Abstract
For small helical membrane proteins, their structures are highly sensitive to their environment, and solid state NMR is a structural technique that can characterize these membrane proteins in native-like lipid bilayers and proteoliposomes. To date, a systematic method by which to evaluate the effect of the solubilizing detergent on proteoliposome preparations for solid state NMR of membrane proteins has not been presented in the literature. A set of experiments are presented aimed at determining the conditions most amenable to dialysis mediated reconstitution sample preparation. A membrane protein from M. tuberculosis is used to illustrate the method. The results show that a detergent that stabilizes the most protein is not always ideal and sometimes cannot be removed by dialysis. By focusing on the lipid and protein binding properties of the detergent, proteoliposome preparations can be readily produced, which provide double the signal-to-noise ratios for both the oriented sample and magic angle spinning solid state NMR. The method will allow more membrane protein drug targets to be structurally characterized in lipid bilayer environments.
Membrane protein isolation and purification requires solubilization of the proteins in detergent micelles. Structure determination by solid state NMR in a native-like lipid bilayer requires the removal of these detergents during reconstitution into liposomes. The membrane protein bilayer or proteoliposome samples can be very sensitive to the presence of residual detergent. To date, a systematic detergent screen has not been described for the reconstitution of small helical membrane protein samples into lipid environments for solid state NMR.
Solid state NMR is responsible for the majority of helical membrane protein structures characterized in lipid bilayers.1−7 Both oriented sample (OS) and magic angle spinning (MAS) samples are sensitive to the detergent used in purification and reconstitution. For OS solid state NMR, mechanically aligned samples rely on planar lipid bilayers, and consequently, the induced curvature from residual detergent molecules can be significant.8 Alternative bicelle samples are very sensitive to the ratio of short to long chain lipids in the sample,9 and an unintended detergent from a purification protocol would alter the phase behavior of bicelles. For MAS, the influence of residual detergent may not be immediately detectable in the spectra. However, recent studies have shown that sensitivity and resolution are both dependent on lipid type10 and require the complete removal of detergent.11 In addition, we have found that residual detergent significantly reduces the stability and hence lifetime of the samples. As the technique is applied to proteins with more than one or two helices, the affinity of detergent for liposomes and for the protein itself must be considered in the preparation of samples. Multispan helical membrane proteins are likely to have more detergent binding capacity in the hydrophobic region of the membrane than single helix membrane proteins.12 It is beneficial then to have a distinct set of experiments by which to optimize the detergent choice for solid state NMR sample preparation.
Many detergent based screens have been performed in the context of membrane protein structural biology. Here, we focus on the use of detergents for reconstitution of purified proteins into liposomes. It is worthwhile noting that different considerations are applicable when solubilizing protein from cells, and these have been discussed in detail.12 Crystallization screens for X-ray13−15 and electron16,17 diffraction have focused on stabilizing proteins in 3D or 2D detergent and lipid lattices. Solution NMR efforts focus on achieving isotropic correlation times in detergent micelle preparations.18−26 In all cases, there should be an emphasis on achieving a fully functional state for the membrane protein, a parameter that can vary greatly depending on the solubilizing conditions used.27−29 Unfortunately, for many membrane proteins, such as ion channels, functional assays require a bilayer preparation. Lipid bilayer and preoliposome samples for solid state NMR permits both structural studies and functional assays in a native-like environment. However, it is often unclear if a residual detergent is present and if present whether or not it interferes with the protein structure and function.30,31 One of the distinguishing characteristics between detergents and lipids is the high monomer concentration of detergents in the presence of micelles that can lead to detergent binding in non-native locations, such as between helices32,33 or at the aqueous surface,34 which could explain the loss of function for proteins in detergent environments.35 Therefore, based on the potential impact on sample preparation and protein structure and function, we have developed a protocol to eliminate the residual detergent from the samples used for solid state NMR spectroscopy.
There have been studies of detergent affinity for liposomes36−39 and of the kinetics of detergent insertion into liposomes.40 Pure liposomes can be prepared with no residual detergent,41 but the introduction of detergent-bound protein into a solution of liposomes can modify the detergent affinity for the liposomes and the kinetics for the detergent removal from the liposomes as a result of the known affinity of detergents for protein molecules.12 While both long and short acyl chain detergents have stabilized a variety of membrane protein structures, long acyl chain detergents provide a better hydrophobic environment.18,23 Unfortunately, detergents, of a given headgroup structure, with longer acyl chains have greater affinity for and increased partitioning into liposomes than those with shorter acyl chains.42 Therefore, a general theme for reconstitution based detergent screens would be to decrease the acyl chain length as much as possible to minimize the residual detergent in the final proteoliposome sample.
Here, we devise a detergent screen that aims to improve the sample quality for solid state NMR. The samples can be used for either OS solid state NMR of mechanically aligned lipid bilayers or MAS solid state NMR of proteoliposomes. Traditional batch purification43 with detergent exchange20 is used to transfer the protein into a new detergent. This can be followed by size exclusion chromatography and circular dichroism (CD) spectroscopy to initially evaluate the protein structure in the new environment. Continued characterization can be achieved with HPLC using evaporative light scattering detection (ELSD) for assessing the residual detergent in the proteoliposomes. We apply the screen here to a small three helix membrane protein, Rv1861 from M. tuberculosis, that hydrolyzes nucleotides when reconstituted into proteoliposomes and participates in the regulation of transglycosylase activity. OS solid state NMR spectra of this protein have been published for samples prepared using two different detergents.44,45 The results of this work indicate that a detergent with moderate affinity for liposomes can sufficiently solubilize the protein and be completely removed during reconstitution allowing for increased sensitivity in the solid state NMR samples. On the basis of the results presented here, we expect that many more helical membrane proteins can be readily prepared in liposomes for solid state NMR studies.
Experimental Procedures
Protein Expression and Batch Purification
The Rv1861 gene from M. tuberculosis strain H37Rv was cloned into a modified pET-16b vector (Novagen, Inc.) modified to include a His6 N-terminal purification tag. The plasmid was transformed into E. coli BL21-RP-Codon Plus cells (Stratagene, Inc.) for expression. Cells were grown in LB media (Ameresco, Inc.) at 37 °C to an OD600 of 1.0 before expression was induced by adding IPTG to 0.4 mM. Cells were harvested, and 10 mL of lysis buffer (75 mM sodium phosphate, pH 7.5, and 500 mM sodium chloride) per unit of OD600 absorbance was added to resuspend the cells. Lysozyme was added to 0.25 mg/mL along with 4 μL benzonase nuclease and cells incubated at room temperature for 30 min prior to French Press at 10,000 PSI three times. Ten milliliters of lysate was centrifuged at 18,000g for 60 min to isolate the inclusion bodies from other cellular components. Six milliliters of solubilization buffer (40 mM sodium phosphate, pH 7.5, 300 mM sodium chloride, 108 mM Empigen-BB) was used to resuspend each pellet, which was then incubated at 4 °C with gentle rocking for 4 h. The suspension was centrifuged at 18,000g for 30 min to remove insoluble material.
Eight purification tests were prepared by adding 700 μL of the supernatant to 100 μL of Qiagen Ni-NTA resin equilibrated with equilibration buffer (40 mM sodium phosphate, pH 7.5, 300 mM sodium chloride, and 25 mM Empigen-BB) in a 1.5 mL Eppendorf tube. The mixtures were incubated at room temperature for 4 h to allow the proteins to bind resin. The tubes were centrifuged at 1,000g for 1 min, and the supernatant was removed without disturbing the resin. Then 500 μL of equilibration buffer was added to each Eppendorf tube and the mixture incubated at room temperature with gentle rocking for 5 min. Next, 500 μL of wash buffer (equilibration buffer with 20 mM imidazole) was added and the tubes incubated and centrifuged as before. The processes was repeated twice more, once with wash buffer, then once with exchange buffer (20 mM sodium phosphate at pH 7.5 with a quantity of detergent expected to yield 0.4 mM concentration of micelles (Tables 1 and 2)). Protein was eluted from the resin by repeating the process three more times with elution buffer (exchange buffer with 500 mM imidazole.)
Table 1. Detergent Concentrations Used for the Batch Purification Assay.
| detergent | concn (mM) |
|---|---|
| sodium dodecylsulfate | 33 |
| Empigen-BB | 25 |
| dodecylphosphocholine | 19 |
| sodium dodecylsarcosine | 41 |
| decyldimethylglycine | 43 |
| Anzergent 3–10 | 55 |
| Anzergent 3–8 | 414 |
| nonylglucoside | 12 |
| nonylmaltoside | 39 |
Table 2. Detergent Parameters in Aqueous Solutiona.
| detergent | molecular weight (Da) | CMC (mM) | aggregation number | micelle mass (kDa) | acyl chain | headgroup charge |
|---|---|---|---|---|---|---|
| SDSb | 289 | 7–10 (0.5)d | 62 | 18 | 12 | negative |
| Empigen-BBb | 272 | 1.6–2.1 | * | * | 12e | zwitterionic |
| DPCc | 351 | 1.5 | 54 | 19 | 12 | zwitterionic |
| Sarcoc | 293 | 14.4 | * | * | 12 | negative |
| DMc | 483 | 1.8 | 69 | 33 | 10 | polar |
| DDGlyc | 243 | 19 | * | * | 10 | zwitterionic |
| A3–10c | 308 | 39 | 41 | 13 | 10 | zwitterionic |
| NMc | 469 | 6 | 25 | 12 | 9 | polar |
| NGc | 306 | 6.5 | 133 | 41 | 9 | polar |
| OGc | 292 | 18–20 | 27–100 | 8–29 | 8 | polar |
| A3–8c | 280 | 390 | * | * | 8 | zwitterionic |
An asterisk indicates that data was not available.
Values obtained from Sigma-Aldrich, Inc.
Values obtained from Anatrace/Affymetrix, Inc.
SDS–CMC in the presence of proteins is reduced to ∼0.5 mM.53
Empigen-BB is primarily 12 carbon but contains 10–16 carbon molecules.
Size Exclusion Chromatography
A Hi-Prep Sephacryl S200 (16/60) 120 mL column (GE Lifesciences, Inc.) and AKTA Xpress (GE Lifesciences, Inc.) system were used to perform size exclusion chromatography at room temperature. For each detergent, ∼20 μL of eluted protein was diluted to 500 μL with exchange buffer (see Protein Expression and Batch Purification section for buffer contents) and concentrated to ∼100 μL using a 3.5 kDa spin column (Millipore, Inc.). The dilution and concentration procedure was repeated once more before harvesting the solubilized protein and diluting it to 5 mL to remove imidazole from the solution that would interfere with the UV detection of the protein and to prepare the sample for injection. The column was equilibrated with two column volumes of size exclusion buffer (20 mM sodium phosphate at pH 7.5, with detergent at the critical micelle concentration (CMC); see Table 2). The protein sample was then injected onto the column, followed by a 5 mL wash with exchange buffer to ensure that the entire sample was removed from the injection loop. The column was run at 0.5 mL/min for 140 mL.
CD Spectroscopy
Purified protein was rinsed three times in 500 μL, 3.5 kDa spin filters (Millipore, Inc.) with 25 mM sodium phosphate at pH 7.5 and a concentration of 1 CMC of the appropriate detergent (Table 2) to remove imidazole. The protein was diluted to between 5 and 10 μM for CD experiments depending on the detergent used. The samples were desalted with data acquired in a 300 μL, 1 mm, Starna quartz cell on an AVIV 202 CD spectrometer. Three sweeps of the 180–260 nm wavelength range in 1.0 nm increments were averaged, and the buffer signal was subtracted from the protein signal before converting the raw data into residual molar ellipticity. The deconvolution of the CD curves was performed in CD-PRO (http://lamar.colostate.edu/∼sreeram/CDPro/main.html) using a basis set of 43 soluble and 13 membrane proteins. The CD data were deposited in the PDCDB with codes CD0004475000, CD0004476000, CD0004477000, CD0004478000, and CD0004479000.
Proteoliposome Preparation
For the reconstitution test and the OS solid state NMR samples, a liposome suspension was prepared by dissolving 50 mg of 1,2-dimyristoyl-sn-glycero-3-phosphocholine/1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DMPC/DMPG) powder at a 4:1 weight ratio in 2 mL of 10 mM HEPES buffer at pH 7.5, in a glass test tube. The mixture was bath sonicated for 30–60 min until the solution became clear indicating the formation of small unilamellar vesicles, which was confirmed by electron microscopy (Figure 1, Supporting Information). The detergent was added to the solution until it was optically clear (0 absorbance at 0.1 cm path length over 300–700 nm), and all lipid vesicles were solubilized into mixed micelles (69 mM for sodium dodecyl sulfate (SDS) and 103 mM for decyl-N,N-dimethylglycine (DDGly)). Then, 4.2 mg (SDS) or 10.5 mg (DDGly) of detergent solubilized protein was added to the lipid detergent solution and the volume adjusted with 10 mM HEPES at pH 7.5 to a final volume of 8 mL. The detergent was added to make concentrations of 69 mM and 82 mM for SDS and DDGly, respectively, and the final lipid concentration was 9 mM. The mixture was incubated at 37 °C for 12 h. During incubation, the solution was mixed by gentle inversion three times at 4 h intervals. The solutions were placed in 6–8 kDa MWCO dialysis tubing and dialyzed against 2 L of 10 mM HEPES at pH 7.5 and 35 °C with daily buffer changes. Proteoliposomes were harvested after 10 d (SDS) or 6 d (DDGly) of dialysis by centrifugation at 228,000g for 1.5 h. The supernatant was discarded and the pellet resuspended with 10 mM HEPES at pH 7.5 to a final volume of 1.2 mL.
For magic angle spinning samples, the same reconstitution procedure was used except the lipid mass was reduced to 20 mg. The protein-to-lipid ratio remained the same for each detergent (1.8 mg for SDS and 4.2 mg for DDGly). After dialysis, the proteoliposomes were was pelleted at 228,000g for 2 h in 3.2 mL thickwall polycarbonate ultracentrifuge tubes (Beckman Coulter, Inc.)
Evaporative Light Scattering Detection
The detergents were separated using HPLC and an Acclaim Surfactant Plus 3.0 μm column (Thermo Scientific, Inc.). An AB linear gradient elution was used (30 °C, 70 to 15% eluent B over 10 min at a flow rate of 0.9 mL/min). Eluent A was HPLC grade acetonitrile, and eluent B was 0.1 M ammonium acetate at pH 5.0. The presence of the detergent was assessed using an ELSD380 system (Agilent Technologies, Inc.). After each daily dialysis buffer change, an aliquot of the detergent–proteoliposome mixture was removed from the dialysis bag. The aliquots were not diluted prior to each 25 μL injection. The N2 flow rate was 1.6 L/min, the nebulizer was 50 °C, the evaporator was 75 °C, and the photomultiplier tube gain was set at 2.5 to ensure a noise level of <0.2 mV peak–peak based on manufacturer recommendations.
Oriented Sample Solid State NMR Samples and Spectroscopy
Proteoliposome solution was deposited on 40 5.7 mm × 12 mm × 60 μm glass slides (35 μL per slide). The slides were dehydrated in a sealed 98% relative humidity chamber at 22 °C for approximately 12 h or until bulk water was visibly removed from the slides. Two microliters of deionized water was added to the center of each slide before stacking 30 of them on top of each other. The stack was incubated at 98% relative humidity at 37 °C for 4 d to remove bulk water and to let the proteoliposome solution between the glass slides become homogenously hydrated. The stack was then inserted into a 5.7 mm × 5.7 mm × 20 mm glass cell and sealed with beeswax after inserting a plastic plug.46 SAMPI447 experiments for measuring 15N chemical shift anisotropy and 1H–15N dipolar couplings were performed on a 21.1 T 105 mm bore magnet using a home-built Low-E 1H-X static probe48 at 310 K. A 4 μs 1H pulse was used, and rf fields were 62.5 kHz on both channels for cross-polarization and 1H decoupling. Thirty-two t1 points were acquired with 2048 or 4096 transients averaged for DDGly and SDS, respectively. A 5 s recycle delay was used for both experiments.
Magic Angle Spinning Solid State NMR Samples and Spectroscopy
Proteoliposome pellets were dehydrated at 37 °C and 16% relative humidity for several hours until the sample was approximately 40% w/w water before packing into a 3.2 mm thin walled rotor (Revolution NMR, Inc.). The detailed packing procedure is described in ref (46). Dipolar assisted rotational resonance (DARR)49,50 experiments were performed on a 14.1 T 89 mm bore magnet using a home-built, Low-E, 1H-13C-15N triple resonance probe51 with 10 kHz MAS at 243 K. One hundred kilo hertz of irradiation on the 1H channel was used for the 90° pulse and SPINAL-64 decoupling.52 During cross-polarization, the 13C rf field was 50 kHz with a ±10% linear 1H ramp. The 13C 90° pulse was 50 kHz. Sixty-four and 128 transients were averaged for each of the 512 t1 points for DDGly and SDS, respectively. Recycle delay was set at 1.5 s.
Results
Detergent Choice
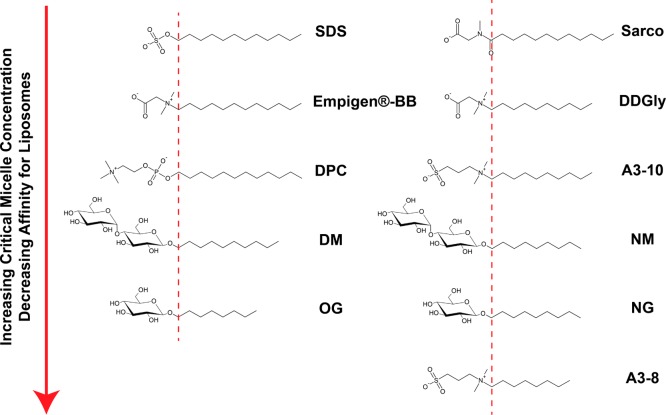
It is important to distinguish between detergents used for isolation and purification and those used for reconstitution. When a preparation of purified protein solubilized in detergent is of interest, the detergent most amenable to the experiment being performed is often chosen. For example, the Rv1861 protein is easily purified in detergents with 12 carbon acyl chains (SDS and DPC) with SDS being the most suitable for solution NMR studies of this protein.20 However, when the protein is reconstituted into liposomes attention must be paid to the detergent affinity for liposomes and protein. Ideally, this affinity should be minimized. The detergent can be exchanged on-column during the purification process once impurities have been removed.20,46 Rv1861 has been studied by OS solid state NMR in DMPC/DMPG lipid bilayers. The first samples were prepared using SDS mediated reconstitution.44 The highest protein-to-lipid molar ratio (P/L) that could be used to prepare high quality mechanically aligned bilayer samples was 1:200. With less lipid per protein, the reconstitutions were incomplete, and the resuspended proteoliposome pellet was too viscous for NMR sample preparation. On the basis of this, we proposed that residual SDS was present in the samples, preventing high quality proteoliposome preparation. The most likely detergent characteristics to induce increased affinity for the proteoliposomes are acyl chain length and headgroup charge. More mild detergents lacking charge and containing shorter acyl chains, such as DM and OG, failed to stabilize the protein long enough for reconstitution into proteoliposomes. The detergent acyl chain length modulates the CMC for a given detergent headgroup42 and is important during dialysis because only detergent monomers exchange with buffer. A higher CMC provides a greater concentration gradient and drives the detergent out of the proteoliposome and into the buffer. Furthermore, acyl chain length also determines relative affinity of the detergents for liposomes42 and dictates how much detergent dissociates from the liposomes during dialysis. On the basis of the structure of SDS, we selected detergents with reduced acyl chain length (9–10 carbons) or a reduction in headgroup hydrophilicity under the premise that such detergents might stabilize the protein adequately and also be readily removed during reconstitution. The commercially available detergents selected for Rv1861 are shown in Figure 1 and Table 2.
Figure 1.
Detergents used in the purification screen for Rv1861. Decreasing the acyl chain length for a given headgroup increases the relative CMC and decreases relative detergent affinity for liposomes. The dashed line is meant to suggest the approximate interface between hydrophilic and hydrophobic moieties for these amphipathic molecules. Molecular structures were drawn with ChemSketch (ACD/Laboratories, Inc.).
Batch Purification Assay
A batch purification assay was performed using the new detergents (Figure 2). Inclusion body fractions were solubilized with the industrial detergent Empigen-BB, bound to a Ni2+ affinity column, and washed in Empigen-BB followed by detergent exchange with the desired detergent and elution at a high concentration of imidazole. While SDS eluted the most protein, the other 8, 9, 10, and 12 carbon detergents eluted considerable protein. The 10 and 12 carbon detergents stabilized the protein for over 2 weeks, while the 8 and 9 carbon detergents all showed visible signs of precipitation within the same period. It is important to note that the >15 kDa bands visible in the InVision stained gel (Figure 2B) are most likely not the Rv1861 protein. The smaller molecular weight band around 16 kDa is not a dimer of Rv1861 whose molecular weight is 11.4 kDa. Also, both bands elute in either the flow through or wash fractions, indicating that these molecules do not have significant affinity for the Ni2+ column. Finally, the detergent A3–8 shows signs of protein precipitation or severe protein aggregation at the top of the gels as well as slightly increased molecular weight for the monomer (Figure 2A).
Figure 2.
Batch purification assay. SDS eluted the most protein, while other detergents eluted slightly less protein. The dashed line indicates detergents that failed to stabilize the protein for longer than two weeks. (A) Coomassie and (B) a UV sensitive poly histidine dye (InVision) were used to stain the gels. Inclusion Body is the resuspended pellet after spinning the lysate at 18,000g, Insoluble is the 18,000g pellet after incubation with the Empigen-BB detergent, Load is the supernatant from the insoluble pellet, Flow Through is the elute from the column with no imidazole present, Wash is the elute from the column with 40 mM imidazole, and all other lanes are the elutes from the column with 250 mM imidazole containing the detergent named at the top of each lane.
Oligomeric State in Micelles
Gel filtration chromatography was performed to assess sample homogeneity and to detect oligomeric states for the protein in the various detergent environments (Figure 3). The data are interpreted based on the standards provided by the manufacturer and the average micellar weight for each detergent. When no data were available for a detergent aggregation number, the value was estimated using data for similar detergents. Here, the detergents tested have micelle sizes spanning ∼7–20 kDa. It should be noted that the detergent contribution to molecular weight per protein molecule for oligomeric complexes can be expected to be somewhat less than that of a monomeric state; therefore, the influence from the detergent is not linear. The SDS sample eluted near the void volume of the column corresponding to a complex of ∼160 kDa. Note that the sample is heterogeneous with small peaks at 85 and 105 mL elution volumes. The DPC sample also eluted primarily as a large oligomeric complex of ∼100 kDa, although there is a small amount of a larger complex for this sample. The A3–10 sample elutes even later from the column and suggests a smaller complex of ∼60 kDa. There is a slight heterogeneity in the sample eluting at ∼55 mL. The DDGly solubilized protein eluted near the column volume (120 mL) and shows a homogeneous sample. This represents a monomeric complex (∼15–20 kDa) based on column standards and the molecular weight of Rv1861 (11.4 kDa). The SDS–PAGE results suggest monomeric states for Rv1861, while the SE results appear to show various oligomeric complexes for SDS, DPC, and A3–10. While the SDS–PAGE samples were not boiled, the higher concentration of SDS in the sample buffer could affect the protein conformation and oligomeric state.
Figure 3.
Size exclusion elution profiles for selected detergents. SDS and DPC indicate the presence of a large oligomeric complex. A3–10 indicates the presence of an intermediate sized oligomeric complex, while DDGly most likely represents a monomeric complex. All detergents except DDGly have some heterogeneity in the sample. The detergent contribution to the molecular weight is most likely similar for all detergents, 7–20 kDa.
Alpha Helical Content in Micelles
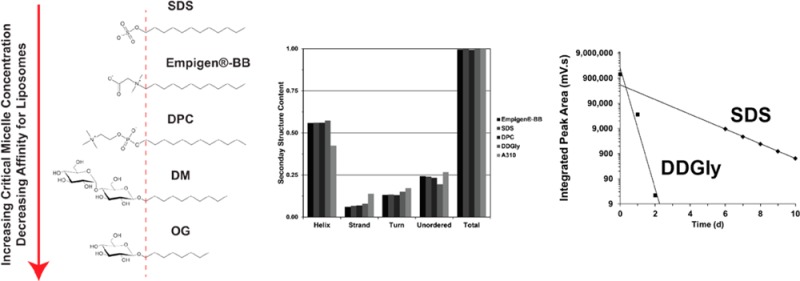
CD spectroscopy was performed on the protein solubilized in detergents to assess helical content in the different environments. Helical prediction from the amino acid sequence using the TMHMM program54 indicates the protein should be roughly 60% helical. The CD spectra all exhibit highly helical profiles (Figure 4) in agreement with TMHMM prediction and published OS solid state NMR data.44,45 The 12 carbon detergents exhibited similar secondary structure content. However, DDGly stabilized slightly more structures presenting slightly less unordered content, while A3–10 stabilized less α-helical content with a marked increase in the β-strand, turn, and unordered content.
Figure 4.
(A) CD profiles for Rv1861 stabilized in various detergents. (B) Secondary structure content analysis of the CD data based on a database of 43 soluble and 13 membrane proteins.
Detergent Removal
Side-by-side reconstitution was performed into DMPC/DMPG lipid bilayers using protein stabilized in SDS or DDGly. Detergent was removed by dialysis with daily buffer changes. Figure 5A shows SDS–PAGE gels monitoring the reconstitution process. For the SDS mediated reconstitution, both detergent solubilized protein and proteoliposomes are present in the sample after extensive dialysis, although the amount of detergent solubilized protein is a small fraction of the total protein shown in Figure 5A. Lanes 1–3 are before dialysis and lanes 4–6 after dialysis with lane 4 being the supernatant after pelleting the proteoliposomes. Lanes 5 and 6 represent the proteoliposome pellet. There must be residual detergent in the sample to solubilize the hydrophobic membrane protein in the supernatant (lane 4). Conversely, the DDGly reconstitution does not show detergent solubilized protein in the supernatant after only three days of dialysis (Figure 5A, lane 4). This suggests that for DDGly virtually all of the detergent is removed by dialysis, while this is not true for SDS. For DDGly, the sample starts with a volume of 8 mL containing ∼80 mM DDGly and is equilibrated with 2 L of buffer for dialysis. After the first buffer change, there should be ∼0.3 mM DDGly remaining in the sample if complete equilibration occurs. By the third dilution, there should only be ∼5 nM of detergent left in the sample. Likewise, for SDS, the sample has an 8 mL volume with ∼70 mM detergent and is dialyzed in 2 L of buffer. Again, after three days the detergent concentration should be ∼5 nM if equilibrium is achieved. In the sample, the protein concentration is ∼0.1 mM, and the lipid concentration is ∼22 mM; therefore, if the detergent is equilibrated with each 2 L buffer change the remaining nanomolar concentration of detergent after 3 days should have no effect on the proteoliposomes because it is so dilute. The presence of detergent during dialysis was monitored by HPLC using an ELSD as shown in Figure 5B. Missing data points for SDS indicate the detector was saturated by large signals resulting from high concentrations of detergent. DDGly is undetectable after the third day of dialysis indicating the concentration is below the limit of detection (a few nanomolar) which verifies that the DDGly completely equilibrates with the buffer within 24 h. SDS, however, was still detected after 10 d indicating that the SDS did not equilibrate completely with buffer in 24 h since it was detectable even after the 10th buffer change. This result and the presence of solubilized protein in the supernatant from the ultracentrifugation suggest that a significant concentration of SDS remains in the sample.
Figure 5.
(A) SDS–PAGE gels monitor the reconstitution process for Rv1861 in SDS and DDGly detergents show the residual soluble protein for the SDS sample. Lane 1 is purified protein in detergent micelles. Lanes 2 and 3 are detergent solubilized protein mixed with proteoliposomes before and after incubation at 37 °C, respectively. Lane 4–6 are samples after extensive dialysis and centrifugation. Lane 4 is the supernatant, and lanes 5 and 6 are the resuspended proteoliposome pellets before and after the removal of any precipitate. (B) Integrated peak volume for the detergent detected by evaporative light scattering as a function of dialysis time. The buffer with a 250-fold larger volume than the sample was changed daily. DDGly (squares) is readily removed but SDS (diamonds) persists in the sample after 10 daily buffer changes. SDS signals saturated the detector until day 6 due to high concentrations of the detergent. DDGly saturates the detector on day 0, but the sample was diluted. Measurement errors are ±5%.
Solid State NMR Sample Quality
OS solid state NMR samples prepared from SDS could only be made at 1:200 P/L, while DDGly can be prepared up to 1:80. The uniformly 15N-labeled SAMPI4 2D spectrum for each liposome preparation is shown in Figure 6. The highly congested spectrum is consistent with a three helix membrane protein.45 A similar intensity pattern, such as the hole at 175 ppm chemical shift and 3.0 kHz dipolar coupling, indicates the protein has similar structure in each sample. However, in the boxed region of the spectrum there are resonances in the SDS spectrum not present in the DDGly sample (Figure 6). The exact structural changes cannot be determined with these spectra. Regardless, the higher P/L ratio available when using DDGly resulted in equivalent signal-to-noise with half of the signal averaging compared to that of the SDS sample.
Figure 6.

SAMPI4 spectra of uniformly 15N-labeled Rv1861 in DMPC/DMPG lipid bilayers prepared using DDGly (A) and SDS (B). The spectra present similar intensity profiles indicating similar helical structure and orientation for both samples. The DDGly detergent allowed higher protein to lipid ratios to be used, which halved the signal averaging time for equivalent signal-to-noise. Contours are drawn at 1.1σ and 1.2σ with the factor between levels set to 1.1 for DDGly and SDS, respectively.
13C–13C magic angle spinning solid state NMR correlation spectra for Rv1861 proteoliposomes prepared using DDGly and SDS are shown in Figure 7. Because of the high content of hydrophobic amino acids, the resonances from these sites are not well resolved. However, both preparations give similar resonance envelopes for valine, leucine, and isoleucine, which are characteristic of helical membrane proteins.55 Importantly, there are significant differences in the spectra. The SDS spectrum has missing threonine and tryptophan Cα/Cβ cross-peaks (Figure 7, red arrows), and the DDGly spectrum has increased resolution (Figure 7, green arrows). The 1D slices shown in Figure 7C and D show narrower lineshapes, twice the signal-to-noise, and extra peaks for the DDGly sample.
Figure 7.
13C–13C DARR spectra (30 ms) for Rv1861 prepared from DDGly (A and C) and SDS (B and D) at 10 kHz MAS and 243 K. The SDS spectrum averaged twice as many transients as the DDGly spectrum but results in less signal. Red arrows indicate resonances missing from the SDS spectrum, and green arrows indicate cross-peaks for threonine, valine, and alanine that are better resolved for DDGly. Contours are drawn at 3.7σ and 7.8σ with the factor between levels set to 1.3 for DDGly and SDS, respectively.
Discussion
Although a given detergent may stabilize a membrane protein, it does not mean that the detergent is the best candidate for the preparation of proteoliposome samples, as exemplified here with Rv1861. Detergents were screened based on the fact that short acyl chain detergents are easily removed from liposomes, while long acyl chain detergents are more difficult to remove from and have more affinity for protein molecules. For Rv1861, we show that reducing the acyl chain length from 12 to 10 allows for a doubling of the protein incorporated into liposomes while forming homogeneous samples without residual detergent. Furthermore, these preparations have been used for the first high resolution structural characterization of the transmembrane domain for a three helix membrane protein using OS and MAS solid state NMR45 (Murray et al., unpublished results). On the basis of the set of experiments presented here, detergents can be screened for their effective removal leading to optimal samples of membrane protein drug targets for structural characterization by solid state NMR.
Every membrane protein interacts differently with each detergent. While some membrane proteins are solubilized very well in long, 12 carbon, acyl chain detergents,56−58 others favor shorter, 8–10 carbon acyl chain detergents.4,12 In general, detergents with acyl chains containing <10 carbons are more easily removed unless there is a highly specific interaction with the protein.12 However, in general, longer, 12 carbon acyl chain detergents are effective for stabilizing membrane proteins.57,59 A directed approach that is aimed at producing a native conformation in a native-like environment for solid state NMR is favored over stabilizing the greatest quantity of protein. As new membrane protein drug targets are studied by solid state NMR, the approach presented here represents an efficient strategy to begin structural characterization efforts. The lipid choice for the reconstitution of a specific protein is dependent on the successful results of a functional assay for each protein. For Rv1861, a GTP hydrolysis assay was used to show the protein is functional in DMPC/DMPG proteoliposomes.
It is completely plausible that the protein under study may be stabilized by a detergent in an inactive state with the wrong tertiary structure. Indeed, some proteins are completely inactivated by short exposure to detergents while remaining soluble,35,60,61 and other proteins require cholesterol or specific lipids to achieve a similar activity in micelles as in liposomes,58 and still others form nonfunctional oligomeric states.62 The focus here is on identifying a detergent that will stabilize the protein for long enough to incorporate it into a native-like liposome environment while also allowing virtually all of the detergent to be removed by dialysis. The intended result is to obtain liposomes for solid state NMR with a high concentration of natively folded protein. It should be stressed that the screen applies to a variety of lipid combinations that can be tailored to the specific needs of a given protein and that the reconstituted proteins can be easily used in a variety of functional assays to validate the native-like character of the sample.
The experiments presented here provide a rapid method to screen detergents to optimize the incorporation of membrane proteins into liposomes. With the advent of solid state NMR technology as a primary technique for investigating membrane protein structure in a native-like environment, these experiments pave the way for many of these important drug targets to be characterized. Through this screening approach, many more membrane protein drug targets will become available for structural characterization.
Acknowledgments
We thank Dr. Claudius Mondoma from the FSU Physical Biochemistry Facility, Institute for Molecular Biophysics for help with the CD experiments. Dr. Huajun Qin assisted with cloning and expression of the Rv1861 protein. Dr. Riqiang Fu, Dr. Bill Brey, and Peter Gor’kov from the NHMFL assisted with the NMR experiments and NMR probe technology.
Glossary
Abbreviations
- A3–8
Anzergent 3–8
- A3–10
Anzergent 3–10
- CD
circular dichroism
- CMC
critical micelle concentration
- DARR
dipolar assisted rotational resonance
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DDGly
decyl-N,N-dimethylglycine
- DMPG
1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- DPC
n-dodecylphosphocholine
- Emp
Empigen-BB
- ELSD
evaporative light scattering detection
- HPLC
high pressure liquid chromatography
- MAS
magic angle spinning
- NM
n-nonly-β-d-glucopyranoside and n-nonly-β-d-maltropyranoside
- OG
n-octyl-β-d-glucopyranoside
- OS
oriented sample
- Sarco
n-dodecanoyl sarcosine
- SDS
sodium dodecyl sulfate
Supporting Information Available
Electron micrographs of the proteoliposome preparations obtained using SDS and DDGly mediated reconstitution. This material is available free of charge via the Internet at http://pubs.acs.org.
The work was made possible by grants from the National Institute of Allergy and Infectious Diseases (R01-AI073891 and P01-AI074805) and the use of the National High Magnetic Field Laboratory, funded by a cooperative agreement between the State of Florida and the National Science Foundation (DMR-1157490).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- De Angelis A. A.; Howell S. C.; Nevzorov A. A.; Opella S. J. (2006) Structure determination of a membrane protein with two trans-membrane helices in aligned phospholipid bicelles by solid-state NMR spectroscopy. J. Am. Chem. Soc. 128, 12256–12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchem R. R.; Hu W.; Cross T. A. (1993) High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science 261, 1457–1460. [DOI] [PubMed] [Google Scholar]
- Park S. H.; De Angelis A. A.; Nevzorov A. A.; Wu C. H.; Opella S. J. (2006) Three-dimensional structure of the transmembrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. Biophys. J. 91, 3032–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M.; Yi M. G.; Dong H.; Qin H. J.; Peterson E.; Busath D. D.; Zhou H. X.; Cross T. A. (2010) Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer. Science 330, 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriot D. S.; Nevzorov A. A.; Zagyanskiy L.; Wu C. H.; Opella S. J. (2004) Structure of the coat protein in Pf1 bacteriophage determined by solid-state NMR spectroscopy. J. Mol. Biol. 341, 869–879. [DOI] [PubMed] [Google Scholar]
- Traaseth N. J.; Shi L.; Verardi R.; Mullen D. G.; Barany G.; Veglia G. (2009) Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc. Natl. Acad. Sci. U.S.A. 106, 10165–10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B. B.; Nothnagel H. J.; Lu G. J.; Son W. S.; Tian Y.; Marassi F. M.; Opella S. J. (2012) Structure determination of a membrane protein in proteoliposomes. J. Am. Chem. Soc. 134, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll F. III; Cross T. A. (1990) Optimizing and characterizing alignment of oriented lipid bilayers containing gramicidin D. Biophys. J. 57, 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis A. A.; Opella S. J. (2007) Bicelle samples for solid-state NMR of membrane proteins. Nat. Protoc. 2, 2332–2338. [DOI] [PubMed] [Google Scholar]
- Wang S.; Munro R. A.; Shi L.; Kawamura I.; Okitsu T.; Wada A.; Kim S. Y.; Jung K. H.; Brown L. S.; Ladizhansky V. (2013) Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat. Methods 10, 1007–1012. [DOI] [PubMed] [Google Scholar]
- Park S. H.; Casagrande F.; Chu M.; Maier K.; Kiefer H.; Opella S. J. (2012) Optimization of purification and refolding of the human chemokine receptor CXCR1 improves the stability of proteoliposomes for structure determination. Biochim. Biophys. Acta 1818, 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Maire M.; Champeil P.; Møller J. V. (2000) Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 1508, 86–111. [DOI] [PubMed] [Google Scholar]
- Low C.; Moberg P.; Quistgaard E. M.; Hedren M.; Guettou F.; Frauenfeld J.; Haneskog L.; Nordlund P. (2013) High-throughput analytical gel filtration screening of integral membrane proteins for structural studies. Biochim. Biophys. Acta 1830, 3497–3508. [DOI] [PubMed] [Google Scholar]
- Ma J. C.; Xia D. (2008) The use of blue native PAGE in the evaluation of membrane protein aggregation states for crystallization. J. Appl. Crystallogr. 41, 1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergis J. M.; Purdy M. D.; Wiener M. C. (2010) A high-throughput differential filtration assay to screen and select detergents for membrane proteins. Anal. Biochem. 407, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Krey I. (2007) Electron crystallography of membrane proteins: Two-dimensional crystallization and screening by electron microscopy. Methods 41, 417–426. [DOI] [PubMed] [Google Scholar]
- Stokes D. L.; Ubarretxena-Belandia I.; Gonen T.; Engel A. (2013) High-throughput methods for electron crystallography. Methods Mol. Biol. 955, 273–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbus L.; Lipfert J.; Jambunathan K.; Fox D. A.; Sim A. Y. L.; Doniach S.; Lesley S. A. (2009) Mixing and matching detergents for membrane protein NMR structure determination. J. Am. Chem. Soc. 131, 7320–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger-Koplin R. D.; Sorgen P. L.; Krueger-Koplin S. T.; Rivera-Torres A. O.; Cahill S. M.; Hicks D. B.; Grinius L.; Krulwich T. A.; Girvin M. E. (2004) An evaluation of detergents for NMR structural studies of membrane proteins. J. Biomol. NMR 28, 43–57. [DOI] [PubMed] [Google Scholar]
- Page R. C.; Moore J. D.; Nguyen H. B.; Sharma M.; Chase R.; Gao F. P.; Mobley C. K.; Sanders C. R.; Ma L.; Sonnichsen F. D.; Lee S.; Howell S. C.; Opella S. J.; Cross T. A. (2006) Comprehensive evaluation of solution nuclear magnetic resonance spectroscopy sample preparation for helical integral membrane proteins. J. Struct. Funct. Genomics 7, 51–64. [DOI] [PubMed] [Google Scholar]
- Shenkarev Z. O.; Lyukmanova E. N.; Paramonov A. S.; Shingarova L. N.; Chupin V. V.; Kirpichnikov M. P.; Blommers M. J. J.; Arseniev A. S. (2010) Lipid-protein nanodiscs as reference medium in detergent screening for high-resolution NMR studies of integral membrane proteins. J. Am. Chem. Soc. 132, 5628–+. [DOI] [PubMed] [Google Scholar]
- Yeliseev A. A.; Wong K. K.; Soubias O.; Gawrisch K. (2005) Expression of human peripheral cannabinoid receptor for structural studies. Protein Sci. 14, 2638–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver R. C.; Lipfert J.; Fox D. A.; Lo R. H.; Doniach S.; Columbus L. (2013) Dependence of micelle size and shape on detergent alkyl chain length and head group. PLoS One 8, e62488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn W. D.; Kim H. J.; Ellis C. D.; Hadziselimovic A.; Sulistijo E. S.; Karra M. D.; Tian C.; Sonnichsen F. D.; Sanders C. R. (2009) Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science 324, 1726–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verardi R.; Shi L.; Traaseth N. J.; Walsh N.; Veglia G. (2011) Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc. Natl. Acad. Sci. U.S.A. 108, 9101–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.; Bobkov A. A.; Plesniak L. A.; Marassi F. M. (2009) Mapping the interaction of pro-apoptotic tBID with pro-survival BCL-XL. Biochemistry 48, 8704–8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu M. L.; Tsang C.; Grihalde N.; MacWilliams M. P. (2008) Over-expression, solubilization, and purification of G protein-coupled receptors for structural biology. Comb. Chem. High Throughput Screening 11, 439–462. [DOI] [PubMed] [Google Scholar]
- Li Q. X.; Mittal R.; Huang L. J.; Travis B.; Sanders C. R. (2009) Bolaamphiphile-class surfactants can stabilize and support the function of solubilized integral membrane proteins. Biochemistry 48, 11606–11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanford M.; Peeples T. L. (2002) Archaeal tetraether lipids - unique structures and applications. Appl. Biochem. Biotechnol. 97, 45–62. [DOI] [PubMed] [Google Scholar]
- Ostuni M. A.; Iatmanen S.; Teboul D.; Robert J. C.; Lacapere J. J. (2010) Characterization of membrane protein preparations: measurement of detergent content and ligand binding after proteoliposomes reconstitution. Methods Mol. Biol. 654, 3–18. [DOI] [PubMed] [Google Scholar]
- Wang L.; Quan C.; Liu B.; Wang J.; Xiong W.; Zhao P.; Fan S. (2013) Functional reconstitution of staphylococcus aureus truncated agrc histidine kinase in a model membrane system. PLoS One 8, e80400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. X.; Cross T. A. (2013) Influences of membrane mimetic environments on membrane protein structures. Ann. Rev. Biophys. 42, 361–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross T.; Murray D.; Watts A. (2013) Helical membrane protein conformations and their environment. Eur. Biophys. J. 42, 731–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoonens M.; Comer J.; Masscheleyn S.; Pebay-Peyroula E.; Chipot C.; Miroux B.; Dehez F. (2013) Dangerous liaisons between detergents and membrane proteins. The case of mitochondrial uncoupling protein 2. J. Am. Chem. Soc. 135, 15174–15182. [DOI] [PubMed] [Google Scholar]
- White J. F.; Grisshammer R. (2010) Stability of the neurotensin receptor NTS1 free in detergent solution and immobilized to affinity resin. PLoS One 5, e12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M.; Kerth A.; Blume A. (1997) Thermodynamics of interaction of octyl glucoside with phosphatidylcholine vesicles: partitioning and solubilization as studied by high sensitivity titration calorimetry. Biochim. Biophys. Acta 1326, 178–192. [DOI] [PubMed] [Google Scholar]
- de la Maza A.; Parra J. L. (1997) Solubilizing effects caused by the nonionic surfactant dodecylmaltoside in phosphatidylcholine liposomes. Biophys. J. 72, 1668–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.; Ziegler A.; Steinbauer B.; Seelig J. (2002) Thermodynamics of sodium dodecyl sulfate partitioning into lipid membranes. Biophys. J. 83, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A.; Blume A. (2004) Solubilization of DMPC-d54 and DMPG-d54 vesicles with octylglucoside and sodium dodecyl sulfate studied by FT-IR spectroscopy. Phys. Chem. Chem. Phys. 6, 1551–1556. [Google Scholar]
- Cócera M.; López O.; Estelrich J.; Parra J. L.; de la Maza A. (2000) Kinetic and structural aspects of the adsorption of sodium dodecyl sulfate on phosphatidylcholine liposomes. Langmuir 16, 4068–4071. [DOI] [PubMed] [Google Scholar]
- López O.; Cócera M.; Coderch L.; Parra J. L.; Barsukov L.; de la Maza A. (2001) Octyl glucoside-mediated solubilization and reconstitution of liposomes: structural and kinetic aspects. J. Phys. Chem. B 105, 9879–9886. [Google Scholar]
- Maza A.; Lopez O.; Baucells J.; Gonzalez P.; Parra J. L. (1998) Solubilization of phosphatidylcholine unilamellar liposomes caused by alkyl glucosides. J. Surfactants Deterg. 1, 381–386. [Google Scholar]
- Feng G.; Winkler M. E. (1995) Single-Step Purifications of His(6)-Muth, His(6)Mutl and His(6)-Muts Repair Proteins of Escherichia-Coli K-12. Biotechniques 19, 956–965. [PubMed] [Google Scholar]
- Li C. G.; Gao P.; Qin H. J.; Chase R.; Gor’kov P. L.; Brey W. W.; Cross T. A. (2007) Uniformly aligned full-length membrane proteins in liquid crystalline bilayers for structural characterization. J. Am. Chem. Soc. 129, 5304–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray D. T.; Hung I.; Cross T. A. (2014) Assignment of oriented sample NMR resonances from a three transmembrane helix protein. J. Magn. Reson. 240, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N.; Murray D. T.; Cross T. A. (2013) Lipid bilayer preparations of membrane proteins for oriented and magic-angle spinning solid-state NMR samples. Nat. Protoc. 8, 2256–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevzorov A. A.; Opella S. J. (2003) A “magic sandwich” pulse sequence with reduced offset dependence for high-resolution separated local field spectroscopy. J. Magn. Reson. 164, 182–186. [DOI] [PubMed] [Google Scholar]
- Gor’kov P. L.; Chekmenev E. Y.; Li C. G.; Cotten M.; Buffy J. J.; Traaseth N. J.; Veglia G.; Brey W. W. (2007) Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. J. Magn. Reson. 185, 77–93. [DOI] [PubMed] [Google Scholar]
- Takegoshi K.; Nakamura S.; Terao T. (2003) 13C–1H dipolar-driven 13C–13C recoupling without 13C rf irradiation in nuclear magnetic resonance of rotating solids. J. Chem. Phys. 118, 2325–2341. [Google Scholar]
- Takegoshi K.; Nakamura S.; Terao T. (2001) 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637. [Google Scholar]
- McNeill S. A.; Gor’kov P. L.; Shetty K.; Brey W. W.; Long J. R. (2009) A low-E magic angle spinning probe for biological solid state NMR at 750 MHz. J. Magn. Reson. 197, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung B. M.; Khitrin A. K.; Ermolaev K. (2000) An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 142, 97–101. [DOI] [PubMed] [Google Scholar]
- Ahmad M. F.; Ramakrishna T.; Raman B.; Rao C. M. (2006) Fibrillogenic and non-fibrillogenic ensembles of SDS-bound human α-synuclein. J. Mol. Biol. 364, 1061–1072. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E. L.; von Heijne G.; Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc.: Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182. [PubMed] [Google Scholar]
- Murray D. T.; Das N.; Cross T. A. (2013) Solid state NMR strategy for characterizing native membrane protein structures. Acc. Chem. Res. 46, 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund S.; Orlowski S.; Deforesta B.; Champeil P.; Lemaire M.; Moller J. V. (1989) Detergent structure and associated lipid as determinants in the stabilization of solubilized Ca-2+-ATPase from sarcoplasmic-reticulum. J. Biol. Chem. 264, 4907–4915. [PubMed] [Google Scholar]
- Rosevear P.; VanAken T.; Baxter J.; Ferguson-Miller S. (1980) Alkyl glycoside detergents: a simpler synthesis and their effects on kinetic and physical properties of cytochrome c oxidase. Biochemistry 19, 4108–4115. [DOI] [PubMed] [Google Scholar]
- Vukoti K.; Kimura T.; Macke L.; Gawrisch K.; Yeliseev A. (2012) Stabilization of functional recombinant cannabinoid receptor CB(2) in detergent micelles and lipid bilayers. PLoS One 7, e46290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowthert L. A.; Ku N. O.; Liao J.; Coulombe P. A.; Omary M. B. (1995) Empigen BB: a useful detergent for solubilization and biochemical analysis of keratins. Biochem. Biophys. Res. Commun. 206, 370–379. [DOI] [PubMed] [Google Scholar]
- Esmann M. (1984) The Distribution of C12e8-Solubilized oligomers of the (Na+ + K+)-ATPase. Biochim. Biophys. Acta 787, 81–89. [DOI] [PubMed] [Google Scholar]
- Esmann M.; Skou J. C.; Christiansen C. (1979) Solubilization and molecular-weight determination of the (Na++K+)-ATPase from rectal glands of Squalus-acanthias. Biochim. Biophys. Acta 567, 410–420. [DOI] [PubMed] [Google Scholar]
- White J. F.; Grodnitzky J.; Louis J. M.; Trinh L. B.; Shiloach J.; Gutierrez J.; Northup J. K.; Grisshammer R. (2007) Dimerization of the class A G protein-coupled neurotensin receptor NTS1 alters G protein interaction. Proc. Natl. Acad. Sci. U.S.A. 104, 12199–12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.