Abstract
The presence of mitochondrial DNA (mtDNA) mutations in human cancer has long been recognized, but their functional significance has remained obscure. Debate persists as to whether the mutations help drive the tumor, or are bystander events. Here, we analyze next-generation mtDNA sequence data from 99 breast cancer patients. High depth coverage enables detection of even low-level heteroplasmic variants, and data from matched normal tissue allow us to distinguish between shifts in heteroplasmy and acquired mutations. Somatic mtDNA mutations are found in 73 (73.7%) of patient tumors, and dramatic shifts from the initial germline allele proportions are observed for many heteroplasmies. Clustering of somatic mutations in promoter and replication regions, and also in genes coding for electron transport chain complex I, suggest selection for mutations affecting critical mitochondrial processes. Furthermore, statistical tests for Darwinian selection reveal evidence for positive and relaxed negative selection for somatic missense mutations. We also observe a dramatic decrease in per-cell mtDNA content in tumor tissues, as well as a surprising positive correlation between somatic mtDNA mutational burden and patient survival. Taken together, our results support the view that somatic mtDNA mutations are not solely bystander events, but have significance in cancer from both biological and clinical perspectives. We also anticipate that the catalog of heteroplasmies and somatic mutations presented here will serve as a reference for future studies of cancer mitochondrial genomes.
Introduction
Mitochondria are the organelles in the cell responsible for generating the majority of energy, in the form of adenosine triphosphate (ATP), through oxidative phosphorylation (OXPHOS) coupled to ATP synthesis. Mitochondria are also involved in other essential, highly regulated processes, such as the production of reactive oxygen species (ROS), intracellular calcium homeostasis and apoptosis (1). Each mitochondrion contains multiple copies of a small (16 569bp) circular DNA molecule [mitochondrial DNA (mtDNA)] that codes for 13 proteins, 22 transfer RNAs (tRNAs) and 2 ribosomal RNAs. In contrast to only two copies of the nuclear genome per cell, thousands of mtDNA copies can be present, although this number varies with tissue type depending on the energy requirements of the cell (1). In the past, it was generally assumed that all copies of mtDNA in a normal cell have the same nucleotide sequence, a state termed homoplasmy. However, once a sporadic mutation has been introduced into mtDNA, it can propagate owing to random drift or positive selection, resulting in a heteroplasmic state (2,3). Minor variants can eventually become dominant after numerous generations or replication cycles and are associated with many human diseases including cancer (4).
A link between mitochondrial defects and cancer was first proposed by Otto Warburg, who established that mitochondria within cancer cells can preferentially undergo glycolysis even in the presence of oxygen (5). Subsequently, researchers have examined the mitochondrial genome in cancer cells for somatic mutations associated with this anomalous behavior, and such mutations have been reported in many tumor types including breast, colon, esophageal, pancreatic, prostate and others (6–11). Although it is yet to be established whether the preferential glycolysis and elevated mutation rate is causal or incidental, clearly the bioenergetic signature of the mitochondria is frequently altered in cancer. For instance, as a consequence of altered metabolism due to respiratory chain disruption, cancer cells harbor elevated levels of ROS (12). Increased concentration of ROS leads to greater oxidative stress, larger genomic instability and defects in DNA repair mechanisms (8). In addition, a critical range of ROS is necessary to regulate cell proliferation and apoptosis, and deviations from ROS homeostasis due to altered metabolism can result in abnormal cell growth, a hallmark of cancer (13). Many chemotherapeutics including cisplatin (Platinol) and doxorubicin (Doxil) attack the tumor by upregulating ROS in cancer cells that already harbor elevated concentrations. These anticancer agents induce excessive ROS accumulation, which in turn induces apoptosis (14,15). Nevertheless, the role of somatic mtDNA mutations in cancer remains a subject of debate, with conflicting reports in the literature (11,16,17). Global profiling and analysis of multiple breast cancer mitochondrial genomes in particular have not, to date, been performed.
Breast cancer is the second most common cancer type in women, with ~12% (1 in 8) of women in USA developing the disease during their lifetime (18). Only 10–15% of breast cancers are due to inherited susceptibility variants and thus the majority of breast cancers arise from sporadic alterations of the genome (19). With the advent of next-generation sequencing (NGS) technology, a number of studies have focused on characterizing somatic mutations in the breast tumor genome (20,21). However, these studies focus almost exclusively on nuclear DNA, leaving the mitochondrial genome largely uncharacterized. Given the well-established mitochondrial dysfunction in cancer and the high rate of somatic mutation in mtDNA, the mitochondrial genome is an under-explored avenue for insight into breast cancer pathogenesis, as well as an attractive candidate source for biomarkers (22).
In this study, we examined the normal and tumor mitochondrial genomes of 99 breast cancer patients, under the hypothesis that the ultra-deep coverage afforded by NGS would help elucidate the relationships between mtDNA mutation, selection and patient outcome. Availability of high-coverage sequence data facilitated accurate detection of somatic variants and quantification of heteroplasmies. In addition, read depth information from the nuclear (two copies per cell) genome served as a calibrator for estimation of mtDNA per-cell copy number in patients for which whole-genome sequencing was available from both the tumor and adjacent normal tissue. The mutational burden in tumor mitochondria was also assessed for its impact on patient survival.
Materials and methods
Data
Whole-genome and/or -exome sequencing.bam files for matched tumor/normal pairs (n = 99; Supplementary Table 1, available at Carcinogenesis Online) were obtained through The Cancer Genome Atlas (TCGA) project via download from Cancer Genomics Hub (https://cghub.ucsc.edu) (20). It has recently been observed that mitochondrial sequences are captured (although they are not explicitly targeted) by whole-exome protocols, and therefore, we were able to detect germline and somatic variants from both whole-genome and whole-exome.bam files (23,24). The numbers of detected variants did not differ between the two protocols (Supplementary Figure 1, available at Carcinogenesis Online). Normal sample data were generated by TCGA using tissue obtained either from non-cancerous breast tissue or from whole blood. Patient clinical information and single nucleotide polymorphism array data were downloaded from TCGA’s Data Portal (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp), as were pathology slides. Herein, each TCGA patient sample is referred to by its (unique) patient ID comprising the 6th through the 12th characters of the barcode (e.g. the patient with barcode TCGA-BH-A0B3 is referred to as A0B3).
Calling and annotating variants
From the .bam sequence files, mitochondrial reads were extracted using samtools (25). The bam2fastq software (http://www.hudsonalpha.org/gsl/information/software/bam2fastq) was used to revert these aligned sequences to fastq format. The sequences were realigned against the revised Cambridge reference sequence using bowtie2 (26,27). PCR duplicate removal and indel realignment were performed using the Genome Analysis Toolkit, and the pileup file was generated using samtools (28). From the pileup file, variants were called using bcftools for both tumor and matched normal samples. Mutations and variants were classified using a series of thresholds designed to filter out false positives while accounting for heteroplasmies. Specifically, any variant present in <2% of the reads was omitted. Those present at ≥2% in the tumor, but <2% in the matched normal were deemed somatic. Germline heteroplasmies were called for those alleles present in normal tissue at levels >2%, unless both tumor and normal carried the allele at levels >90%. All resulting putative variants were manually inspected in the Integrative Genome Viewer (29) for quality control purposes.
Impact (synonymous versus non-synonymous) and pathogenicity assessment of protein-coding variants were performed using snpEff, which is able to account for the mitochondrial genetic code (30). Tumor-specific tRNA variants were compared with the MITOMAP and Mamit-tRNA databases (31,32). tRNA structures were produced using the tRNA scan software (33).
Mutational enrichment computations
Testing difference in proportions of transitions/transversions between germline and somatic mutations was performed using a standard two-sided Fisher’s exact test, as was testing the significance of mutational base context. Enrichment in control region mutations impacting functional sites was assessed by comparing the number of such mutations with the (upper tail) null binomial distribution Binom(N, p), with parameters N = number of mutations in control region and p = proportion of control region bases in functional sites. Differences in distribution of mutations in complexes I, II, IV and V between germline and somatic mutations were assessed by applying Fisher’s exact test to the 2 × 4 contingency table of mutation counts, with rows indicating germline and somatic categories, and columns indicating the four complexes. A similar calculation was performed to compare the distributions, across regions, of the numbers of base positions with the numbers of somatic mutations.
K a /K s computation
The K a /K s ratios were computed using the seqinr package (34). For a given transcribed region, K a denotes the average number non-synonymous mutations per non-synonymous site. Similarly, K s denotes the average number synonymous mutations per synonymous site. To assess the effect of mutations (germline and somatic separately) on the mitochondrial genome, all mutations were first added, in silico, to the reference revised Cambridge reference sequence genome. For each protein-coding gene, the reference and mutated amino acid sequences were generated using the ‘translate’ command in seqinr with the mitochondrial genetic code argument, then the resulting amino acid sequences (revised Cambridge reference sequence and mutated) were aligned to one another using Clustal X (35). The corresponding aligned DNA sequences were then generated using the ‘reverse.align’ command (again specifying the mitochondrial genetic code) in seqinr, and then used the ‘kaks’ command to compute K a, K s and their variances. Two-sided P-values were calculated, testing the null hypothesis K a – K s = 0, using the normal approximation.
Quantification of mtDNA content via normalized read depth
To perform a well-controlled quantification of relative mtDNA quantity between tumor and normal samples, we designed a novel approach to calibrate depth coverage using the known (two copy) nuclear genome per-cell content. The gene HMOX2 on chromosome 16p13.3 was deemed to have two copies in virtually all whole-genome samples (both tumor and normal) from analysis of single nucleotide polymorphism array data. For each sample, the ratio of median read depth across the mitochondrial genome (D m) to the median read depth across the two copy nuclear HMOX2 gene (D n) was computed as
For each patient sample, the ratios r (n) and r (t) were thereby obtained for the normal and tumor tissues, respectively, with estimated mtDNA copy numbers computed as 2 × r (n) and 2 × r (t). To assess tumor-specific mtDNA copy number changes, attention was initially restricted to three patients for which whole-genome sequencing was available (to avoid exome-capture artifacts) and for which the normal DNA was derived from matched tissue (as opposed to blood). This latter requirement is necessary because different normal tissues are known to have different quantities of mitochondria and the goal was to assess aberrant DNA quantities in the tumor.
Survival analysis
For each sample, the number of somatic mtDNA mutations was used as a survival prediction variable. The mutation number was initially treated as a numerical variable and then dichotomized. A Cox proportional hazards model was fit to overall survival, initially adjusting for patient age. Subsequently, tumor stage was added as a model term. Because there were only 10 stage I patients, stages I and II were combined, yielding two stage categories (I/II and III). Next, hormone receptor status was added as a binary term. Here, a tumor was deemed hormone receptor positive if it was either estrogen or progesterone receptor positive (or both). All model fitting and hazard ratio computations were performed using the survival package in R (36).
Results
Patient cohort and accompanying data
Our sample set comprised of 99 patients with mean age 60.0 (range 34–88). Patient details may be found in Supplementary Table 1, available at Carcinogenesis Online. Briefly, all tumors were breast carcinomas, with 89 distinguished as ductal or having ductal features. Thirteen of the tumors were confirmed to be triple-negative, and overall, there were 10 stage I, 60 stage II and 28 stage III patients (staging was unavailable for one patient). Of the 99 patients, whole-genome sequencing was performed on 37 tumor-normal pairs and whole-exome sequencing on 62 pairs (median mitochondrial genome depth coverage 3532X for whole genome and 184X for whole exome, ranges 57–13676X and 33–1331X, respectively). The normal DNA samples were obtained from matched breast tissue for 21 of the patients, with the remaining 78 from whole blood. For three of the patients, sequence data were available for both types of normal tissue.
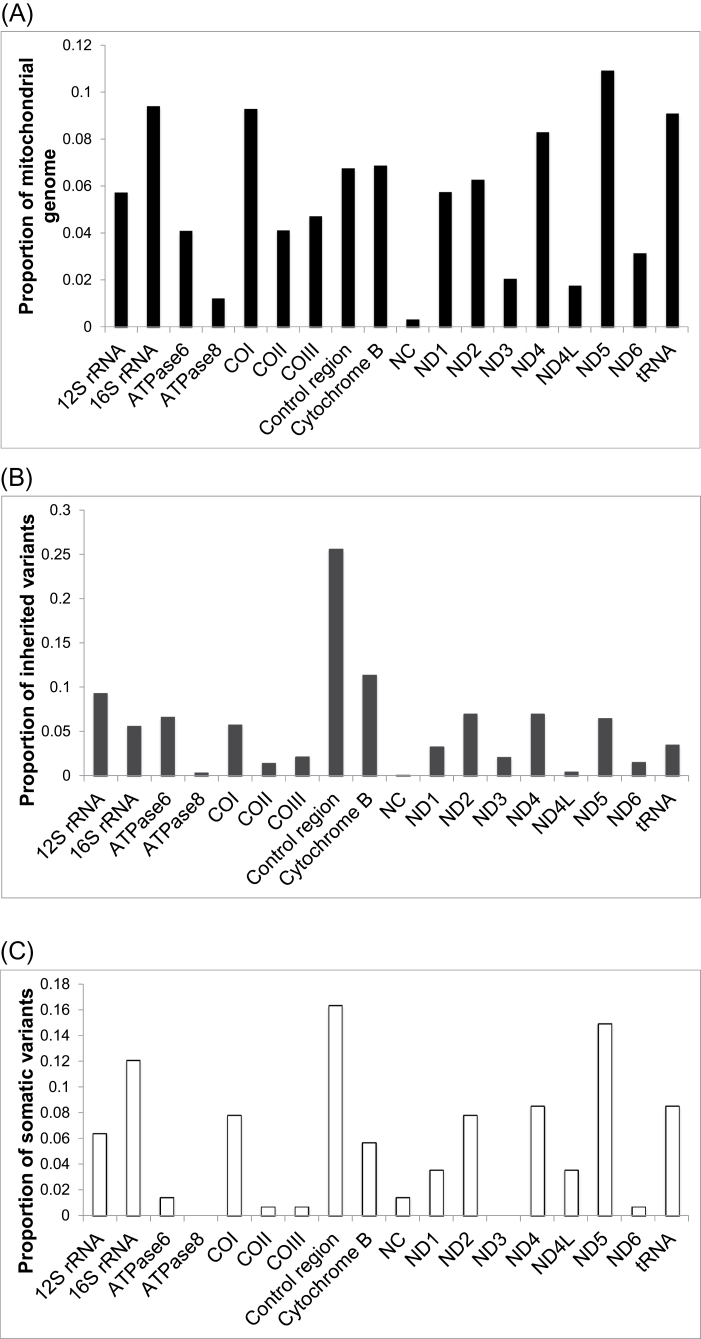
Germline variants and heteroplasmies
Normal tissue sequencing showed an average of 27 germline variants per individual. In the patients for whom data from both normal tissue types were available, no sequence differences were observed between breast tissue and matched blood DNA. This indicates that, in our setting, tissue-specific mutations are not likely to arise and be misidentified as germline variants. Also, the normal breast tissue samples do not appear to be contaminated with tumor cells. Overall, 55.9% of germline variants were located in protein-coding regions, a smaller proportion than the ~68% of the mitochondrial genome that is protein coding (Figure 1A and B). A total of 30 germline heteroplasmies were detected, arising in 27 individual patients. Heteroplasmy occurs when a mixture of reference and non-reference alleles is present at the same base position in an individual’s germline. Non-reference allele abundance here ranged between 2.17% and 97.5% (median 21.9%; Supplementary Table 2, available at Carcinogenesis Online). Two of the heteroplasmic sites arose as such in multiple individuals—positions 16 093 (heteroplasmic in five individuals) and position 16 325 (heteroplasmic in two individuals). Five of the sites have been previously reported as being associated with disease (Supplementary Table 2, available at Carcinogenesis Online). There were two nominally significant associations between heteroplasmy level and patient characteristics. First, women of African ancestry show a larger heteroplasmy shift toward reference allele dominance in the tumor than do white women (P = 0.004). However, the number of patients in the former group is relatively small (N = 4). Second, HER2-positive tumors show larger shifts in heteroplasmy (P = 0.035) than HER2-negative tumors.
Fig. 1.
Mutational distribution across the mitochondrial genome in breast cancer patients. (A) The proportion of the 16.6kb mitochondrial genome that lies within each region/gene. (B) The distribution of germline variants (2632 total) across the mitochondrial regions/genes. (C) The distribution of somatic mutations (141 total) across the mitochondrial regions/genes. (NC, non-coding regions outside of the control region).
Tumor-specific mtDNA mutational overview
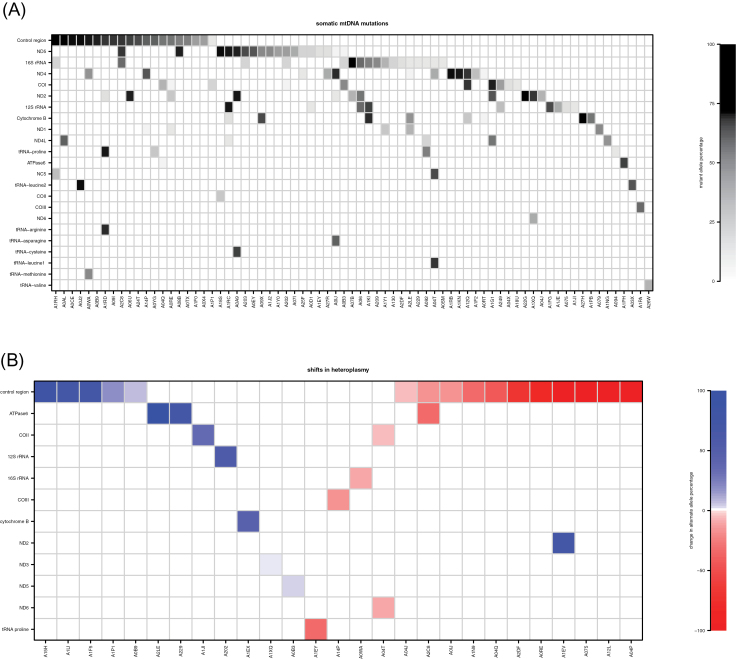
Somatic mutations were found in 73 of 99 patients (73.7%), and in total, there were 141 such mutations (Figure 2A). The vast majority of somatic mutations were singletons, arising in only one patient (Supplementary Table 3, available at Carcinogenesis Online). Exceptions to this were at positions 186, 13 069, 16 114 (two patients each) and 16 390 (three patients). Twelve of the somatic mutations were previously reported as arising in various diseases (Supplementary Table 3, available at Carcinogenesis Online). There was no statistical association observed between mutational presence or abundance and patient characteristics. Several of these have previously been reported in various diseases, including cancer (Supplementary Table 3, available at Carcinogenesis Online). Interestingly, all of the somatic mutations were present in a heteroplasmic state (mutant allele abundance 5.6–97.4%, median 44.1%). However, it is possible that this apparent variable heteroplasmy is due to differing levels of normal cell content or tumor heterogeneity rather than a heteroplasmic state within each tumor cell. We cannot distinguish among these possibilities with the available data. One recent study suggested that many putative somatic mutations are in fact low-level heteroplasmies undetected in the germline that have undergone clonal expansion in the tumor (37). Careful inspection of germline reads covering the sites of somatic mutations reported in this study showed no evidence of the mutations’ presence in normal cells, and therefore, the mutations reported here seem to be truly somatic.
Fig. 2.
Somatic changes and their genomic locations in each sample. (A) Each shaded rectangle corresponds to a somatic mutation, with shading indicating mutant allele percentage (white = 0%, black = 100%). The mutations are presented in tabular form in Supplementary Table 3, available at Carcinogenesis Online. (B) Each colored rectangle corresponds to a heteroplasmic germline variant that shifts variant allele percentage in tumor (white = no heteroplasmy present or no shift, red = variant allele decrease from normal to tumor, blue = variant allele increase from normal to tumor). The variants are presented in tabular form in Supplementary Table 2, available at Carcinogenesis Online.
The majority of somatic mutations were transitions, although there was a significant enrichment of transversions among somatic as compared with germline variants (Fisher’s exact P = 0.047; Supplementary Figure 2, available at Carcinogenesis Online). Closer examination of the somatic base substitutions showed that a disproportionate number—65 of 141 (~46%)—occur at guanines on the reference (‘light’ or L-) strand, and 62 of these are G>A changes. We cannot determine whether the mutations originally arose on the light strand G or on the complementary heavy-strand cytosine, but the enrichment of mutations at guanines is particularly striking given their relative scarcity on the light strand. Only 13% of bases are guanines, yet they account for 46% of the mutations (P = 2.47 × 10– 21).
Distribution of somatic alterations across genomic regions
Overall, 8.5% of somatic mutations were within tRNAs, 17.7% within ribosomal RNAs, 55.3% in protein-coding regions, 17.0% in the control region and 1.5% in other non-coding regions (Figures 1C and 2A; Supplementary Table 3, available at Carcinogenesis Online). There were also a number of substantial shifts in variant allele proportions from the normal to the tumor tissues among the heteroplasmic germline variants (Figure 2B; Supplementary Table 2, available at Carcinogenesis Online). Among protein-coding genes, the number of somatic mutations correlated strongly with transcript size (Pearson r = 0.89; Figure 1A and C; Supplementary Figure 3, available at Carcinogenesis Online). ND5 harbored the most somatic mutations, consistent with another recent breast cancer study (38).
Given the role of mitochondrial genes in the OXPHOS cascade, we examined the distribution of mutations within the complexes of the cascade. These complexes are involved in a series of redox reactions that ultimately result in the production of ATP. This series of reactions begins when electron carriers from the citric acid cycle transfer electrons at sites located in complexes I and III of the OXPHOS cascade. The redox reactions result in the production of cellular ROS, which are known to cause DNA damage. Our analysis indicates that the distribution of somatic mutations across the mitochondrial-encoded components of the OXPHOS cascade (complexes I, III, IV and V) differed from that of inherited variants (omnibus Fisher P = 5.5 × 10– 4) and did not correspond to what would be expected from transcript sizes (P = 0.034), with complex I particularly showing a significant enrichment of somatic mutations (Supplementary Figure 4, available at Carcinogenesis Online).
Across the mitochondrial genome as a whole, it has been well established that the control region (including the displacement loop) contains a disproportionate number of germline and somatic mutations (39). We sought to determine whether the rate of somatic mutations is particularly elevated in functional sites within the control region. These annotated sites include heavy- and light-strand promoters, as well as the heavy-strand origin of replication. We found that, although these functional sites make up less than one-third (~32%) of control region base positions, mutations in them comprise nearly half (~48%) of those in the control region (binomial test P = 0.035). The two promoter regions together also showed an enrichment in mutation rate (P = 0.018).
mtDNA copy number in normal and tumor cells
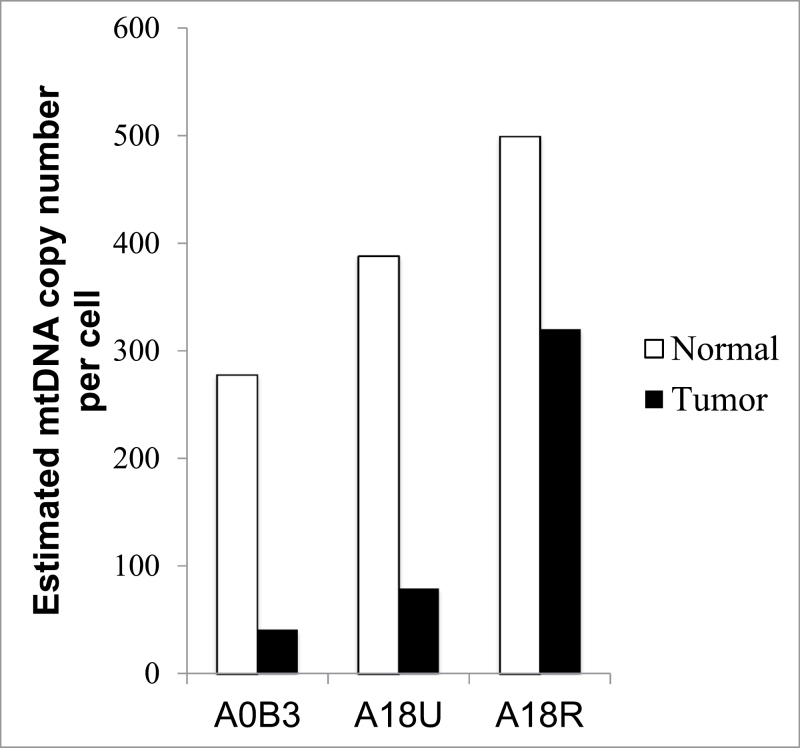
Previous studies have reported either an increase or a decrease of mtDNA content in tumor cells relative to matched normal tissue (40,41). To investigate this in the current data set, we developed a novel method to estimate mtDNA content per cell using whole-genome NGS data. Local depth of coverage in NGS experiments is used as a proxy for DNA content by several methods that aim to find genomic copy number aberrations (42,43). Here, we calibrated the mitochondrial depth of coverage using a region of the nuclear genome that is confirmed to be unaffected by copy number aberrations, i.e. has two copies per cell (see Materials and methods). Our method requires whole-genome (as opposed to exome) sequence to avoid biases induced by exome-capture technologies. Furthermore, the per-cell mtDNA content in the tumor should be compared with that of matched normal cells of the same tissue type. This ensures that any observed copy changes can be attributed to malignancy and are not instead due to variable energy requirements in disparate tissue types that would affect numbers of mitochondria per cell (44). Therefore, we focused our analysis to patients from whom whole-genome sequence was available for both tumor and matched normal breast tissue (as opposed to blood).
In each of these cases, we found a dramatically lower mtDNA content in cancer cells compared with the normal cells (Figure 3), though the degree of mtDNA depletion varied from tumor to tumor. This variation is not due to differences in amount of normal cell infiltration into the tumor, as all three tumor samples have between 75% and 85% purity according to the pathology reports (pathology slides images provided in Supplementary Figure 5, available at Carcinogenesis Online), and there is no correlation between purity and mtDNA content. The same analysis performed on matched tumor-blood pairs showed a lower copy number in tumor cells (Supplementary Table 4, available at Carcinogenesis Online), but here, it is impossible to determine whether the copy number differences are attributable to the cancer/normal distinction, or rather to the differences in cell type.
Fig. 3.
Estimated per-cell mtDNA copy number in normal and cancer cells. MtDNA content was determined using the nuclear DNA-normalized read count-based method described in the Materials and methods. TCGA patient IDs are indicated on the horizontal axis.
Impact of somatic mutations on RNA
To further evaluate the impact of somatic mutations on the mitochondrial genome, we examined their distribution within mitochondrial-encoded tRNAs and proteins. There were 12 somatic tRNA mutations (Supplementary Table 5 and Figure 6, available at Carcinogenesis Online), two of which are known polymorphisms, and five of which have been previously reported as pathogenic (32).The remaining five somatic tRNA mutations have not been previously reported in the MITOMAP database (31). Although there were no mutations within the tRNA anticodons, variants located within the stem or loop regions can result in instability and thus affect the synthesis of mitochondrial proteins (45).
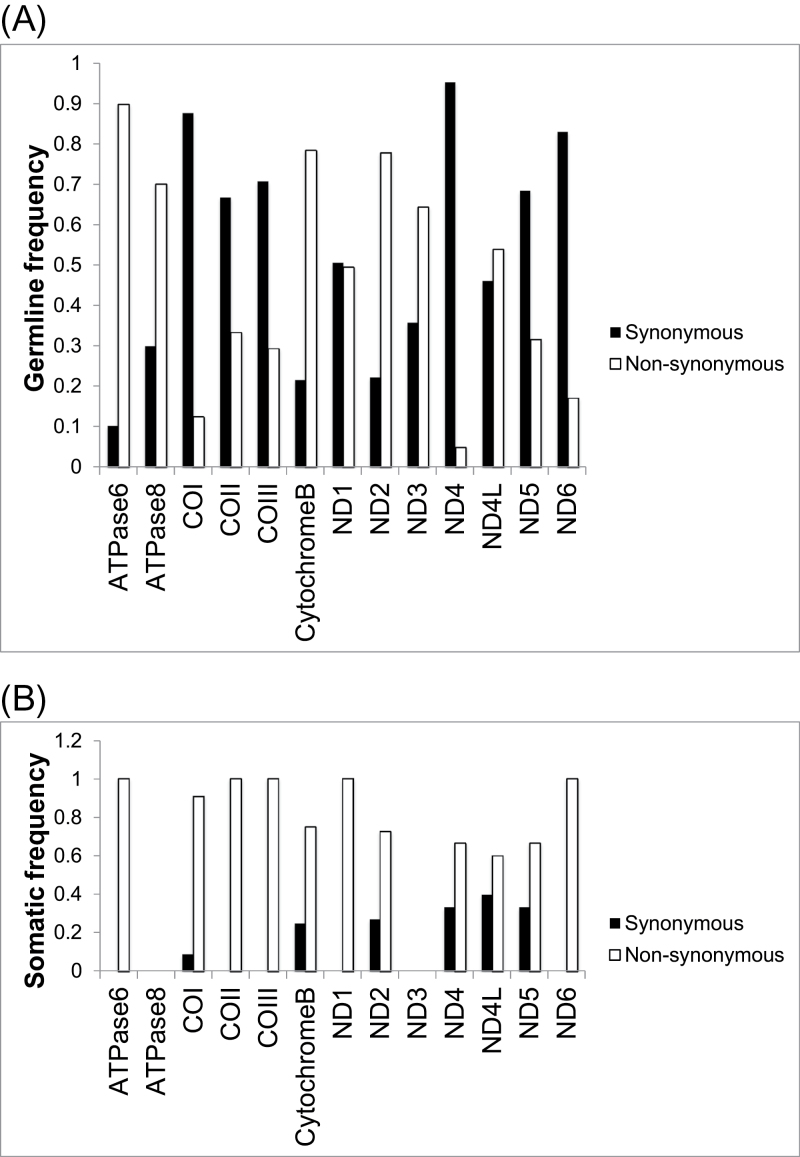
In protein-coding genes, the somatic mutations were more likely than germline variants to be non-synonymous amino acid changes (Figure 4). This suggests the possibility that some of the genes are under positive selection in the tumor environment. To investigate further, we computed the rates of non-synonymous mutations per non-synonymous site (K a) and synonymous mutations per synonymous site (K s) for each gene, as well as for the mitochondrial transcriptome as a whole (Table I). The K a /K s ratio is commonly used in evolutionary genetics to measure the degree to which a gene is under Darwinian selection, whether positive (K a /K s > 1), negative (K a /K s < 1) or neutral (K a /K s ≈ 1). Overall, the mitochondrial transcriptome showed very strong purifying selection against germline protein-coding changes (K a /K s = 0.329, P = 5.64 × 10– 21). All genes trended in this direction (K a /K s < 1) save ND6, which showed nominal statistical significance for positive selection for missense mutations (K a /K s = 3.0, P = 0.04). In contrast, the pattern of somatic mutations in the transcriptome as a whole trended toward positive selection for non-synonymous mutations (K a /K s = 1.59, P = 0.082). Although the relative sparseness of somatic mutations rendered our test somewhat underpowered, the majority of genes with sufficient mutations had K a /K s values >1, and positive selection for somatic amino acid changes in COI attained nominal significance (K a /K s = 5.36, P = 0.038). The majority (six of nine) of these mutations were within transmembrane helical segments, with the remainder located within domains found in the mitochondrial matrix and inner membrane space (Supplementary Figure 7, available at Carcinogenesis Online).
Fig. 4.
Relative frequencies of non-synonymous and synonymous mutations across mitochondrial protein-coding regions. (A) Germline mutations; (B) somatic mutations.
Table I.
K a / K s calculations for germline and somatic mutations
| Gene | K a | K s | K a variance | K s variance | K a/K s | z-score | P-value |
|---|---|---|---|---|---|---|---|
| Germline | |||||||
| ND1 | 0.023 | 0.090 | 4.40E-05 | 3.50E-04 | 0.256 | −3.38 | 7.25E-04 |
| ND2 | 0.024 | 0.077 | 4.12E-05 | 2.91E-04 | 0.306 | −2.94 | 3.28E-03 |
| ND3 | 0.043 | 0.082 | 2.53E-04 | 7.28E-04 | 0.526 | −1.24 | 2.15E-01 |
| ND4 | 0.010 | 0.083 | 1.39E-05 | 1.99E-04 | 0.122 | −4.97 | 6.54E-07 |
| ND4L | 0.010 | 0.037 | 5.47E-05 | 3.90E-04 | 0.282 | −1.25 | 2.10E-01 |
| ND5 | 0.025 | 0.076 | 2.44E-05 | 1.62E-04 | 0.327 | −3.73 | 1.92E-04 |
| ND6 | 0.051 | 0.017 | 1.63E-04 | 1.15E-04 | 3.000 | 2.05 | 4.05E-02 |
| Cytochrome b | 0.029 | 0.079 | 5.07E-05 | 2.93E-04 | 0.371 | −2.69 | 7.09E-03 |
| COI | 0.016 | 0.090 | 1.61E-05 | 2.48E-04 | 0.177 | −4.54 | 5.72E-06 |
| COII | 0.020 | 0.098 | 4.58E-05 | 7.62E-04 | 0.205 | −2.73 | 6.35E-03 |
| COIII | 0.020 | 0.068 | 3.95E-05 | 3.01E-04 | 0.290 | −2.61 | 9.04E-03 |
| ATPase6 | 0.060 | 0.065 | 1.53E-04 | 3.59E-04 | 0.923 | −0.22 | 8.24E-01 |
| ATPase8 | 0.044 | 0.077 | 4.37E-04 | 1.34E-03 | 0.571 | −0.79 | 4.30E-01 |
| Transcriptome | 0.025 | 0.076 | 3.86E-06 | 2.59E-05 | 0.329 | −9.40 | 5.64E-21 |
| Somatic | |||||||
| ND1 | 0.287 | 0.340 | 1.38E-03 | 4.54E-03 | 0.846 | −0.68 | 0.497 |
| ND2 | 0.012 | 0.008 | 1.87E-05 | 2.20E-05 | 1.500 | 0.63 | 0.525 |
| ND3 | 0.000 | 0.000 | NA | NA | NA | NA | NA |
| ND4 | 0.009 | 0.006 | 1.07E-05 | 1.21E-05 | 1.534 | 0.67 | 0.502 |
| ND4L | 0.017 | 0.018 | 9.63E-05 | 1.75E-04 | 0.918 | −0.09 | 0.927 |
| ND5 | 0.011 | 0.012 | 1.01E-05 | 3.18E-05 | 0.911 | −0.17 | 0.863 |
| ND6 | 0.000 | 0.006 | NA | 3.20E-05 | NA | NA | NA |
| Cytochrome b | 0.008 | 0.005 | 1.19E-05 | 1.85E-05 | 1.565 | 0.51 | 0.611 |
| COI | 0.010 | 0.002 | 1.01E-05 | 5.22E-06 | 5.357 | 2.08 | 0.038 |
| COII | 0.002 | 0.000 | 5.27E-06 | NA | NA | NA | NA |
| COIII | 0.002 | 0.000 | 3.84E-06 | NA | NA | NA | NA |
| ATPase6 | 0.005 | 0.000 | 1.12E-05 | NA | NA | NA | NA |
| ATPase8 | 0.000 | 0.000 | NA | NA | NA | NA | NA |
| Transcriptome | 0.008 | 0.005 | 1.10E-06 | 1.64E-06 | 1.585 | 1.74 | 0.082 |
NA refers to insufficient number of mutations to perform calculation.
Here, positive z-scores are consistent with positive selection and negative z-scores with negative selection.
Association with patient outcome
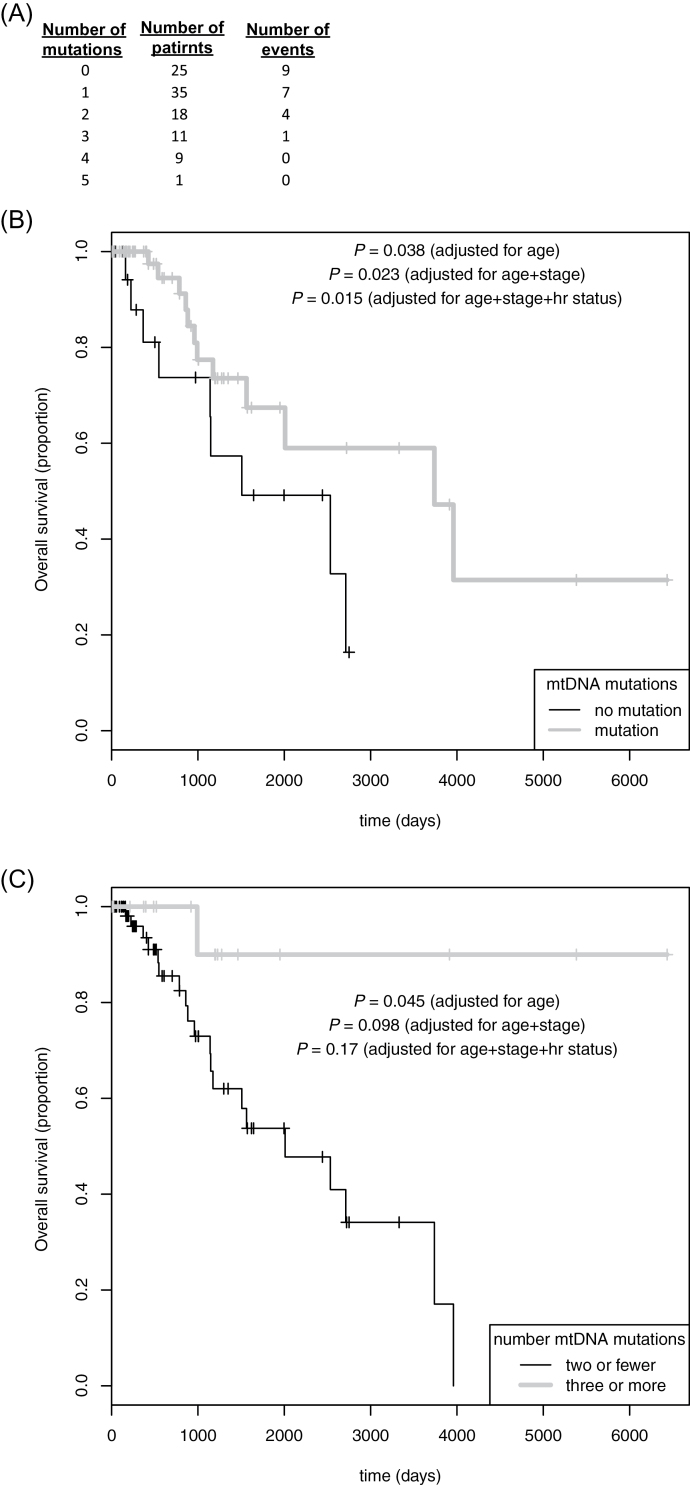
By incorporating clinical survival data, we sought to determine whether there is an association between somatic mtDNA mutations and patient outcome (overview in Figure 5A). Treating number of mutations as a quantitative predictor, survival analysis revealed that a higher mutational burden was significantly associated with better overall survival (hazard ratio for each additional mutation 0.59, 95% confidence interval 0.39–0.90, age-adjusted P = 0.015). Dichotomizing patients by mutational presence or absence (Figure 5B) similarly showed better outcomes for patients with mutations (hazard ratio 0.38, 95% confidence interval 0.15–0.95, age-adjusted P = 0.038). Furthermore, patients having a mutational burden above the median among those carrying mutations (≥3) fared better than other patients (hazard ratio 0.12, 95% confidence interval 0.02–0.95, age-adjusted P = 0.045; Figure 5C).
Fig. 5.
Overall survival by mitochondrial somatic mutation status. (A) Counts of patients by mutational burden, with number of events (deaths) in each category. In addition, Kaplan–Meier curves were generated for patients dichotomized by (B) presence or absence of somatic mtDNA mutations and (C) high (≥3) or low (≤2) somatic mutation burden. Log rank test P-values are given, adjusted for age, age plus stage, and age plus stage and hormone receptor (hr, estrogen and/or progesterone) status.
Additionally adjusting for tumor stage in the models slightly alters the P-values for mutation (adjusting P-values from above yields 0.035, 0.023 and 0.098, respectively), though stage itself is not a statistically significant term in any of models. Similarly, additionally incorporating hormone receptor status as a model term moderately alters mutation P-values (0.055, 0.015 and 0.017, respectively), though receptor status itself is not significantly predictive in the models.
Discussion
We have performed the first, to our knowledge, NGS-based study of the mitochondrial genome in breast cancer. A substantial majority of tumors harbor somatic mtDNA mutations, and many tumors show shifts in allele frequencies from their corresponding germline heteroplasmies. Our results provide evidence for selection of somatic mutations within key regulatory and coding regions. Furthermore, mtDNA depletion and overall increased survival of breast cancer patients harboring somatic mutations suggest that altered metabolism in mitochondria has a fundamental role in tumorigenesis and treatment response. The relationship between mutation load and patient outcome seems to be largely independent of tumor stage and hormone receptor status (Figure 5B and C), but larger cohorts are necessary for additional evidence of this association.
A recent report by Larman et al. (17) analyzing NGS data from the mitochondrial genomes of brain, colorectal, ovarian and myeloid cancers hypothesized a link between the presence of somatic mutations and metabolic deregulation in tumors. Their results support Warburg’s hypothesis that tumor progression is driven by mtDNA alterations, resulting in an aberrant metabolic signature. Although the Larman et al. (17) study focused exclusively on protein-coding regions, our analysis showed that sequence elements controlling transcription and replication were preferentially mutated in tumor samples. Somatic mutations in these sequences may affect the production of mitochondrial-encoded respiratory chain components. Indeed, a recent study (46) showed that some mutations cause impairment of complex I, resulting in a compensatory metabolic shift. Interestingly, one of the mutations implicated in that report—an A to G change at position 3243—was among the somatic mutations found in our study.
Examination of mitochondrial genes revealed a significant enrichment of somatic mutations in complex I of the electron transport chain, and indeed the gene with the most somatic mutations was ND5, a component of complex I. A study performed by Mayr et al. (47) suggested that complex I is an important factor in apoptosis signaling, and therefore, reduced activity of complex I would favor tumor formation and growth. Our own results, therefore, suggest that tumors may select for mutations within key regions of the mitochondrial genome, thereby inducing altered metabolism and supporting tumor growth. Additionally, the elevated ratio of non-synonymous to synonymous somatic substitutions, K a /K s, supports the hypothesis that protein-altering mutations within tumor mtDNA may confer a selective advantage to the cell.
We observed a very strong preference for L-strand G>A substitutions. Furthermore, G>A mutations on the H-strand (indistinguishable from C>T mutations on the L-strand) are much less common in our sample set, despite the H-strand being far more G-rich. As with many of the signatures emerging from recent studies of somatic mutational base context in the nuclear genome, the actual mechanisms underlying our observation remain unknown (48,49). In contrast to cancers arising as a result of smoking or UV damage, most of the tumors in our study were probably not initiated by exogenous carcinogenic exposures that can result in specific mutational signatures. Nor is there an enrichment in cytosines immediately 5ʹ to the mutated guanines, as would be expected if these G>A mutations were the result of deamination of a methylated complementary cytosine on the H-strand. This puzzling phenomenon is worthy of further investigation.
In addition to acquired mutations in the tumor, we also detected large expansions in the cancer cell of some low-level germline variants. For instance, the T15394C mutation in cytochrome b is present in patient A1EX at 2.17% in the germline, but 42.64% in the tumor (Supplementary Table 2, available at Carcinogenesis Online; Figure 2B). This particular example is consistent with our analysis of the K a /K s statistic, which showed a signal of purifying selection from germline variants in cytochrome B, but then neutrality or positive selection from somatic mutations. It is important to note that, using traditional Sanger sequencing, this variant would likely have appeared to be absent in the germline but present in the tumor, and would have therefore been classified as a somatic mutation. This underscores the importance of ultra-deep sequencing to accurately classify mutations and heteroplasmies in mtDNA. It is also interesting to note that we observed significantly larger shifts in heteroplasmy in HER2-positive tumors than in HER2-negative tumors, perhaps reflecting the more aggressive nature of the latter class.
We demonstrated a positive association between somatic mtDNA mutational burden and overall survival in breast cancer patients, consistent with a previous study of acute myeloid leukemia where patients with mutated ND4 showed greater overall survival than patients with wild-type ND4 (50). We hypothesize that many acquired mutations may be disruptive to OXPHOS, causing a repetitive cycle of increased ROS and more mtDNA mutations. However, if deleterious mutations are acquired that completely shift energy metabolism in the tumor from OXPHOS to glycolysis, then there will likely be a decrease in ROS. A decrease in ROS would support tumor growth by inhibiting apoptosis. Therefore, tumors with more mutations might be those that have not gained a deleterious mutation that shuts down OXPHOS. These tumors would, therefore, be subject to severe oxidative stress, thereby making them more susceptible to anticancer agents that are capable of inducing apoptosis in the presence of excess oxidative stress.
As mentioned previously, studies in larger cohorts of breast and other tumor types will be necessary to draw firm conclusions regarding the importance and function of mtDNA mutations. However, the work presented here, and the mutations that we catalog, may serve as a basis for further study regarding the role of acquired variants in the cancer mitochondrial genome, which has been under-studied as compared with the nuclear genome in recent high-profile reports.
Supplementary material
Supplementary Tables 1–5 and Figures 1–7 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute (R01 CA131341 to T.L.); American Cancer Society Research Scholar grant (ACS-RSG-12-159-01DMC to T.L.).
Supplementary Material
Acknowledgements
The authors wish to thank Mr M.Ruffalo for expert help with the figures.
Conflicts of Interest Statement: None declared.
Glossary
Abbreviations:
- ATP
adenosine triphosphate
- mtDNA
mitochondrial DNA
- NGS
next-generation sequencing
- OXPHOS
oxidative phosphorylation
- ROS
reactive oxygen species
- TCGA
The Cancer Genome Atlas
- tRNA
transfer RNA.
References
- 1. Malik A.N., et al. (2013). Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion, 13, 481–492 [DOI] [PubMed] [Google Scholar]
- 2. Chinnery P.F., et al. (2000). The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends Genet., 16, 500–505 [DOI] [PubMed] [Google Scholar]
- 3. Gasparre G., et al. (2008). Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum. Mol. Genet., 17, 986–995 [DOI] [PubMed] [Google Scholar]
- 4. Li H., et al. (2012). Physiology and pathophysiology of mitochondrial DNA. Adv. Exp. Med. Biol., 942, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Warburg O., et al. (1927). The metabolism of tumors in the body. J. Gen. Physiol., 8, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fendt L., et al. (2011). Accumulation of mutations over the entire mitochondrial genome of breast cancer cells obtained by tissue microdissection. Breast Cancer Res. Treat., 128, 327–336 [DOI] [PubMed] [Google Scholar]
- 7. Polyak K., et al. (1998). Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat. Genet., 20, 291–293 [DOI] [PubMed] [Google Scholar]
- 8. Tan D.J., et al. (2006). Significance of somatic mutations and content alteration of mitochondrial DNA in esophageal cancer. BMC Cancer, 6, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones J.B., et al. (2001). Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass”-ive advantage over detection of nuclear DNA mutations. Cancer Res., 61, 1299–1304 [PubMed] [Google Scholar]
- 10. Kloss-Brandstätter A., et al. (2010). Somatic mutations throughout the entire mitochondrial genome are associated with elevated PSA levels in prostate cancer patients. Am. J. Hum. Genet., 87, 802–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhidkov I., et al. (2009). MtDNA mutation pattern in tumors and human evolution are shaped by similar selective constraints. Genome Res., 19, 576–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carew J.S., et al. (2002). Mitochondrial defects in cancer. Mol. Cancer, 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lenaz G. (2012). Mitochondria and reactive oxygen species. Which role in physiology and pathology? Adv. Exp. Med. Biol., 942, 93–136 [DOI] [PubMed] [Google Scholar]
- 14. Gogvadze V., et al. (2009). Mitochondria as targets for cancer chemotherapy. Semin. Cancer Biol., 19, 57–66 [DOI] [PubMed] [Google Scholar]
- 15. Trachootham D., et al. (2009). Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov., 8, 579–591 [DOI] [PubMed] [Google Scholar]
- 16. Zhidkov I., et al. (2001). High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat. Genet., 2, 147–150 [DOI] [PubMed] [Google Scholar]
- 17. Larman T.C., et al. (2012). Spectrum of somatic mitochondrial mutations in five cancers. Proc. Natl Acad. Sci. USA, 109, 14087–14091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siegel R., et al. (2012). Cancer statistics, 2012. CA. Cancer J. Clin., 62, 10–29 [DOI] [PubMed] [Google Scholar]
- 19. Ellsworth R.E., et al. (2010). Breast cancer in the personal genomics era. Curr. Genomics, 11, 146–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. The Cancer Genome Atlas Research Network. (2012). Comprehensive molecular portraits of human breast tumours. Nature, 490, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yost S.E., et al. (2012). Identification of high-confidence somatic mutations in whole genome sequence of formalin-fixed breast cancer specimens. Nucleic Acids Res., 40, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu J., et al. (2009). Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res., 19, 802–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo Y., et al. (2013). MitoSeek: extracting mitochondria information and performing high-throughput mitochondria sequencing analysis. Bioinformatics, 29, 1210–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Picardi E., et al. (2012). Mitochondrial genomes gleaned from human whole-exome sequencing. Nat. Methods, 9, 523–524 [DOI] [PubMed] [Google Scholar]
- 25. Li H., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andrews R.M., et al. (1999). Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet., 23, 147. [DOI] [PubMed] [Google Scholar]
- 27. Langmead B., et al. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DePristo M.A., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson J.T., et al. (2011). Integrative genomics viewer. Nat. Biotechnol., 29, 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cingolani P., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin), 6, 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ruiz-Pesini E., et al. (2007). An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res., 35, D823–D828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pütz J., et al. (2007). Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA, 13, 1184–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schattner P., et al. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res., 33, W686–W689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Charif D., et al. (2007). SeqinR 1.0–2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. Structural approaches to sequence evolution: molecules, networks, populations. In Bastolla U., et al. (ed.) Biological and Medical Physics, Biomedical Engineering. Springer-Verlag, New York, pp. 207–232 [Google Scholar]
- 35. Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 36. R Development Core Team. (2011). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 37. Payne B.A., et al. (2013). Universal heteroplasmy of human mitochondrial DNA. Hum. Mol. Genet., 22, 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen L., et al. (2011). Evaluating mitochondrial DNA in patients with breast cancer and benign breast disease. J. Cancer Res. Clin. Oncol., 137, 669–675 [DOI] [PubMed] [Google Scholar]
- 39. Stoneking M. (2000). Hypervariable sites in the mtDNA control region are mutational hotspots. Am. J. Hum. Genet., 67, 1029–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y., et al. (2005). The increase of mitochondrial DNA content in endometrial adenocarcinoma cells: a quantitative study using laser-captured microdissected tissues. Gynecol. Oncol., 98, 104–110 [DOI] [PubMed] [Google Scholar]
- 41. Fan A.X., et al. (2009). Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. J. Cancer Res. Clin. Oncol., 135, 983–989 [DOI] [PubMed] [Google Scholar]
- 42. Abyzov A., et al. (2011). CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res., 21, 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chiang D.Y., et al. (2009). High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods, 6, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Veltri K.L., et al. (1990). Distinct genomic copy number in mitochondria of different mammalian organs. J. Cell. Physiol., 143, 160–164 [DOI] [PubMed] [Google Scholar]
- 45. Schmeing T.M., et al. (2011). How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat. Struct. Mol. Biol., 18, 432–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iommarini L., et al. (2013). Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment. Hum. Mol. Genet., in press. [DOI] [PubMed] [Google Scholar]
- 47. Mayr J.A., et al. (2008). Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin. Cancer Res., 14, 2270–2275 [DOI] [PubMed] [Google Scholar]
- 48. Alexandrov L.B., et al. (2013). Signatures of mutational processes in human cancer. Nature, 500, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lawrence M.S., et al. (2013). Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature, 499, 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Damm F., et al. (2012). Prognostic implications and molecular associations of NADH dehydrogenase subunit 4 (ND4) mutations in acute myeloid leukemia. Leukemia, 26, 289–295 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.