Abstract
The centrosome consists of a pair of centrioles and surrounding pericentriolar material (PCM). Many vertebrate cells also have an array of granules, termed centriolar satellites, that localize around the centrosome and are associated with centrosome and cilium function. Centriole duplication occurs once per cell cycle and is effected by a set of proteins including PLK4, CEP192, CEP152, CEP63 and CPAP. Information on the relationships between these components is limited due to the difficulty in assaying interactions in the context of the centrosome. We use proximity-dependent biotin identification (BioID) to identify proximity interactions among centriole duplication proteins. PLK4, CEP192 and CEP152 BioID identified known physically interacting proteins, and a new interaction between CEP152 and CDK5RAP2 consistent with a function of CEP152 in PCM recruitment. BioID for CEP63 and its paralog CCDC67 revealed extensive proximity interactions with centriolar satellite proteins. Focusing on these satellite proteins identified two new regulators of centriole duplication, CCDC14 and KIAA0753. Both proteins co-localize with CEP63 to satellites, bind to CEP63, and identify other satellite proteins by BioID. KIAA0753 positively regulates centriole duplication and CEP63 centrosome localization, whereas CCDC14 negatively regulates both processes. These results suggest that centriolar satellites have a previously unappreciated function in regulating centriole duplication.
Results and Discussion
A combination of approaches has identified many proteins that reside at the centrosome of mammalian cells [1]. A limitation in identifying interactions at the centrosome is that standard techniques such as co-immunoprecipitation cannot access proteins within the centrosome without disrupting its structure. To overcome this limitation, we used BioID proximity labeling [2–4] to identify spatial and temporal relationships proximal to the site of centriole duplication. In the BioID approach, the protein of interest is tagged with a promiscuous mutant of E. coli BirA biotin ligase BirA(R118G). Cells expressing this protein are incubated with excess biotin, and biotinylated proteins are affinity-purified and identified by mass spectrometry. Due to the strong affinity of the biotin-streptavidin interaction, purification can take place under denaturing conditions to solubilize centrosome proteins while preserving, in the form of covalent biotinylation, information about proximal relationships. We will refer to such relationships as proximity interactions, as opposed to physical interactions derived from traditional approaches.
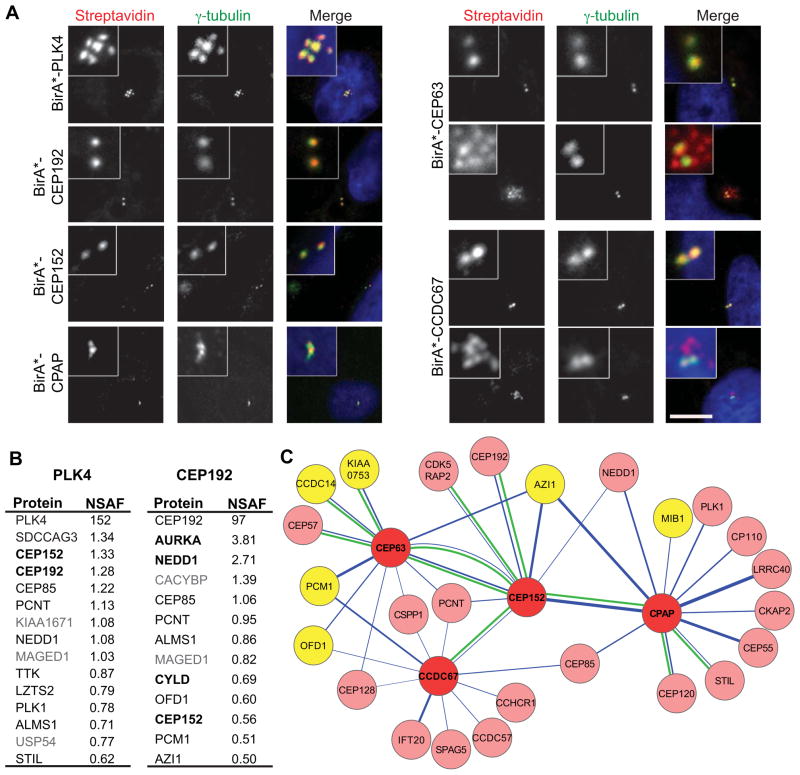
To test the feasibility of using BioID in the context of the centrosome, we applied this approach to human PLK4 and CEP192, key proteins in centriole duplication and maturation [5]. CEP192 interacts with PLK4, but we used an isoform of CEP192 that does not interact [6] to maximize the opportunity to observe differential spatial labeling. Myc-BirA(R118G) (hereafter BirA*) was fused to the N-termini of PLK4 and CEP192 and the constructs were transfected into human U2OS cells. Both fusion proteins localized to the centrosome (Fig. S1A), and also stimulated biotinylation at the centrosome (Fig. 1A); similar results were observed in HEK293T cells (not shown). Expression of BirA*-PLK4 caused formation of multiple centrioles adjacent to the parental centrioles (Fig. 1A, S1A), as observed for wild-type PLK4 [7], indicating that the fusion protein is functional.
Figure 1. Localization, activity and proximity interactors of BirA*-tagged centriole duplication and maturation proteins.
(A) U2OS cells were transfected with Myc-BirA*-tagged PLK4, CEP192, CEP152, CPAP, CEP63 and CCDC67. After 18 h incubation with biotin, cells were fixed and stained for biotinylated proteins with fluorescent streptavidin and centrosomes with anti-γ-tubulin antibody. DNA was stained with DAPI. Two panels are shown for CEP63 and CCDC67 to reflect the observed centrosome (upper panel) and centriole satellite (lower panel) staining. Satellite labeling was observed in 19±2.4 percent of cells (n=300) for CEP63 and 22±3.2 percent of cells (n=300) for CCDC67 and. Scale bar, 10 μm, all insets show 4x enlarged centrosomes. (B) Mass spectrometry analysis of proximity interactors of Myc-BirA*-PLK4 and Myc-BirA*-CEP192. Proximity interactors are ranked in the order of their NSAF values (average of three independent experiments). Proteins in black were previously shown to localize to the centrosome and proteins in bold were previously shown to interact physically with the indicated BirA*-tagged protein. (C) A proximity-based interaction map of centriole duplication proteins. The map was constructed with selected proteins from mass spectrometry analysis of Myc-BirA*-tagged CEP63, CCDC67, CEP152 and CPAP, and published data, using Cytoscape software. Nodes representing individual Myc-BirA*-fusion proteins are connected by edges (in blue) to the identified proximity interactors. The width of each edge is proportional to the NSAF value of the interaction. A green edge between nodes indicates physical interactions validated in this study or previous studies: CEP152-CPAP [9, 24], CEP63-CEP152 [17, 18], CEP63-CEP57 [25], CCDC67-CEP152 [19], CPAP-STIL [12, 26], CPAP-CEP120 [4, 27] and CEP152-CEP192 [6], CCDC14-CEP63, KIAA0753-CEP63, CEP152-CDK5RAP2. The nodes are color-coded to represent Myc-BirA*-fusion proteins (red) and centriolar satellite proteins (yellow).
To identify proximity interactors for PLK4 and CEP192, HEK293T cells were transfected with BirA*-PLK4 or BirA*-CEP192, supplemented with 50 μM biotin 24 h post-transfection, and incubated for 18 h. Centrosome-enriched fractions were prepared by sucrose gradient fractionation, proteins were solubilized, and biotinylated proteins were precipitated by streptavidin beads and analyzed by mass spectrometry. Cells transfected with BirA*, or mock-transfected, were processed in parallel as controls. Relative signals from mass spectrometry were compared as normalized spectral abundance factors (NSAF), which takes into account protein length and run-to-run variation [8]. Results were filtered as described in the Methods.
The top proximity interactors for PLK4 and CEP192 are listed in Fig. 1B, with the complete list in Table S1. In both cases the fusion protein itself has the highest NSAF value, presumably due to self-biotinylation or trans-biotinylation within a dimer. Indeed, some peptides identified with BirA*-CEP192 corresponded to peptides not in the isoform used in the BirA* construct, indicating that they derive from trans-biotinylation, and suggesting that at least some CEP192 is multimeric in the centrosome. Several of the PLK4 proximity interactors stand out by their known relationship to PLK4, or involvement in centriole duplication. CEP152 and CEP192 both function in centriole duplication and have been reported to interact with PLK4 by independent approaches [6, 9, 10]. STIL is a core centriole duplication protein, which, like PLK4, causes centriole amplification when overexpressed [11, 12], but has not been shown to interact with PLK4. The PLK4 BioID hits also include proteins not previously implicated in centriole duplication. SDCCAG3, the highest non-PLK4 hit, localizes to the centrosome, endosomes, and the midbody during cytokinesis, and regulates cytokinesis [13].
The proximity interactors for CEP192 (Fig. 1B, Table S1) comprise a different, but overlapping set of proteins. CEP192 interacts with PLK4 and is required for centriole duplication, but is also required for the recruitment of PCM proteins. Since we used the isoform of CEP192 that does not bind PLK4, we expected that many of the proximity interactors would be involved in recruitment. Accordingly, two of the top hits, NEDD1, a component of the pericentriolar material, and AURKA, an Aurora kinase, physically interact with CEP192 and are involved in mitotic PCM recruitment [14, 15]. Other proximity interactors include CEP152, which together with CEP192 recruits PLK4 to the centrosome [6], and the K63-deubiquitinase CYLD, which functions with CEP192 in mitotic spindle assembly [16].
The results with PLK4 and CEP192 show that combining BioID labeling with centrosome enrichment identified proximity interactors consistent with known physical interactions and functions. We next applied this approach to CEP152, CEP63 and CPAP [9, 17, 18]. CEP152 scaffolds procentriole formation by promoting centrosomal accumulation of CPAP and PLK4 [9, 10]. CEP152 and CEP63 co-localize in a ring around the proximal end of the parental centriole, and are dependent on each other for centrosomal localization [17, 18]. CEP63 is ubiquitously expressed but has a paralog, CCDC67/Deup1, which we also included; CCDC67 functions specifically in de novo centriole amplification in multiciliated cells [19].
N-terminal BirA* fusions of CEP152, CEP63, CPAP and CCDC67 each localized to the centrosome and stimulated centrosomal biotinylation (Fig. 1A, S1A). CEP152 was also tagged at the C-terminus because super-resolution imaging suggests that it has an elongated structure within the PCM [20], and thus that the two termini might sample different spatial domains (Fig. S1A). Overexpression of BirA*-CPAP caused centriole elongation, indicating that this protein fusion is functional (Fig. 1A, S1A) [21–23]. Proximity interactions for CEP152, CEP63, CCDC67 and CPAP are represented graphically in Fig. 1C and listed in Table S1. As for PLK4 and CEP192, many previously reported physical interactions were identified by BioID (Fig. 1C).
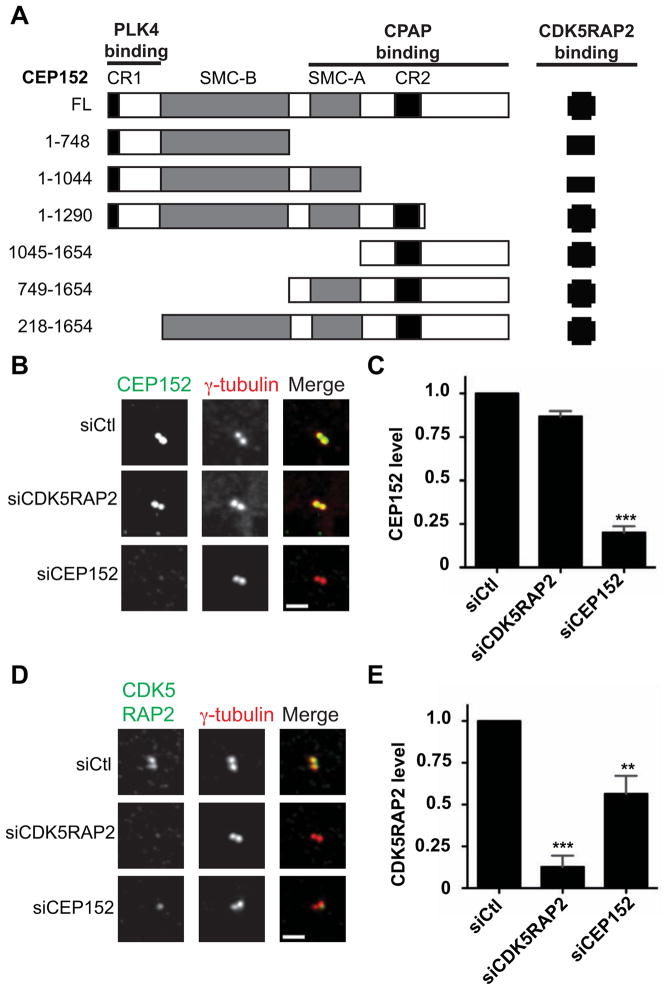
The proximity interaction map revealed a previously uncharacterized relationship between CEP152 and CDK5RAP2, a protein important in PCM recruitment and centrosome organization [28, 29]. This interaction was shown to reflect a physical association by co-immunoprecipitation in HEK293T cells (Fig. 2A, S2A). The C-terminal 608 residues of CEP152 were sufficient to bind CDK5RAP2 (Fig. S2B). The proximity interaction between CEP152 and CDK5RAP2 was identified with CEP152-BirA*, but not with BirA*-CEP152, showing that this approach yields sub-protein level spatial information reflecting actual interactions (Table S1). CEP152 localized normally to the centrosome in cells depleted of CDK5RAP2 (Fig. 2B, C), however centrosome-associated CDK5RAP2 was significantly reduced in cells depleted of CEP152 (Fig. 2D, E). This suggests that recruitment of CDK5RAP2 is dependent on CEP152, consistent with results for the Drosophila orthologs Centrosomin and Asterless [30], and with super-resolution imaging of human centrosomes, in which CEP152 and CDK5RAP2 form adjacent concentric rings [20, 31].
Figure 2. CEP152 interacts with and recruits CDK5RAP2 to the centrosome.
(A) Schematic of CEP152 full length (FL) and deletion constructs and summary of interactions with CDK5RAP2. Interactions were determined by co-immunoprecipitation of GFP-CEP152 constructs from HEK293T lysates with Myc-CDK5RAP2. Numbers indicate amino acid positions, CR1 and CR2 indicate conserved domains in CEP152 orthologs, and SMC-A and SMC-B indicate coiled-coil regions. (B–E) CDK5RAP2 depends on CEP152 for centrosome localization. (B, D) U2OS cells were transfected with control (siCtl), siCDK5RAP2, or siCEP152 siRNA for 72 h, fixed, and stained for the indicated proteins. Images represent centrosomes in cells from the same coverslip taken with the same camera settings. (C, E) Quantification of fluorescence intensities from experiments in B and D, respectively. CEP152 and CDK5RAP2 fluorescence intensities at the centrosome were measured for cells treated with siRNA as in B and D. Control siRNA-transfected levels were normalized to 1.0. Data represent mean value from three experiments per condition, +/− SEM; n≥50 cells per experiment. Scale bars = 2 μm.
A striking feature of the proximity map for the centriole duplication proteins is that several components of centriolar satellites are among the most prominent hits for CEP63 and CCDC67 (Fig. 1C, Table S1). Centriolar satellites are linked to centrosome and cilium protein trafficking, cellular stress response, autophagy and are implicated in human disease [32–36]. However, little is known about the structure and function of centriolar satellites, and they have not been previously linked to centriole duplication proteins. We found that BirA*-CEP63 and BirA*-CCDC67 localized to punctate structures resembling centriolar satellites in addition to their prominent centrosome localization (Fig. 1A; S1A). We demonstrated that these puncta colocalize with a subset of structures labeled by PCM1, a marker centriolar satellites (Fig. S1B) and that these BirA*-CEP63/PCM1-positive puncta dispersed throughout the cell upon depolymerization of microtubules (Fig. S1C), as expected for centriolar satellites.
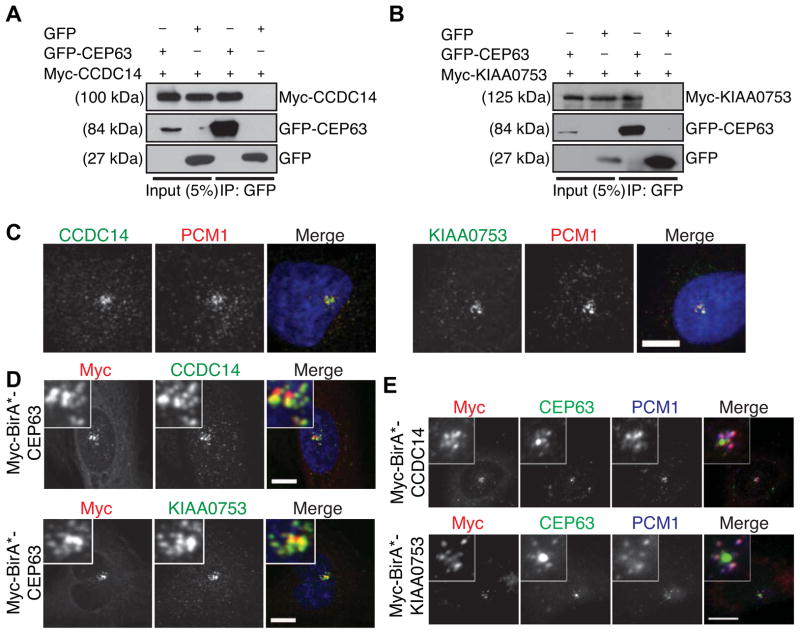
We examined the functional significance of the proximity interactions between duplication proteins and centriolar satellites by focusing on two proteins that were identified by BirA*-CEP63, KIAA0753 and CCDC14. Both proteins were identified previously as centrosome components by mass spectrometry [1], and CCDC14 was reported to localize to centriolar satellites in that study. Co-immunoprecipitation in HEK293T cells revealed that both CCDC14 and KIAA0753 physically interact with CEP63 (Fig. 3A,B; S3). Endogenous CCDC14 and KIAA0753 localized to centriolar satellites in U2OS cells, colocalizing with PCM1 (Fig. 3C). BioID proximity interactors for both BirA*-CCDC14 and BirA*-KIAA0753 included several previously characterized centriolar satellite proteins (i.e. PCM1, OFD1, CEP90, CDK1) with high NSAF values (Table S2), consistent with these proteins being satellite components. CEP63 was also identified as a proximity interactor for both CCDC14 and KIAA0753. Consistent with the reciprocal proximity relationship of CEP63 to CCDC14 and KIAA0753 we found that BirA*-CEP63 colocalized with CCDC14 and KIAA0753 at satellites (Fig. 3D), and that endogenous CEP63 localized to centriolar satellites in some cells expressing BirA*-CCDC14 or BirA*-KIAA0753 (Fig. 3E).
Figure 3. KIAA0753 and CCDC14 are components of centriolar satellites and interact with CEP63.
Co-immunoprecipitation of (A) Myc-CCDC14 and GFP-CEP63, and (B) Myc-KIAA0753 and GFP-CEP63, after co-expression in HEK293T cells. Complexes were immunoprecipitated with anti-GFP antibody and co-precipitated proteins were detected with anti-GFP and anti-Myc antibodies. (C) Localization of CCDC14 and KIAA0753 to centriolar satellites. U2OS cells were fixed and stained for CCDC14 or KIAA0753, and PCM1, which marks centriolar satellites. DNA was stained with DAPI. (D) Co-localization of expressed CEP63 with CCDC14 and KIAA0753 at satellites. U2OS cells expressing Myc-BirA*-CEP63 were stained with anti-Myc, and anti-CCDC14 or anti-KIAA0753 antibodies. DNA was stained with DAPI. (E) Overexpression of CCDC14 or KIAA0753 drives CEP63 to satellites. U2OS cells expressing Myc-BirA*-CCDC14 or Myc-BirA*-KIAA0753 were stained with anti-Myc, anti-CEP63 and anti-PCM1 antibodies. Scale bars = 5 μm, all insets show 4x enlarged centrosomes.
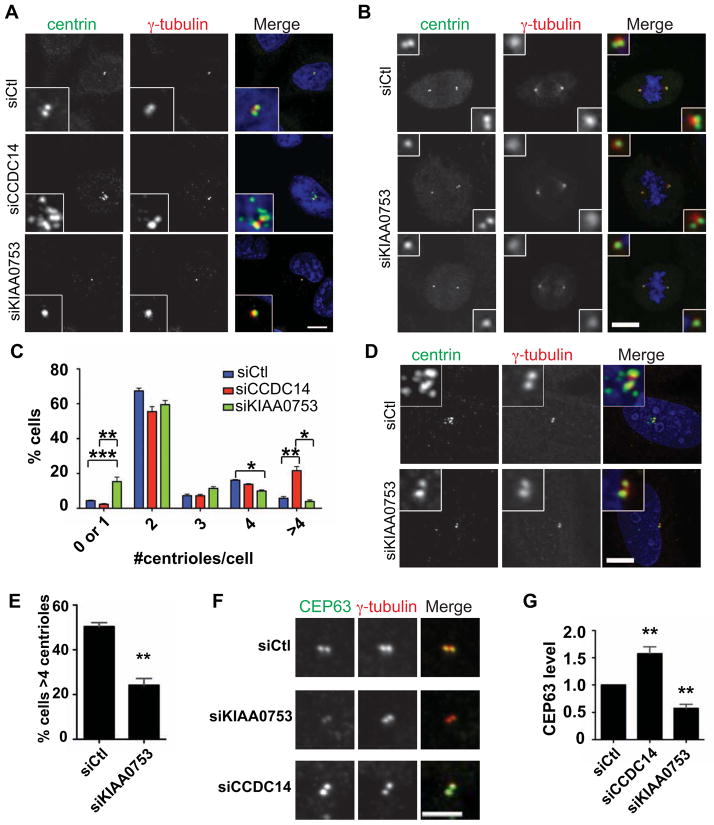
We tested the function of CCDC14 and KIAA0753 by depleting them from U2OS cells. Transfection with siRNAs against CCDC14 or KIAA0753 resulted in depletion after 48 h (Fig. S4A). Strikingly, cycling KIAA0753-depleted cells had a significant decrease in centriole number relative to control cells (Fig 4A-C). This phenotype was also apparent in mitotic cells, evidenced as spindle poles with single centrioles (Fig. 4B), and thus is not due to failure to traverse the G1/S transition. KIAA0753 depletion also compromised centriole reduplication during prolonged S phase arrest [37] in U2OS cells (Fig. 4D, E). These phenotypes were rescued by expression of RNAi-resistant GFP-KIAA0753 (Fig S4B-E).
Figure 4. KIAA0753 and CCDC14 regulate centriole duplication and CEP63 level at the centrosome.
(A, B) Effect of CCDC14 and KIAA0753 depletion on centriole duplication. U2OS cells were fixed 48 h after transfection with control siRNA or siRNAs directed against CCDC14 or KIAA0753, and centriole number was determined by staining for centrin and γ-tubulin. (C) Quantification of A and B. n≥100 cells per experiment. (D) KIAA0753 depletion compromises S phase-arrest overduplication of centrioles. U2OS cells were transfected with control siRNA or KIAA0753 siRNA, and arrested in S phase by hydroxyurea treatment for 48 h. (E) Quantification of E. n≥100 cells per experiment. (F) Effect of CCDC14 and KIAA0753 depletion on CEP63 level at the centrosome. U2OS cells were fixed 48 h after transfection with control siRNA or siRNAs directed against CCDC14 or KIAA0753, and stained for endogenous CEP63 and γ-tubulin. Images represent centrosomes in cells from the same coverslip taken with the same camera settings. Scale bar = 1 μm. (G) Quantification of F. Control siRNA-transfected levels were normalized to 1.0; n≥50 cells per experiment. Data in B, D, F represent mean value from three experiments per condition, +/− SEM. Scale bars = 5 μm, all insets show 4x enlarged centrosomes.
In contrast to KIAA0753, depletion of CCDC14 caused a significant increase in the percentage of cells with more than four centrioles (Fig 4A, C). The overduplication phenotype was not due to a prolonged S phase-arrest, since the percentage of S-G2 cells was similar in CCDC14-depleted cells (36±2.8 percent, n=300) and controls (39±3.6 percent, n=300), assayed by nuclear-localized cyclin A [38]. This phenotype could be rescued by expression of RNAi-resistant GFP-CCDC14 (Fig S4F, G).
The phenotypes of CCDC14 and KIAA0753 depletion suggest that these centriolar satellite proteins regulate centriole duplication, with CCDC14 acting negatively and KIAA0753 positively. Based on the interaction of CCDC14 and KIAA0753 with CEP63, we tested whether localization of CEP63 to the centrosome was altered upon depletion of either protein. Remarkably, the CEP63 immunofluorescence signal at the centrosome increased in CCDC14-depleted cells and decreased in KIAA0753-depleted cells (Fig. 4F, G). This result suggests that these two centriolar satellite proteins achieve their opposing functions, at least in part, by antagonistically regulating CEP63 levels at the centrosome.
Our experiments demonstrate the power of BioID labeling combined with centrosome enrichment in identifying functionally important relationships among centrosome proteins. The BioID proximity interactions included most known physical interactions, as well as previously uncharacterized interactions. Interestingly, some centrosome proteins, such as PCNT, CEP85 and ALMS1, were common to the BioID lists for several of the BirA* fusions. It is likely that these common proteins neither physically interact with, nor are specifically proximal to, all of these fusion proteins. Rather, these proteins might be characterized by high abundance with dispersed localization within the centrosome, or high mobility through the centrosome domain. These types of interactions might only be accessible to BioID or other proximity labeling approaches.
The detection of a proximity interactor by BioID relies on the spatial relationship of that interactor to the BirA* fusion protein, the amount of time spent in the vicinity of the fusion protein, and the abundance relative to other nearby proteins. For the centrosome proteins studied here, these parameters are not known, and it is not yet possible to derive a detailed spatial map from the BioID data. However, BioID yielded an overlapping but distinct set of proximity interactors for PLK4 and CEP192, which occupy similar domains in the centrosome, and for the N- and C-termini of CEP152 [20], consistent with spatial relationships being a critical factor in BioID proximity labeling.
We identified two new regulators of centriole duplication, CCDC14 and KIAA0753, and showed that they are components of centriolar satellites, interact with CEP63, and regulate CEP63 level at the centrosome. CCDC14 interacts with the C-terminus of CEP63 (Fig. S3B–D), which is also required for interaction of CEP63 with CEP152 [17], providing a potential mechanism by which CCDC14 could negatively affect centriole duplication. Centriolar satellites are known to have activities relevant to determining protein level and activity [33, 35], but we have not established whether the relevant effects are occurring at satellites or elsewhere. We propose that centriolar satellite proteins affect centriole duplication by regulating the localization of important duplication proteins, similar to their role in primary cilium formation [34]. We demonstrated this type of regulation for CEP63 but note that our BioID data for other centriole duplication proteins also reveals proximity interactions with satellite proteins (see Fig. 1). Thus, centriolar satellites might have a broader function in regulating centriole duplication in vertebrates.
Supplementary Material
HIGHLIGHTS.
Proximity-labeling identifies interactions among centriole duplication proteins.
CEP152 interacts with CDK5RAP2 and recruits it to the centrosome.
The centriolar satellite proteins CCDC14 and KIAA0753 interact with CEP63.
CCDC14 and KIAA0753 have opposing effects on centriole duplication.
Acknowledgments
We thank Erkang Ai for preliminary work on CCDC67, and Andrew Kodani and Jeremy Reiter for sharing unpublished data. This work was supported by NRSA grant 5 F32 GM106620 to ENF, NIH grants P41 GM103533 and R01 MH067880 to JRY, and NIH grant R01 GM52022 to TS.
Footnotes
Supplemental Information includes Experimental Procedures, four figures, and two tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, Anrather D, Kostan J, Djinovic-Carugo K, Roux KJ, et al. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell. 2013;12:356–367. doi: 10.1128/EC.00326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comartin D, Gupta GD, Fussner E, Coyaud E, Hasegan M, Archinti M, Cheung SW, Pinchev D, Lawo S, Raught B, et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol. 2013;23:1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Hatch E, Stearns T. The life cycle of centrioles. Cold Spring Harb Symp Quant Biol. 2010;75:425–431. doi: 10.1101/sqb.2010.75.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnen KF, Gabryjonczyk AM, Anselm E, Stierhof YD, Nigg EA. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J Cell Sci. 2013;126:3223–3233. doi: 10.1242/jcs.129502. [DOI] [PubMed] [Google Scholar]
- 7.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 8.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 9.Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arquint C, Sonnen KF, Stierhof YD, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci. 2012;125:1342–1352. doi: 10.1242/jcs.099887. [DOI] [PubMed] [Google Scholar]
- 12.Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, Liu F, Bestvater F, Raab MS, Zentgraf H, Izraeli S, et al. STIL is required for centriole duplication in human cells. J Cell Sci. 2012;125:1353–1362. doi: 10.1242/jcs.104109. [DOI] [PubMed] [Google Scholar]
- 13.Hagemann N, Ackermann N, Christmann J, Brier S, Yu F, Erdmann KS. The serologically defined colon cancer antigen-3 interacts with the protein tyrosine phosphatase PTPN13 and is involved in the regulation of cytokinesis. Oncogene. 2013;32:4602–4613. doi: 10.1038/onc.2012.485. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Ferreria MA, Bashkurov M, Helbig AO, Larsen B, Pawson T, Gingras AC, Pelletier L. Novel NEDD1 phosphorylation sites regulate gamma-tubulin binding and mitotic spindle assembly. J Cell Sci. 2012;125:3745–3751. doi: 10.1242/jcs.105130. [DOI] [PubMed] [Google Scholar]
- 15.Joukov V, De Nicolo A, Rodriguez A, Walter JC, Livingston DM. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc Natl Acad Sci U S A. 2010;107:21022–21027. doi: 10.1073/pnas.1014664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Ferreria MA, Bashkurov M, Mullin M, Gingras AC, Pelletier L. CEP192 interacts physically and functionally with the K63-deubiquitinase CYLD to promote mitotic spindle assembly. Cell Cycle. 2012;11:3555–3558. doi: 10.4161/cc.21574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown NJ, Marjanovic M, Luders J, Stracker TH, Costanzo V. Cep63 and cep152 cooperate to ensure centriole duplication. PLoS One. 2013;8:e69986. doi: 10.1371/journal.pone.0069986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sir JH, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D’Santos C, Woods CG, Gergely F. A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet. 2011;43:1147–1153. doi: 10.1038/ng.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H, Zhu L, Zhu Y, Cao J, Li S, Huang Q, Xu T, Huang X, Yan X, Zhu X. The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat Cell Biol. 2013;15:1434–1444. doi: 10.1038/ncb2880. [DOI] [PubMed] [Google Scholar]
- 20.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 24.Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 25.Lukinavicius G, Lavogina D, Orpinell M, Umezawa K, Reymond L, Garin N, Gonczy P, Johnsson K. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr Biol. 2013;23:265–270. doi: 10.1016/j.cub.2012.12.030. [DOI] [PubMed] [Google Scholar]
- 26.Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011;30:4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YN, Wu CT, Lin YC, Hsu WB, Tang CJ, Chang CW, Tang TK. CEP120 interacts with CPAP and positively regulates centriole elongation. J Cell Biol. 2013;202:211–219. doi: 10.1083/jcb.201212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barr AR, Kilmartin JV, Gergely F. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J Cell Biol. 2010;189:23–39. doi: 10.1083/jcb.200912163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fong KW, Choi YK, Rattner JB, Qi RZ. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol Biol Cell. 2008;19:115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conduit PT, Brunk K, Dobbelaere J, Dix CI, Lucas EP, Raff JW. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr Biol. 2010;20:2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 31.Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 32.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stowe TR, Wilkinson CJ, Iqbal A, Stearns T. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol Biol Cell. 2012;23:3322–3335. doi: 10.1091/mbc.E12-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, Zhong Q. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013;502:254–257. doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villumsen BH, Danielsen JR, Povlsen L, Sylvestersen KB, Merdes A, Beli P, Yang YG, Choudhary C, Nielsen ML, Mailand N, et al. A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J. 2013;32:3029–3040. doi: 10.1038/emboj.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 38.Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.