Summary
Srx induced by tumor promoter reduces hyperoxidized Prxs, increases cells’ ability to survive through oxidative stress by inhibition of TPA-induced apoptosis and facilitates skin tumor development.
Abstract
Sulfiredoxin (Srx), the exclusive enzyme that reduces the hyperoxidized inactive form of peroxiredoxins (Prxs), has been found highly expressed in several types of human skin cancer. To determine whether Srx contributed to skin tumorigenesis in vivo, Srx null mice were generated on an FVB background. Mouse skin tumorigenesis was induced by a 7,12-dimethylbenz[α]anthracene/12-O-tetradecanoylphorbol-13-acetate (DMBA/TPA) protocol. We found that the number, volume and size of papillomas in Srx−/− mice were significantly fewer compared with either wild-type (Wt) or heterozygous (Het) siblings. Histopathological analysis revealed more apoptotic cells in tumors from Srx−/− mice. Mechanistic studies in cell culture revealed that Srx was stimulated by TPA in a redox-independent manner. This effect was mediated transcriptionally through the activation of mitogen-activated protein kinase and Jun-N-terminal kinase. We also demonstrated that Srx was capable of reducing hyperoxidized Prxs to facilitate cell survival under oxidative stress conditions. These findings suggested that loss of Srx protected mice, at least partially, from DMBA/TPA-induced skin tumorigenesis. Therefore, Srx has an oncogenic role in skin tumorigenesis and targeting Srx may provide novel strategies for skin cancer prevention or treatment.
Introduction
Sulfiredoxin (Srx), also known as neoplastic progression 3, has been demonstrated as the exclusive enzyme that catalyzes the ATP-dependent reduction of the hyperoxidized form of peroxiredoxins (Prxs) (1,2). Prxs are the major cellular antioxidants that scavenge peroxides and also mediate hydrogen peroxide-induced intracellular signaling. In the peroxidase reaction, the cysteine residue (Cys-SH) of Prxs is oxidized to cysteine-sulfenic acid (Cys-SOH) and then forms a disulfide bond with another Prx. The disulfide bond is subsequently reduced by thioredoxin and completing a catalytic cycle (3,4). Under oxidative stress conditions, however, the Cys-SOH intermediate of Prxs is further oxidized to cysteine-sulfinic acid (Cys-SO2/3H), leading to the loss of peroxidase function and facilitating the formation of oligomer that bears protein chaperone activity (5–7). In yeast and other eukaryotes, oxidative stress stimulates expression of Srx, whose primary biochemical function is to reduce the hyperoxidized Prxs to its active form, and this process involves ATP, magnesium and other cofactors (1,6). Srx has also been reported to catalyze the deglutathionylation of protein phosphatase and Prx II (8,9).
Compared with its well-documented redox function, the biological significance of Srx in development and disease, such as cancer development, was not fully explored. The first indication of a potential role of Srx in cancer was emerged from the discovery that Srx transcript was one of the three messengers that were differentially expressed in the transformation-sensitive mouse skin epithelial cells (10). Our subsequent study further demonstrated that Srx was highly expressed in human skin squamous cell carcinoma, sweat gland carcinoma, basal cell carcinoma and melanoma, whereas it was weakly expressed or absent in normal skin or patients with skin non-malignant tumors (11). Skin cancer is the most common cancer with millions of new patients diagnosed each year worldwide. Chemically induced skin tumorigenesis in mice provides an important experimental strategy to study the molecular basis of multiple stage carcinogenesis and has contributed to the discovery and validation of numerous agents for cancer prevention and treatment. Whether Srx is required for skin tumorigenesis in vivo has not been investigated. In this study, we have used Srx null mice and a well-characterized 7,12-dimethylbenz[α]anthracene (DMBA)- and 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced carcinogenesis model to explore the functional significance of Srx in skin cancer development in vivo. Our data suggest that loss of Srx protects mice from DMBA/TPA-induced skin tumorigenesis.
Materials and methods
Srx null mice and genotyping
Mouse breeding and animal protocols were performed according to the guidelines of the Animal Care and Use Committee (ACUC) of the National Cancer Institute at Frederick. All mice were exposed to a 12:12 h light/dark cycle, fed with an AIN-96G purified diet (Harlan Teklad, Madison, WI) and supplied with drinking water ad libitum. Srx nulls on an FVB background were generated using Srx−/− B6/129 mice backcrossed onto an FVB strain. After seven generations, the offspring of Srx+/− sibling breeding was used for experiments. For mouse genotyping, the genomic DNA was extracted from tail clip using genomic DNA extraction kit (Qiagen, Valencia, CA). PCR-based genotyping was performed as previously reported (12).
DMBA/TPA protocol and tumor measurement
A randomized, double-blind experimental design was applied to eliminate potential subjective bias (13). Briefly, mice at 7 weeks of age, including wild-type (Wt, n = 17), heterozygous (Het, n = 24) and nulls (n = 14), were used in the study. The shaved dorsal skin was painted with 400 nmol of DMBA (Sigma–Aldrich, St Louis, MO) in 200 µl of acetone. Ten days later, mice were treated with 10 nmol of TPA (Sigma–Aldrich) in 200 µl of acetone twice weekly for a period of 20 weeks. Skin papillomas/carcinomas were examined following previously established guidelines (13,14). Briefly, a skin lesion was recorded as a papilloma when it reached a diameter >1mm and was present for 2 consecutive weeks. Suspected carcinomas characterized by cratering, ulceration, invasion and downward growing were verified histologically by standard pathological criteria (15). Every other day, the number of tumors was counted and the length of perpendicular axes of tumors was measured using a caliper. Tumor volume was calculated as previously reported (16). Mouse tissues were fixed in 4% paraformaldehyde and stored in 70% ethanol before being processed with standard paraffin embedding, sectioning, hematoxylin and eosin (H&E) or immunohistochemistry (IHC) staining.
IHC staining and in situ apoptosis assay
IHC with hematoxylin counterstaining was performed using Dako LSAB2-HRP Kit (Dako, Carpinteria, CA). Antibodies used included anti-Srx (1:100; Proteintech, Chicago, IL) and anti-Ki67 (1: 50; Dako). To analyze apoptosis, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay was performed using TACS 2TdT-DAB In Situ Apoptosis Detection Kit (Trevigen, Gaithersburg, MD). Samples were counter stained with methyl green, dehydrated and mounted before microscopic visualization following the manufacturer suggested protocol. Images of IHC staining were taken using a digital slide scanner (Aperio ScanScope XT System; Aperio Technologies, Vista, CA). Samples were analyzed by quantification of staining intensity using the ImageScope software. Additionally, samples were also analyzed by calculating the proliferation index or apoptotic index, which was defined as the percentage of proliferating or apoptotic cells, respectively, in each sample (the number of positive nuclei divided by the total number of cells in each microscopic field) (17). For every slide, data from five representative fields were collected.
Cell culture and western blot
Mouse skin epithelial JB6 (RT101) cells were maintained in minimum essential medium with l-glutamine (2mM), penicillin (100 units/ml)/streptomycin (100 µg/ml), gentamicin (5 µg/ml) and 5% fetal bovine serum. Lentiviral particles encoding either a ShRNA sequence targeting the coding region of mouse Srx (ShSrx) or a ShRNA containing a non-target sequence (ShNT) were generated using the MISSION® ShRNA lentiviral construct and packaging mix (Sigma–Aldrich) as previously established (11). Cells were infected with lentiviral particles and stable selection was accomplished using 1 µg/ml of puromycin. All chemicals and inhibitors, including actinomycin D (AMD), N-acetyl cysteine (NAC), PD98059 (MEK1/2 inhibitor), SP600125 [Jun-N-terminal kinase (JNK)1/2 inhibitor], GO6983 (pan-PKC inhibitor) and Wortmannin (PI3K inhibitor), were commercially obtained (Sigma–Aldrich). For TPA and/or AMD treatment, cells were plated at the same time and chemicals (dimethyl sulfoxide for control) were added at different time. Cells were then harvested at the same time after indicated time of treatment. For specific kinase inhibition, cells were preincubated with a chemical inhibitor for 1 h and then treated with TPA or dimethyl sulfoxide (control) for another 6 h before being harvested. For western blot, cells were lysed in radioimmunoprecipitation assay buffer containing protease inhibitors (Santa Cruz Biotech, Santa Cruz, CA). Protein was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blotting was performed following standard protocol. The primary antibodies used included anti-Srx (Proteintech), Prx I (Abcam), Prx II and Prx III (Santa Cruz), Prx IV (Abcam), caspase 3 (Santa Cruz), phospho-C-Jun (Cell Signaling), total C-Jun (Santa Cruz), phospho-p44/p42 and total p44/p42 (Cell Signaling) and β-actin (Sigma–Aldrich).
Quantitative real-time PCR
Total cellular RNA was extracted using RNeasy Kit (Qiagen) and 300ng of total RNA was used to synthesize the first strand cDNA using random hexamers. The quantitative real-time PCR was performed using SYBR Green Master Mix (Roche Applied Science) with following primers: mSRX forward 5′- AAAGTGCAGAGCCTGGTGG-3′, reverse 5′- CTTGGCAGGAATGGTCTCTC-3′; mPrx1 forward 5′- ACCTCTTCCTGCG TTCTCAC-3′, reverse 5′-TGTCCATCTGGCATAACAGC-3′; mPrx2 forward 5′- GCTGAGGACTTCCGAAAGCT-3′, reverse 5′- AGTCACGTCAGCAAG CAGAG-3′; mPrx3 forward 5′- CATCTTGCCTGGATCAACAC-3′, reverse 5′- CTTTCCAACAGCACTCCGTA-3′; mPrx4 forward 5′- ATCA GTGGACGAGACACTGC-3′, reverse 5′- CCAGCTGGATCTGGGATTAT-3′; mGAPDH forward 5′- ACAACTTTGGCATTGTGGAA-3′, reverse 5′- GATGCAGGGATGATGTTCTG-3′. The quantitative results from eight replicates were normalized by level of GAPDH mRNA.
Measurement of reactive oxygen species
Cellular reactive oxygen species (ROS) levels were examined using 2′,7′-dichlorofluorescein diacetate (DCFDA) as previously described (18). Briefly, cells cultured in 96-well plates were treated with 5mM of NAC, 10nM of TPA alone or in combination for 5 h. Each treatment was repeated in eight wells. Cells were then stained for 30min in the presence of 25 µM of DCFDA in Krebs-Ringer solution (119mM NaCl, 2.5mM KCl, 1mM NaH2PO4, 2.5mM CaCl2, 1.3mM MgCl2, 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4). After washing with phosphate-buffered saline, absorption was measured using a fluorescence spectrometer at the wavelength of 488/528 nanometer (Ex/Em). To visualize the intracellular ROS, cells were cultured in chamber slide and stained with H2DCFDA and examined by fluorescence microscope.
Statistical analysis
Quantitative data were presented as means ± SD (± SD). Data were analyzed with the indicated statistical method using GraphPad Prism (Version 5.04) or Microsoft Excel (Version 2010). For calculation of the P value, parameters of two-tailed, 95% confidence interval were used for all analyses. A P value of <0.05 was considered statistically significant.
Results
Srx null mice were resistant to DMBA/TPA-induced skin tumorigenesis
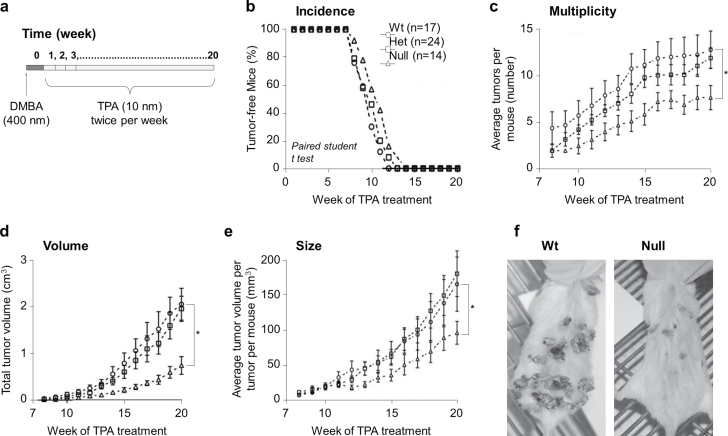
Srx null mice, either on B6/129 or FVB background, were completely normal under laboratory conditions. To study the role of Srx in skin tumorigenesis in vivo, a well-established DMBA/TPA-induced mouse skin tumorigenesis protocol was applied (Figure 1a). Mouse skin papilloma typically began to be visible at the 7th week of TPA application, and all mice developed skin papillomas by the 14th week (Figure 1b). This process of papilloma development was consistent with previously reported features of the DMBA/TPA skin tumorigenesis model on the FVB strain (13,14). We found no significant difference in the overall incidence of skin tumors (including papilloma and carcinoma) between groups of Wt (+/+), Het (−/+) or nulls (−/−) (Figure 1b). If we only examined the incidence of invasive carcinomas, there were also no significant difference (two cases each in Wt and null and three cases in Het). However, the overall tumor multiplicity (as reflected by the average tumor number per mouse) and the average tumor volume per mouse in the Srx−/ − group were significantly fewer compared with either the −/+ or +/+ group (both P < 0.05) (Figure 1c and 1d). The size of each tumor (as reflected by the average tumor volume per tumor per mouse) in the Srx−/− group was also significantly smaller than either Srx(−/+) or (+/+) group (Figure 1e). In all of these measurements, we found no statistically significant differences between the Srx(−/+) and (+/+) group (P > 0.05 in these comparisons). Representative images of skin tumors from Srx+/+ and Srx−/− mouse were shown (Figure 1f). Our data indicated that Srx null mice had a significant reduction in tumor multiplicity, overall tumor volume and individual tumor size compared with their Wt or Het counterparts. Therefore, these data suggested that genomic depletion of Srx rendered mice resistant, at least partially, to DMBA/TPA-induced skin tumorigenesis.
Fig. 1.
Srx null mice were resistant to DMBA/TPA-induced skin tumorigenesis. (a) Schematic presentation of the DMBA/TPA protocol. (b–e) The average rate/value of Srx Wt, Het and null mice in skin tumor incidence (b), multiplicity (c), volume (d) and size (e). Tumors were macroscopically examined and tumor size was measured with a caliper. Data were presented as means ± SD ( ± SD). Statistical methods used were paired Student’s t-test (b) and Fisher’s exact test (c–e). *P < 0.05. (f) Representative gross images of skin tumors in Wt or Srx null mice.
DMBA/TPA-induced Srx expression in mouse skin epithelium and tumors
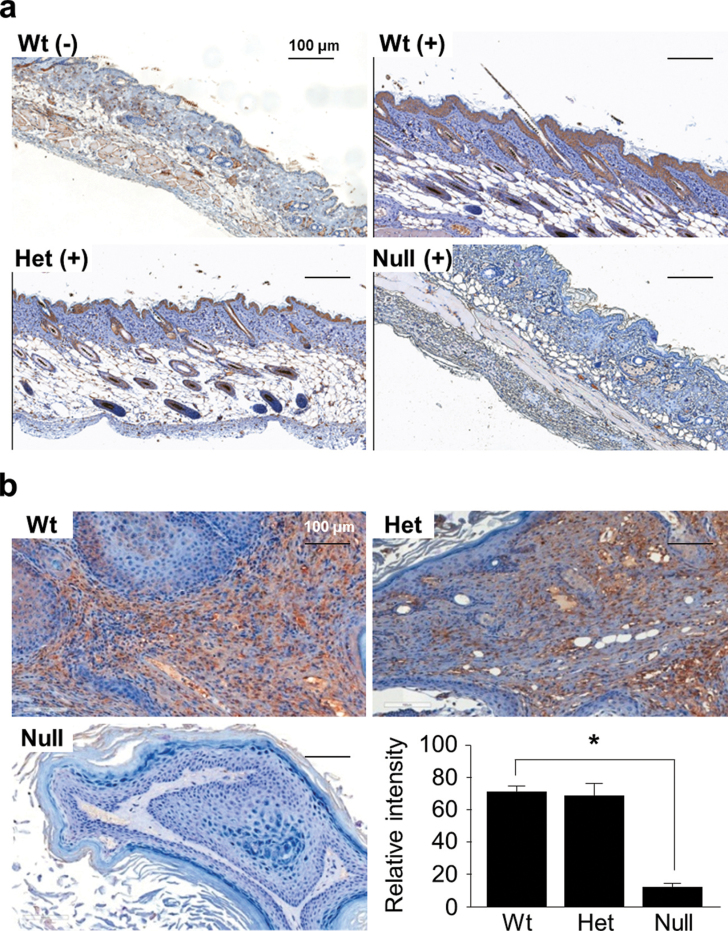
To investigate why depletion of Srx in mice led to reduction of skin tumorigenesis, we examined the effect of DMBA/TPA treatment on the expression of Srx in mouse skin and tumors. An IHC method that specifically detected Srx in formaldehyde-fixed tissue was applied as previously reported (17). Without DMBA/TPA treatment, Srx was barely detectable in normal mouse skin epidermis, except that a few positive cells were identified in the basal cell layer and hair follicles (Figure 2a). After 20 weeks of DMBA/TPA treatment, the thickened epidermis with features of hyperplasia was obvious in tumor-adjacent normal skin, regardless of mouse genotypes (Figure 2a). The vast majority of those proliferated cells in the epidermis and hair follicles of Srx(+/+) or (−/+) mice were Srx-staining positive (Figure 2a). We also examined the expression of Srx in DMBA/TPA-induced skin tumors and found that Srx was expressed in tumor cells as well as stroma cells (Figure 2b). Lack of positive staining in the epithelium and tumors of Srx knockout mice further validated the specificity of the anti-Srx staining (Figure 2a and b). The intensities of Srx staining in tumors from Srx(+/+) and (−/+) mice were quantitated, and there were no statistical differences (Figure 2b). These data suggested that application of DMBA/TPA led to increased expression of Srx in mouse skin and tumors.
Fig. 2.
Expression of Srx in mouse skin and tumors. (a) Expression of Srx in normal mouse skin (−) or DMBA/TPA-treated, hyperproliferated skin epithelium (+). (b) Expression of Srx in DMBA/TPA-induced skin tumors from Wt, Het or null mice. Bar = 100 µm. In the bar graph, data were presented as means ± SD ( ± SD) (n = 6). *P < 0.05 (t-test).
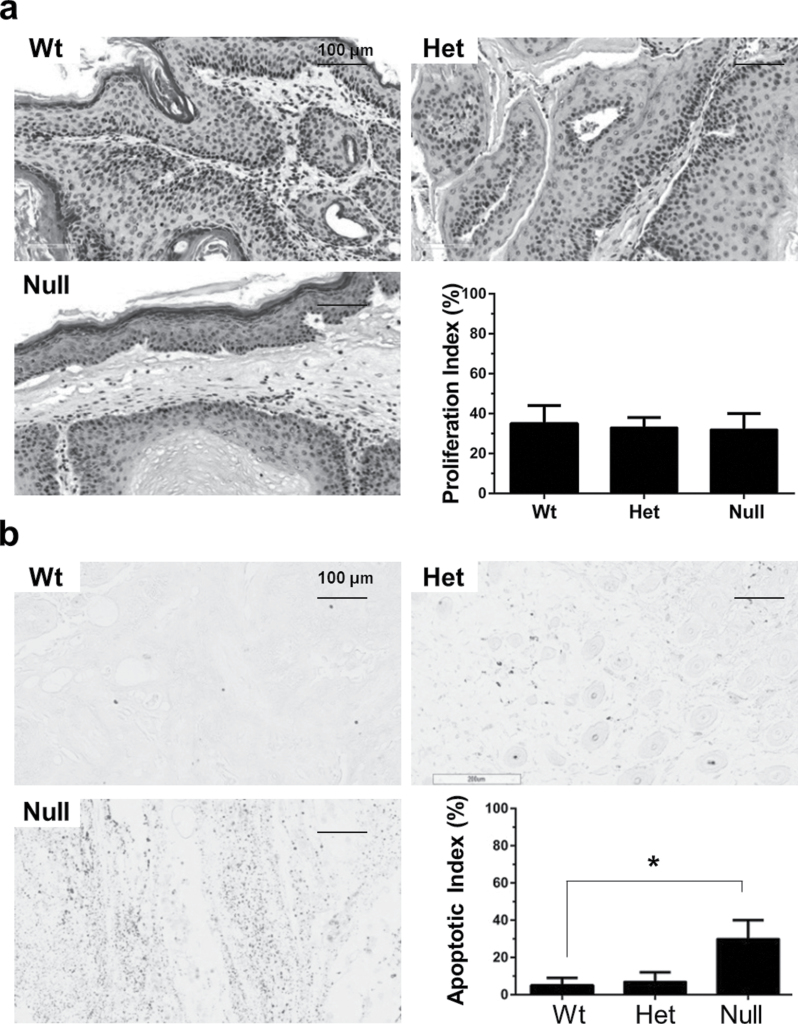
Depletion of Srx did not affect the rate of cell proliferation but increased intratumoral apoptosis
To investigate whether the reduction of tumor multiplicity and volume in Srx−/− mice was resulted from alterations in cell proliferation and/or apoptosis, mouse skin tumors were stained for Ki67, an intracellular marker for cell proliferation. The proliferation index as calculated by the percentage of Ki67+ cells in tumors from either Srx(+/+), (−/+) or (−/−) mice were comparable (Figure 3a). These data may suggest that depletion of Srx in vivo did not affect the rate of cell proliferation, which was consistent with our previous findings indicating that depletion of Srx in mouse skin epithelial cells had no effect on cell cycle progression and proliferation in cell culture (11). Therefore, the differences in tumor multiplicity and volume between Srx null and Wt mice may not be resulted from changes in cell proliferation. Next, we examined the levels of intratumoral apoptosis in these tumors using TUNEL staining. A significantly increased staining of TUNEL+ cells was demonstrated in tumors from Srx−/− compared with those from either +/+ or −/+ mice either by the quantification of staining intensity or calculation of apoptotic index (Figure 3b). These data suggested that loss of Srx was associated with increased intratumoral apoptosis, which may contribute to the reduced tumor multiplicity and volume observed in Srx−/− mice.
Fig. 3.
Tumors from Srx null mice had comparable levels of cell proliferation but increased rate of intratumoral apoptosis. (a) Similar levels of cell proliferation were found in tumors from Wt, Het and null mice by staining of nuclei Ki67. (b) Increased rate of apoptosis was found in tumors from Srx null mice by TUNEL assay. Bar = 100 µm. In the bar graph, data were presented as means ± SD ( ± SD) (n = 6). *P < 0.05 (t-test).
TPA stimulated Srx protein expression through transcriptional activation
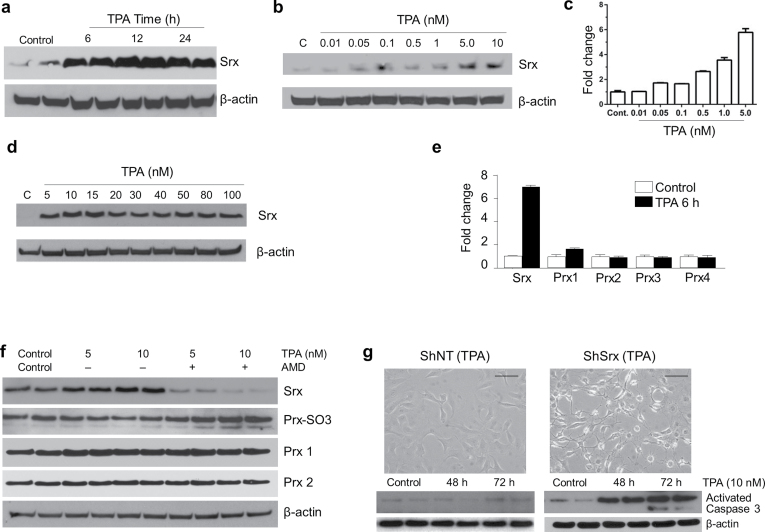
To understand the molecular basis of Srx expression in DMBA/TPA-induced skin tumorigenesis, we asked whether the expression of Srx found in mice was directly caused by TPA treatment. Mouse skin epithelial JB6 cells were treated with TPA in culture and then harvested for western blot. As early as 6 h after TPA treatment at a dosage of 10nM, the levels of Srx protein expression were dramatically increased in these cells (Figure 4a). Within this time frame, a dose-dependent increased expression of Srx at the protein and transcript levels was observed (Figure 4b and c). Treatment of JB6 cells with higher concentrations of TPA (>10nM) did not cause further increase in Srx protein expression (Figure 4d). Furthermore, the effect of TPA on Srx was relatively specific in that 10nM of TPA treatment did not activate transcription of Prxs (Figure 4e). To further confirm that TPA-induced Srx protein expression occurred through activation of transcription, AMD was added to the medium to inhibit transcription. The effect of TPA to stimulate Srx expression was completely abolished in the presence of AMD (Figure 4d). Interestingly, the levels of hyperoxidized Prxs were accumulated when Srx was inhibited (Figure 4f). This observation was consistent with the primary function of Srx to reduce hyperoxidized Prxs as previously reported in yeast and mammalian cells (1,2). Taken together, these data suggested that TPA stimulated Srx protein expression through activation of transcription, and increased levels of Srx was required to reduce the hyperoxidized Prxs in mouse epithelial cells.
Fig. 4.
TPA directly stimulated Srx transcription and Srx-knockdown cells were more sensitive to apoptosis. (a) Western blot shown time-dependent induction of Srx protein by TPA in cultured JB6 cells. (b and c) TPA activated Srx expression dose dependently at the protein and the transcript levels as indicated by western blot (b) and quantitative real-time PCR (c), respectively. (d) Higher concentration of TPA (>10nM) did not further stimulate Srx expression. (e) 10nM of TPA stimulated Srx but not Prxs transcription as measured by quantitative real-time PCR. (f) TPA-induced Srx protein expression was abolished by AMD, and Srx reduced hyperoxidized Prxs. (g) Srx-knockdown cells were more sensitive to TPA-induced apoptosis. Microscopic images of cell in culture were taken after 72h of TPA treatment. Results from duplicated samples in each treatment were shown in (a), (f) and (g). Bar = 100 µm. Control cells were treated with equal amounts of dimethyl sulfoxide (solvent for TPA and AMD) for 24h (a), 6h (b–e), 6h (f) and 72h (g), respectively.
Previous study demonstrated that TPA also induced apoptosis of mouse skin epithelial cells (19). Next, we asked whether Srx had a role in TPA-induced apoptosis. With a lentivirus-based ShRNA knockdown strategy, we successfully depleted/inhibited endogenously expressed as well as TPA-induced Srx protein in JB6 cells as previously reported (11). We treated these cells with 10nM of TPA for 2–3 days and examined for signs of apoptosis. We found that Srx-knockdown cells showed obvious morphological signs of apoptosis and the levels of activated caspase 3 were significantly increased (Figure 4g); whereas control cells under same conditions were completely normal and the levels of activated caspase 3 were barely detectable (Figure 4g). These data suggested that depletion/inhibition of Srx may sensitize mouse skin epithelial cells to TPA-induced apoptosis, whereas enhanced expression of Srx may promote DMBA/TPA-induced skin tumorigenesis by antagonizing the apoptotic effect of TPA and thus facilitate tumor cell growth and survival.
TPA-induced Srx expression was independent of cellular redox status
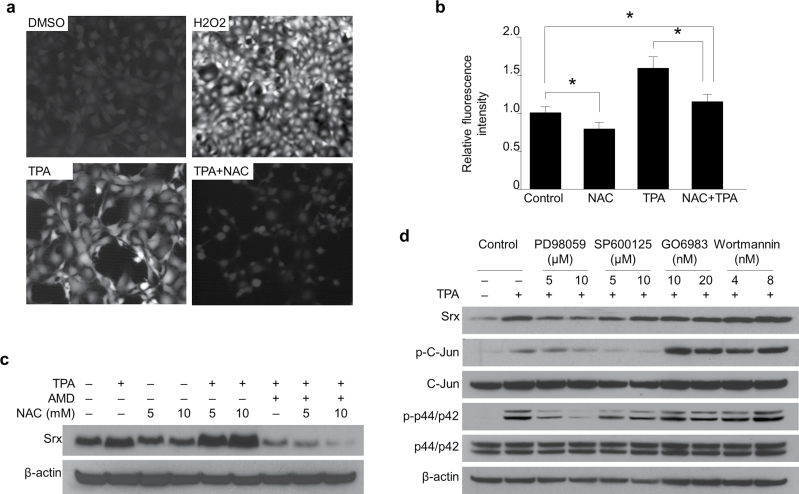
Oxidative stress has been identified as one of the major factors that stimulated Srx gene expression in yeast and mammalian cells through activation of the antioxidant response elements located in its promoter (1,20,21). TPA has been known to induce accumulation of ROS that function as essential mediators to stimulate expression of a number of genes (22,23). Therefore, we asked whether TPA-induced oxidative stress contributed to the expression of Srx in mouse skin epithelial cells. We used DCFDA and fluorescent microscope imaging to measure the levels of intracellular ROS. JB6 cells were treated with TPA in the presence or absence of an exogenous antioxidant, NAC. We found that TPA treatment led to the accumulation of ROS in JB6 cells, and addition of NAC successfully reduced the level of ROS in these cells (Figure 5a and b). In the presence of NAC, TPA was still fully capable of stimulating Srx expression in these cells (Figure 5c). These data suggested that TPA-induced Srx expression in mouse skin epithelial cells was most likely independent of cellular redox status.
Fig. 5.
TPA-induced Srx expression was independent of cellular redox status and was dependent on the activation of JNKs and MAPK signaling pathways. (a) TPA induced ROS accumulation in JB6 cells and this effect was abolished by treatment with NAC. The levels of ROS were measured by DCFDA-based fluorescence imaging. (b) Quantitative data from (a) were presented as means ± SD ( ± SD) (n = 8). *P < 0.05 (t-test). (c) NAC was not sufficient to reduce TPA-induced Srx expression. (d) TPA-induced Srx expression was abolished by inhibition of JNKs and MAPK signaling pathways.
TPA-induced Srx expression was dependent on the activation of mitogen-activated protein kinase and JNKs
To further understand the molecular mechanism of TPA-induced Srx expression, JB6 cells were treated with TPA alone or together with an inhibitor of mitogen-activated protein kinase (MAPK; PD98059), AP-1 (SP600125), PKC (GO6983) or PI3K (Wortmannin) signaling pathway. Coincident with Srx induction, TPA stimulated AP-1 and MAPK signaling as indicated by the induction of phosphorylation on c-Jun and Erk1/2 (p44/42) (Figure 5d). When the AP-1 or MAPK signaling was blocked by PD98059 or SP60015, TPA was unable to induce Srx expression; however, block of PKC or PI3K signaling by GO6983 and Wortmannin, respectively, had no effect on TPA-induced Srx expression in these cells (Figure 5d). These data suggested that TPA-induced Srx expression was activated, at least partially, through the activation of AP-1 and MAPK signaling pathways but was not likely through the activation of PKC or PI3K pathway.
Discussion
Srx is a critical enzyme whose primary biochemical function is to reduce the hyperoxidized Prxs in yeast, plants and mammalians (1,2,24). It also has alternative functions to reverse protein gluta- thionylation (8,9) and to hydrolyze single- or double-stranded nucleic acids (25). In addition to Srx, elevated expression of cellular antioxidants, such as glutaredoxin, Prx and thioredoxin, has been identified in a wide range of human cancers (26,27). The capability of Srx to support cell survival through oxidative stress, such as hydrogen peroxide, has been previously demonstrated in cell lines including embryonic fibroblasts (28), neurons (29,30) and mouse skin epithelial cells (11). To study the role of Srx in skin tumorigenesis, we used the DMBA/TPA model to mimic skin cancer development in humans. We established Srx nulls on an FVB background and demonstrated that depletion of Srx rendered mice resistant to DMBA/TPA-induced skin carcinogenesis. In mechanistic studies, we found that Srx was strongly expressed in DMBA/TPA-treated epidermis and induced tumors; we also demonstrated that depletion of Srx led to increased intratumoral apoptosis, which may contribute to tumor-resistant phenotype of Srx null mice.
Skin cancer is the most common type of cancer in the USA and worldwide. Identification of novel therapeutic target for skin cancer is of social and economic significance. Srx is found to be highly expressed in human skin malignant tumors including melanoma (11). Induction of Srx has also been confirmed in human melanoma cells treated with cinnamic aldehyde (31), liver cells treated with D3T (32) and lung epithelial cells exposed to cigarette smoke extracts (33). Expression of Srx is critical for the pathogenesis of a variety of human disease, in particular, cancer development. In lung cancer, Srx is indispensable for cancer cell proliferation, migration and invasion (34–37). In pancreatic adenocarcinoma, expression of Srx is associated with poor patient survival (38). In breast cancer, genetic polymorphism and expression of Nrf2 and Srx predict patient survival outcomes (39). All these studies and our current data clearly demonstrate that Srx plays a significant pathogenic role in human cancer development.
Although less studied, the cellular levels of Srx may be regulated through mechanisms of transcriptional and/or translational activation. For example, the transcription and translation of Srx in yeast are stimulated by oxidative stress and can be further enhanced by activation of the Ras-cAMP-PKA pathway (40). It has been demonstrated in many mammalian cells, such as neurons, pancreatic β cells and lung epithelial cells, that activation of AP-1 is one of the major transcription factors contributing to Srx expression (21,41,42). Additionally, activation of Nrf2 may also stimulate Srx expression (21,33). Given the oncogenic importance of AP-1 activation and Nrf2 signaling in cancer (43,44), it is not surprising that tumor promoter-induced Srx plays an oncogenic role in skin tumorigenesis. In this study, we demonstrate that TPA directly stimulates the transcription of Srx through a redox-independent mechanism, and we further demonstrate that activation of JNK and MAPK pathways are required for its expression.
Our findings of the tumor-resistant phenotype of Srx null mice may reflect an intrinsic, long-term accumulative effect of DMBA/TPA treatment. Despite their normal phenotype, Srx null mice appear to have a delayed/prolonged inflammatory response that is characterized by the upregulation of genes involved in adaptive and innate immunity under stress conditions such as severe endotoxic shock (12). It is still unknown how the defect in adaptive and/or innate immunity may affect the process of skin tumorigenesis. In addition, whether loss of Srx has any effect on the mutagenic consequence of DMBA remains to be investigated. Nevertheless, our findings suggest that Srx is one of the critical components that contribute to mouse skin tumorigenesis in vivo. Targeting Srx may thus be used as a novel strategy for skin cancer prevention and treatment in the future.
Funding
National Cancer Institute, National Institutes of Health (R00CA149144 to Q.W.).
Acknowledgements
We thank Ms Jen Wise, Ms Lisa Dodge and Mr Dan Logsdon (Frederick National Laboratory for Cancer Research) for mouse maintenance. We also thank Ms Donna A.Gilbreath (Markey Cancer Center, University of Kentucky) for professional language editing.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- AMD
actinomycin D
- DCFDA
2′,7′-dichlorofluorescein diacetate
- DMBA
7,12-dimethylbenz[α]anthracene
- Het
heterozygous
- IHC
immunohistochemistry
- JNK
Jun-N-terminal kinase
- MAPK
mitogen-activated protein kinase
- NAC
N-acetyl cysteine
- Prxs
peroxiredoxins
- ROS
reactive oxygen species
- Srx
sulfiredoxin
- TPA
12-O-tetradecanoylphorbol-13-acetate
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- Wt
wild-type.
References
- 1. Biteau B., et al. (2003). ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature, 425, 980–984 [DOI] [PubMed] [Google Scholar]
- 2. Chang T.S., et al. (2004). Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem., 279, 50994–51001 [DOI] [PubMed] [Google Scholar]
- 3. Fourquet S., et al. (2008). The dual functions of thiol-based peroxidases in H2O2 scavenging and signaling. Antioxid. Redox Signal., 10, 1565–1576 [DOI] [PubMed] [Google Scholar]
- 4. Rhee S.G., et al. (2005). Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr. Opin. Cell Biol., 17, 183–189 [DOI] [PubMed] [Google Scholar]
- 5. Jacob C., et al. (2004). The sulfinic acid switch in proteins. Org. Biomol. Chem., 2, 1953–1956 [DOI] [PubMed] [Google Scholar]
- 6. Lowther W.T., et al. (2011). Reduction of cysteine sulfinic acid in eukaryotic, typical 2-Cys peroxiredoxins by sulfiredoxin. Antioxid. Redox Signal., 15, 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jang H.H., et al. (2004). Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell, 117, 625–635 [DOI] [PubMed] [Google Scholar]
- 8. Findlay V.J., et al. (2006). A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer Res., 66, 6800–6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park J.W., et al. (2009). Deglutathionylation of 2-Cys peroxiredoxin is specifically catalyzed by sulfiredoxin. J. Biol. Chem., 284, 23364–23374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun Y., et al. (1994). Molecular cloning of five messenger RNAs differentially expressed in preneoplastic or neoplastic JB6 mouse epidermal cells: one is homologous to human tissue inhibitor of metalloproteinases-3. Cancer Res., 54, 1139–1144 [PubMed] [Google Scholar]
- 11. Wei Q., et al. (2008). Sulfiredoxin is an AP-1 target gene that is required for transformation and shows elevated expression in human skin malignancies. Proc. Natl Acad. Sci. USA, 105, 19738–19743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Planson A.G., et al. (2011). Sulfiredoxin protects mice from lipopolysaccharide-induced endotoxic shock. Antioxid. Redox Signal., 14, 2071–2080 [DOI] [PubMed] [Google Scholar]
- 13. Abel E.L., et al. (2009). Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat. Protoc., 4, 1350–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hennings H., et al. (1993). FVB/N mice: an inbred strain sensitive to the chemical induction of squamous cell carcinomas in the skin. Carcinogenesis, 14, 2353–2358 [DOI] [PubMed] [Google Scholar]
- 15. Knutsen G.L., et al. (1986). Gross and microscopic lesions in the female SENCAR mouse skin and lung in tumor initiation and promotion studies. Environ. Health Perspect., 68, 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Euhus D.M., et al. (1986). Tumor measurement in the nude mouse. J. Surg. Oncol., 31, 229–234 [DOI] [PubMed] [Google Scholar]
- 17. Henery S., et al. (2008). Quantitative image based apoptotic index measurement using multispectral imaging flow cytometry: a comparison with standard photometric methods. Apoptosis, 13, 1054–1063 [DOI] [PubMed] [Google Scholar]
- 18. Wardman P. (2007). Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free Radic. Biol. Med., 43, 995–1022 [DOI] [PubMed] [Google Scholar]
- 19. Wang F., et al. (2010). Mitochondrial uncoupling inhibits p53 mitochondrial translocation in TPA-challenged skin epidermal JB6 cells. PLoS One, 5, e13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calvo I.A., et al. (2012). The transcription factors Pap1 and Prr1 collaborate to activate antioxidant, but not drug tolerance, genes in response to H2O2 . Nucleic Acids Res., 40, 4816–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soriano F.X., et al. (2009). Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol. Cells, 27, 279–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parfett C., et al. (1993). Oxidative stress mediates tumor promoter-induced proliferin gene-expression in c3h10t1/2 cells. Int. J. Oncol., 3, 917–925 [DOI] [PubMed] [Google Scholar]
- 23. Durán H.A., et al. (1991). Differential oxidative stress induced by two different types of skin tumor promoters, benzoyl peroxide and 12-O-tetradecanoylphorbol-13-acetate. Carcinogenesis, 12, 2047–2052 [DOI] [PubMed] [Google Scholar]
- 24. Rey P., et al. (2007). The Arabidopsis thaliana sulfiredoxin is a plastidic cysteine-sulfinic acid reductase involved in the photooxidative stress response. Plant J., 49, 505–514 [DOI] [PubMed] [Google Scholar]
- 25. Chi Y.H., et al. (2012). Dual functions of Arabidopsis sulfiredoxin: acting as a redox-dependent sulfinic acid reductase and as a redox-independent nuclease enzyme. FEBS Lett., 586, 3493–3499 [DOI] [PubMed] [Google Scholar]
- 26. Kang S.W., et al. (2005). 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol. Med., 11, 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arnér E.S., et al. (2006). The thioredoxin system in cancer. Semin. Cancer Biol., 16, 420–426 [DOI] [PubMed] [Google Scholar]
- 28. Baek J.Y., et al. (2012). Sulfiredoxin protein is critical for redox balance and survival of cells exposed to low steady-state levels of H2O2 . J. Biol. Chem., 287, 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu C.L., et al. (2012). c-Jun-dependent sulfiredoxin induction mediates BDNF protection against mitochondrial inhibition in rat cortical neurons. Neurobiol. Dis., 46, 450–462 [DOI] [PubMed] [Google Scholar]
- 30. Nagashima S., et al. (2011). CRMP5-associated GTPase (CRAG) protein protects neuronal cells against cytotoxicity of expanded polyglutamine protein partially via c-Fos-dependent activator protein-1 activation. J. Biol. Chem., 286, 33879–33889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cabello C.M., et al. (2009). The cinnamon-derived Michael acceptor cinnamic aldehyde impairs melanoma cell proliferation, invasiveness, and tumor growth. Free Radic. Biol. Med., 46, 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kwak M.K., et al. (2003). Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem., 278, 8135–8145 [DOI] [PubMed] [Google Scholar]
- 33. Singh A., et al. (2009). Nrf2-dependent sulfiredoxin-1 expression protects against cigarette smoke-induced oxidative stress in lungs. Free Radic. Biol. Med., 46, 376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lei K., et al. (2008). Protein cysteine sulfinic acid reductase (sulfiredoxin) as a regulator of cell proliferation and drug response. Oncogene, 27, 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei Q., et al. (2011). Sulfiredoxin-Peroxiredoxin IV axis promotes human lung cancer progression through modulation of specific phosphokinase signaling. Proc. Natl Acad. Sci. USA, 108, 7004–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bowers R.R., et al. (2012). Sulfiredoxin redox-sensitive interaction with S100A4 and non-muscle myosin IIA regulates cancer cell motility. Biochemistry, 51, 7740–7754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim Y.S., et al. (2011). Nuclear factor E2-related factor 2 dependent overexpression of sulfiredoxin and peroxiredoxin III in human lung cancer. Korean J. Intern. Med., 26, 304–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soini Y., et al. (2014). Nuclear Nrf2 expression is related to a poor survival in pancreatic adenocarcinoma. Pathol. Res. Pract., 210, 35–39 [DOI] [PubMed] [Google Scholar]
- 39. Hartikainen J.M., et al. (2012). Genetic polymorphisms and protein expression of NRF2 and Sulfiredoxin predict survival outcomes in breast cancer. Cancer Res., 72, 5537–5546 [DOI] [PubMed] [Google Scholar]
- 40. Molin M., et al. (2011). Life span extension and H(2)O(2) resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae . Mol. Cell, 43, 823–833 [DOI] [PubMed] [Google Scholar]
- 41. Papadia S., et al. (2008). Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nat. Neurosci., 11, 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Glauser D.A., et al. (2007). Transcriptional response of pancreatic beta cells to metabolic stimulation: large scale identification of immediate-early and secondary response genes. BMC Mol. Biol., 8, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lau A., et al. (2008). Dual roles of Nrf2 in cancer. Pharmacol. Res., 58, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Colburn N.H., et al. (2008). Targeting transcription factors for cancer prevention–the case of Nrf2. Cancer Prev. Res. (Phila)., 1, 153–155 [DOI] [PubMed] [Google Scholar]