Abstract
Understanding the microtubule-dependent behaviors of viruses in live cells is very meaningful for revealing the mechanisms of virus infection and endocytosis. Herein, we used a quantum dots-based single-particle tracking technique to dynamically and globally visualize the microtubule-dependent transport behaviors of influenza virus in live cells. We found that the intersection configuration of microtubules can interfere with the transport behaviors of the virus in live cells, which lead to the changing and long-time pausing of the transport behavior of viruses. Our results revealed that most of the viruses moved along straight microtubules rapidly and unidirectionally from the cell periphery to the microtubule organizing center (MTOC) near the bottom of the cell, and the viruses were confined in the grid of microtubules near the top of the cell and at the MTOC near the bottom of the cell. These results provided deep insights into the influence of entire microtubule geometry on the virus infection.
Many viruses hijack the endocytic pathway to enter host cells and utilize the microtubule-dependent transport to deliver their genomes to specific compartments for replication.1−4 Understanding the microtubule-dependent behaviors of viruses in live cells is thus critical for revealing the mechanisms of virus infection and endocytosis. Extensive efforts have been devoted to deciphering virus infection pathways, and several reports have indicated that the viruses move along microtubules from the cell periphery to the perinuclear region in a rapid and unidirectional way.5−8 However, detailed microtubule-dependent transport behaviors of viruses remain poorly investigated. Microtubule is a component of cytoskeleton and essential for the intracellular transport of cargos based on molecular motors.9−11 Kinesin and dynein are both intracellular motor proteins that move unidirectionally in opposite directions along microtubules, which may lead to the complex movements along microtubules in live cells.10,12−14 In vitro and in vivo experiments indicated that the intersection of microtubules, which is a tethering point for cargos, can influence the cargo movements.14−17 Such observations raise the question whether the microtubule intersections or other microtubule configurations can influence the microtubule-dependent transport behaviors of viruses during their infection.
Here, we chose avian influenza A H9N2 virus as a model to dissect the microtubule-dependent transport behaviors of influenza viruses in live cells. Influenza A virus is an enveloped virus, consisting of eight segmented single-stranded negative-sense RNA, and the genome segmentation enables influenza viruses to own the advantage of genetic reassortment.18,19 Due to the reassortment among viruses, new types of influenza viruses that are more dangerous to human and animals can arise easily. In the past decades, the outbreaks of several disastrous pandemics have confirmed that the influenza A virus is a very significant risk to public health.20−23 The recent human infection with avian influenza A H7N9 virus has again proven that it is urgent to investigate the infection mechanism of influenza viruses in order to fight the virus infection.24,25
In this work, we used quantum dots (QDs) to label the viruses and tracked the individual viruses in live cells by the single-particle tracking technique, which allowed us to globally visualize the microtubule-dependent motion behaviors of viruses in live cells for a long time. Our single-virus studies showed that the virus moved along microtubules via six types of motion behaviors, including the previously reported unidirectional rapid movement in live cells, and the distribution of the motion behaviors was related to the distribution of the complex microtubule configuration. These results indicated that the movement of the virus along microtubules was a complex process and influenced by the complex configuration of microtubules.
Experimental Section
Cell Culture and Virus Propagation
Madin-Darby canine kidney (MDCK) cells were cultured with Dulbecco’s modified Eagle medium (DMEM) containing 100 μg/mL streptomycin sulfate, 100 U/mL penicillin G, and 10% fetal bovine serum (FBS, Gibco). For transfection and fluorescence imaging, MDCK cells were planted onto a 20 mm Petri dish and 35 mm glass-bottomed Petri dish (NEST Corp), respectively, for 24 h before experiments. Avian influenza A virus (H9N2) strain was propagated in the allantoic cavity of 10-day-old embryonated eggs. After purification by ultracentrifugation and density gradient centrifugation, the viruses were harvested, aliquoted, and stored at −70 °C before use.6
Labeling Virus Envelope with QDs
To label the virus with QDs, we used the strong interaction of biotin–streptavidin to link biotinylated viruses with streptavidin-modified QDs (SA-QDs). To modify viruses with biotin, 100 μL of purified viruses was incubated with 0.1 mg of EZ-Link Sulfo-NHS-LC-Biotin (Thermo) at room temperature for 2 h. A NAP-5 column (GE Healthcare) was used to remove unbound biotin and 0.22 μm pore size filters removed virus aggregates before fluorescence imaging. A two-step method was used to label the biotinylated viruses with SA-QDs (Wuhan Jiayuan Quantum Dots Co., Ltd., China) as reported.6 First, biotinylated viruses were added to the MDCK cells at 4 °C for 10 min. After being washed with PBS containing 0.1% BSA, the MDCK cells were incubated with SA-QDs (2 nm) under the same conditions.
Labeling Microtubule of MDCK Cell
To label the microtubules of MDCK cells, the cells were transfected with the plasmid expressing GFP-microtubule-associated protein 4 (GFP-MAP4).26 To transfect cells in a 20 mm Petri dish, 0.5 μg of DNA and 1 μL of lipofectamine LTX reagent were mixed in 100 μL of Opti-MEM I reduced serum medium (Gibco). After 30 min, the lipofectamine LTX-DNA mixture was added to the cell culture. After incubation at 37 °C for 4 h, the medium was changed and the cells were then plated on a 35 mm glass-bottomed Petri dish for 6 h before fluorescence imaging.
To label microtubules with immunofluorescence, the cells were fixed in 4% (w/v) paraformaldehyde for 20 min at room temperature and exposed in PBS containing 5% (w/v) BSA and 0.1% (w/v) Triton-X 100 for 30 min at 37 °C. After being washed with PBS containing 1% (w/v) BSA, the cells were incubated with tubulin-beta-monoclonal antibody (Abnova) for 1.5 h and Dylight 649-conjugated goat antimouse IgG (Thermo) for 40 min at 37 °C to stain microtubules, and the cells were then incubated with 5 μg/mL Hoechst 33342 for 30 min at 37 °C.
Fluorescence Imaging
Fluorescence images were acquired with a spinning-disk confocal microscope (Andor Revolution XD), which was equipped with an Olympus IX 81 microscope, an EMCCD (Andor iXon DV 885K single photon detector), a Nipkow disk type confocal unit (CSU 22, Yokogawa), and a cell culture system (INUBG2-PI). Hoechst 33342, GFP, 605 nm QDs, and Dylight 649 were excited with 405, 488, 561, and 605 nm lasers, respectively. The fluorescence signals were separated with 447/60, 525/50, 617/73, and 685/40 nm band-pass emission filters, respectively. For multicolor imaging, fluorescence signals were detected separately with the EMCCD by the corresponding different channels.
Image Analysis
Each frame of the movies was denoised with a gauss filter. Kymograph image and orthogonal slice view were both obtained by Andor IQ software (Andor technology). To track the QDs-labeled virus, Imaging-Pro-Plus software (Media Cybernetics Inc. USA) was utilized. The MSD of each trajectory was calculated for each time interval by the user-written program with Matlab.27
Results and Discussion
Distribution of Microtubules and Influenza Viruses in Live Cells
First, we labeled the influenza virus with QDs by biotin–streptavidin interaction as reported previously.6 The specificity and efficiency of QDs to label influenza viruses were confirmed by incubating cells only with streptavidin-modified QDs and immunolabeling the hemagglutinin of the virus with Dylight 649, respectively (Figure S1 in Supporting Information).
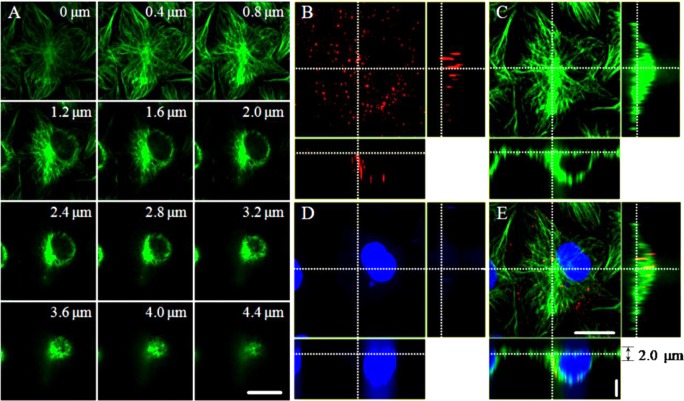
Earlier studies have shown that influenza virus can be transported via microtubules to the perinuclear region of the host cell for RNA release.6,7 To investigate the relationship between microtubules and viruses during virus infection, we immunolabeled the microtubules of Madin-Darby canine kidney (MDCK) cells, which were infected by the QDs-labeled influenza virus for 40 min and analyzed the distribution of virus in live cells. Using three-dimensional fluorescence imaging, we observed that the configurations of microtubules differ from near the bottom, i.e., close to the dish which cell is attached to, to near the top of the cell (Figure 1A). The obvious straight microtubules mainly existed in the range of 0–2 μm from the bottom of the cell, while the microtubules mostly intersected with each other in the upper part of the cell. Furthermore, we observed different microtubule configurations from the perinuclear region to the cell periphery. The microtubules in the region from the cell periphery to the perinuclear region were mostly linear but intersected with each other in the perinuclear region of the cell. Figure 1B–E showed that the QDs signals were accumulated in the perinuclear region of the cell, where the microtubules intersected with each other. Three-dimensional imaging, e.g., Movie S1 in Supporting Information, showed that the viruses were accumulated to the microtubule organizing center (MTOC) of the cell. Interestingly, we found that viruses moved unidirectionally in the range of 0–2 μm from the bottom of the cells. These results raised the questions of how the virus moved in different regions of the cell and how the various configurations of microtubules influenced the movement behaviors of viruses in live cells.
Figure 1.
Distributions of microtubules and influenza viruses in MDCK cells. (A) Snapshots of microtubules in a cell from the bottom to the top. The number in each panel indicates the distance from the cell bottom, close to the dish (Scale bar: 20 μm. The gap of Z: 0.4 μm). (B–E) Orthogonal slice views of QDs-labeled viruses (red), Dylight 649-labeled microtubules (green), Hoechst 33342-labeled nucleus (blue), and the overlapped image (Horizontal scale bar: 20 μm. Vertical scale bar: 3 μm).
Six Types of the Microtubules-Related Motion Behaviors of Viruses
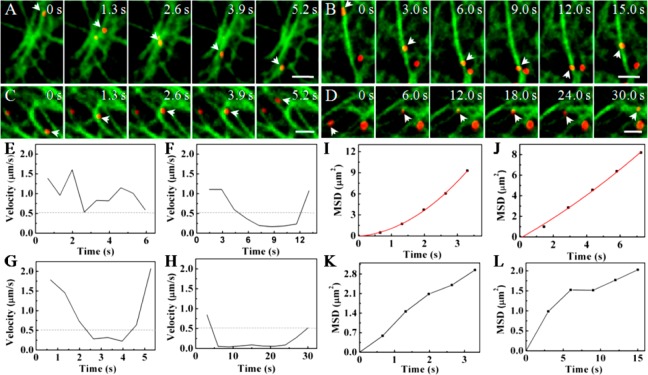
To investigate the influence of microtubule configuration on the movement behaviors of the virus, we monitored the movement of QDs-labeled viruses along microtubules in live cells by single-particle tracking in real time. We observed that many viruses moved along microtubules rapidly in a directed and regular motion mode, similar to that reported previously.6,7 In addition, we monitored the movement of viruses in cells treated with nocodazole, a microtubule-disrupting drug (Figure S2 in Supporting Information). In such cells, the movement of the virus was limited near the cytomembrane, indicating that the rapid transport of the virus was indeed dependent on microtubules. However, we also found the motion behaviors of viruses were complex frequently when the viruses encountered the intersections of microtubules. Statistical analysis revealed two distinct motion behaviors of viruses: movement along microtubules and movement confined to the certain microtubule regions/configurations. For those viruses that moved along the microtubules, there were four types of motion behaviors (Figure 2A–D): moving unidirectionally along microtubules (Type 1); decelerating near an intersection of microtubules and subsequently moving back along the same microtubule (Type 2); decelerating near an intersection of microtubules and then continuing to move along the same microtubule in the same direction (Type 3); and decelerating near an intersection of microtubules and then moving along another microtubule (Type 4).
Figure 2.
Influenza viruses moving along different configurations of microtubules. (A) Snapshots of a virus moving unidirectionally along microtubules (Scale bar: 2 μm). (B) Snapshots of a virus decelerating near an intersection of microtubules and moving back along the same microtubule (Scale bar: 2 μm). (C) Snapshots of a virus decelerating near an intersection of microtubules and moving along the same microtubule sequentially (Scale bar: 2 μm). (D) Snapshots of a virus decelerating near an intersection of microtubules and moving along another microtubule sequentially (Scale bar: 2 μm). (E–H) Velocity vs time plots of the movements shown in A–D, respectively. The dotted lines indicate the velocity of 0.5 μm/s. (I–L) MSD vs time plots of the movements shown in A–D, respectively. The red lines are the fits to MSD = 4Dτ + (Vτ)2 + constant (D and V are the diffusion coefficient and mean speed of the particle, the constant term was due to noise), and the black lines indicate the plots cannot be fitted due to the abnormalities of the movements.
To investigate the dynamic features of the different motion behaviors, we analyzed the typical trajectories of viruses in detail. It has been reported that actin filaments and microtubules are required for cellular delivery and transport by motor proteins and involved in the infection of influenza viruses. Myosin is the molecular motor that transports cargos along microfilaments at a speed of about 0.1–0.4 μm/s,28−30 while kinesin and dynein, the molecular motors traveling along microtubules, move faster than myosin, at a speed of several μm/s.31 Additionally, cellular movements can also be characterized in terms of the dependence of mean square displacement (MSD) on time. The linear, upward, and downward relationships in MSD indicate the movements are in normal diffusion, directed motion with diffusion, and anomalous diffusion, respectively. Therefore, the speed and motion mode of the particles moving in cells are often thought of as the important criteria to determine whether they move along actin filaments or microtubules. As motors travel along microtubules normally at a speed of several μm/s, if the speed of viruses moving along microtubules is below 0.5 μm/s, this would suggest that the microtubule-dependent movement is interfered by some mechanisms.31
Figure 2A showed the typical Type 1 virus movement. The virus kept on moving rapidly along the microtubule (Movie S2 in Supporting Information) with a speed higher than 0.5 μm/s (Figure 2E). On the basis of the relationship of the MSD vs time, we found that the virus moved in a directed motion mode, with the diffusion coefficient (D) and fitting velocity (V) of 0.043 μm2/s and 0.95 μm/s, respectively (Figure 2I). The result is consistent with the microtubule-dependent movements reported previously.6
A typical Type 2 movement was shown in Figure 2B. The virus moved along the microtubule rapidly and then slowed down near an intersection, followed by a sudden return to the opposite direction along the same microtubule (Movie S3 in Supporting Information). The speed vs time plot showed that the virus was decelerating at the intersection to a speed below 0.5 μm/s (Figure 2F). The MSD vs time plot also suggested that the movement was in a directed motion mode with D and V of 0.229 μm2/s and 0.18 μm/s, respectively (Figure 2J). These results indicated that the D was in the range of the microtubule-dependent movement, while the fitting velocity is very low and similar to the actin filaments-dependent movement reported previously.7 Thus, the deceleration of the virus movement may be caused by the redistribution of the force exerted on vesicles by molecular motors at the intersection and resulted in the complexity of the dynamic information about intracellular transport.
We also observed that, in some cases, when reaching the intersection of microtubules, the virus slowed down and then moved forward in the same direction along the same microtubule (Type 3). The snapshots of a typical movement of this kind were shown in Figure 2C (Movie S4 in Supporting Information). The speed vs time plot suggested that the virus slowed down to below 0.5 μm/s at the intersection and then moved rapidly along the same microtubule again (Figure 2G). On the basis of the MSD vs time plot, we found that the movement did not belong to any type of motion modes mentioned above (Figure 2K). The results suggested that the intersection significantly influenced the movement of the virus and hindered the analysis of motion mode under the conditions.
In some other cases, when the virus reached the intersection, the virus was found to abandon the original microtubule and move along another microtubule (Type 4). The snapshots and a movie for this type of motion were shown in Figure 2D and Movie S5 in Supporting Information, respectively. Here, we found that the virus slowed down to below 0.5 μm/s at the intersection and subsequently moved rapidly along another microtubule (Figure 2H). The MSD vs time plot was also irregular (Figure 2L), similar to that of Type 3.
Unlike the movement along the simple configuration of microtubules as described above, there were two types of movement with confined motion behaviors related to the complex configurations of microtubules. Figure 3A showed a virus confined within a grid formed with several microtubules (Type 5 motion behavior) (Movie S6 in Supporting Information). Analyzing the speed and MSD vs time plots, we found that the speed of the virus was below 0.25 μm/s (Figure 3E) and the motion behavior was consistent with the anomalous diffusion mode with the D and α value of 0.002 μm2/s and 0.59, respectively (Figure 3F), indicating that the virus was confined by the grid of microtubules. In addition, another confined movement was observed at the intersection of several microtubules, where the virus was confined to the intersection, the Type 6 motion behavior (Figure 3B and Movie S7 in Supporting Information). The speed and MSD vs time plots suggested that the virus moved slowly with the speed below 0.25 μm/s (Figure 3G) and the movement was in anomalous diffusion mode with D and α value of 0.001 μm2/s and 0.74, indicating that the virus was confined by the intersection (Figure 3H). Kymograph images further confirmed that the movements were slow and confined by the two kinds of microtubule configuration (Figure 3C,D). Our analyses indicated that these two confined motion behaviors were similar to each other except the types of microtubule configurations where the virus was confined.
Figure 3.
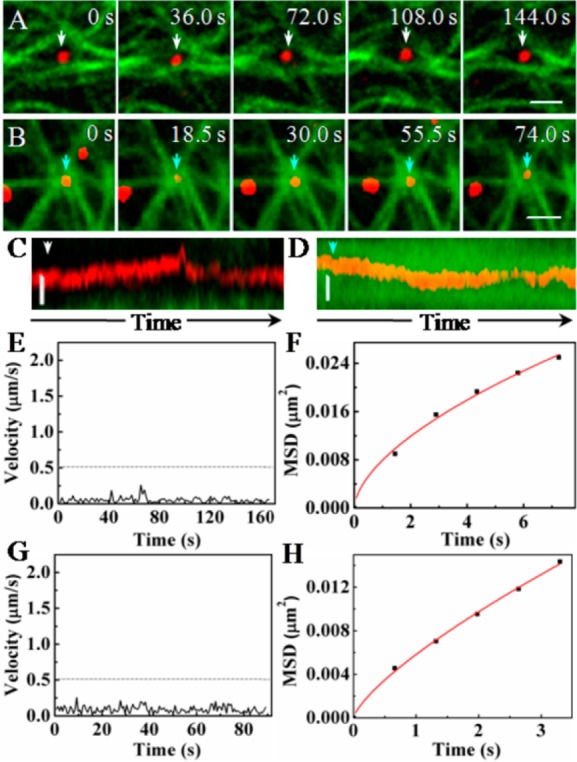
Influenza viruses moving at two typical configurations formed by several microtubules. (A) Snapshots of a virus being confined by a grid formed by microtubules (Scale bar: 2 μm). (B) Snapshots of a virus being confined by an intersection of microtubules (Scale bar: 2 μm). (C, D) Kymograph images of the movements of the viruses shown in A and B, respectively (Scale bar: 0.5 μm). (E, G) Velocity vs time plots of the movements shown in A and B, respectively. The dotted lines indicate the velocity of 0.5 μm/s. (F, H) MSD vs time plots of the movements shown in A and B, respectively. The red lines are the fits to MSD = 4Dτα (α is a coefficient and α < 1).
Taken together, the complex configurations of microtubules appeared to bring about different types of motion behaviors of the virus. When a virus underwent the various motion behaviors mentioned above successively, the infection pathway of the virus could be full of twists and turns (Figure S3A and Movie S8 in Supporting Information). The speed vs time plot showed that the virus moved very irregularly along the microtubules (Figure S3B in Supporting Information). Analyzing the MSD vs time plot, we found that the movement was still in directed motion mode with the D and V values of 0.070 μm2/s and 0.081 μm/s, respectively (Figure S3C in Supporting Information). This result demonstrated that the D value could reflect the characteristic of the microtubule-dependent movement, even though the complexity of microtubules led to the irregular fitting speed. The result confirmed that the D value could be used to estimate whether the movement of viruses was related to microtubule or actin filament.
Intracellular Distribution of Microtubule-Related Motion Behaviors
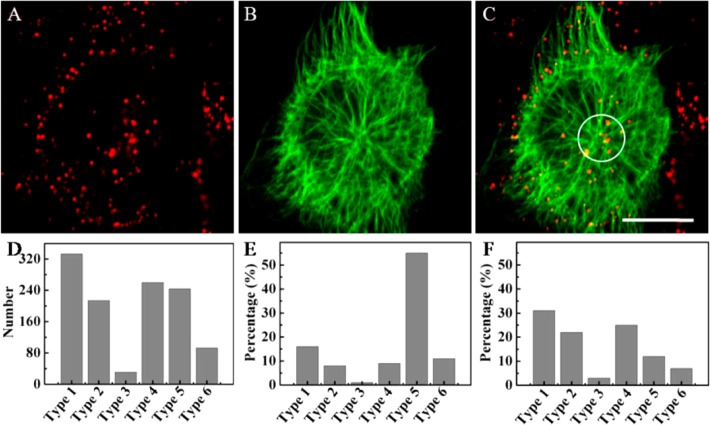
As mentioned above, the configuration of microtubules was different in different regions of the cell. Herein, we chose ten cells randomly from six parallel experiments to further investigate the distribution of the six types of motion behavior mentioned above in live cells. We statistically analyzed the motion behaviors of viruses related to microtubules and obtained 1183 trajectories of the viruses. The percentages of the six types of motion behavior were about 28%, 19%, 2%, 22%, 21%, and 8%, respectively (Figure 4D), suggesting that the directed rapid motion mode (Type 1) as reported previously6,7 was just the main motion behavior of viruses moving along microtubules.
Figure 4.
Tracking the movements of viruses in the bottom of the cells. (A–C) Fluorescence images of QDs-labeled viruses (red), Dylight 649-labeled microtubules (green), and the overlapped image of panels A and B (Scale bar: 20 μm). (D–F) Distributions of six types of the movements in the whole cells, near the Microtubule organizing center (MTOC), and in the region from the cell periphery to the MTOC region, respectively.
Given the different patterns of microtubule configuration in the upper and lower halves of the cells, we first studied the motion behaviors of the virus in the lower part of cells. Considering the morphological differences of microtubules between the MTOC region (the white circle in Figure 4C) and the region from the cell periphery to the MTOC region, we studied the distribution of motion behaviors in the two regions. At the MTOC region, the percentages of the six types of motion behavior were 16%, 8%, 1%, 9%, 55%, and 11%, respectively (Figure 4E), indicating that Type 5 movement was the main motion behavior of viruses at the MTOC, i.e., the viruses were mainly confined by the grids formed by microtubules here. We further used the QDs-labeled virus to infect the DiO-labeled MDCK cells (DiO is a membrane dye) (Figure S4 in Supporting Information). The signals of QDs always colocalized with DiO signals in the cytoplasm and accumulated in the perinuclear region, suggesting that the viruses were trapped in vesicles and transported to the MTOC region. Thus, it was speculated that the grid of microtubules might be the support structure of vesicles at the MTOC. In contrast to the MTOC, we found that the percentages for the different motion types in the region from the cell periphery to the MTOC region were 31%, 22%, 3%, 25%, 12%, and 7%, respectively (Figure 4F), indicating that the directed rapid motion mode (Type 1) was the main motion behavior of the virus in this region. These two contrasting distribution patterns of the motion types explained why the viruses converged to the perinuclear region in a directed rapid motion mode. The percentages of Type 2 and Type 4 behaviors were also large in the region from the cell periphery to the MTOC, suggesting that the intersection of microtubules indeed interfered with the movement of viruses along microtubules.
We next studied the motion behaviors of the virus in the upper part of cells by the same method. As shown in Figure 5, four viruses represented by four arrows with different colors, respectively, were essentially motionless on the complexly crossed microtubules (Figure 5A and Movie S9 in Supporting Information). Kymograph images showed that the four viruses were confined to the regions (Figure 5B). The speeds of viruses were below 0.5 μm/s, and the motion modes were anomalous diffusion modes with small values of D based on the speed and MSD vs time plots, respectively (Figure 5C,D). These results suggested that viruses in the upper part of cells were mainly confined by the grids or intersections of microtubules. The motion behaviors of the virus were relatively simple, mostly the fifth and sixth types of motion behaviors, owing to the grids and intersections of microtubules.
Figure 5.
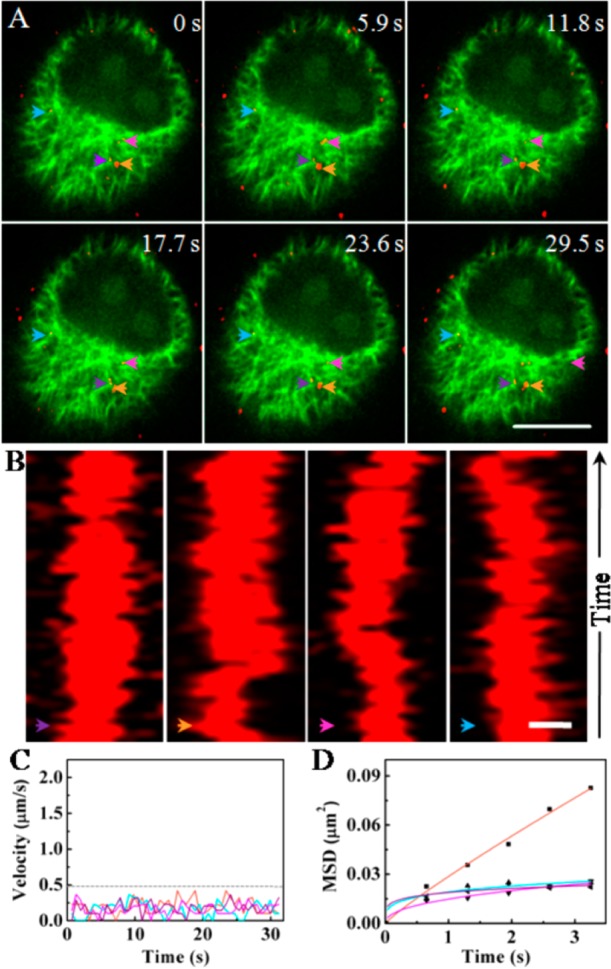
Tracking the movements of the viruses in the upper part of the cells. (A) Snapshots of four viruses moving in the upper part of the cell (Scale bar: 10 μm). The colored arrows indicate the viruses. (B) Kymograph images of the movements of the four viruses shown in (A) (Scale bar: 0.5 μm). (C) Velocity vs time plots of the movements of the four viruses shown in (A). The dotted line indicates the velocity of 0.5 μm/s. (D) MSD vs time plots of the movement shown in (A). The lines are the fits to MSD = 4Dτα with D and α values of 0.0072 μm2/s and 0.895 (orange), 0.0037 μm2/s and 0.432 (blue), 0.0048 μm2/s and 0.248 (pink), and 0.0047 μm2/s and 0.177 (purple), respectively. The four different colored lines in C and D refer to the four different viruses shown in (A).
Conclusions
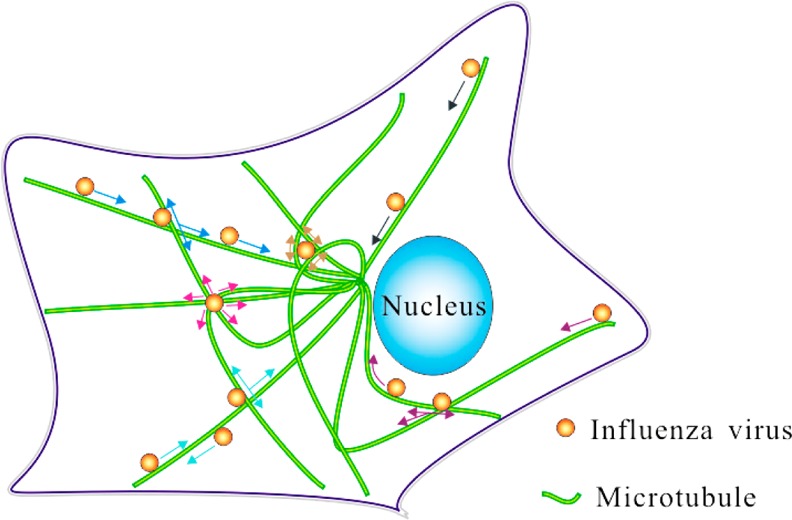
Given the obvious importance of understanding virus infection, there have been extensive studies on the movement of viruses in cells. These earlier studies revealed that the movement of viruses in cells is dependent on microtubules and further suggested that the movement along microtubules is a simple directed rapid process.7 Our live-cell imaging of the virus infection process for the first time revealed that the infection process was much more complicated. By studying the behaviors of viruses moving along microtubules by the real-time and long-term SPT technique based on the superior optical properties of QDs, we showed that there were many types of complicated motion behaviors of viruses in addition to the previously reported directed rapid motion mode (Figure 6). On the basis of statistical analyses, we found that the distribution of virus motion behaviors was different in different regions of the cell and that the grid of microtubules may be the support structure of vesicles in live cells. The complex transport behaviors of viruses may be caused by the interactions of multiple motor proteins brought about by the complexity of microtubule configurations. Earlier studies have suggested that it is important to monitor the process of viruses converging rapidly from cell membrane to perinuclear region in the study of the transport dynamics of viruses in cells.6,7 Our research shows that only by choosing the appropriate locations within a cell can we observe the process of rapid converging movement of viruses in cells. For MDCK cells, the most appropriate cell level to track viruses is the region from the bottom to 2 μm of cells. Our findings have not only revealed the importance of intracellular structures for cellular transport by endocytosis but also raised cautions about interpreting the data about intracellular virus movement without carefully considering the cellular locations and distribution of different types of virus movement.
Figure 6.
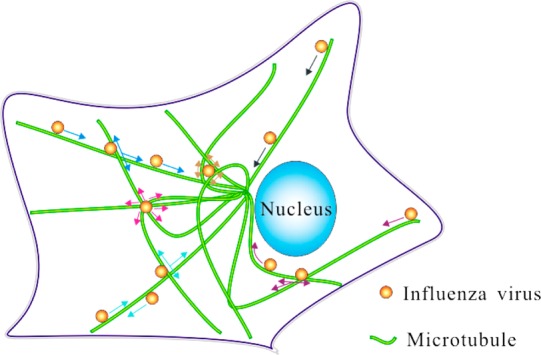
Schematic diagram of the microtubule-related behaviors of viruses. The black arrow indicates that the virus is moving unidirectionally along microtubules (Type 1). The light blue arrow indicates the virus is decelerating near an intersection of microtubules and moving back along the same microtubule (Type 2). The blue arrow indicates that the virus is decelerating near an intersection of microtubules and moving along the same microtubule sequentially (Type 3). The purple arrow indicates that the virus is decelerating near an intersection of microtubules and moving along another microtubule sequentially (Type 4). The yellow arrow indicates that the virus is moving confinedly in a grid formed by microtubules (Type 5). The pink arrow indicates that the virus is moving confinedly at an intersection of microtubules (Type 6).
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program, No. 2011CB933600), the Science Fund for Creative Research Groups of NSFC (No. 20921062), the 111 Project, and the 3551 Talent Program of the Administrative Committee of East Lake Hi-Tech Development Zone ([2011]137). Y.-B.S. was supported by the Intramural Research Program of NICHD, National Institutes of Health.
Supporting Information Available
Figures S1–S4 and Movies S1–S9 (ac500640u_si_002.avi–ac500640u_si_010.avi, respectively). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
‡ S.-L.L. and L.-J.Z. contributed equally.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Suomalainen M.; Nakano M. Y.; Keller S.; Boucke K.; Stidwill R. P.; Greber U. F. J. Cell Biol. 1999, 144, 657–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J. J.; Ng M. L. J. Virol. 2004, 78, 10543–10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderheit A.; Helenius A. PLoS Biol. 2005, 3, 1225–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L.; Kartenbeck J.; Helenius A. Nat. Cell Biol. 2001, 3, 473–483. [DOI] [PubMed] [Google Scholar]
- Sodeik B.; Ebersold M. W.; Helenius A. J. Cell Biol. 1997, 136, 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L.; Zhang Z. L.; Tian Z. Q.; Zhao H. S.; Liu H.; Sun E. Z.; Xiao G. F.; Zhang W.; Wang H. Z.; Pang D. W. ACS Nano 2011, 6, 141–150. [DOI] [PubMed] [Google Scholar]
- Lakadamyali M. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 9280–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger G.; Ried M. U.; Endress T.; Buning H.; Hallek M.; Brauchle C. Science 2001, 294, 1929–1932. [DOI] [PubMed] [Google Scholar]
- Gundersen G. G. Nat. Rev. Mol. Cell Biol. 2002, 3, 296–304. [DOI] [PubMed] [Google Scholar]
- Döhner K.; Nagel C.-H.; Sodeik B. Trends Microbiol. 2005, 13, 320–327. [DOI] [PubMed] [Google Scholar]
- Dodding M. P.; Way M. EMBO J. 2011, 30, 3527–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Science 1998, 279, 519–526. [DOI] [PubMed] [Google Scholar]
- Welte M. A. Curr. Biol. 2004, 14, R525–R537. [DOI] [PubMed] [Google Scholar]
- Ross J. L.; Ali M. Y.; Warshaw D. M. Curr. Opin. Cell Biol. 2008, 20, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. L.; Shuman H.; Holzbaur E. L.; Goldman Y. E. Biophys. J. 2008, 94, 3115–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershinin M.; Carter B. C.; Razafsky D. S.; King S. J.; Gross S. P. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint S.; Verdeny Vilanova I.; Sandoval Alvarez A.; Lakadamyali M. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R. A.; Garcia-Sastre A. Nat. Rev. Microbiol. 2011, 9, 590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K.; Joseph T. Nat. Rev. Immunol. 2007, 7, 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G.; Noda T.; Kawaoka Y. Nature 2009, 459, 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H. L.; Webster R. G. Curr. Top. Microbiol. Immunol. 2009, 333, 3–24. [DOI] [PubMed] [Google Scholar]
- Claas E. C. J.; Osterhaus A. D. M. E.; van Beek R.; De Jong J. C.; Rimmelzwaan G. F.; Senne D. A.; Krauss S.; Shortridge K. F.; Webster R. G. Lancet 1998, 351, 472–477. [DOI] [PubMed] [Google Scholar]
- Capua I.; Alexander D. J. Acta Trop. 2002, 83, 1–6. [DOI] [PubMed] [Google Scholar]
- Gao R.; Cao B.; Hu Y.; Feng Z.; Wang D.; Hu W.; Chen J.; Jie Z.; Qiu H.; Xu K.; et al. N. Engl. J. Med. 2013, 368, 1888–1897. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Liang W.; Yang S.; Wu N.; Gao H.; Sheng J.; Yao H.; Wo J.; Fang Q.; Cui D.; et al. Lancet 2013, 381, 1916–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J.; Chua N. H. Plant Cell 2000, 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton M. J.; Jacobson K. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 373–399. [DOI] [PubMed] [Google Scholar]
- Dunn A. R.; Spudich J. A. Nat. Struct. Mol. Biol. 2007, 14, 246–248. [DOI] [PubMed] [Google Scholar]
- Sakata S.; Watanabe Y.; Usukura J.; Mabuchi I. J. Biochem. 2007, 142, 481–490. [DOI] [PubMed] [Google Scholar]
- Nagy S.; Ricca B. L.; Norstrom M. F.; Courson D. S.; Brawley C. M.; Smithback P. A.; Rock R. S. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 9616–9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan J. C.; Brandenburg B.; Hogle J. M.; Zhuang X. Biophys. J. 2009, 97, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.