Summary
We show that the E3 ubiquitin ligase EDD/UBR5 regulates both survival and cisplatin resistance in ovarian cancer cells and is a novel therapeutic target for ovarian cancer.
Abstract
The E3 ubiquitin ligase EDD is overexpressed in recurrent, platinum-resistant ovarian cancers, suggesting a role in tumor survival and/or platinum resistance. EDD knockdown by small interfering RNA (siRNA) induced apoptosis in A2780ip2, OVCAR5 and ES-2 ovarian cancer cells, correlating with loss of the prosurvival protein myeloid cell leukemia sequence 1 (Mcl-1) through a glycogen synthase kinase 3 beta-independent mechanism. SiRNA to EDD or Mcl-1 induced comparable levels of apoptosis in A2780ip2 and ES-2 cells. Stable overexpression of Mcl-1 protected cells from apoptosis following EDD knockdown, accompanied by a loss of endogenous, but not exogenous, Mcl-1 protein, suggesting that EDD regulated Mcl-1 synthesis. Indeed, EDD knockdown induced a 1.87-fold decrease in Mcl-1 messenger RNA and EDD transfection enhanced murine Mcl-1 promoter-driven luciferase expression 5-fold. To separate EDD survival and potential cisplatin resistance functions, we generated EDD shRNA stable cell lines that could survive initial EDD knockdown and showed that these cells were 4- to 21-fold more sensitive to cisplatin. Moreover, transient EDD overexpression in COS-7 cells was sufficient to promote cisplatin resistance 2.4-fold, dependent upon its E3 ligase activity. In vivo, mouse intraperitoneal ES-2 and A2780ip2 xenograft experiments showed that mice treated with EDD siRNA by nanoliposomal delivery [1,2-dioleoyl-sn-glycero-3-phophatidylcholine (DOPC)] and cisplatin had significantly less tumor burden than those treated with control siRNA/DOPC alone (ES-2, 77.9% reduction, P = 0.004; A2780ip2, 75.9% reduction, P = 0.042) or control siRNA/DOPC with cisplatin in ES-2 (64.4% reduction, P = 0.035), with a trend in A2780ip2 (60.3% reduction, P = 0.168). These results identify EDD as a dual regulator of cell survival and cisplatin resistance and suggest that EDD is a therapeutic target for ovarian cancer.
Introduction
Initial therapy for ovarian cancer involves surgical debulking combined with chemotherapy, which consists of platinum and paclitaxel; however, resistance to chemotherapy often occurs in recurrent tumors. Identifying mechanisms of acquired drug resistance is important to developing novel therapeutics. One indicator of poor prognosis in recurrent ovarian cancer is the E3 ubiquitin ligase EDD (E3 ligase identified by differential display), a 300kDa nuclear phosphoprotein that we previously identified as a direct substrate of the MAP kinase extracellular signal-regulated kinase 2 (1–4). E3 ubiquitin ligases modify proteins through the addition of ubiquitin, most often resulting in protein degradation (5,6). EDD contains a C-terminal HECT (Homologous to the E6-AP Carboxyl Terminus) ubiquitin ligase domain and is the human homolog of the Drosophila tumor suppressor hyperplastic discs (hyd), which regulates imaginal disk formation (7). EDD has a reported role in the DNA damage response and has been implicated in the S phase and G2/M DNA damage checkpoints (2,8,9). EDD enhances activation of the DNA damage response kinase Chk2 in response to ionizing radiation or the radiomimetic phleomycin (10). EDD also acts as a transcriptional coactivator for the progesterone and vitamin D receptors, dependent upon its middle domain and independent of its E3 ligase activity (2).
EDD protein is overexpressed or mutated in several solid tumors including ovarian, breast, hepatocellular, tongue, gastric and melanoma (11–13). EDD protein levels are low in benign ovarian tissue and borderline tumors, but overexpression is observed in 47% of ovarian cancer tumors overall, 73% of serous ovarian tumors and was associated with a 2-fold increased risk of recurrence and death in patients who had a favorable response to initial chemotherapy (1,11). The edd gene is on chromosome 8q22.3 and amplification of this chromosomal region is associated with cisplatin resistance (7,14). Knockdown of EDD with small interfering RNA (siRNA) decreased colony formation in A2780-cp70 ovarian cancer cells, a derivative selected for cisplatin resistance in vitro, when cotreated with cisplatin (1). Collectively, these results suggest that EDD may play a role in tumor maintenance and/or cisplatin resistance.
Altered expression of many genes and proteins has been reported in tumor tissue and in isogenic cell lines that have been selected for cisplatin resistance. However, many of these studies failed to demonstrate that changes in expression of a particular protein were sufficient to induce cisplatin resistance, raising the possibility that the observed overexpression of EDD in ovarian tumors may not be directly responsible for acquired cisplatin resistance. In this article, we demonstrate that EDD directly contributes to both cell survival through myeloid cell leukemia sequence 1 (Mcl-1) upregulation and cisplatin resistance through its E3 ubiquitin ligase activity in ovarian cancer cells and we provide evidence for EDD as a therapeutic target for the treatment of epithelial ovarian cancer.
Materials and methods
Cell lines and antibodies
ES-2 and TOV21G cells were from Runzhao Li, OVCAR3 cells were from Kristen Atkins, A2780 cells were from Andrew Godwin, A2780ip2 cells were from Charles Landen, OVCAR5 cells were from Thomas Hamilton and IOSE cells were from Nelly Auersperg. COS-7, HeLa, COS-1 and SKOV-3 cells were from American Type Culture Collection (ATCC, Manassas, VA). Stable EDD shRNA cells were generated by retroviral transduction: control shRNA (5′GCTGCAAGACCA TACACTTAT), EDD-shRNA1 (5′GCTGTAGA TTTCAACTTAGAT), EDD-shRNA2 (5′GCCATTAGA AAGAACCAC AAA) and EDD-shRNA3 (5′TGACAGCAGAA CA ACATAATT). Puromycin-resistant clones (ES-2 and A2780ip2) or populations (OVCAR5) were selected. Mcl-1 stable cells were generated by transduction with pBabe or pBabe-Flag-Mcl-1 (Addgene) and puromycin-resistant clones (A2780ip2) or populations (ES-2) were selected. Cisplatin was from Sigma–Aldrich (St Louis, MO). Antibodies [poly(ADP-ribose) polymerase (PARP), Bcl2 family proteins, actin] were from Cell Signaling (Danvers, MA) and the EDD (M19) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA).
siRNA transfection
Cell lines were transfected with 45 nmol of control or EDD siRNA (Sigma–Aldrich). siRNA1: SASI_Hs01_00175227 (5′CCAUUUACCCUGGC UAGUA); siRNA2: SASI_Hs02_00348492 (5′GCGACUCUCCAUG GUUUCU). Mcl-1 siRNA: SASI_Hs01_00162656 (5′GUAAUAGA ACUA UGACUGU). Bcl-xL siRNA: SASI_Hs01_00165963 (5′CUGAUUGGU GCAACCCUUA). Glycogen synthase kinase 3 beta (GSK-3β) siRNA1: SASI_ Hs01_00192106 (5′GGACUAUGU UCCGGAAA CA) and GSK-3β siRNA2: SASI_Hs01_00192105 (5′CACUCAA GAACUGUCAAGU). Twenty nanomoles of Mcl-1, Bcl-xL and GSK-3β siRNA were used. Control siRNA was Universal Negative Control #1 (Sigma–Aldrich).
Western blotting
Floating and adherent cells were lysed with M2 lysis buffer containing 0.5% sodium dodecyl sulfate (15). Typically, 65 μg of protein lysate was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (7–12% gradient gel) and immunoblotted proteins were visualized using enhanced chemiluminescence (Pierce). GSK-3β inhibitors used were LiCl (20mM, Sigma–Aldrich), TDZD-8 (10 μM) and L803-mts (20 μM, EMD Chemicals, Gibbstown, NJ). Cycloheximide (Sigma–Aldrich) was used at 50 μg/ml. For caspase inhibition, cells were cotreated with siRNA and either 25 μM pan caspase inhibitor Q-VD-OPH (R&D Systems, Minneapolis, MN) or the negative control Z-FA-FMK (BD Pharmingen, Franklin Lakes, NJ).
Crystal violet staining
Cells were fixed with 4% paraformaldehyde, stained with 0.05% crystal violet in 2% ethanol for 15 min, washed five times with phosphate-buffered saline and dried. Stained cells were solubilized with 2% sodium dodecyl sulfate in phosphate-buffered saline and absorbance was measured at 550 nm.
Quantitative real-time PCR
RNA was extracted using the Qiagen (Valencia, CA) RNeasy Plus Mini Kit and cDNA was synthesized using the Bio-Rad (Hercules, CA) iScript™ Advanced cDNA Synthesis Kit for RT-qPCR. Bio-Rad’s SsoAdvanced™ SYBR® Green Supermix was used for quantitative real-time PCR on an Eppendorf (Hauppage, NY) Mastercycler Realplex 2. The average fold change of the test sample over control sample was determined for each experimental condition with normalization to two housekeeping genes, actin and glyceraldehyde 3-phosphate dehydrogenase. The Mcl-1 primer was from Integrated DNA Technologies (Coralville, IA) (forward: 5′-AAA GAGGCTGGG ATGGGTTT-3′, reverse: 5′-CAAAA GCAAGC AGCACATTC-3′). The actin primer used was from Real-Time Primers (forward: 5′-GGACTTCGAGCA AGAGATGG-3′, reverse: 5′-AG CACTGTGT TGGCGTACAG-3′) along with glyceraldehyde 3-phosphate dehydrogenase (forward: 5′-GAGTCAACGGATTTGGTCG T-3′, reverse: 5′-TTGATTTTGG AGGGATCTCG-3′).
Flow cytometry
Floating and adherent cells were fixed in ethanol and stained with propidium iodide (Molecular Probes, Eugene, OR). DNA content was determined by flow cytometry and sub-2n cells were counted as apoptotic. The Student’s t-test was performed on three independent experiments done in duplicate.
MTS assay
Stable ES-2 shRNA cell lines were plated in quadruplicate onto 96-well dishes and treated with cisplatin or saline for 72 h. MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) reagent (Promega, Madison, WI) was added for the last 2 h and absorbance measured. The results are a combination of three independent experiments.
Apoptosis assay
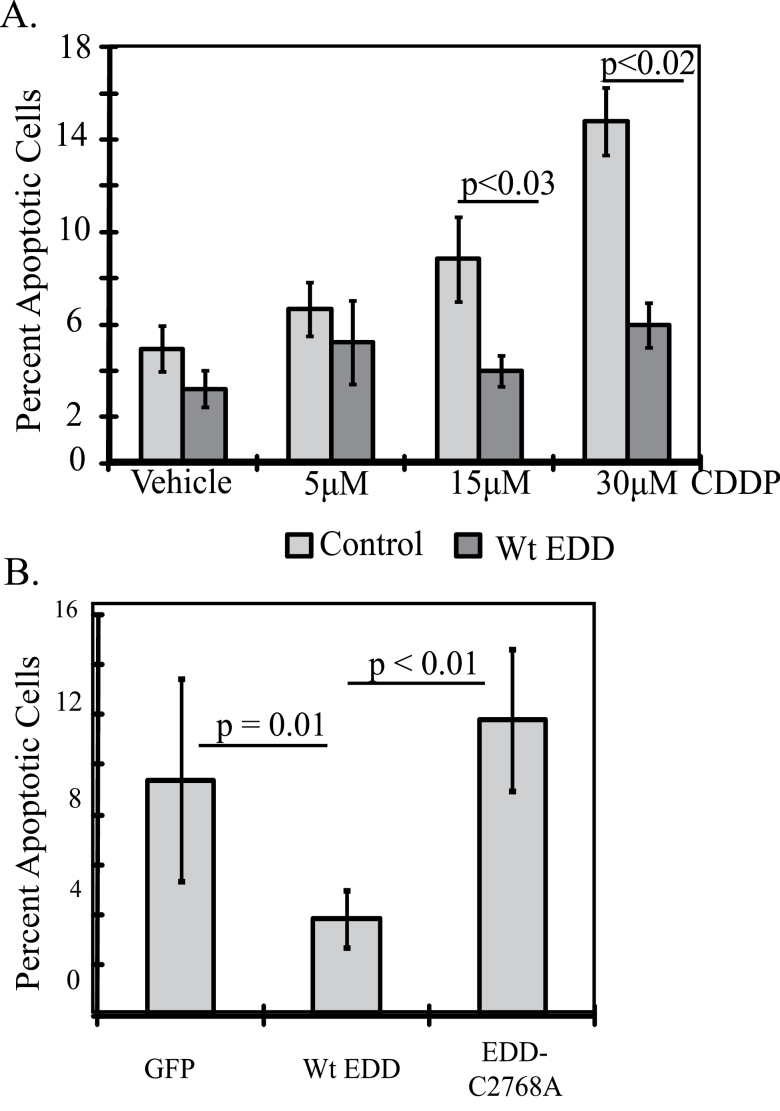
COS-7 cells on coverslips were transfected with 2 µg of Flag-EDD, Flag-EDD-C2768A or green florescent protein (GFP). After 24 h, the cells were treated with cisplatin for 24 h and fixed with 4% paraformaldehyde. Apoptotic cells were labeled using the TACS® 2 Tdt-Blue Label In Situ Apoptosis Detection Kit (Trevigen, Gaithersburg, MD). Flag-EDD-transfected cells were immunostained with M2 anti-Flag antibody (Sigma–Aldrich), followed by fluorescein isothiocyanate-labeled secondary antibody (Jackson ImmunoResearch, West Grove, PA). At least 500 transfected cells per coverslip were counted and the percentage of transfected apoptotic cells was determined. Four independent experiments were performed for the cisplatin dose experiment. For the EDD-C2768A experiment, three independent experiments were performed comparing GFP, EDD and EDD-C2768A at a single dose of 15 μM cisplatin. The data for GFP compared with EDD included the data from the 15 μM group in the cisplatin dose experiment, for an n = 7. Two-sample t-tests were conducted to determine significance.
Luciferase assays
HeLa cells were transfected with 40 ng TK Renilla luciferase, 400 ng of firefly luciferase plasmid p(−2389/+10)mcl-luc (16) and 2 μg of either wild-type or mutant Flag-EDD or empty vector. Luciferase assays were performed at 48 h using the Dual Luciferase Reporter Assay (Promega) on a Monolight 2010 Luminometer (Analytical Luminescence, Ann Arbor, MI). Firefly luciferase activity was normalized to Renilla luciferase. The results are a combination of four independent experiments done in triplicate. After averaging over experimental replicates, a two-sample t-test was conducted for each luciferase plasmid testing the effect of EDD or EDD mutant versus vector. Cell lysates were immunoblotted for EDD and actin.
Intraperitoneal ovarian cancer model and in vivo delivery of siRNA
Female athymic nude mice (NCr-nu) were purchased from the National Cancer Institute (Frederick, MD) after Institutional Animal Care and Use Committee approval of protocols and cared for in accordance with guidelines of the American Association for Accreditation of Laboratory Animal Care. ES-2 and A2780ip2 cells were suspended in serum-free Hanks' balanced salt solution at a concentration of 5 × 106 cells/ml, and 1 × 106 cells were injected intraperitoneally in 200 µl into 40 mice per experiment. After 1 week, mice (n = 10 per group) were randomized to treatment with (i) 5 µg control siRNA (sense sequence: 5′-UUCUCCGAAC GUGUCACGU-3′, Sigma) in 1,2-dioleoyl-sn-glycero-3-phophatidylcholine (DOPC), (ii) 5 µg anti-human EDD siRNA (Sigma product SASI_Hs01_00175227), (iii) 5 µg control siRNA plus cisplatin or (iv) 5 µg EDD-targeting siRNA in DOPC plus cisplatin. siRNA constructs were incorporated in DOPC nanoparticles (DOPC) as described previously (17,18) and the lyophilized product was stored at −4°C for <4 weeks. Prior to treatment, the siRNA/DOPC complex was reconstituted in 0.9% saline and administered intraperitoneally twice per week in a volume of 100 µl. Cisplatin was administered intraperitoneally at a dose of 40 µg weekly. Mice were treated for 4 weeks before killing and tumor collection. Tumors were excised and total tumor weight recorded. Statistical analysis comparisons of tumor weights were made using a two-tailed Student’s t-test, if assumptions of data normality were met. Those represented by alternate distribution were examined using a non-parametric Mann–Whitney U-test. Differences between groups were considered statistically significant at P < 0.05. Error bars represent standard error. Number of mice per group (n = 10) was chosen as directed by a power analysis to detect a 50% decrease in tumor growth with beta error of 0.2. Immunohistochemistry was performed using anti-EDD antibody.
Results
EDD knockdown induces apoptosis in ovarian cancer cells
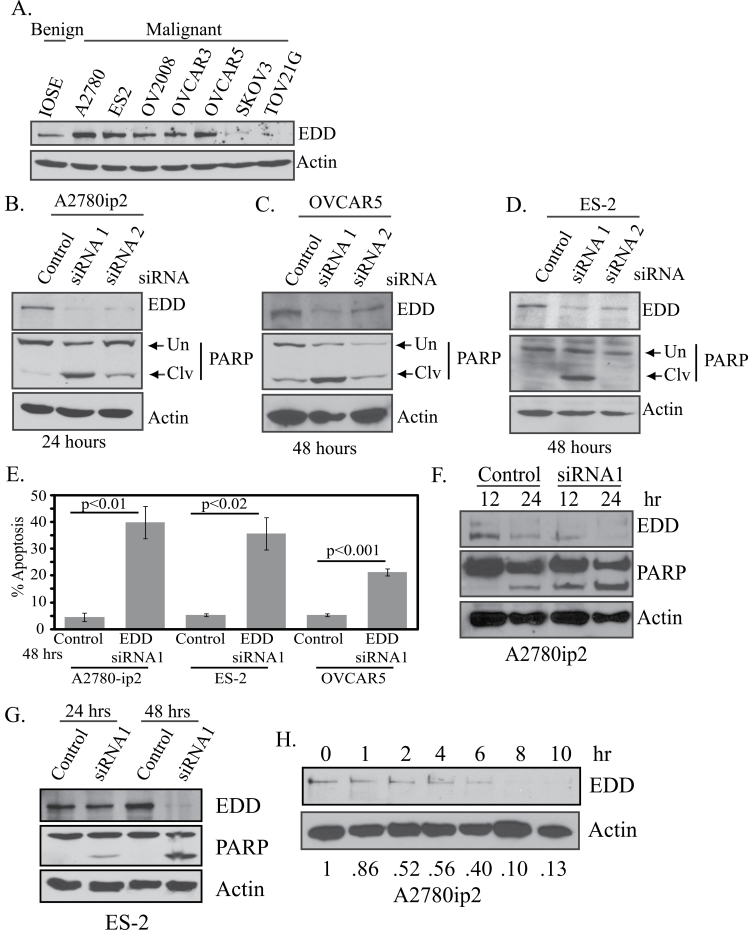
Immunoblotting lysates from ovarian cell lines showed high EDD expression in five of seven ovarian cancer cell lines compared with the preneoplastic IOSE398 cell line, with the highest expression in ES-2, OVCAR5 and A2780 cells (Figure 1A). To determine the effect of EDD knockdown, we transfected A2780ip2 (Supplementary Figure 1A, available at Carcinogenesis Online), ES-2 (Supplementary Figure 1B, available at Carcinogenesis Online) and OVCAR5 (Supplementary Figure 1C, available at Carcinogenesis Online) cells with control siRNA or one of two EDD siRNAs. EDD siRNAs knocked down EDD protein expression, with siRNA1 having the strongest effect. Interestingly, cells transfected with EDD siRNA showed a significant reduction in cell number in all three cell lines within 48 h, as measured by quantitation of crystal violet staining, with the exception of siRNA2 in ES-2 cells (Supplementary Figure 1D, available at Carcinogenesis Online). Loss of cell viability after EDD siRNA1 transfection increased from 24 to 72 h (Supplementary Figure 1E, available at Carcinogenesis Online). To determine whether EDD knockdown induced apoptosis, lysates from floating and adherent siRNA-transfected cells were immunoblotted for cleavage of PARP, a substrate of caspases and an indicator of apoptosis. Enhancement of cleaved PARP relative to total PARP (cleaved plus uncleaved) was observed in all three cell lines after EDD siRNA transfection (Figure 1B–D), with siRNA1 having a greater effect, coinciding with greater EDD knockdown, especially in ES-2 cells. A2780ip2 cells showed enhanced apoptotic sensitivity to EDD knockdown at earlier time points (Figure 1B). In addition, propidium iodide staining followed by flow cytometry showed significant apoptosis, measured by <2n DNA content, after 48 h of EDD knockdown in A2780ip2 (control = 5.8%; EDD = 44.6%), ES-2 (control = 5.8%; EDD = 42.6%) and OVCAR5 (control = 5.9%; EDD = 22.6%) cells (Figure 1E). The induction of apoptosis showed a temporal increase in both A2780ip2 (Figure 1F) and ES-2 cells (Figure 1G). The relatively rapid induction of apoptosis suggested a short EDD half-life and strong requirement for cell survival. Cycloheximide experiments demonstrated the half-life of EDD protein was ~4 h in A2780ip2 cells (Figure 1H).
Fig. 1.
EDD is overexpressed in ovarian cancer cell lines and EDD knockdown induces apoptosis. (A) EDD expression was determined by immunoblotting lysates from ovarian cell lines. (B) A2780ip2, (C) OVCAR5 and (D) ES-2 cells were transfected with control siRNA or one of two siRNAs to EDD. After transfection for the indicated time, floating and adherent cells were harvested and cell lysates were immunoblotted for EDD expression and PARP. Uncleaved (Un) and cleaved (Clv) PARP are indicated with arrows. (E) Cells were transfected with control siRNA or EDD siRNA1 for 48h and floating and adherent cells were stained with propidium iodide. Flow cytometry was used to determine the percentage of cells with sub-2n DNA content, an indicator of apoptosis. The results are from three independent experiments. (F) A2780ip2 cells were transfected with control siRNA or EDD siRNA1 for 12 or 24h and lysates from floating and adherent cells were immunoblotted for EDD expression and PARP cleavage. (G) ES-2 cells were transfected with control siRNA or EDD siRNA1 for 24 or 48h and lysates from floating and adherent cells were immunoblotted for EDD expression and PARP cleavage. (H) A2780ip2 cells were treated with 50 μg/ml of cycloheximide for the indicated time. Cell lysates were immunoblotted for EDD and actin. The number under each lane indicates the relative intensity of the EDD band compared with actin, with the amount in time zero set at 1.
EDD knockdown causes loss of Mcl-1 through a degradation- independent mechanism
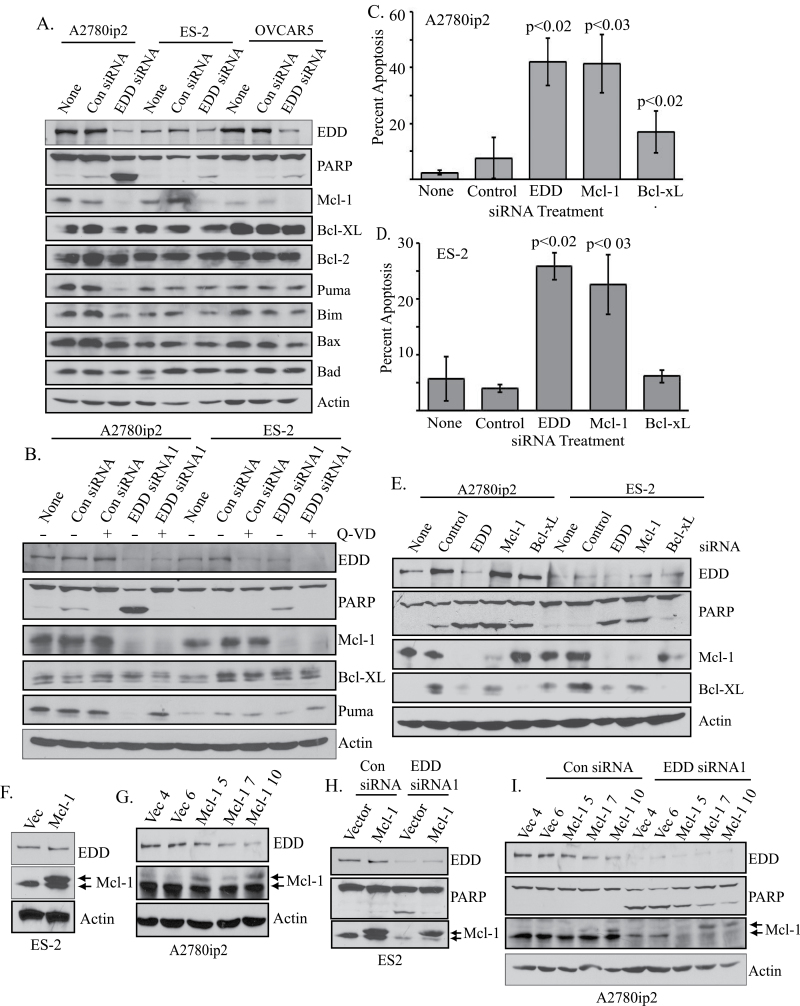
To identify a potential mechanism of apoptosis induction, we immunoblotted siRNA-transfected cell lysates with antibodies to Bcl2 family members, which have both prosurvival and proapoptotic functions (19,20). EDD knockdown resulted in specific downregulation of the prosurvival protein Mcl-1 in all three cell lines, correlating with increased PARP cleavage (Figure 2A), and Mcl-1 loss was detected using either EDD siRNA1 or siRNA2 (Supplementary Figure 2A, available at Carcinogenesis Online). Pretreatment of the cells with the pan caspase inhibitor Q-VD inhibited PARP cleavage and the loss of the proapoptotic caspase 3 substrate p53 upregulated modulator of apoptosis (Puma) upon EDD knockdown, but did not inhibit loss of Mcl-1, suggesting that Mcl-1 loss was not a consequence of caspase action or apoptosis induction (Figure 2B) (21). To compare the requirements for EDD and Mcl-1 in cell survival, we transfected cells with siRNA against EDD, Mcl-1, the prosurvival protein Bcl-xL or control siRNA. Apoptotic cells were identified by propidium iodide staining. EDD or Mcl-1 siRNA induced equal and significant induction of apoptosis in A2780ip2 (control = 7.6%; EDD siRNA1 = 42%; Mcl-1 = 41.4%; Bcl-xL = 16.9%) and ES-2 cells (control = 4%; EDD siRNA1 = 25.8%; Mcl-1 = 22.6%; Bcl-xL = 6.1%), whereas Bcl-xL knockdown induced less apoptosis that was only significantly different from control in A2780ip2 cells and much less than that induced by EDD or Mcl-1 siRNA (Figure 2C and D). Immunoblotting demonstrated knockdown of the targeted proteins and levels of PARP cleavage that corresponded to the relative level of apoptosis observed by propidium iodide staining (Figure 2E). These data show that these ovarian cancer cell lines have the same survival requirement for EDD and Mcl-1.
Fig. 2.
EDD downregulation decreases Mcl-1 protein levels, whereas Mcl-1 overexpression inhibits apoptosis upon EDD knockdown. (A) Cells were either untreated (none) or transfected with control or EDD siRNA1 for 24h. Lysates from floating and adherent cells were immunoblotted for EDD, PARP and Bcl2 family members as indicated. (B) A2780ip2 and ES-2 cells were untreated (none) or transfected with control siRNA or EDD siRNA1 and simultaneously treated with either Q-VD-OPH pan caspase inhibitor (+) or the negative control Z-FA-FMK (−). After 24h, floating and adherent cells were collected, lysed, run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotted for EDD, PARP, Mcl-1, Bcl-xL, p53 upregulated modulator of apoptosis and actin. (C) A2780ip2 and (D) ES-2 cells were either untreated (none) or were transfected with the control siRNA, EDD siRNA1 or siRNA to Mcl-1 or Bcl-xL for 24h. Floating and adherent cells were fixed, stained with propidium iodide and the percentage of sub-2n cells determined by flow cytometry. P values represent significance compared with the control siRNA-transfected cells. (E) Cells were transfected with siRNA as in (D) for 24h. Lysates from floating and adherent cells were immunoblotted to confirm knockdown and to determine PARP cleavage. (F) Stable populations of ES-2 cells and (G) stable clones of A2780ip2 cells expressing either pBabe vector (Vec) or pBabe-Flag-Mcl-1 (Mcl-1) were generated by retroviral transduction. Cell lysates were immunoblotted as indicated. Arrows indicate endogenous Mcl-1 and the slower-migrating Flag-Mcl-1. (H) Stable ES-2 or (I) A2780ip2 cells were transfected with control or EDD siRNA1 for 24h and cell lysates immunoblotted as indicated. Arrows indicate endogenous Mcl-1 and Flag-Mcl-1.
To determine if EDD regulated survival by promoting Mcl-1 levels, we generated stable cell lines expressing either Flag-Mcl-1 or empty vector. Stable ES-2 populations (Figure 2F) and A2780ip2 clones with varying levels of Flag-Mcl-1 (Figure 2G) were selected with puromycin. Flag-Mcl-1 migrated slower than endogenous Mcl-1 on sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Flag-Mcl-1 overexpression inhibited PARP cleavage upon EDD knockdown in both ES-2 (Figure 2H) and A2780ip2 (Figure 2I) stable lines compared with the vector control lines, with a dose-dependent effect of exogenous Mcl-1 expression on inhibition of PARP cleavage in the A2780ip2 clones. Interestingly, EDD knockdown induced loss of endogenous Mcl-1, but not expression of the exogenous Flag-Mcl-1 expressed from a cytomegalovirus promoter. Collectively, these results show that Mcl-1 overexpression protects cells from apoptosis upon EDD knockdown.
Mcl-1 protein stability is controlled in part through phosphorylation by GSK-3β, stimulating Mcl-1 ubiquitination by β-transducin repeat-containing protein, followed by proteosomal degradation (22). EDD binds to GSK-3β and stimulates its nuclear accumulation (23). To determine if EDD binding to GSK-3β ‘protects’ Mcl-1 from GSK-3β-induced degradation, which would be lost upon EDD knockdown, we transfected parental A2780ip2 cells with EDD siRNA1 and treated the cells with the GSK-3β inhibitors TZDZ, lithium chloride or L803-mts (24–28). GSK-3β inhibitors did not inhibit Mcl-1 downregulation or PARP cleavage upon EDD knockdown (Supplementary Figure 2B, available at Carcinogenesis Online). Furthermore, GSK-3β knockdown for 24 h prior to EDD knockdown with siRNA1 did not prevent the loss of Mcl-1 protein or inhibit PARP cleavage (Supplementary Figure 2C, available at Carcinogenesis Online), suggesting that Mcl-1 downregulation after EDD knockdown is GSK-3β independent.
EDD enhances Mcl-1 expression at the messenger RNA level
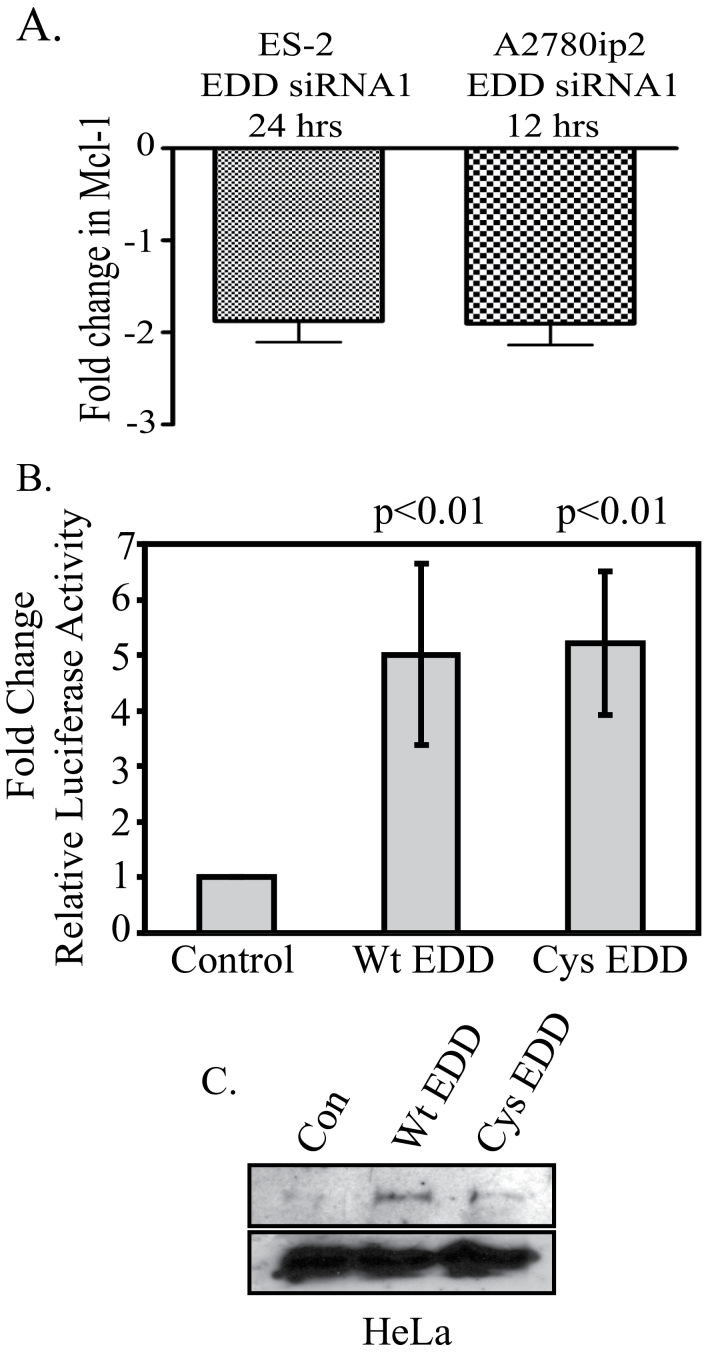
The above results suggest that EDD may regulate Mcl-1 synthesis, not its degradation. Indeed, quantitative real-time PCR analysis demonstrated that EDD knockdown inhibited Mcl-1 messenger RNA (mRNA) expression by 1.87-fold in both A2780ip2 and ES-2 cells at 12 and 24 h, respectively, compared with transfection with control siRNA, demonstrating that EDD downregulation inhibits Mcl-1 transcription (Figure 3A).
Fig. 3.
EDD regulates Mcl-1 levels through transcriptional regulation. (A) EDD knockdown inhibits Mcl-1 mRNA expression. ES-2 and A2780ip2 cells were transfected with EDD siRNA1 for 24 or 12h, respectively, and RNA was harvested. Quantitative real-time PCR was performed using Mcl-1-specific primers. The y-axis represents the fold change in Mcl-1 mRNA in EDD siRNA1-transfected cells compared with that in control siRNA-transfected cells. The results are a combination of three independent experiments. (B) EDD activates the Mcl-1 promoter. HeLa cells were transfected with p(−2389/+10)mcl-luc, an Mcl-1 promoter-driven firefly luciferase plasmid (16), TK Renilla luciferase and either Flag-EDD, Flag-EDD-C2768A or empty vector. Cells were harvested at 48h and firefly luciferase activity was normalized to Renilla luciferase activity in each sample. P values indicate significance (P < 0.05) within a group between Flag-EDD- and vector-transfected cells. These results are a combination of four independent experiments. (C) Western blot of Flag-EDD from (B).
EDD has been shown to act as a transcriptional coactivator for the progesterone and vitamin D receptors, independent of the C-terminal ubiquitin ligase domain (2). Flag-EDD cotransfection in HeLa cells enhanced transcription from an Mcl-1 promoter-driven luciferase reporter p(−2389/+10)mcl-luc by 5-fold when normalized to cotransfected TK Renilla luciferase (Figure 3B) (16). Transfection of the ubiquitin ligase-deficient point mutant, Flag-EDD-C2768A, also induced luciferase expression 5-fold. Western blotting confirmed equal EDD expression (Figure 3C). These data suggest that EDD positively regulates Mcl-1 transcription, independent of its ubiquitin ligase activity.
EDD is sufficient to induce cisplatin resistance
O’Brien et al. (1) showed that EDD siRNA reduced colony formation after cisplatin treatment in the cisplatin-resistant A2780-cp70 cell line. However, although 72 h cisplatin treatment induced dose-dependent cell death in ES-2 and A2780ip2 cells transfected with control siRNA (Supplementary Figure 3A and B, available at Carcinogenesis Online), the catastrophic apoptosis induced by EDD siRNA obscured any cisplatin effect. At 24 h of cotreatment, EDD knockdown in ES-2 cells conferred cisplatin sensitivity (Supplementary Figure 3C, available at Carcinogenesis Online), whereas the strong apoptotic response of EDD knockdown alone in A2780ip2 cells masked any potential effects on cisplatin sensitization (Supplementary Figure 3D, available at Carcinogenesis Online). Although EDD knockdown induced apoptosis in A2780-cp20 cisplatin-resistant cells, it did not enhance cell death in response to cisplatin in these cells (Supplementary Figure 3E, available at Carcinogenesis Online).
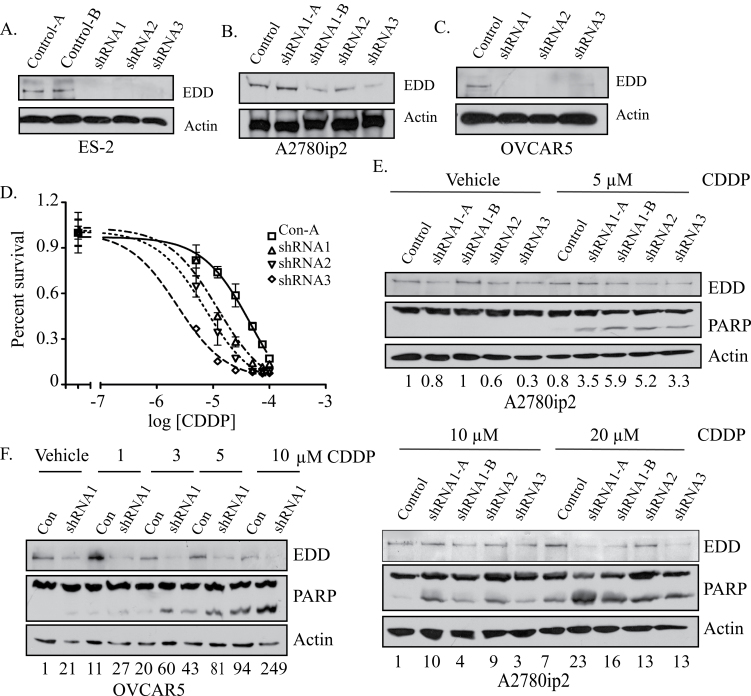
In order to separate the basic cell survival function of EDD from a potential role in cisplatin resistance, we generated ES-2 (Figure 4A), A2780ip2 (Figure 4B) and OVCAR5 (Figure 4C) cell lines with constitutive knockdown of EDD using retroviral transduction of three separate shRNAs. These cells represent the small portion of the population that can survive initial EDD knockdown, as the majority of the cells undergo apoptosis. Immunoblotting showed that these pools of cells survive because they are not dependent upon EDD for Mcl-1 expression (Supplementary Figure 4, available at Carcinogenesis Online). Cellular clones of ES-2 and A2780ip2 cells and a population of OVCAR5 cells were selected. MTS assays demonstrated that ES-2 clones expressing EDD shRNA were 4- to 21-fold more sensitive to cisplatin than cells expressing control shRNA, with EC50 values of 48.8 μM for the control-1 (clone 1) shRNA line, 12.0 μM for EDD shRNA1, 7.4 μM for EDD shRNA2 and 2.3 μM for the EDD shRNA3 cell lines (Figure 4D). In addition, A2780ip2 (Figure 4E) and OVCAR5 (Figure 4F) EDD shRNA cells were more sensitive to cisplatin after 24 h of treatment compared with the control shRNA cells, as measured by increased induction of PARP cleavage. These results demonstrate that stable loss of EDD sensitizes cells to cisplatin.
Fig. 4.
Stable EDD knockdown increases cisplatin sensitivity. (A) ES-2, (B) A2780ip2 and (C) OVCAR5 cells were retrovirally transduced with control or one of three EDD shRNAs and clones (ES-2 and A2780ip2) or populations (OVCAR5) were selected. Cell lysates were immunoblotted for EDD expression. Multiple clones from the same shRNA are designated as A or B. (D) ES-2 control shRNA or EDD shRNA cells were treated with cisplatin for 72h and cell viability measured by MTS assay. Percent survival was plotted against the log of the cisplatin concentration. The results are from three independent experiments performed in quadruplicate. (E) A2780ip2 and (F) OVCAR5 shRNA cells were treated with cisplatin for 24h and cell lysates from floating and adherent cells were immunoblotted for EDD and PARP cleavage. The numbers underneath the blot represent the relative intensity of cleaved PARP in each lane compared with the first lane of each blot.
To determine if EDD is sufficient to induce cisplatin resistance, COS-7 cells were transfected with Flag-EDD or GFP for 24 h and then treated with cisplatin for an additional 24 h. Cells were immunostained for Flag-EDD and costained with the TACS® 2 Tdt-Blue Label In Situ Apoptosis Detection Kit, staining apoptotic nuclei black under brightfield microscopy (Supplementary Figure 5A, available at Carcinogenesis Online). The percentage of transfected cells that were apoptotic after cisplatin treatment was determined by counting. Cells transfected with Flag-EDD had significantly less apoptosis at the higher cisplatin doses of 15 μM (GFP = 8.9%, EDD = 4.0%, P < 0.03) and 30 μM (GFP = 14.6%, EDD = 6.0%, P < 0.02) compared with the GFP-transfected cells, demonstrating that EDD overexpression was sufficient to induce cisplatin resistance (Figure 5A). To determine if EDD ubiquitin ligase activity was required, cells were transfected with GFP, Flag-EDD or Flag-EDD-C2768A, a ubiquitin ligase-deficient mutant, and treated with 15 μM cisplatin for 24 h. EDD-C2768A did not induce cisplatin resistance compared with the GFP control, whereas EDD caused 2.4-fold protection (GFP = 9.4%, EDD = 3.8%, EDD-C2768A = 11.8%) (Figure 5B). Statistical significance was seen between EDD and GFP and EDD and EDD-C2768A. These results show that EDD-induced cisplatin resistance is dependent upon its E3 ubiquitin ligase activity. EDD localizes to the nucleus, where cisplatin induces DNA damage, and mutation of EDD at Cys2768 did not affect nuclear localization (Supplementary Figure 5B, available at Carcinogenesis Online) (2,10).
Fig. 5.
EDD overexpression is sufficient to induce cisplatin resistance, dependent upon its ubiquitin ligase activity. (A) COS-7 cells on coverslips were transfected with Flag-EDD or GFP for 24h and then treated with cisplatin for an additional 24h. Fixed cells were stained for transfected and apoptotic cells and 4′,6-diamidino-2-phenylindole stained as shown in Supplementary Figure 5, available at Carcinogenesis Online. The percentage of apoptotic transfected cells was determined by cell counting. At least 500 cells were counted per condition in each of four experiments. (B) Same as in (A), but cells were transfected with GFP, Flag-EDD or Flag-EDD-C2768A, a ubiquitin ligase-deficient mutant. Cells were treated with 15 μM cisplatin for 24h on the day following transfection and the percentage of apoptotic transfected cells was determined by cell counting 24h later.
EDD knockdown in vivo enhances cisplatin efficacy
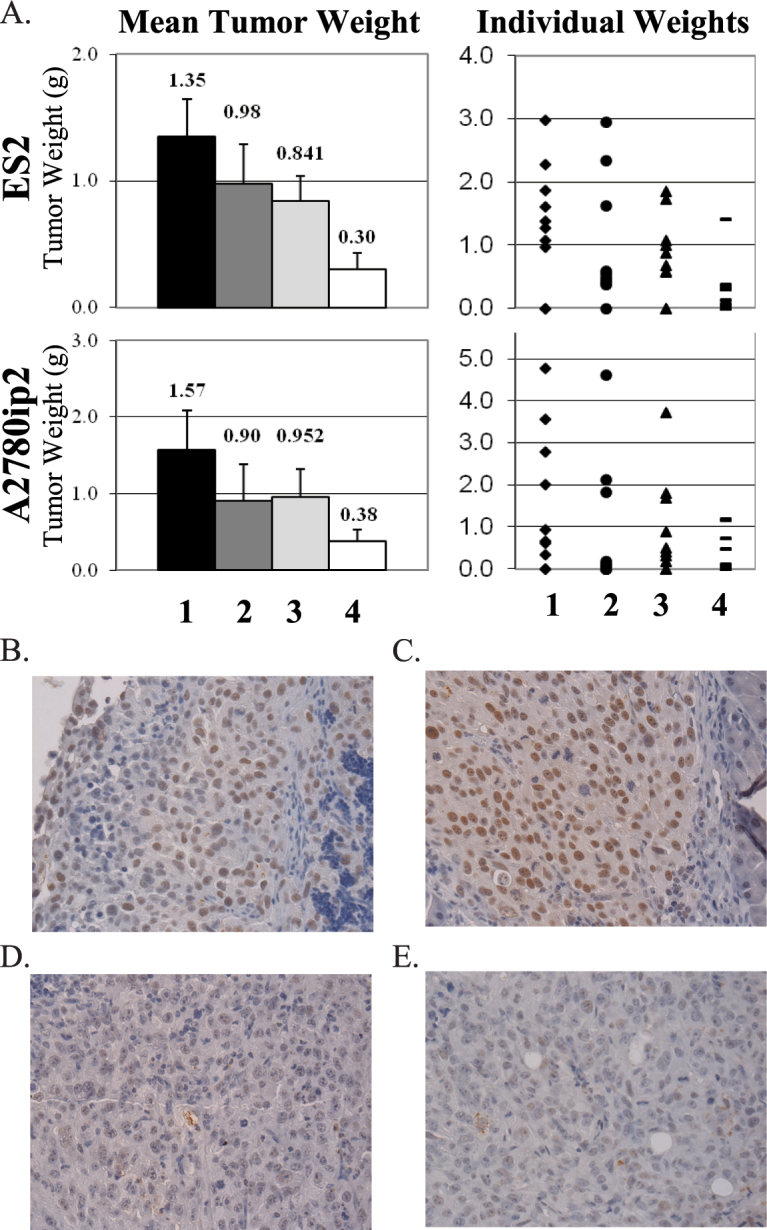
Our previous work has demonstrated in vivo delivery of siRNA to ovarian tumors via DOPC liposomal nanoparticles, resulting in knockdown of the target protein and a reduction in tumor burden (18,29–35). To determine if EDD is a viable target for the treatment of ovarian cancer, we generated intraperitoneal xenografts of ES-2 and A2780ip2 cells in female athymic nude mice. One week later, 10 mice per group were treated intraperitoneally twice per week with either control or EDD siRNA1 in DOPC liposomes, in combination with either cisplatin or saline treatment once weekly. After 4 weeks, mice were killed and tumor tissue was harvested. When compared to control siRNA treatment alone, cisplatin combined with control siRNA/DOPC showed a trend toward significance in ES-2 xenografts when measuring tumor weight (37.7% reduction, P = 0.167) but became statistically significant when cisplatin was combined with EDD siRNA1/DOPC (77.9% reduction, P = 0.004) (Figure 6A). In A2780ip2 xenografts, cisplatin plus control siRNA/DOPC treatment was not significantly different compared with control siRNA/DOPC alone (39.2% reduction, P = 0.349), but cisplatin plus EDD siRNA1/DOPC was significantly better than control siRNA/DOPC alone (75.9% reduction, P = 0.042). In those mice treated with cisplatin, cotreatment with EDD siRNA1/DOPC was significantly better than cotreatment with control siRNA/DOPC in ES-2 (64% reduction, P = 0.035) and showed a trend toward significance in A2780ip2 (60.3% reduction, P = 0.168). Immunohistochemistry of A2780ip2 tumors with EDD antibody showed EDD expression in tumors treated with control siRNA, with a possible enhancement of EDD expression in tumors following cisplatin treatment (Figure 6B and C). EDD siRNA1 treatment in vivo decreased EDD expression in tumors (Figure 6D and E). Collectively, these results suggest that therapies targeting EDD expression might be an attractive treatment for ovarian cancer patients.
Fig. 6.
DOPC nanoparticle delivery of EDD siRNA in vivo reduces tumor burden. (A) ES-2 or A2780ip2 cells were injected intraperitoneally into 40 female athymic nude mice per cell line. Mice were either treated with control siRNA in DOPC (lane 1), EDD siRNA1 in DOPC (lane 2), control siRNA in DOPC plus cisplatin (lane 3) or EDD siRNA1 in DOPC plus cisplatin (lane 4). Mice were treated for 4 weeks before killing and tumor collection. Tumors were excised and total tumor weight determined. The number above each lane represents the mean tumor weight in grams. Immunohistochemistry demonstrates EDD knockdown in vivo. A2780ip2 tumors from mice treated with (B) control siRNA in DOPC, (C) control siRNA in DOPC plus cisplatin, (D) EDD siRNA1 in DOPC and (E) EDD siRNA1 in DOPC plus cisplatin were immunostained with EDD antibody followed by horseradish peroxidase secondary antibody.
Discussion
This study illustrates the potential for the E3 ubiquitin ligase EDD as a therapeutic target for the treatment of epithelial ovarian cancer, particularly in those patients who develop platinum resistance. We demonstrate a dual role for EDD overexpression in ovarian cancer cells, regulating both cell survival and cisplatin resistance (Supplementary Figure 6, available at Carcinogenesis Online). We show that EDD enhances cell survival through the prosurvival protein Mcl-1, an important mediator of survival in ovarian cancer cells (36–38). EDD knockdown inhibited Mcl-1 mRNA and endogenous protein expression, whereas EDD overexpression increased Mcl-1 transcriptional expression in luciferase assays using the murine Mcl-1 promoter. Induction of the Mcl-1 promoter was independent of EDD’s ubiquitin ligase activity, as mutation of the critical cysteine residue in the E3 ligase domain still allowed for induction of the Mcl-1 promoter. This is in agreement with a previous study that showed that EDD acted as a transcriptional coactivator through the middle third of the protein, independent of the C-terminal ubiquitin ligase domain (2).
Several transcription factors have been demonstrated to regulate Mcl-1 expression, some of which have links to EDD. Platelet-derived growth factor stimulation of prostate cancer cells enhances Mcl-1 expression via a β-catenin and hypoxia-inducible factor 1 alpha subunit-dependent pathway and EDD ubiquitinates β-catenin to promote its stabilization, nuclear localization and activity (23,39). E2F transcription factor 1 represses Mcl-1 expression and knockdown of EDD induces E2F transcription factor 1 protein levels in HeLa cells (8,40,41). Transcription factor software analysis (TFSEARCH, www.cbrc.jp/research/db/ TFSEARCH.html) of the human Mcl-1 promoter (accession no. DQ088966) identified potential binding sites for other transcription factors, including GATAs 1–3, heat shock factors 1 and 2, nuclear factor kappa B and activator protein 1. The progesterone receptor cooperates with GATA-2 in transcriptional activation in breast cancer cells, suggesting that EDD–progesterone receptor interactions may regulate Mcl-1 expression through a GATA-2-dependent pathway (2,42). We have not ruled out translational control of Mcl-1 expression by EDD as an additional mechanism of regulation.
We further show that EDD directly regulates cisplatin sensitivity. A previous study has shown that EDD overexpression correlates with poor survival for patients with recurrent ovarian cancer and that knockdown of EDD with siRNA in cisplatin-resistant A2780-cp70 cells decreases colony formation by 40% after cisplatin treatment (1). However, a portion of this effect may be due to the cell survival functions of EDD described here. To separate these functions, we generated stable knockdown cells to select for those cells that could survive initial EDD knockdown. These cells showed normal levels of Mcl-1, demonstrating that this small portion of the initial cell population was not dependent upon EDD for Mcl-1 expression. By separating these functions, we demonstrated that loss of EDD sensitizes cells to cisplatin. Expression of EDD in ovarian cancer cell lines does not directly correlate with reported cisplatin sensitivity, as some ovarian cancer cell lines with high EDD expression have low cisplatin IC50s and some of those with higher resistance express lower levels of EDD (Figure 1 and Supplementary Table 1, available at Carcinogenesis Online) (43–46). This is likely due to the multiple mechanisms of cisplatin resistance in cells and tumors (47). Indeed, A2780-cp70 cells selected in vitro for cisplatin resistance after long-term exposure did not have higher levels of EDD expression than parental A2780 cells (1). Importantly, we show that overexpression of EDD was sufficient to induce resistance to cisplatin and was dependent upon EDD ubiquitin ligase activity. EDD has been suggested to play a role in the DNA damage response, particularly in response to double strand breaks. EDD and the E3 ubiquitin ligase TRIP12 regulate levels of RNF168, a regulator of histone ubiquitination after DNA damage, resulting in controlled spread of histone ubiquitination from the area of double strand breaks (9); however, no reports have linked RNF168 to cisplatin resistance. EDD is important in activation of the DNA damage response kinase Chk2, as EDD-depleted cells show reduced activation of Chk2 in response to double strand breaks (10). EDD knockdown increased sensitivity of HeLa cells to phleomycin, regulating both the S phase and the G2/M phase checkpoints in treated cells (8,10). In the presence of DNA damage, EDD knockdown cells underwent radio-resistant DNA synthesis and premature entry into mitosis, leading to mitotic catastrophe (8).
Both Bcl-xL and Mcl-1 have been implicated to protect ovarian cancer cells from chemotherapy-induced apoptosis, suggesting that EDD upregulation of Mcl-1 expression may also contribute to cisplatin resistance; however, the requirement for ubiquitin ligase activity for cisplatin resistance, but not for induction of the Mcl-1 promoter, strongly suggests that the regulation of Mcl-1 by EDD is distinct from the induction of cisplatin resistance (38). Interestingly, EDD itself appeared to be upregulated in xenografts from mice treated with cisplatin, which may be clinically important in regards to a study showing EDD overexpression in recurrent ovarian tumors from patients who had a favorable response to initial chemotherapy (1).
Small molecule inhibitors of ubiquitin ligases have had little success due to the lack of a defined catalytic domain and the utilization of protein–protein interactions in order to ubiquitinate targets. Members of our group have previously demonstrated that DOPC nanoparticles can be utilized to efficiently deliver siRNA to ovarian tumor tissue to inhibit tumor growth and metastasis and to enhance chemosensitivity (18,29–35). Our in vivo data demonstrated that EDD is a valid target for treating epithelial ovarian cancer in combination with chemotherapy. EDD siRNA showed enhanced efficacy over cisplatin treatment alone in ES-2 xenografts and a trend toward significance in A2780ip2 xenografts. This effect of EDD siRNA was likely due to both the positive effects of EDD on cell survival and the enhancement of cisplatin resistance. Upon knockdown in vivo, loss of EDD likely enhances both cell death and cisplatin sensitivity. Our findings that EDD regulates survival Mcl-1 regulation independent of its ubiquitin ligase activity and cisplatin resistance through its ubiquitin ligase domain suggest that therapies targeting EDD expression, such as EDD siRNA in nanoparticles, may prove to be a more beneficial therapeutic approach than a chemical inhibitor of EDD ubiquitin ligase activity, although the latter alone may have some beneficial role in enhancing cisplatin sensitivity.
Supplementary material
Supplementary Figures 1– 6 and Table 1 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (CA131200 to S.T.E.); Medical University of South Carolina (to S.T.E.); National Institutes of Health (P30 CA138313 to MUSC, U54 CA151668 and P50 CA083639 to MD Anderson); Ovarian Cancer Research Fund, the RGK Foundation (to A.K.S. and G.L.B.); J.B.B. was supported by GM086510; South Carolina EPSCoR/IDeA Postdoctoral Academic Career Development Program (NIH-NIGMS SC-INBRE P20GM103499 to M.B.O.).
Supplementary Material
Acknowledgements
We thank R.Li (Emory University), K.Atkins (University of Virginia), A.Godwin (University of Kansas), T.Hamilton (Fox Chase Cancer Center) and N.Auersperg (University of British Columbia) for cell lines and H.F.Yang-Yen (Academia Sinica, Taiwan) for luciferase plasmids.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- Bcl
B cell leukemia
- Chk2
checkpoint kinase 2
- DOPC
1,2-dioleoyl-sn-glycero-3-phophatidylcholine
- EDD
E3 ubiquitin ligase identified by differential display
- GATA-2
GATA-binding protein 2
- GFP
green florescent protein
- GSK-3β
glycogen synthase kinase 3 beta
- Mcl-1
myeloid cell leukemia sequence 1
- mRNA
messenger RNA
- MTS
(3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)
- PARP
poly(ADP-ribose) polymerase
- siRNA
small interfering RNA
- TRIP12
thyroid hormone receptor interactor 12
- UBR5
ubiquitin protein ligase E3 component n-recognin 5.
References
- 1. O’Brien P.M., et al. (2008). The E3 ubiquitin ligase EDD is an adverse prognostic factor for serous epithelial ovarian cancer and modulates cisplatin resistance in vitro . Br. J. Cancer, 98, 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henderson M.J., et al. (2002). EDD, the human hyperplastic discs protein, has a role in progesterone receptor coactivation and potential involvement in DNA damage response. J. Biol. Chem., 277, 26468–26478 [DOI] [PubMed] [Google Scholar]
- 3. Bethard J.R., et al. (2011). Identification of phosphorylation sites on the E3 ubiquitin ligase UBR5/EDD. J. Proteomics, 75, 603–609 [DOI] [PubMed] [Google Scholar]
- 4. Eblen S.T., et al. (2003). Identification of novel ERK2 substrates through use of an engineered kinase and ATP analogs. J. Biol. Chem., 278, 14926–14935 [DOI] [PubMed] [Google Scholar]
- 5. Rechsteiner M., et al. (2005). Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol., 15, 27–33 [DOI] [PubMed] [Google Scholar]
- 6. Wolf D.H., et al. (2004). The proteasome: a proteolytic nanomachine of cell regulation and waste disposal. Biochim. Biophys. Acta, 1695, 19–31 [DOI] [PubMed] [Google Scholar]
- 7. Callaghan M.J., et al. (1998). Identification of a human HECT family protein with homology to the Drosophila tumor suppressor gene hyperplastic discs. Oncogene, 17, 3479–3491 [DOI] [PubMed] [Google Scholar]
- 8. Munoz M.A., et al. (2007). The E3 ubiquitin ligase EDD regulates S-phase and G(2)/M DNA damage checkpoints. Cell Cycle, 6, 3070–3077 [DOI] [PubMed] [Google Scholar]
- 9. Gudjonsson T., et al. (2012). TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell, 150, 697–709 [DOI] [PubMed] [Google Scholar]
- 10. Henderson M.J., et al. (2006). EDD mediates DNA damage-induced activation of CHK2. J. Biol. Chem., 281, 39990–40000 [DOI] [PubMed] [Google Scholar]
- 11. Clancy J.L., et al. (2003). EDD, the human orthologue of the hyperplastic discs tumour suppressor gene, is amplified and overexpressed in cancer. Oncogene, 22, 5070–5081 [DOI] [PubMed] [Google Scholar]
- 12. Fuja T.J., et al. (2004). Somatic mutations and altered expression of the candidate tumor suppressors CSNK1 epsilon, DLG1, and EDD/hHYD in mammary ductal carcinoma. Cancer Res., 64, 942–951 [DOI] [PubMed] [Google Scholar]
- 13. Mori Y., et al. (2002). Instabilotyping reveals unique mutational spectra in microsatellite-unstable gastric cancers. Cancer Res., 62, 3641–3645 [PubMed] [Google Scholar]
- 14. Wasenius V.M., et al. (1997). Comparative genomic hybridization analysis of chromosomal changes occurring during development of acquired resistance to cisplatin in human ovarian carcinoma cells. Genes Chromosomes Cancer, 18, 286–291 [PubMed] [Google Scholar]
- 15. Eblen S.T., et al. (2001). Biochemical and biological functions of the N-terminal, noncatalytic domain of extracellular signal-regulated kinase 2. Mol. Cell. Biol., 21, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chao J.R., et al. (1998). mcl-1 is an immediate-early gene activated by the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathway and is one component of the GM-CSF viability response. Mol. Cell. Biol., 18, 4883–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landen C.N., Jr, et al. (2005). Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res., 65, 6910–6918 [DOI] [PubMed] [Google Scholar]
- 18. Landen C.N., et al. (2006). Intraperitoneal delivery of liposomal siRNA for therapy of advanced ovarian cancer. Cancer Biol. Ther., 5, 1708–1713 [DOI] [PubMed] [Google Scholar]
- 19. Ola M.S., et al. (2011). Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol. Cell. Biochem., 351, 41–58 [DOI] [PubMed] [Google Scholar]
- 20. Burlacu A. (2003). Regulation of apoptosis by Bcl-2 family proteins. J. Cell. Mol. Med., 7, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hadji A., et al. (2010). Caspase-3 triggers a TPCK-sensitive protease pathway leading to degradation of the BH3-only protein puma. Apoptosis, 15, 1529–1539 [DOI] [PubMed] [Google Scholar]
- 22. Ding Q., et al. (2007). Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol. Cell. Biol., 27, 4006–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hay-Koren A., et al. (2011). The EDD E3 ubiquitin ligase ubiquitinates and up-regulates beta-catenin. Mol. Biol. Cell, 22, 399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martinez A., et al. (2002). First non-ATP competitive glycogen synthase kinase 3 beta (GSK-3beta) inhibitors: thiadiazolidinones (TDZD) as potential drugs for the treatment of Alzheimer’s disease. J. Med. Chem., 45, 1292–1299 [DOI] [PubMed] [Google Scholar]
- 25. Klein P.S., et al. (1996). A molecular mechanism for the effect of lithium on development. Proc. Natl Acad. Sci. U. S. A., 93, 8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phiel C.J., et al. (2001). Molecular targets of lithium action. Annu. Rev. Pharmacol. Toxicol., 41, 789–813 [DOI] [PubMed] [Google Scholar]
- 27. Kaidanovich-Beilin O., et al. (2006). Long-term treatment with novel glycogen synthase kinase-3 inhibitor improves glucose homeostasis in ob/ob mice: molecular characterization in liver and muscle. J. Pharmacol. Exp. Ther., 316, 17–24 [DOI] [PubMed] [Google Scholar]
- 28. Rao R., et al. (2007). Glycogen synthase kinase 3 inhibition improves insulin-stimulated glucose metabolism but not hypertension in high-fat-fed C57BL/6J mice. Diabetologia, 50, 452–460 [DOI] [PubMed] [Google Scholar]
- 29. Landen C.N., et al. (2005). EphA2 as a target for ovarian cancer therapy. Expert Opin. Ther. Targets, 9, 1179–1187 [DOI] [PubMed] [Google Scholar]
- 30. Halder J., et al. (2006). Focal adhesion kinase targeting using in vivo short interfering RNA delivery in neutral liposomes for ovarian carcinoma therapy. Clin. Cancer Res., 12, 4916–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merritt W.M., et al. (2008). Effect of interleukin-8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J. Natl Cancer Inst., 100, 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin Y.G., et al. (2008). Targeting aurora kinase with MK-0457 inhibits ovarian cancer growth. Clin. Cancer Res., 14, 5437–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mangala L.S., et al. (2009). Liposomal siRNA for ovarian cancer. Methods Mol. Biol., 555, 29–42 [DOI] [PubMed] [Google Scholar]
- 34. Nick A.M., et al. (2011). Silencing of p130cas in ovarian carcinoma: a novel mechanism for tumor cell death. J. Natl Cancer Inst., 103, 1596–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chakravarty D., et al. (2011). Therapeutic targeting of PELP1 prevents ovarian cancer growth and metastasis. Clin. Cancer Res., 17, 2250–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shigemasa K., et al. (2002). Increased MCL-1 expression is associated with poor prognosis in ovarian carcinomas. Jpn. J. Cancer Res., 93, 542–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brotin E., et al. (2010). Bcl-XL and MCL-1 constitute pertinent targets in ovarian carcinoma and their concomitant inhibition is sufficient to induce apoptosis. Int. J. Cancer, 126, 885–895 [DOI] [PubMed] [Google Scholar]
- 38. Simonin K., et al. (2009). Mcl-1 is an important determinant of the apoptotic response to the BH3-mimetic molecule HA14-1 in cisplatin-resistant ovarian carcinoma cells. Mol. Cancer Ther., 8, 3162–3170 [DOI] [PubMed] [Google Scholar]
- 39. Iqbal S., et al. (2012) PDGF upregulates Mcl-1 through activation of β-catenin and HIF-1α-dependent signaling in human prostate cancer cells. PLoS One, 7, e30764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Croxton R., et al. (2002). Direct repression of the Mcl-1 promoter by E2F1. Oncogene, 21, 1359–1369 [DOI] [PubMed] [Google Scholar]
- 41. Croxton R., et al. (2002). Differences in DNA binding properties between E2F1 and E2F4 specify repression of the Mcl-1 promoter. Oncogene, 21, 1563–1570 [DOI] [PubMed] [Google Scholar]
- 42. Magklara A., et al. (2009). A composite intronic element directs dynamic binding of the progesterone receptor and GATA-2. Mol. Endocrinol., 23, 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saran U., et al. (2012). Secreted frizzled-related protein 4 expression is positively associated with responsiveness to cisplatin of ovarian cancer cell lines in vitro and with lower tumour grade in mucinous ovarian cancers. BMC Cell Biol., 13, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsumura N., et al. (2011). Epigenetic suppression of the TGF-beta pathway revealed by transcriptome profiling in ovarian cancer. Genome Res., 21, 74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ye G., et al. (2011). MicroRNA 376c enhances ovarian cancer cell survival by targeting activin receptor-like kinase 7: implications for chemoresistance. J. Cell Sci., 124, 359–368 [DOI] [PubMed] [Google Scholar]
- 46. Smith J.A., et al. (2005). An evaluation of cytotoxicity of the taxane and platinum agents combination treatment in a panel of human ovarian carcinoma cell lines. Gynecol. Oncol., 98, 141–145 [DOI] [PubMed] [Google Scholar]
- 47. Galluzzi L., et al. (2012). Molecular mechanisms of cisplatin resistance. Oncogene, 31, 1869–1883 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.