;193:6020–6031. doi: 10.1128/JB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs. Method Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. Highly Potent Inhibitors of Quorum Sensing in Staphylococcus aureus Revealed Through a Systematic Synthetic Study of the Group-III Autoinducing Peptide. J Am Chem Soc. 2013;135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide Signaling in the Staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SA, Brewster JA, Bourret RB. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol. 2008;69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmen M, Bassler BL, Jung K. AI-1 influences the kinase activity but not the phosphatase activity of LuxN of Vibrio harveyi. J Biol Chem. 2006;281:24398–24404. doi: 10.1074/jbc.M604108200. [DOI] [PubMed] [Google Scholar]
- Tran QH, Unden G. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur J Biochem. 1998;251:538–543. doi: 10.1046/j.1432-1327.1998.2510538.x. [DOI] [PubMed] [Google Scholar]
- Wang C, Sang JY, Wang JW, Su MY, Downey JS, Wu QG, Wang SD, Cai YF, Xu XZ, Wu J, et al. Mechanistic Insights Revealed by the Crystal Structure of a Histidine Kinase with Signal Transducer and Sensor Domains. Plos Biol. 2013;11:e1001493. doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YZ, Ng WL, Cong JP, Bassler BL. Ligand and antagonist driven regulation of the Vibrio cholerae quorum-sensing receptor CqsS. Mol Microbiol. 2012;83:1095–1108. doi: 10.1111/j.1365-2958.2012.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schaffer AA, Miller W, Madden TL, Lipman DJ, Koonin EV, Altschul SF. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebandt AK, Becher D, Ohlsen K, Hacker J, Hecker M, Engelmann S. The influence of agr and sigma(B) in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics. 2004;4:3034–3047. doi: 10.1002/pmic.200400937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.