Summary
Acriflavine, a known HIF inhibitor, limits murine colorectal cancer progression through HIF-specific effects on both colonic epithelial cells and recruited macrophages.
Abstract
Hypoxia-inducible factors (HIFs) accumulate in both neoplastic and inflammatory cells within the tumor microenvironment and impact the progression of a variety of diseases, including colorectal cancer. Pharmacological HIF inhibition represents a novel therapeutic strategy for cancer treatment. We show here that acriflavine (ACF), a naturally occurring compound known to repress HIF transcriptional activity, halts the progression of an autochthonous model of established colitis-associated colon cancer (CAC) in immunocompetent mice. ACF treatment resulted in decreased tumor number, size and advancement (based on histopathological scoring) of CAC. Moreover, ACF treatment corresponded with decreased macrophage infiltration and vascularity in colorectal tumors. Importantly, ACF treatment inhibited the hypoxic induction of M-CSFR, as well as the expression of the angiogenic factor (vascular endothelial growth factor), a canonical HIF target, with little to no impact on the Nuclear factor-kappa B pathway in bone marrow-derived macrophages. These effects probably explain the observed in vivo phenotypes. Finally, an allograft tumor model further confirmed that ACF treatment inhibits tumor growth through HIF-dependent mechanisms. These results suggest pharmacological HIF inhibition in multiple cell types, including epithelial and innate immune cells, significantly limits tumor growth and progression.
Introduction
Chronic inflammation increases an individual’s risk of cancer, as exemplified by the well-established relationship between ulcerative colitis and the development of colorectal cancer (1–7). Inflammatory lesions and solid tumors are similar in that both contain regions of varying oxygen (O2) levels and are composed of complex, highly heterogeneous cell populations (8,9). Hypoxic domains within tumors are characterized by the infiltration of certain bone marrow-derived cells that act to promote disease progression (10). In particular, tumor-associated macrophages (TAMs) have been implicated in promoting tumorigenesis, often as a result of chronic inflammation (7,11,12). Hypoxia-driven inflammatory intracellular and cytokine signaling and macrophage infiltration clearly enhance tumor progression (7,13). Because both tumor cells and infiltrating TAMs must adapt to the unique stress of survival and proliferation under low O2 concentrations, hypoxic responses in these cell types directly impact tumor growth, local invasion and metastasis (8,14,15). As such, targeting the hypoxic response in either or both population(s) could have a beneficial effect on cancer therapy (16,17).
The transcriptional response to O2 deprivation is mediated, in large part, by hypoxia-inducible factors (HIFs) (18,19). HIFs are composed of an O2-sensitive α subunit and a constitutively expressed β (HIF-1β/ARNT) subunit (19). The α subunit is regulated by the von Hippel–Lindau (VHL) E3 ligase complex and degraded by the 26S proteasome under elevated O2 tensions (20). Low O2 levels stabilize HIF-α subunits by inhibiting prolyl hydroxylases that modify HIF-α proteins and promote their degradation (21). Once stabilized, HIF-α subunits translocate to the nucleus, form heterodimers with ARNT and bind hypoxia-response elements (HREs) to promote gene expression devoted to adaptation to hypoxic stress (18,22–25). Three α subunits (HIF-1α, HIF-2α and HIF-3α) have been identified; however, HIF-1α and HIF-2α appear to account for the majority of HIF-mediated transcriptional responses (26,27). The HIF-1α subunit is expressed in virtually all cells, whereas HIF-2α has a more restricted expression profile, including components of the liver, kidney, lung, intestine and brain (28). HIF-1α and HIF-2α possess distinct and occasionally overlapping roles; however, both have been suggested to actively promote the progression of a variety of cancers, including clear cell renal carcinoma, neuroblastoma, hepatocellular carcinoma and colorectal cancer (26,29). HIFs play an important role in neoplastic and inflammatory cells within the tumor microenvironment, and cross talk between these populations has clear effects on tumor growth (10,22,30–33). Both HIF-α isoforms are expressed in TAMs but have different downstream functions. For example, in the setting of nitric oxide (NO) metabolism, HIF-1α and HIF-2α elicit differential effects on arginase and inducible nitric oxide synthase (iNOS) activity, respectively (26,34). Both isoforms have been implicated in bone marrow-derived macrophages (BMDMs), mature macrophages and the protumorigenic and proangiogenic signaling observed in TAMs (14,35). Importantly, HIF-1α expression in macrophages has been implicated in modulating the switch from aerobic to anaerobic metabolism, as well as classical activation via Th1 cytokines, whereas HIF-2α expression in TAMs has been associated with an alternative activation via Th2 cytokines (14,22,34,36). It is becoming increasingly apparent that HIFs are a common link between hypoxia, chronic inflammation and tumorigenesis through their activity in macrophages during cancer development.
Pharmacological HIF inhibition as a novel therapeutic strategy is an active area of ongoing research (37–39). In particular, targeting HIF is well suited to colorectal cancer, as the HIF pathway has been repeatedly implicated in colorectal cancer pathogenesis (16). Acriflavine (ACF), a mixture of trypaflavin and proflavine, inhibits HIF-α:ARNT dimerization, has shown promise in xenograft models of human cancers and may be a viable source for future therapeutic interventions aimed at targeting HIF-1α and HIF-2α (40). Recently, ACF has also been shown to inhibit the recruitment of CD11b+ bone marrow-derived cells to the tumor microenvironment in an orthotopic model of breast cancer (41). Importantly, ACF does not appear to elicit any adverse side effects when administered to patients for extended periods of time (42). Whereas ACF has proven effective in subcutaneous and orthotopic models, it has yet to be evaluated in an autochthonous tumor model in immunocompetent mice, which more accurately mimics the cellular complexity observed in clinical disease. Here we demonstrate that ACF limits tumor progression in murine models of colitis-associated colon cancer (CAC) and use in vitro cellular assays to assess underlying mechanisms in both macrophages and malignant colonic epithelial cells.
Materials and methods
Autochthonous and subcutaneous colorectal cancer models
Eight-week-old female Balb/C mice were purchased from Jackson Laboratory. Briefly, mice received a single intraperitoneal (i.p.) injection of 12.5 mg/kg azoxymethane (AOM) at 8 weeks of age followed by four cycles of 2% dextran sulfate sodium (DSS) in their drinking water (cycle 1: 5 days, cycle 2: 4 days, cycle 3: 4 days, cycle 4: 4 days) with 2 weeks of regular water between each cycle for autochthonous induction of CAC. For subcutaneous experiments, 1 × 106 CT26 cells containing either shSCR or shARNT were injected subcutaneously into the left or right flank of 8-week-old female Balb/C mice, respectively. For all in vivo experiments, mice received ACF (Sigma M.W. 259.7) via daily i.p. injections at 2mg/kg dissolved in phosphate-buffered saline (PBS) or an equivalent volume of PBS alone for the control cohort. The laboratory animal program is accredited by the American Association for Accreditation of Laboratory Animal Care. Animal health, well-being and comfort were monitored constantly by certified veterinary staff. Every effort was made to minimize discomfort, stress, pain and injury to the mice and the mice was maintained in accordance with the Animal Welfare Act and the DHHS Guide for the care and use of laboratory animals. These procedures were performed according to the protocols reviewed by the Institutional Animal Care and Use Committee (IACUC).
Cell lines and cell culture
CMT93 (ATCC CCL-223) and CT26 (ATCC CRL-2638) cell lines were purchased from ATCC and cultured according to instructions. Cells were cultured under normoxia (21% O2) or hypoxia (0.5% O2) using a Ruskinn InvivO2 400 workstation. ACF (Sigma M.W. 259.7) was administered at 5μM in dimethyl sulfoxide (DMSO).
Bone marrow-derived macrophages
Generation of VavCre and Arnt fl/fl mice has been previously discussed (43,44). VavCre;Arnt mice were created by crossing VavCre mice (obtained as a gift from Speck lab) with Arnt fl/fl mice on a mostly C57BL/6 background. Macrophages were isolated from C57BL/6, VavCre;Arnt fl/+ or VavCre;Arnt fl/fl (littermates) mice by removing the long bones and flushing the marrow followed by red blood cell lysis. BMDMs were cultured in Dulbecco’s Modified Eagle’s Medium containing 20% Hyclone serum, 30% LCM, 1% l-glutamine, 1% Anti-Anti and 0.1% beta-mercaptoethanol and stimulated with 5ng/ml lipopolysaccharides (Sigma L3024) and 20ng/ml interferon-γ (R&D 485-MI). For hypoxia induction, BMDMs were cultured under normoxia (21% O2) or hypoxia (0.5% O2 or 3% O2). ACF (Sigma M.W. 259.7) was administered at 1μM in DMSO.
Luciferase assay
CMT93 and CT26 cells were transfected according to Fugene protocols (Roche) with PGL3 plasmids containing firefly luciferase under control of a wild-type (WT) HRE promoter from the human PGK gene or a mutant HRE promoter along with renilla control, or an pGL4.32[luc2p/NF-κB-RE/Hygro] vector (Promega E849A) also with renilla control. Twenty-four hours after transfection, media were changed to either DMSO or ACF and then cells were placed under normoxia or hypoxia. Luciferase activity was read on a luminometer 16 h after addition of DMSO or ACF (Promega E1960). Firefly activity from the WT HRE plasmid was normalized to renilla and mutant HRE activity.
Reverse transcription–quantitative polymerase chain reaction
RNA was isolated from tumor tissue or cells using the RNAeasy minikit (Qiagen #74106). RNA concentration was quantified using the Nanodrop with equal amounts of mRNA used for reverse transcription to cDNA using the High-Capacity RNA-to-cDNA kit (ABI #4387406). Expression was determined by quantitative PCR of synthesized cDNA using the Applied Biosystems 7900HT system and ΔΔCT program settings. Target cDNA amplification was measured using the following TaqMan primers: vascular endothelial growth factor (VEGF) (Mm00437304_m1), interleukin (IL)-1β (Mm00434228_m1), IL-6 (Mm00446190_m1), CXCR4 (Mm01292123_m1), COX-2 (Mm00478377_g1), SDF-1 (Mm0044552_m1), ARNT (Mm00507836_m1), PGK (Mm00435617_m1), HPRT (Mm01318743_m1), HIF-1α (Mm01283758_g1), HIF-2α (EPAS Mm00438717_m1), iNOS (Mm00440502_m1), ANG4 (Mm03647554_g1) and RETNLB (Mm00445845_m1). Results were analyzed with HPRT as an endogenous control.
Production of shRNA containing lentiviruses and transduction
HEK-293T cells were used for lentiviral production using the following constructs: pLKO.1 scrambled shRNA (Addgene 1864), pLKO.1 ARNT shRNA (ThermoScientific TRCN0000079931), pLKO.1 HIF1α shRNA (ThermoScientific TRCN0000054448), and pLKO.1 HIF-2α shRNA (ThermoScientific TRCN0000082307), G protein of the vesicular stomatitis virus (VSV-G), pMDLG and pRSV-rev. 293T cells were transfected according to the Fugene (Roche) protocol. Twenty-four and 48 h after transfection, supernatant was collected and concentrated using Amicon centrifugal filter units (Millipore). As the pLKO.1 shRNA constructs contain a puromycin resistance gene, transduction was followed by puromycin selection. CT26 cells were transduced with lentiviral particles containing copGFP in the form of the pCDH-CMV-EF1-copGFP vector (System Biosciences).
Immunostaining and imaging
Immunohistochemistry was performed using enzymatic Avidin-Biotin Complex–diaminobenzidine staining (Vector Labs) with hematoxylin used for counterstaining of nuclei. Stained sections were visualized using an Olympus IX81 microscope. CD68 1:100 (Abcam ab955) used according to instructions (Vector PK-2200), CD31 1:50 (Abcam ab28364) and Ki67 (Novocastra NCL-Ki67-MM1) used according to instructions (Vector PK-2200), terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining done according to instructions (Millipore ApopTag S7111) and copGFP staining performed using anti-TurboGFP antibody (Evrogen AB514). Staining was quantified using ImageJ software.
Immunoblot assays
Whole cell extracts were isolated in sodium dodecyl sulfate/Tris pH 7.6 lysis buffer. Subcellular fractionation was performed as previously described (45). Protein was quantified using BCA and equal protein amounts were run on an 8% or 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel, transferred to nitrocellulose and probed with the following antibodies: HIF-1α 1:1000 (Cayman 10006421), ARNT 1:1000 (Cell Signaling #5537), GAPDH 1:1000 (Cell Signaling #2118), nuclear factor-kappa B (NF-κB; Cell Signaling #3034), p-NF-κB (Cell Signaling #3039), IκBα (Cell Signaling #4814) p-IκBα (Cell Signaling #2859), M-CSFR (Cell Signaling #3152), DNMT1 (Cell Signaling #5032), AKT (Cell Signaling #9272) and histone deacetylase 1 (HDAC1; (Cell Signaling #5356). Representative western blots from multiple independent experiments are presented.
Flow cytometry and sorting
Subcutaneous tumors were grossly dissected, minced, collagenase treated and run through a 70 μm cell strainer to generate a single cell suspension. Live cells were run on a FACSVantage SE and sorted based on GFP staining. GFP negative parent cells were run to set up GFP+ and GFP− gates. Acquired data were analyzed using FlowJo software.
Statistical analysis
Unless otherwise indicated, data are shown as mean ± SEM. GraphPad Prism software was used to conduct statistical analyses and graph data. Unless otherwise indicated, unpaired two-tailed Student’s t-test was performed to evaluate statistical differences between control and experimental groups. In situations where more than two groups were compared, a one-way Anova was used followed by posttest Tukey analysis. Significance is demonstrated by ‘#’ indicating P > 0.05, ‘*’ representing P < 0.05, ‘**’ representing 0.001 < P < 0.01 and ‘***’ representing P < 0.001.
Results
ACF limits tumor burden in an autochthonous murine model of CAC
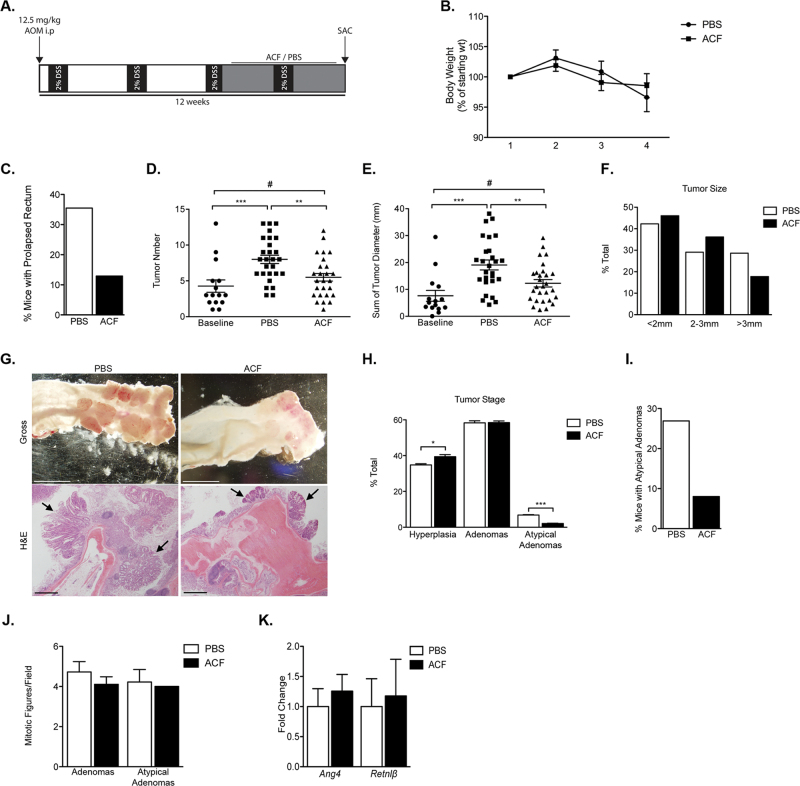
To analyze the effect of ACF treatment in the setting of CAC, 8-week-old female Balb/C mice were subjected to a single intraperitoneal (i.p.) injection of the procarcinogen AOM, followed by repeated treatments of 2% DSS to induce autochthonous CAC (46–48) (Figure 1A). Prior to initiating ACF treatment, we confirmed that mice exhibited gross intestinal polyp formation, with hyperplastic lesions making up approximately 60% of tumor burden and adenomas making up the remaining 40%, based on histologic examination of mice killed at baseline (Supplementary Figure 1A–C, available at Carcinogenesis Online). The AOM/DSS-treated mice were then separated into two cohorts: an experimental group that received daily i.p. injection of 2mg/kg ACF for 4 weeks and a control group that received daily injections of PBS. Over the course of treatment, no significant difference in mouse weight was observed between experimental and control groups (Figure 1B, Supplementary Figure 2A, available at Carcinogenesis Online); however, the control cohort exhibited increased incidence of prolapsed rectum, indicative of underlying pathology (Figure 1C).
Fig. 1.
ACF treatment limits disease progression in an autochthonous model of colitis-induced colon cancer. (A) Schematic diagram of AOM/DSS treatment followed by 28 days of i.p. ACF or PBS administration. (B) Weight change in mice over course of treatment, shown as percent change from weight at the beginning of daily injections—measured weekly. (C) Percentage of each cohort that exhibited prolapsed rectum based on visual observation. (D and E) Tumor number and burden upon gross examination either at the end of the third DSS administration (baseline) or PBS versus ACF treatment. (F) Tumor size as percent of total. (G) Gross and H&E pictures of representative samples from PBS and ACF treatment cohorts—images acquired on dissecting microscope. Arrows indicate individual hyperplastic or adenomatous lesions. (H) Tumor staging based on H&E slides from each mouse for either PBS- or ACF-treated cohorts. (I) Percent of each treatment cohort with at least one atypical adenoma, based on tumor staging. (J) Number of mitotic figures between either cohort. (K) RNA analysis of PBS- and ACF-treated tumors evaluating the expression of Ang4 and Retnlβ. (mean ± SEM, PBS: n = 27, ACF: n = 28, # P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001).
After 1 month, mice were euthanized, colons were dissected, and tumor burden was analyzed. The control cohort exhibited significant disease progression compared with the baseline group, whereas the ACF-treated cohort did not (Figure 1D–F). Mice in the control group, on average, developed increased numbers of colorectal tumors (Figure 1D). Similarly, there was an overall greater tumor burden, with a greater proportion of large tumors, in the control cohort compared with ACF-treated or baseline groups (Figure 1E–G). The control cohort also displayed a larger fraction of high-grade lesions, based on nuclear pleiomorphism and atypia, than the corresponding ACF-treated group (Figure 1H). In particular, the most highly pleiomorphic lesions in this model (atypical adenomas) were observed at nearly 3-fold higher frequency in the control group than in the ACF-treated cohort (Figure 1I). Although increased nuclear atypia were detected, no significant decreases in number of mitotic figures were noted in the control group compared with the ACF-treated group (Figure 1J). Lastly, we observed a trend toward decreased expression of multiple HIF-associated inflammatory molecules (Supplementary Figure 1D, available at Carcinogenesis Online) in RNA isolated from unstaged and unmatched individual polyps from ACF- and PBS-treated mice, respectively. Resistin like beta (Retnlβ) and Angiogenin 4 (Ang4) expression corresponds with colonic inflammation and bacterial influx (49,50). Importantly, expression of both Ang4 and Retnlβ was unchanged, suggesting ACF does not alter bacterial influx into the colonic epithelial cells (Figure 1K). As such, any observations in ACF-treated mice are unlikely to be due to the antimicrobial effects of ACF. Although previous studies have shown that intestinal microflora may contribute to the pathogenesis of colitis, and antibiotics are effective at minimizing disease in acute models of DSS-induced colitis, antibiotic treatment appears to be ineffective in models of chronic DSS-induced colitis (49–51). We concluded that ACF limits tumor progression in an autochthonous model of CAC and may be doing so through HIF-dependent mechanisms.
Effects on tumor vascularity and proliferation corresponding to ACF treatment
Previous work has demonstrated an effect on tumor growth along with decreased vascularity and infiltrating CD11b+ cells in mice treated with ACF (40,41). Similarly, although a trend toward decreased mitotic figures was detected upon H&E analysis, further investigation revealed that tumors from ACF-treated mice exhibited approximately 30% fewer proliferating (Ki67-positive) cells in stage-matched sections (Figure 2A). In contrast, no difference in apoptotic cell numbers was noted between the two cohorts, based on TUNEL staining (Figure 2A). Moreover, ACF-treated tumors exhibited significantly reduced vessel density and size, with vessel area nearly 3 times greater in PBS-treated mice, as assessed by CD31 staining (Figure 2B). In the setting of an inflammation-driven cancer, such as the AOM/DSS model of colitis-induced colorectal cancer, ACF appears to limit tumor burden through effects on tumor growth and progression, correlated with lower rates of tumor cell proliferation and decreased angiogenesis. ACF may also have an effect on the inflammatory component of this model.
Fig. 2.
Decreased tumor vascularity and cellular proliferation upon ACF treatment. (A) Representative examples of Ki67 and TUNEL staining. Ki67+ and TUNEL+ cells quantified from stage-matched samples. Images taken at ×200 magnification. (B) Representative images and quantification of CD31+ cells in both adenomas and atypical adenomas for stage-matched samples. Arrows indicate individual vessels. (mean ± SEM, PBS: n = 6, ACF: n = 6 for adenomas and PBS: n = 4, ACF: n = 2 for atypical adenomas, # P > 0.05, *P < 0.05, **P < 0.01).
ACF-treated tumors exhibit decreased macrophage infiltration
We hypothesized that the effects of ACF on the growth and progression of AOM/DSS-induced colorectal tumors were caused by altered HIF activity in TAMs, transformed colonic epithelial cells or both. To investigate ACF-mediated effects on macrophage recruitment, CD68+ cells were counted in stage-matched tumor sections from PBS- and ACF-treated mice, respectively. Adenomas and atypical adenomas in control animals exhibited significantly greater numbers of infiltrating macrophages than corresponding tumors from the ACF-treated cohort (Figure 3A, Supplementary Figure 3A, available at Carcinogenesis Online). These results confirm previous work in different non-inflammation-driven tumor models and are unlikely to be a consequence of general myelosuppressive effects of ACF treatment, as experimental animals displayed no discernible changes in the number of B220+, F4/80+ and Gr1+ cells in bone marrow or spleen, following 1 month of treatment (Supplementary Figure 2B–D, available at Carcinogenesis Online) (41). Because regulatory T cells possess anti-inflammatory functions and have been implicated in CAC, we stained for CD3 and Foxp3 in sections from ACF-treated and control mice (52). However, no differences were observed in Foxp3+ or CD3+ T lymphocyte numbers among control and ACF-treated cohorts, indicating that ACF treatment primarily affects innate immune cells associating with colonic tumors in this setting (Figure 3B). Collectively, these findings, along with previously published data, underscore the importance of infiltrating macrophages in inflammation-driven cancers and provide a possible mechanism to explain the less aggressive CAC observed in ACF-treated mice.
Fig. 3.
Tumors treated with ACF exhibit decreased macrophage infiltration. (A) CD68 staining in PBS- and ACF-treated tumors, with representative images and quantification of both adenomas and atypical adenomas, respectively. Arrows indicate individual CD68+ cells. (B) CD3 and Foxp3 staining with respective quantifications of PBS- and ACF-treated tumors from stage-matched tumors; images taken at ×400 and ×200 magnification. (mean ± SEM, PBS: n = 6, ACF: n = 6 for adenomas and PBS: n = 4, ACF: n = 2 for atypical adenomas, # P > 0.05, *P < 0.05).
ACF inhibits HIF signaling in macrophages
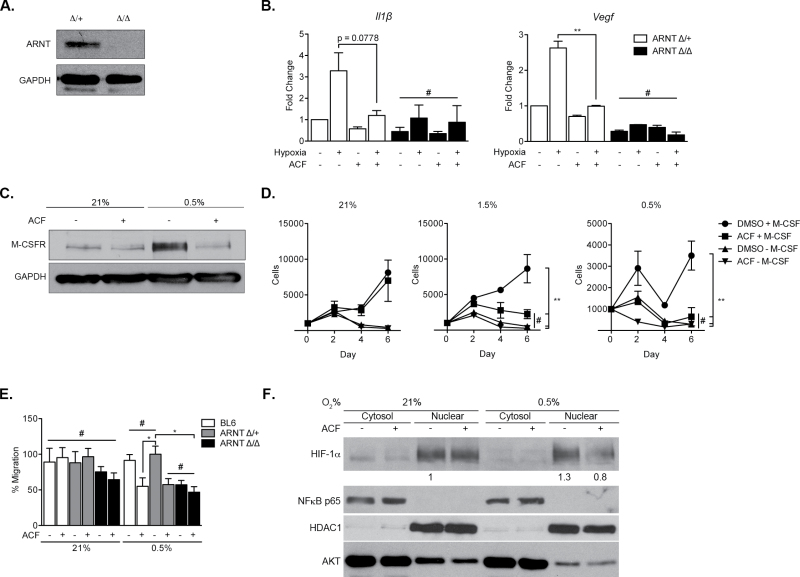
To evaluate the general impact of ACF treatment on macrophages, we investigated HIF-dependent responses in BMDMs obtained from WT C57BL/6, VavCre;Arnt fl/+ or VavCre;Arnt fl/fl mice (also on a C57BL/6 background). To confirm efficacy of Arnt deletion, BMDMs were purified and whole cell lysates analyzed for ARNT protein levels. As shown in Figure 4A, no detectable ARNT protein was observed in macrophages isolated from VavCre;Arnt fl/fl mice (Figure 4A) (Krock,B.L. et al., submitted for publication). Arnt ∆ /∆ macrophages are therefore deficient in the obligate HIF-α dimerization partner and, consequently, fail to engage HIF-1α and HIF-2α responses. We propose that this genetic model is similar to pharmacological HIF inhibition by ACF. To mimic the hypoxic microenvironment, macrophages were cultured in complete media at 3% O2 and growth compared with that at 21% O2. Macrophage proliferation was unaffected by ACF treatment under normoxia or hypoxia in complete media (21 or 3% O2, respectively, Supplementary Figure 3B, available at Carcinogenesis Online), suggesting that the observed decrease in CAC infiltrating macrophages (Figure 3A) is secondary to decreased recruitment rather than an effect on resident macrophage numbers. In contrast, ACF treatment significantly inhibited the expression of genes encoding IL-1β and VEGF, both HIF targets, in Arnt ∆ /+ macrophages (Figure 4B). Interestingly, Il-1β and Vegf transcript levels in ACF-treated Arnt ∆ /+ macrophages are similar to those observed in HIF-deficient Arnt ∆ /∆ macrophages. Furthermore, ACF treatment failed to substantially further reduce the expression of either gene in Arnt ∆ /∆ macrophages (Figure 4B), confirming that ACF is acting via a predominantly HIF-dependent pathway. We also observed that hypoxic stimulation of M-CSFR, a principal receptor for the macrophage growth factor and chemoattractant M-CSF, was ablated upon ACF treatment (Figure 4C). This is consistent with our previous observation that macrophage M-CSFR expression is regulated by HIF-2α and suggests a mechanism by which ACF treatment limits macrophage recruitment to, and infiltration of, inflammation-associated tumors (22). Interestingly, when macrophages are cultured without M-CSF under 21, 1.5 or 0.5% O2 and treated with DMSO or ACF, there appeared to be a specific proliferative effect on macrophages treated with ACF under hypoxic conditions. This may be secondary to decreased M-CSFR expression, as the proliferative defect is recapitulated with macrophages cultured in the absence of M-CSF (Figure 4D). We concluded that ACF treatment limits hypoxic induction of M-CSFR expression, minimizing macrophage stimulation by M-CSF under low O2. This effect was also detected during macrophage motility, as demonstrated by M-CSF-mediated migration of seeded macrophages in a modified Boyden chamber migration assay (Figure 4E). Although no difference between WT, Arnt ∆ /+ or Arnt ∆ /∆ cells was noted under normoxia, migration under 0.5% O2 was limited in ACF-treated WT and Arnt ∆ /+ cells and in both DMSO-treated and ACF-treated Arnt ∆ /∆ macrophages. These results indicated ACF treatment acts upon macrophages in a hypoxia and HIF-dependent manner, in large part through the expression of M-CSFR.
Fig. 4.
Effects of ACF on macrophages are largely HIF dependent. (A) Western blot validation of ARNT ablation in macrophages purified from VavCre;Arnt fl/fl mice. (B) Expression of proinflammatory (Il-1β) or proangiogenic (Vegf) genes upon ACF treatment or Arnt deletion. (C) M-CSFR protein levels based on immunoblot of primary bone marrow-derived macrophages from C57BL/6 mice cultured under 21% O2 or 0.5% O2 and in the presence of either DMSO or ACF. (D) Proliferation of either control or ARNT-deficient macrophages under 21 or 3% O2 over a 4-day period. (E) Normoxic and hypoxic migration of BL6, Arnt ∆ /+ or Arnt ∆ /∆ macrophages treated with DMSO or ACF. Shown as percent total. (F) Cytoplasmic/nuclear subcellular fractionation of macrophage lysates under normoxia (21% O2) and hypoxia (0.5% O2) with or without ACF treatment and immunoblotting for HIF-1α, NF-κB p65, HDAC1 and AKT. (mean ± SEM, PBS: n = 3, ACF: n = 3, # P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001).
NF-κB (Nuclear factor kappa-light-chain-enhancer of activated B cells) is a central regulator of the inflammatory response and has an established role in inflammation-associated cancers (53). Moreover, ACF has been suggested to have possible effects on this pathway (40,53). NF-κB transcription complexes are generally maintained in an inactive state in the cytoplasm and only translocate to the nucleus when dissociated from inhibitors such as IκBα (nuclear factor of kappa light polypeptide gene enhancer in B-cell inhibitor alpha) (54). As such, NF-κB nuclear localization is a strong indicator of transcriptional activity. We investigated whether ACF treatment alters NF-κB nuclear transit in BMDMs cultured under normoxia or hypoxia. Of note, ACF treatment had no appreciable effect on NF-κB subcellular localization or IκBα phosphorylation (Figure 4F and Supplemental Supplementary Figure 3C, available at Carcinogenesis Online), a necessary step to release NF-κB and allow nuclear entry. However, the same lysates clearly demonstrate decreased hypoxia-induced nuclear localization of HIF-1α (Figure 4F). AKT and HDAC1 immunoblotting indicate the purity of cytosolic and nuclear fractions, respectively. Similarly, ACF treatment had no detectable effect on the expression of transcripts encoding COX-2, an inflammatory protein regulated by multiple stimuli, including NF-κB (Supplementary Figure 3D, available at Carcinogenesis Online). Taken together, these findings indicate ACF inhibits macrophage recruitment and signaling through HIF-α specific mechanisms, with little to no impact on the NF-κB pathway.
HIF signaling is inhibited by ACF treatment in murine colorectal cells
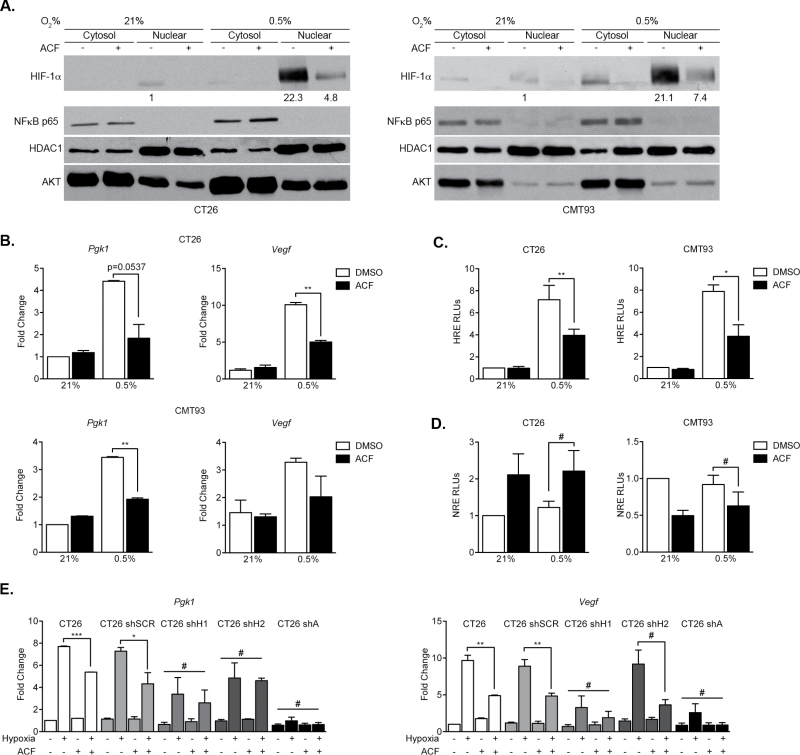
In addition to its effects on macrophages, ACF treatment likely inhibits CAC progression by inhibiting HIF responses in transformed colonic epithelial cells. To address this hypothesis, we analyzed ACF effects on murine CT26 cells (derived from Balb/C colon carcinoma) and CMT93 cells (derived from C57BL/6 polypoid carcinoma of the rectum). As expected, ACF administration did not impact HIF-1α stabilization under 0.5% O2 in either CT26 or CMT93 cells (Supplementary Figure 4A, available at Carcinogenesis Online), consistent with its proposed role in blocking HIF-α/ARNT dimerization rather than α subunit accumulation (40). Furthermore, ACF treatment had no effect on Hif1α or Arnt transcript levels (Supplementary Figure 4C, available at Carcinogenesis Online). Importantly, ACF treatment instead limits nuclear localization of HIF-1α in both CT26 and CMT93 cells under 0.5% O2 (Figure 5A) with AKT and HDAC1 immunoblotting demonstrating cytosolic and nuclear fractions, respectively. In contrast, ACF administration had no effect on the cellular localization of NF-κB (Figure 5A and Supplemental Supplementary Figure 4B, available at Carcinogenesis Online), or expression of Cox-2 (Supplementary Figure 4F, available at Carcinogenesis Online), indicating that ACF is unlikely to be affecting the NF-κB pathway. As noted for macrophages, Cox-2 transcription is likely regulated by multiple hypoxia-dependent, HIF-independent factors and does not appear to be impacted by ACF treatment. Instead, HIF targets Vegf and phosphoglycerate kinase 1 (Pgk1) were markedly reduced in both CT26 and CMT93 cells upon ACF exposure (Figure 5B). Additionally, ACF administration resulted in decreased HRE-driven luciferase reporter gene expression in both CT26 and CMT93 cells under hypoxia (Figure 5C), demonstrating decreased HIF transcriptional activity. Unlike that observed with HRE-driven luciferase assays, ACF had no effect (hypoxic or otherwise) on an NF-κB-response element luciferase reporter assay in either cell line (Figure 5D). Importantly, in the absence of HIF-1α or HIF-2α in CT26 cells, ACF treatment resulted in a modest decrease in Vegf or Pgk1 expression, whereas in the absence of ARNT, no further reduction in HIF target gene expression was observed (Figure 5E and Supplemental Supplementary Figure 4D and E, available at Carcinogenesis Online). These findings reinforce the notion that ACF is acting through the HIF transcriptional pathway.
Fig. 5.
Effects of ACF on colorectal cell lines are HIF dependent. (A) Cytoplasmic/nuclear fractionation of CT26 and CMT93 lysates under normoxia (21% O2) and hypoxia (0.5% O2) with or without ACF treatment and immunoblotting for HIF-1α, NF-κB p65, HDAC1 and AKT. (B) Expression of HIF targets Pgk1 and Vegf under hypoxia in both CT26 and CMT93 cell lines upon treatment with DMSO or ACF. (C) CT26 and CMT93 cells were transfected with a plasmid containing firefly luciferase under control of the PGK promoter (with three consecutive HRE sequences) and cultured under normoxia (21% O2) or hypoxia (0.5% O2) in the presence or absence of ACF. (D) CT26 and CMT93 cells were transfected with a plasmid containing firefly luciferase under control of an NRE and cultured under normoxia (21% O2) or hypoxia (0.5% O2) in the presence or absence of ACF. (E) Pgk1 and Vegf expression levels in control, shSCR, shHIF-1α, shHIF-2α or shARNT CT26 cells when cultured under normoxia or hypoxia in the presence or absence of ACF. (mean ± SEM, PBS: n = 3, ACF: n = 3, # P > 0.05, *P < 0.05, **P < 0.01).
ACF slows allograft tumor growth, dependent on HIF-α/ARNT activity
To further investigate the HIF specificity of ACF treatment, we employed a lentiviral shRNA construct to inhibit ARNT expression, and thus HIF-1α- and HIF-2α-mediated responses, in CT26 cells (Figure 6A). GFP-expressing CT26 cells transduced with either control (shSCR) or ARNT-specific (shARNT) lentiviruses were injected into the left and right flanks, respectively, of syngeneic Balb/C mice (Supplementary Figure 5A and B, available at Carcinogenesis Online). Mice then received daily i.p. injection of PBS or ACF for 3 weeks. Over the course of treatment, shSCR tumors in mice receiving PBS grew significantly larger than shSCR tumors in mice administered ACF (Figure 6B). Interestingly, there was minimal effect on growth rate in shARNT tumors as a result of ACF treatment, and shSCR tumors in mice treated with ACF grew at a similar rate as the shARNT tumors. The fact that shARNT tumors in mice receiving ACF were nearly identical in size to shARNT tumors in PBS-treated mice strongly suggests that ACF is primarily targeting the HIF pathway. Moreover, an appreciable decrease in tumor weight was observed in ACF-treated shSCR tumors; however, there was no difference in the weight of shARNT tumors (Figure 6C). These observations indicate that a majority of the antitumorigenic effects of ACF are directly related to HIF inhibition. RNA analysis of FACS-sorted tumor cells (based on GFP+ staining) confirmed that suppression of ARNT expression was maintained in shARNT cells throughout the experiment (Figure 6D). Similarly, the expression of canonical HIF targets Vegf and Pgk was reduced in shSCR tumors from ACF-treated mice compared with controls (although they do not achieve statistical significance), and no additional decrease was observed in shARNT tumors treated with ACF (Figure 6D). Immunohistochemical staining of subcutaneous tumors revealed little inflammatory infiltration as evidenced by a lack of CD68 staining (data not shown), demonstrating that the bulk of each tumor was composed of CT26 cells, as suggested by relatively prolific GFP staining (Supplementary Figure 4C, available at Carcinogenesis Online). Of note, reduced CD31+ positive cells were detected in sections from ACF-treated shSCR tumors, similar to values in PBS- and ACF-treated shARNT tumors (Figure 6E). The decrease in vascularity was not as pronounced as in the autochthonous CAC model (Figure 3B), which exhibited dramatically higher numbers of infiltrating macrophages. TAMs are known to influence tumor angiogenesis (15,55,56). As such, although ACF treatment has clear HIF-dependent effects in transformed colorectal cell lines, its antitumorigenic properties may be magnified by the changes in TAM activity in the setting of an inflammation-driven tumor model.
Fig. 6.
ACF slows allograft tumor growth, dependent on HIF-α/ARNT activity. (A) Immunoblot of ARNT protein in shSCR and shARNT cells, respectively. (B) Tumor volume changes in each cohort of mice over time—measured with calipers every 3 days. Two mice in the PBS cohort had to be euthanized on day 18 due to advanced disease. (C) Average tumor weight of each cohort at culmination of experiment. (D) Relative Arnt expression in FACS-sorted CT26 cells. (E) Vegf and Pgk1 RNA expression in isolated tumor cells from each treatment cohort. (F) CD31 staining of shSCR and shARNT tumors treated with PBS and ACF, respectively, with quantification on the right. Arrows indicate individual CD31+ vessels. (mean ± SEM, n = 5 for each group, # P > 0.05, *P < 0.05, **P < 0.01).
Discussion
HIFs are important transcription factors involved in cellular adaptation to low O2, a common feature of solid tumors, and thus represent attractive potential therapeutic targets (25,57). Additionally, many tumors exhibit extensive leukocytic infiltration—especially those occurring as a result of chronic inflammation (4). Therefore, therapies designed to target specific features of the tumor microenvironment may be impacted by naturally occurring O2 gradients, as well as hypoxic adaptations in both tumor parenchyma and stroma, including recruited inflammatory cells. The HIFs function in hypoxic responses of both tumor compartments, making HIF inhibition in tumor cells, TAMs or both likely to mitigate tumor progression. One known HIF inhibitor, digoxin, is currently in phase II clinical trials for breast cancer (http://clinicaltrials.gov/). Importantly, ACF is an example of a HIF inhibitor that has already been proven safe in patients for up to 5 months, with very few side effects (42). Although previous reports demonstrated HIF inhibition by ACF in xenograft and orthotopic models (40,41), both used severe combined immunodeficiency mice. This study represents the first time the effects of ACF-mediated HIF inhibition have been explored in fully immunocompetent mice, mimicking the complexity of HIF activity in tumor microenvironments within an autochthonous setting. Despite the utility of xenograft (or allograft) tumor models, they typically cannot recapitulate the cellular complexity and natural history of autochthonous tumors in immunocompetent hosts, and treatments that eradicate xenograft tumors have often proved ineffective in patients. Similarly, the use of tissue- or cell-type-specific genetic deletion can provide important insights into the role of specific genes in tumor initiation and progression but may be formally distinct from using pharmacological compounds to target a particular molecular target that is expressed in extant tumors.
The work described here investigates ACF treatment of autochthonous tumors and suggests that pharmacological HIF inhibition in multiple cell types, including epithelial and innate immune cells, reduces tumor growth and progression. Mice treated with ACF consistently developed fewer and smaller colonic lesions with a marked decrease in vascularity and number of recruited macrophages. We demonstrated that ACF acts largely on HIF-dependent responses in macrophages, without effect on the NF-κB pathway. Because HIFs have been shown to be important in multiple components of the tumor microenvironment, inhibiting HIF activity in any single cell population may effect tumor progression with increased efficacy observed when multiple compartments are targeted simultaneously (26). Previous studies have demonstrated the importance of HIFs in TAMs. We have now shown that ACF limits macrophage infiltration and signaling in the tumor microenvironment in a HIF-dependent manner. A likely mechanism for the reduced macrophage infiltration detected in ACF-treated mice is decreased hypoxic induction of M-CSFR expression—a finding similar to loss of HIF-2α in these cells (22). There may also be HIF-dependent effects on resident macrophages of the colon as a result of ACF treatment, contributing to reduced tumor progression. ACF exhibits clear effects on multiple colorectal cancer cell lines in vitro and in vivo in HIF-dependent mechanisms and is very likely acting on the tumor parenchyma. In future work, it will be important to employ Cre-mediated recombination to delete ARNT, and thereby both HIF-1α and HIF-2α activity, in both colonic epithelial cells and macrophages, to assess the effects of pan-HIF ablation during tumor initiation and progression.
These observations are clinically relevant, as increasingly specific HIF inhibitors will likely have a more significant effect on tumor progression. Whereas our work has focused extensively on the HIF-dependent effects of ACF treatment on the tumor microenvironment, it is possible that ACF has effects that are partially independent of HIF transcriptional activity (42,58,59). However, as transcription factors are effectively targeted for cancer therapeutics in the future, HIF inhibition in the tumor microenvironment by a safe, naturally occurring compound, in the setting of inflammation-driven cancer, represents an important finding. Targeting HIFs may be a viable therapeutic strategy in a myriad of cancers, as the data collectively indicate HIF inhibition can slow advancement of established tumors. Finally, the observations of HIF inhibition in both colorectal cancer cells and recruited macrophages provide insight into the usefulness of future genetic models for studying effects of HIF activity in the setting of inflammation-driven cancers.
Supplementary material
Supplementary Figures 1–5 can be found at http://carcin.oxfordjournals.org/
Funding
United States National Cancer Institute (1F30CA167949-01 to J.E.S.S.).
Supplementary Material
Acknowledgements
The authors thank H. Yu of the Abramson Family Cancer Research Institute Histopathology Core for assistance with histology preparation, as well as T. Richardson-Metzger for assistance with mouse experiments, and B. Keith for helpful advice regarding manuscript preparation. J.E.S.S. conceived the study, performed experiments and wrote the manuscript. H.Z.I. performed experiments; S.S. performed experiments; A.C.D. gave technical support and advice; N.S. gave technical support and advice; S.H. performed experiments; V.M. gave technical support and advice; T.S.K.E.M. gave technical support and advice; B.L.K. gave technical support and advice; D.N.G. gave technical support and advice; M.C.S. conceived the study, gave conceptual and technical advice, edited the manuscript and administered the project.
Conflict of Interest Statement: M.C.S. is an investigator of the Howard Hughes Medical Institute.
Glossary
Abbreviations:
- ACF
acriflavine
- AOM
azoxymethane
- BMDMs
bone marrow-derived macrophages
- CAC
colitis-associated colon cancer
- DMSO
dimethyl sulfoxide
- DSS
dextran sulfate sodium
- HDAC1
histone deacetylase 1
- HIF
hypoxia-inducible factor
- HRE
hypoxia-response element
- IL
interleukin
- NF-κB
nuclear factor-kappa B
- PBS
phosphate-buffered saline
- TAMs
tumor-associated macrophages
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- VEGF
vascular endothelial growth factor
- WT
wild type.
References
- 1. Virchow R. (1863). Aetologie der neoplastichen Geshwulste.Pathogenie der neoplastischen Geschwustle. Die Krankhafter Geschwulste, 1, 57–101 [Google Scholar]
- 2. Danese S., et al. (2010). Inflammatory bowel disease and intestinal cancer: a paradigm of the Yin-Yang interplay between inflammation and cancer. Oncogene, 29, 3313–3323 [DOI] [PubMed] [Google Scholar]
- 3. Vakkila J., et al. (2004). Inflammation and necrosis promote tumour growth. Nat. Rev. Immunol., 4, 641–648 [DOI] [PubMed] [Google Scholar]
- 4. Balkwill F., et al. (2001). Inflammation and cancer: back to Virchow? Lancet, 357, 539–545 [DOI] [PubMed] [Google Scholar]
- 5. Rhodes J.M., et al. (2002). Inflammation and colorectal cancer: IBD-associated and sporadic cancer compared. Trends Mol. Med., 8, 10–16 [DOI] [PubMed] [Google Scholar]
- 6. Eaden J.A., et al. et al. (2001). The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut, 48, 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Connor P.M., et al. (2010). Mechanisms by which inflammation may increase intestinal cancer risk in inflammatory bowel disease. Inflamm. Bowel Dis., 16, 1411–1420 [DOI] [PubMed] [Google Scholar]
- 8. Bertout J.A., et al. (2008). The impact of O2 availability on human cancer. Nat. Rev. Cancer, 8, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruan K., et al. (2009). Role of hypoxia in the hallmarks of human cancer. J. Cell. Biochem., 107, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 10. Murdoch C., et al. (2004). Mechanisms regulating the recruitment of macrophages into hypoxic areas of tumors and other ischemic tissues. Blood, 104, 2224–2234 [DOI] [PubMed] [Google Scholar]
- 11. Saleh M., et al. (2011). Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat. Rev. Immunol., 11, 9–20 [DOI] [PubMed] [Google Scholar]
- 12. Solinas G., et al. (2009). Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J. Leukoc. Biol., 86, 1065–1073 [DOI] [PubMed] [Google Scholar]
- 13. Tanner A.R., et al. (1984). Macrophage activation, chronic inflammation and gastrointestinal disease. Gut, 25, 760–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang H.Y., et al. (2009). Hypoxia-inducible factors 1 and 2 are important transcriptional effectors in primary macrophages experiencing hypoxia. Blood, 114, 844–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crowther M., et al. (2001). Microenvironmental influence on macrophage regulation of angiogenesis in wounds and malignant tumors. J. Leukoc. Biol., 70, 478–490 [PubMed] [Google Scholar]
- 16. Waldner M.J., et al. (2010). The molecular therapy of colorectal cancer. Mol. Aspects Med., 31, 171–178 [DOI] [PubMed] [Google Scholar]
- 17. Bingle L., et al. (2002). The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J. Pathol., 196, 254–265 [DOI] [PubMed] [Google Scholar]
- 18. Semenza G.L. (2007). Life with oxygen. Science, 318, 62–64 [DOI] [PubMed] [Google Scholar]
- 19. Majmundar A.J., et al. (2010). Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell, 40, 294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cockman M.E., et al. (2000). Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem., 275, 25733–25741 [DOI] [PubMed] [Google Scholar]
- 21. Jaakkola P., et al. (2001). Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science, 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 22. Imtiyaz H.Z., et al. (2010). Hypoxia-inducible factor 2alpha regulates macrophage function in mouse models of acute and tumor inflammation. J. Clin. Invest., 120, 2699–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Talks K.L., et al. (2000). The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol., 157, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. White J.R., et al. (2004). Genetic amplification of the transcriptional response to hypoxia as a novel means of identifying regulators of angiogenesis. Genomics, 83, 1–8 [DOI] [PubMed] [Google Scholar]
- 25. Semenza G.L. (2007). Evaluation of HIF-1 inhibitors as anticancer agents. Drug Discov. Today, 12, 853–859 [DOI] [PubMed] [Google Scholar]
- 26. Keith B., et al. (2012). HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer, 12, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keith B., et al. (2007). Hypoxia-inducible factors, stem cells, and cancer. Cell, 129, 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiesener M.S., et al. (2003). Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J., 17, 271–273 [DOI] [PubMed] [Google Scholar]
- 29. Mucaj V., et al. (2012). Effects of hypoxia and HIFs on cancer metabolism. Int. J. Hematol., 95, 464–470 [DOI] [PubMed] [Google Scholar]
- 30. Burke B., et al. (2003). Hypoxia-induced gene expression in human macrophages: implications for ischemic tissues and hypoxia-regulated gene therapy. Am. J. Pathol., 163, 1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burke B., et al. (2002). Expression of HIF-1alpha by human macrophages: implications for the use of macrophages in hypoxia-regulated cancer gene therapy. J. Pathol., 196, 204–212 [DOI] [PubMed] [Google Scholar]
- 32. Lewis J.S., et al. (1999). Macrophage responses to hypoxia: relevance to disease mechanisms. J. Leukoc. Biol., 66, 889–900 [DOI] [PubMed] [Google Scholar]
- 33. Murdoch C., et al. (2005). Macrophage migration and gene expression in response to tumor hypoxia. Int. J. Cancer, 117, 701–708 [DOI] [PubMed] [Google Scholar]
- 34. Takeda N., et al. (2010). Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes Dev., 24, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cramer T., et al. (2003). HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell, 112, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shay J.E., et al. (2012). Hypoxia-inducible factors: crosstalk between inflammation and metabolism. Semin. Cell Dev. Biol., 23, 389–394 [DOI] [PubMed] [Google Scholar]
- 37. Semenza G.L. (2006). Development of novel therapeutic strategies that target HIF-1. Expert Opin. Ther. Targets, 10, 267–280 [DOI] [PubMed] [Google Scholar]
- 38. Semenza G.L. (2012). Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol. Sci., 33, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Semenza G.L. (2012). Hypoxia-inducible factors in physiology and medicine. Cell, 148, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee K., et al. (2009). Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl. Acad. Sci. USA, 106, 17910–17915 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41. Wong C.C., et al. (2012). Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis. J. Mol. Med. (Berl)., 90, 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wainwright M. (2001). Acridine-a neglected antibacterial chromophore. J. Antimicrob. Chemother., 47, 1–13 [DOI] [PubMed] [Google Scholar]
- 43. Stadtfeld M., et al. (2005). Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development, 132, 203–213 [DOI] [PubMed] [Google Scholar]
- 44. Tomita S., et al. (2000). Conditional disruption of the aryl hydrocarbon receptor nuclear translocator (Arnt) gene leads to loss of target gene induction by the aryl hydrocarbon receptor and hypoxia-inducible factor 1alpha. Mol. Endocrinol., 14, 1674–1681 [DOI] [PubMed] [Google Scholar]
- 45. Pan Y., et al. (2004). p53 cannot be induced by hypoxia alone but responds to the hypoxic microenvironment. Oncogene, 23, 4975–4983 [DOI] [PubMed] [Google Scholar]
- 46. Okayasu I., et al. (1990). A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology, 98, 694–702 [DOI] [PubMed] [Google Scholar]
- 47. Okayasu I., et al. (1996). Promotion of colorectal neoplasia in experimental murine ulcerative colitis. Gut, 39, 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Okayasu I., et al. (2002). Dysplasia and carcinoma development in a repeated dextran sulfate sodium-induced colitis model. J. Gastroenterol. Hepatol., 17, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 49. Hogan S.P., et al. (2006). Resistin-like molecule beta regulates innate colonic function: barrier integrity and inflammation susceptibility. J. Allergy Clin. Immunol., 118, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hooper L.V., et al. (2003). Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol., 4, 269–273 [DOI] [PubMed] [Google Scholar]
- 51. Hans W., et al. (2000). The role of the resident intestinal flora in acute and chronic dextran sulfate sodium-induced colitis in mice. Eur. J. Gastroenterol. Hepatol., 12, 267–273 [DOI] [PubMed] [Google Scholar]
- 52. Ullman T.A., et al. (2011). Intestinal inflammation and cancer. Gastro enterology, 140, 1807–1816 [DOI] [PubMed] [Google Scholar]
- 53. Barnes P.J., et al. (1997). Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med., 336, 1066–1071 [DOI] [PubMed] [Google Scholar]
- 54. Pahl H.L. (1999). Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene, 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- 55. Dirkx A.E., et al. (2006). Monocyte/macrophage infiltration in tumors: modulators of angiogenesis. J. Leukoc. Biol., 80, 1183–1196 [DOI] [PubMed] [Google Scholar]
- 56. Murdoch C., et al. (2008). The role of myeloid cells in the promotion of tumour angiogenesis. Nat. Rev. Cancer, 8, 618–631 [DOI] [PubMed] [Google Scholar]
- 57. Onnis B., et al. (2009). Development of HIF-1 inhibitors for cancer therapy. J. Cell. Mol. Med., 13, 2780–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hassan S., et al. (2011). Novel activity of acriflavine against colorectal cancer tumor cells. Cancer Sci., 102, 2206–2213 [DOI] [PubMed] [Google Scholar]
- 59. Lim M.J., et al. (2012). Acriflavine enhances radiosensitivity of colon cancer cells through endoplasmic reticulum stress-mediated apoptosis. Int. J. Biochem. Cell Biol., 44, 1214–1222 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.