Abstract
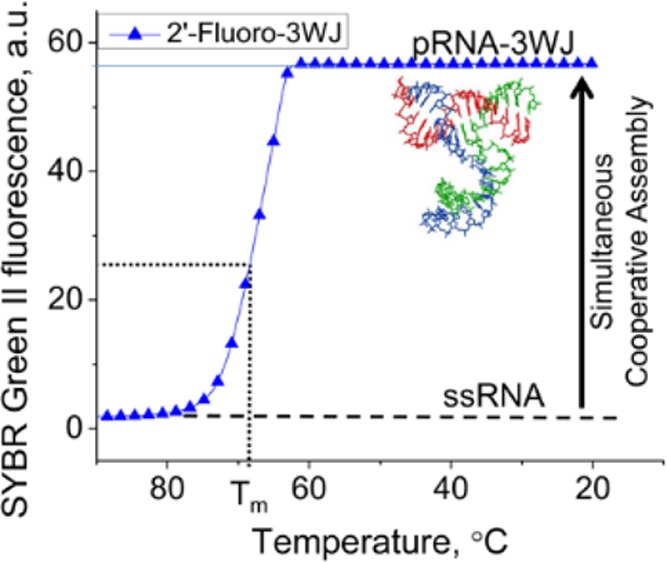
The emerging field of RNA nanotechnology necessitates creation of functional RNA nanoparticles but has been limited by particle instability. It has been shown that the three-way junction of bacteriophage phi29 motor pRNA has unusual stability and can self-assemble from three fragments with high efficiency. It is generally believed that RNA and DNA folding is energy landscape-dependent, and the folding of RNA is driven by enthalpy. Here we examine the thermodynamic characteristics of the 3WJ components as 2′-fluoro RNA, DNA, and RNA. It was seen that the three fragments existed either in 3WJ complex or as monomers, with the intermediate of dimers almost undetectable. It seems that the three fragments can lead to the formation of the 3WJ complex efficiently within a rapid time. A low dissociation constant (apparent KD) of 11.4 nM was determined for RNA, inclusion of 2′-F pyrimidines strengthened the KD to 4.5 nM, and substitution of DNA weakened it to 47.7 nM. The ΔG°37, were −36, −28, and −15 kcal/mol for 3WJ2′-F, 3WJRNA, and 3WJDNA, respectively. It is found that the formation of the three-component complex was governed by entropy, instead of enthalpy, as usually found in RNA complexes.
Since the proof-of-concept in 1998,1 RNA nanotechnology has been emerged as a popular field.2−10 RNA has the simplistic chemical characteristics of DNA with the complex folding and functionality of proteins.2 These attributes make RNA an ideal candidate for creating nanoparticles with diverse functionalities for targeting and treating cancer tumors and viral infections, as well as other applications in nanodevices.1,5,11−21 RNA nanotechnology in therapeutics provides many advantages over current technologies:2,22 (1) RNA can be produced with a defined shape and stoichiometry as well as high reproducibility.1,5,12,23 There are many modified nucleosides within RNA, but it is primarily composed of only four nucleic acid bases, allowing for simplicity in structure and predictable interactions between molecules and formation of structures. (2) RNA can target specific cell groups by targeting cell surface receptors through the use of RNA aptamers that function like protein or chemical ligands.24−26 These structures do not induce antibody production, allowing for repeated delivery and therapy.24 (3) RNA nanoparticles that have been produced have a size range of 10–50 nm,27−29 the perfect size to be retained within the body and pass through leaky blood vessels in cancer tumors,30,31 as well as cell membranes by cell surface receptor endocytosis.29 (4) RNA can be created to harbor multiple therapeutic elements by utilizing branch scaffolds21,32−35 and bottom-up construction.5,36 Even with these advantages, RNA nanotechnology has been hindered because of the instability of RNA itself, specifically in vivo. Dissociation of complex without covalent bonds is an intrinsic property of molecules, for example, RNA molecules, that are thermodynamically unstable in nature.37−45
When RNA nanoparticles are delivered systemically to the body, the particles will exist in low concentrations due to dilution by circulating blood. Only those RNA particles that do not dissociate at low concentrations are feasible for therapeutic purposes that require systemic delivery. Furthermore, RNA can easily be degraded and cleaved by RNases found throughout the human body.22 Chemically modified nucleotides have been developed to combat the nuclease degradation. Specifically, 2′-F modified nucleotides have been shown to keep the original folding and functionality of the RNA molecules while significantly increasing the half-life.46 To overcome the instability issues and push the RNA nanotechnology field to progress further, a stable platform needs to be produced that can remain stable at low concentrations and high temperatures while resisting RNase degradation.12−14
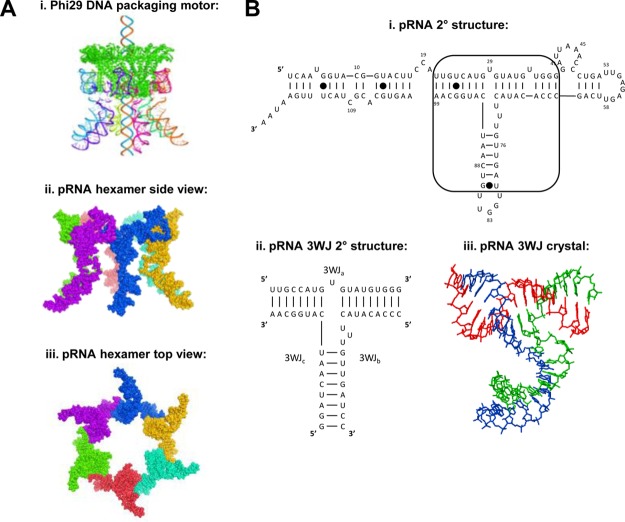
Recently, a three way junction motif (3WJ) in the packaging RNA (pRNA) of the bacteriophage phi29 dsDNA packaging motor was found to be ultrastable.12 The pRNA-3WJ produced a melting curve indicative of a low Gibbs free energy (ΔG°),47,48 as well as a high melting temperature (Tm). It has also been elucidated that the 3WJ is stable in ultralow concentrations, as well as in 8 M urea. This junction serves as the central core of the pRNA linking the helical domain49 to the interlocking looped regions50 and allows for intermolecular interactions with other pRNA molecules (Figure 1). The core can be formed from three individual RNA oligos with a high efficiency without the presence of metal ions. It has been found that this junction can incorporate RNA functional moieties, such as receptor-binding aptamer,51,52 siRNA,33,36,53 ribozyme,54−56 miRNA,57−59 or riboswitch.60,61 Additionally, the resulting structures have the ability to keep the strong folding of the core, while retaining the functionality of the conjugated RNA moieties.12 Even with the strong stability of the 3WJ, it is still very much susceptible to degradation by RNases; therefore, chemical modifications to the RNA are required.40,62,63 The effects on the thermodynamic stability of the pRNA-3WJ core, though, are still unknown.
Figure 1.
Background information for motor pRNA, pRNA three-way junction (3WJ), and the application of their thermodynamic stability to build RNA nanoparticles. (A) The side-view of the hexametric structure of phi29 DNA packaging motor.79 Two bottom panels are side-view and top-view of the hexameric pRNA derived from X-ray crystallography.80 (B) Predicted secondary structure of the phi29 packaging RNA (pRNA) with the 3WJ motif outlined and the pRNA-3WJ and pRNA-3WJ secondary structure with solved crystal structure.
Previously, thermodynamics of nucleic acids and their folding properties have been studied; however, the majority of the studies have been completed on RNA and DNA duplex sequences.38,41−43,64−67 An estimation of thermodynamic parameters for RNA 3WJ and 4WJ have been surmised68 from studies completed using two piece designs by incorporating looped regions between helical branches. A gap has remained within the thermodynamic studies of nucleic acids regarding elucidating thermodynamic characteristics of multibranched structures, that is, multistranded motifs. Additionally, the understanding of chemical modifications to the RNA backbone on RNA junctions has remained a mystery, and untouched. To understand the thermodynamic characteristics of such structures and comprehend the governing laws of motif folding, new methods must be developed to be able to see multistranded interactions.
Here we report the measured thermodynamic parameters for 3WJ complexes containing DNA, RNA, and 2′-F U/C modified RNA strands (Figure 1), as well as hybrid complexes by means of comparison of their stability using a real-time polymerase chain reaction (rtPCR) machine and temperature gradient gel electrophoresis (TGGE). Results concluded that use of DNA strands weakened the structure of the pRNA-3WJ, while 2′-F modifications strengthened the RNA motif by elevating the transition temperatures and lowering ΔG°. More importantly, the data appear to show the 3WJ formed in a single step without displaying the presence of dimer species, showing that all three strands of the pRNA-3WJ formed together into a complex rapidly with the intermediate product undetectable. The assembly was also revealed to be entropy driven.
Materials and Methods
Oligonucleotides and Assembly of 3WJs
RNA and DNA oligonucleotides were obtained from Integrated DNA Technologies (IDT). RNA oligonucleotides containing 2′-F U/C modifications were ordered from Trilink BioTechnologies. Assembly of 3WJs was performed by mixing equimolar concentration of corresponding strands in TMS buffer (50 mM TRIS pH = 8.0, 100 mM NaCl, 10 mM MgCl2), heating to +80 °C, and slowly cooling to +4 °C at a rate of 2.0 °C/min for a total of 37 min. The 3WJ formations were assayed on a 12% native PAGE TBM running buffer (89 mM Tris, 200 mM borate acid, and 5 mM MgCl2) ran at 100 V at 4 °C for 90 min. Gels were stained with ethidium bromide (EB) and imaged using a Typhoon (GE).
KD Measurements
Apparent equilibrium dissociation constants (KD) for 3WJ formations were determined by titration over a range of concentrations from 0.1 nM to 512 nM, as previously described.23,69 Concentrations of the 3WJs were calculated using the absorbance of UV light at 260 nm using a Nanodrop 2000 (Thermo Scientific) using an optical density equaling one as 40 μg/mL and 50 μg/mL for RNA and DNA, respectively. Briefly, fixed amounts of [32P] ATP 5′-end labeled 3WJb strands of RNA, DNA, and 2′-F modified RNA (0.1 nM final) were mixed with variable amounts of unlabeled 3WJa and 3WJc strands RNA to make the indicated final concentration (0.1 nM to 512 nM) of each. The resulting 3WJs were then heated to 80 °C for 3 min in TMS buffer and slowly cooled to 4 °C. The resulting gel shifts were measured using Image J software and interpreted with the program Origin 8.0. The fractions (f) for each trimer forming complex was calculated by dividing the corresponding quantified values for trimers by the total sum of values for all complexes (monomer, dimer, and trimer) presented in the corresponding lane. The combined data from several independent measurements were subjected to nonlinear curve fitting using the equation:
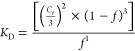 |
1 |
where Ct is the total concentration of RNA strands in each lane, and f is the fraction of 3WJ complex to total concentration.
In calculating the Ct, the concentration of the labeled 3WJb strand was not included, as it was in trace amounts and negligible to the concentrations of 3WJa and 3WJc. The Ct at equilibrium was calculated by interpolation of the fraction (f) at 50%. These values were then in turn used to calculate the equilibrium KDs.48 Marky et al. described this calculation using changing temperature to reach equilibrium, thus requiring equal concentrations of each strand; however, equilibrium was reached here by varying concentrations, as previous performed,23,69 thus not requiring equal concentrations of each strand.
Real-Time PCR Annealing Curves of 3WJ Complexes
Each synthesized pRNA-3WJ strand (3WJa, 3WJb, 3WJc) was mixed at equimolar concentrations in the presence of 1× SYBR Green II dye (Invitrogen) at equal molar ratios of 10 μM to create a 3WJRNA, 3WJDNA, and 3WJ2′-F. SYBR Green II dye is a reporter dye with higher specificity for RNA but shows binding to both DNA and RNA bases.70 Using a Roche Lightcycler 480 real-time PCR machine, with accurate readings of temperatures to ± >0.25 °C, samples were heated to 95 °C for 5 min for an initial denaturing period and then slowly cooled at a rate of 0.11 °C/s until 20 °C. The Roche Lightcycler 480 was able to detect the fluorescence levels and thus monitor the formation of the 3WJ cores. All samples were completed in triplicate to ensure accuracy of the annealing temperatures and profiles. Samples were then subjected to electrophoresis on a 12% polyacrylamide gel to ensure the formation of the 3WJs. All hybrid structures of the pRNA-3WJ core were created using a mix of RNA/DNA, RNA/2′-F RNA, or DNA/2′-F RNA in the same method described as above.
TGGE and Thermodynamic Parameters
All 3WJs were assembled as described above. The TGGE system from BiometraHmbGh, Germany, was used in this study. Temperature was varied perpendicular or parallel to the electrical current. All experiments were performed in TMS running buffer. The RNA bands were detected in native 15% gel by total RNA stain using EB. The concentrations of 3WJs were typically 10 μM. All varieties were run a minimum of three times to ensure accuracy of measurement.
Various concentrations of preassembled pRNA-3WJs covering 1000-fold dilutions from 10 μM to 0.016 μM were subjected to the perpendicular TGGE for thermodynamic parameter calculations. 3WJ bands were detected with radiolabeled 3WJc strands at the 5′-end using γ-32P ATP (Perkin-Elmer). A linear temperature gradient was applied perpendicular to the electric field. The gels were run for 1 h at constant 80 V, dried, and exposed to phosphoroimaging screen overnight. Gels were then visualized on a Typhoon (GE), and Image J software was used to quantify the area of bands on each lane by plotting intensity of each band intensity and integrating the area under the curve for each band. Background signal of each lane immediately surrounding the band of interest from the gel was subtracted and removed from the band intensity and not included in calculations. The fractions of 3WJ bands were obtained by subtracting the area of melted bands from the trimer band. Melting temperatures were then calculated from a plot of 3WJ fraction (f) versus temperature, with f = 0.5 (50%) corresponding to the Tm.
Calculation of Thermodynamic Parameters
Tm values at different concentration of 3WJs were used to calculate thermodynamic parameters. van’t Hoff plots were generated by plotting the Tm versus the concentrations of non-self-complementary three molecular strands according to a previous method:48
| 2 |
where R is the gas constant 1.987 cal mol–1 K–1, Ct is the total concentration of each 3WJs, and ΔH° and ΔS° are enthalpy and entropy changes, respectively. Under this method of calculation, the assumption is made that the heat capacity remains unchanged throughout the entire melting profile.
Fluorescence of pRNA 3WJ-MG Aptamer
The assembled unmodified and 2′-F A/C modified 3WJs (1 μM) were mixed with MG (2 μM) in binding buffer containing 100 mM KCl, 5 mM MgCl2, and 10 mM HEPES (pH 7.4) and incubated at room temperature for 30 min. The fluorescence was measured using a fluorospectrometer (Horiba Jobin Yvon; SPEX Fluolog-3), excited at 615 nm, and scanning from 625 to 800 nm for emission.71
Results and Discussion
Assembly of the Three-Way Junctions
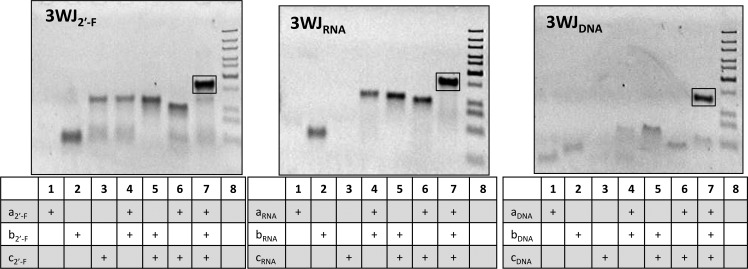
pRNA three way junctions composed of RNA (3WJRNA), DNA (3WJDNA), and 2′-F U/C modified RNA (3WJ2′-F) were assembled using equimolar concentrations of each strand of the 3WJ. The formation of the pRNA-3WJs in each of the species was confirmed by 12% native PAGE (Figure 2). A stepwise assembly through the polyacrylamide gel was observed between the monomer to dimer and finally to the fully assembled trimer band indicating the formation of 3WJs. The assembled RNA, DNA, and 2′-F 3WJs products resulted in high yield (>90%). Any side product is assumed to be contributed to a slight mismatch in strand concentrations loaded during assembly, preventing all nucleic acid strands from forming into the 3WJ complex. The data are in agreement with previously reported results of RNA-3WJ assembly.12
Figure 2.
Assembly of pRNA-3WJ core structures. 12% polyacrylamide gels in native conditions displaying the stepwise assembly of the 3WJ2′-F, 3WJRNA, and 3WJDNA.
Dissociation Constants (KD) Measurements of the 3WJs
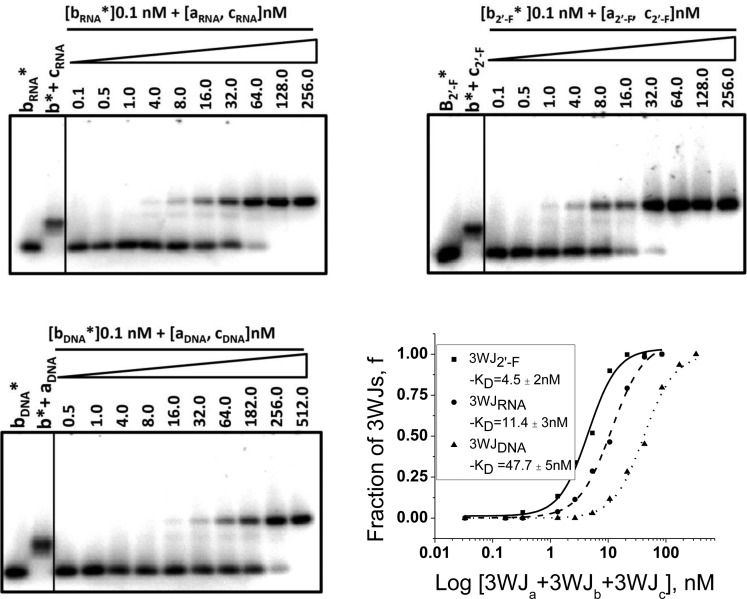
The KD values of nucleic acids formation are directly related to their stabilities as more stable complexes require lower concentrations of the components for self-assembly resulting in a lower KD value. We measured the apparent dissociation equilibrium constants for RNA, DNA, and 2′-F modified RNA 3WJ complexes (Figure 3). For the 3WJRNA and 3WJ2′-F, the KD values were found to be 11.4 and 4.5 nM, respectively. For the 3WJDNA complex, this value was about five times higher (47.7 nM). This indicates that 3WJ2′-F and 3WJRNA were the most stable complexes, and the least stable was 3WJDNA.
Figure 3.
Dissociation constant measurements. Electrophoeretic mobility shift assay (EMSA) of 32P-labeled 3WJb (constant concentration) assembling with increasing concentrations of 3WJa and 3WJc for RNA, 2′-F RNA, and DNA on 12% polyacrylamide gels in native conditions. Plot of percentage of 3WJ formed versus total concentration to solve for KD of each 3WJ.
These results demonstrated that the incorporation of enzymatically resistant DNA or 2′-F modified RNA strands into the RNA 3WJ motif could increase the resistance of the complex to RNases. However, while the DNA strand incorporation decreases the stability, the 2′-F modified RNA increases the stabilization of the 3WJ complex, as shown by the lower dissociation constant compared to the 3WJRNA. Therefore, the stability and properties of 3WJRNA motif can be potentially tuned by alternating the ratio of 2′-F modified RNA and DNA strands.
Determination of the Formation of Complex and Ta of DNA, RNA, and 2′-F RNA-3WJs by rtPCR Machine
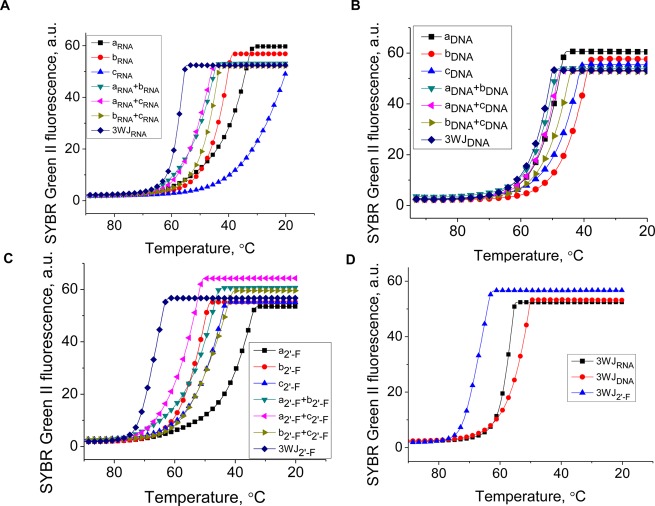
The thermostability of 3WJs complexes were compared by measuring their fluorescence intensities as a function of temperature in the presence of SYBR Green II dye on a Roche 480 Lightcycler to obtain Ta.12 The annealing transitions in Figure 4A–C show that the mixture of three strands (3WJa, 3WJb, and 3WJc) produced the highest annealing temperature compared to any monomer or dimer formation. Within the assembling profile, the slope of the transitions directly correlates with ΔG°, as the steeper slope results in more negative ΔG° values.47,48 These results showed that the assembly of the 3WJ was preferred over any dimer formation of any two strands in each of the three species: RNA, DNA, and 2′-F modified RNA.
Figure 4.
Assembly curves produced from the Roche 480 Lightcycler using SYBR green II reporter dye. (A) 3WJRNA, (B) 3WJDNA, (C) 3WJ2′-F, and each of the components of the 3WJs. (D) The three 3WJ species directly compared, showing the differences in annealing temperatures (Ta).
From each of the transitions obtained by the Roche 480 Lightcycler, the curves of the each of the completed 3WJ structures were compared (Figure 4D). The 3WJ2′-F was the most stable with Ta = 69.8 °C, the 3WJRNA was the next stable at 59.3 °C, and the 3WJDNA had the lowest where Ta = 48.9 °C. These results correlated with literature reported values of overall thermostability for nucleic acids and followed the order of stability: 2′-F RNA > RNA > DNA.43,64,72
Surprisingly, from the assembly profiles of the pRNA-3WJ species, a single annealing temperature, not two, was seen. This is evidenced by the single slope to the transition profile and hinting that the 3WJ forms rapidly with all three strands producing no byproducts, whereas a two-step association would display a plateau within the melting curve itself or two individual sloped regions, resulting in two annealing temperatures. These results show the three-stranded motif forming together, and a dimer species was undetectable due to the rapid 3WJ formation. This rapid formation from three fragments to form the pRNA-3WJ is highly beneficial because it allows a high yield of assembly while permitting the construction and assembly of RNA nanoparticles without creating side products as a result of the dimer formation.
Comparison of Stability between DNA, RNA, and 2′-F RNA-3WJs by TGGE
TGGE is common technique to measure Tm of large and complex nucleic acids.23,73,74 This approach has an advantage over the real-time PCR in that it can be applied to directly measure the Tm of RNA complexes as fractions of RNA versus temperature with no intercalation dye required.
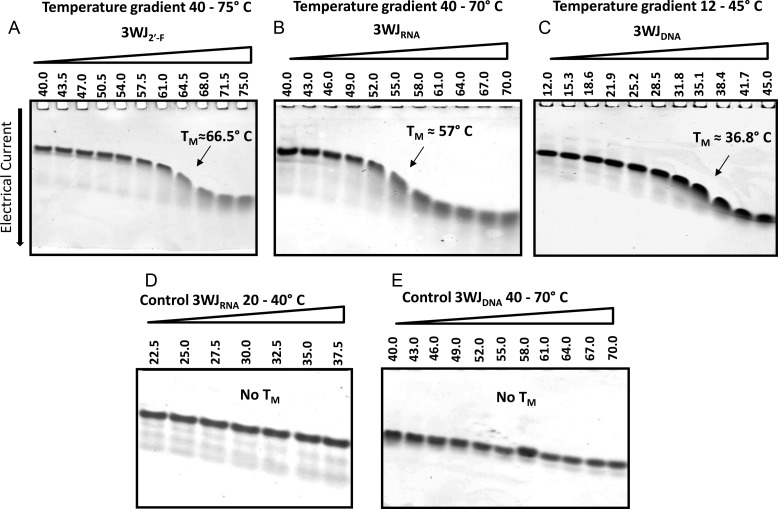
Melting temperatures of 3WJ complexes were determined by measuring the decrease in the yield of 3WJs versus temperature (Figure 5). A temperature gradient was applied perpendicular to the electrical current, with an increasing temperature that resulted in the melting of the structures in the later lanes. Here, the percent 3WJ complex was compared to dimer and monomer formations. Tm values were determined as 50% of 3WJ remaining. Any remaining dimers were not considered as a complex, as it was not complete 3WJ formations.
Figure 5.
Representative 15% native TGGE with temperature gradient perpendicular to the electrical current. (A) 3WJ2′-F, (B) 3WJRNA, and (C) 3WJDNA; total 3WJ concentration in each lane = 10 μM. Panels (D) and (E) are control gels showing migration of 3WJRNA (D) before reaching its Tm in the temperature range of 20–40 °C and migration of 3WJDNA (E) in the temperature range of 40–70 °C that is over its Tm. The 3WJ bands were detected by total nucleic acid stain with EB.
From the resulting gels, melting temperatures were derived for the 3WJ2′-F, 3WJRNA, and 3WJDNA as 66.5 °C, 57 °C, and 35.2 °C, respectively. The melting temperatures for the two RNA species were within the range of error from the Tm found by the fluorescence melting curves, but there was a large difference in Tm between the two methods for the 3WJDNA. This difference between these two methods was presumably due to differing affinities of SYBR Green II to DNA and RNA stacking bases.70 Nevertheless, both methodologies produced data that allowed direct comparison of the stability among 3WJ complexes and demonstrated the trend that 2′-F RNA had a higher melting temperature than RNA, and RNA had a higher Tm than DNA.
Within the TGGE melting gels, the dimer species were again undetected in the melting curves in the 2′-F RNA and RNA 3WJ species, further pointing to the possibility of the unusual assembly pathway. However, the 3WJDNA seemed to display a dimer intermediate, which was previously not seen from the PCR profiles. It is believed that this difference is because the TGGE showed the melting profile, while the PCR curves showed the association of the molecules. Therefore, the TGGE system of the DNA species was dissociation down to a dimer then a monomer, possibly because DNA was not the natural species of the pRNA-3WJ and forced the motif into an unnatural conformation. Furthermore, any dimer species that is seen in radiolabeled gel analysis can be attributed to the presence of extreme excess unlabeled strands allowing for a mismatch in concentrations between each individual strand and were therefore ignored.
Thermodynamic Parameters for 3WJs Formation
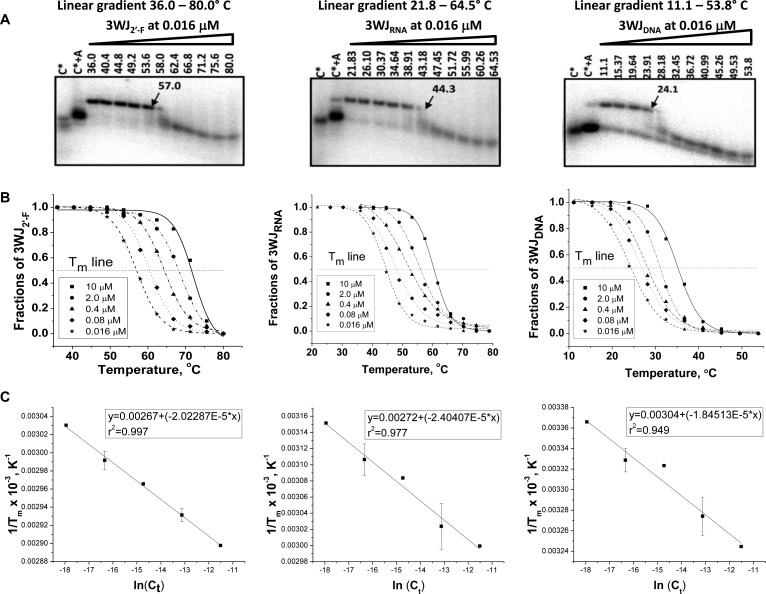
Non-self-complementary nucleic acids usually exhibit a linear correlation Tm and RNA concentration, as the Tm increased with the increase in nucleic acid strands concentration.38,68,75 The TGGE approach was further applied to measure Tm of 3WJ species as a function of concentration using ∼1000 fold dilution of the 3WJRNA, 3WJ2′-F, and 3WJDNA complexes from 10 μM to 0.016 μM (Figure 6A). Tm’s were calculated by finding the temperature at which 50% of the nucleic acids were in trimer formation compared to the total concentration of the bands observed. 3WJ Tms were used to calculate thermodynamic parameters for three components of nonself complementary sequences, according to Marky et al.48 The typical van’t Hoff plots for 3WJ2′-F, 3WJRNA, and 3WJDNA are represented in Figure 6C. From the obtained ΔH° and ΔS° parameters, the ΔG°37 was calculated using eq 3:
| 3 |
where ΔH° and ΔS° are enthalpy and entropy, respectively, and K is the abbreviation for Kelvin. All thermodynamic parameters are summarized in Table 1. Here the data produced a near linear trend lines with r2 values very near 1.00. Because this little error is seen in calculating melting temperatures, samples were repeated in a selective fashion due to the high number of gels that would be needed. The repeated experiments allowed for calculation of error in the thermodynamic parameters which remained low. The results indicated that the most thermodynamically stable was the 3WJ2′-F (ΔG°37 = −36 kcal/mol) complex, followed by 3WJRNA (ΔG°37 = −28 kcal/mol), and the 3WJDNA (ΔG°37 = −15 kcal/mol). On the basis of the parameters of free energy change for RNA and 2′-F RNA-3WJ assemblies, it can be determined that the energy for these complexes were favored 2-fold compared to the 3WJDNA. The 3WJDNA displayed a more favorable decreased enthalpy value of ΔH° = −220 kcal/mol, compared to 3WJ2′-F (ΔH° = −200 kcal/mol) and 3WJRNA (ΔH° = −170 kcal/mol). However, the comparison of entropy parameters resulted in a significant increase for the RNA and 2′-F RNA complexes. The two RNA species resulted in more negative free energy changes, yet had higher enthalpies when compared to DNA. This data combined with the increased entropy values of the RNA species indicated that the thermodynamic stabilities of the 3WJRNA and 3WJ2′-F were entropy-driven (Table 1).
Figure 6.
Calculation of thermodynamic parameter for 3WJs formation. (A) Representation of native 15% TGGE for the 3WJ complexes at the lowest concentration (0.016 μM in each lane). The radiolabeled C strand of corresponding 3WJ complexes indicated with asterisk “*”. Lanes 3WJc* and 3WJc*+3WJa served as controls for monomer and dimer location. (B) Melting temperature profiles of RNA, 2′-F RNA, and DNA 3WJs obtained after quantification of the corresponding band from the perpendicular TGGE at various concentrations. (C) Plots of Tm versus 3WJ concentrations (van’t Hoff analysis) evaluated from melting curves of the 3WJ complexes obtained at different strand concentrations.
Table 1. Thermodynamic Parameters for 3WJs Formationa.
| 1/Tm vs log (Ct) |
||||
|---|---|---|---|---|
| 3WJs | ΔG°37 (kcal/mol) | ΔH° (kcal/mol) | ΔS°37(e.u.) | Tmb (°C) |
| 2′-F RNA | –36 ± 0.45 | –200 ± 5.7 | –520 ± 17 | 72.1 |
| RNA | –28 ± 0.58 | –170 ± 13 | –440 ± 39 | 60.4 |
| DNA | –15 ± 0.71 | –220 ± 25 | –650 ± 83 | 35.2 |
Parameters derived from 15% native TGGE.
Tm values for 3WJ strand concentrations of 10–6 M.
The findings of more negative ΔG° values for 2′-F RNA and RNA are consistent with other studies comparing helical DNA, RNA, and 2′-F RNA.43,64,65,72,76 However, the notion that was RNA entropically driven, compared to DNA in helical structures, appeared less unanimous as the majority report RNAs to be less entropically favored and that folding is normally driven by a lower enthalpy value.43,64,65,76 Here it is believed that the 3WJDNA provided a more rigid structure, producing a lower internal energy or ΔH compared to the RNAs; however, the flexibility of the RNAs provided more disorder, thus giving strong stability, ease of folding, and a more negative ΔG°. This entropy-driven assembly combined with the one-step assembly expresses the unusual thermodynamic characteristics of the pRNA-3WJ.
Analysis of 3WJ Hybrid Formations and Thermostability
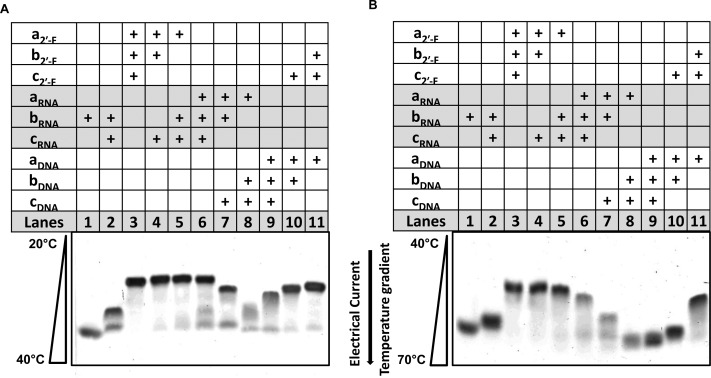
The hybrid composition (2′-F RNA/RNA; RNA/DNA; DNA/2′-F RNA) within the RNA 3WJs are of great interest due to their ability to maintain the diverse functionality of structured RNA molecules, while incorporating the chemical stability of 2′-F RNA and DNA. To test for hybrid 3WJ viability, the 2′-F RNA/RNA; RNA/DNA; DNA/2′-F RNA hybrids of the 3WJs complexes were characterized by parallel TGGE and fluorescence annealing temperature experiments. Using the TGGE, a temperature gradient was applied in parallel to the electrical current (Figure 7). As the samples migrated through the gel, the temperature increased from 20 to 70 °C, therefore melting the hybrid structures as they migrated further into the gel. A less stable 3WJ complex migrates further as the elevated temperature melts the structure to smaller single strands, thus causing an increased rate of migration and separating the stable hybrids from unstable hybrids.
Figure 7.
Native 15% TGGE of some hybrids 3WJs with temperature gradient in parallel of the electrical current. As the 3WJs migrated into the gels, weaker structures melted due to the elevating temperatures, resulting in a more rapid migration; stable structures migrated at slower rates. Concentration of hybrids in each lane = 10 μM; the bands were detected by total nucleic acid stain with EB. (A) Hybrids analyzed in a linear temperature gradient of 20–40 °C and (B) the same samples but the temperature range was 40–70 °C.
The TGGE analysis demonstrates that each hybrid structure forms correctly and is stable at lower temperature ranges 20–40 °C (A). However, at a higher range of 40–70 °C, the RNA/DNA and DNA/2′-F RNA-3WJ hybrid structures melted, resulting in a more rapid migration rate compared to 2′-F RNA/RNA hybrids (Figure 7B). This direct comparison between hybrid stability demonstrates weakness in the thermostability of hybrids involving DNA strands. In addition, the 3WJs hybrids followed the general trend that more strands with 2′-F modifications equate to a higher stability overall. These results were further confirmed by the annealing temperatures produced for each hybrid on the Roche 480 Lightcycler as shown in Figure 8 and Table 2. Combining the TGGE gels along with the annealing temperatures provided from the fluorescence annealing curves, the data further support the findings that 2′-F modifications strengthen the thermostability of the pRNA-3WJ, while DNA substitutions only weaken the 3WJ complex. In this case, even limited modifications lead to a difference in the thermostability.
Figure 8.
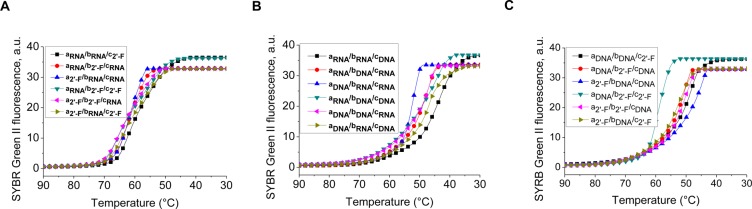
Comparison of pRNA-3WJ hybrid structures. Assembly curves produced from the Roche 480 Lightcycler of the pRNA-3WJ (A) RNA/DNA hybrids, (B) RNA/2′-F RNA hybrids, and (C) DNA/2′-F RNA hyrbids.
Table 2. Annealing Temperature for 3WJ Hybrid Formationa.
| 2′-F RNA to RNA | Ta (°C) | RNA to DNA | Ta (°C) | DNA to 2′-F RNA | Ta (°C) |
|---|---|---|---|---|---|
| a2′-F/b2′-F/c2′-F | 69.8 ± 2.0 | aRNA/bRNA/cRNA | 59.3 ± 1.7 | aDNA/bDNA/cDNA | 48.9 ± 3.2 |
| a2′-F/b2′-F/cRNA | 65.4 ± 0.1 | aRNA/bRNA/cDNA | 42.6 ± 2.2 | aDNA/bDNA/c2′-F | 48.4 ± 1.6 |
| a2′-F/bRNA/c2′-F | 64.1 ± 0.2 | aRNA/bDNA/cRNA | 48.6 ± 1.5 | aDNA/b2′-F/cDNA | 51.6 ± 0.4 |
| aRNA/b2′-F/c2′-F | 65.5 ± 0.2 | aDNA/bRNA/cRNA | 53.1 ± 0.1 | a2′-F/bDNA/cDNA | 47.2 ± 1.5 |
| a2′-F/bRNA/cRNA | 62.1 ± 0.1 | aRNA/bDNA/cDNA | 44.5 ± 2.6 | aDNA/b2′-F/c2′-F | 59.5 ± 0.2 |
| aRNA/b2′-F/cRNA | 62.7 ± 0.2 | aDNA/bRNA/cDNA | 45.9 ± 2.4 | a2′-F/bDNA/c2′-F | 52.4 ± 0.6 |
| aRNA/bRNA/c2′-F | 61.9 ± 0.4 | aDNA/bDNA/cRNA | 47.3 ± 0.4 | a2′-F/b2′-F/cDNA | 51.2 ± 1.8 |
Annealing temperatures calculated at 10 μM total strand concentration in TMS buffer.
MG-Aptamer Functionality Assay and Stability
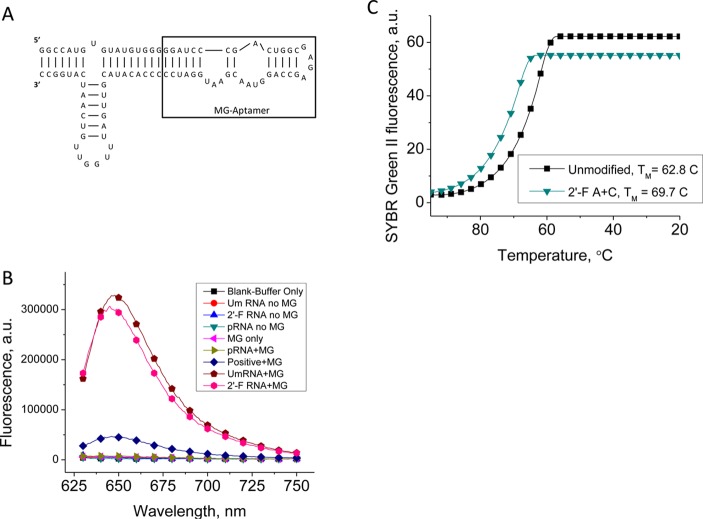
To ensure that the added stability of the 2′-F modifications to the pRNA-3WJ was true for a functional, more complex RNA nanoparticle, a fluorescence assay was performed. The pRNA-3WJ used in this study harbored the Malachite Green (MG) RNA aptamer that binds to Malachite green triphenylmethane dye causing the chemical to fluoresce.77 Malachite green itself emits very low fluorescence; therefore, a change in the fluorescence can be used to confirm binding and complexation between the RNA aptamer and chemical.30,71,77,78
It has been previously shown that the MG aptamer remains active in binding when nucleotides remain unmodified, as well as when using 2′-F cytosine and adenine modified nucleotides.71 Therefore, we constructed the pRNA-3WJ-MG using unmodified RNA strands and 2′-F A/C modified RNA strands (see Figure 9A for 2D structure). The resulting particles were then tested for binding to the MG to show the folding and functionality of the MG aptamer by measuring its fluorescence emissions (Figure 9B). Both nanoparticles showed almost identical fluorescence values indicating the formation of correctly folded structures.
Figure 9.
Thermostability of functional pRNA-3WJ nanoparticle. (A) Secondary structure and sequence of the 3WJ-MG aptamer. (B) Fluorescence emission of 3WJ-MG aptamer, demonstrating binding of the nanoparticle to MG in both RNA and 2′-F species. (C) Melting curves for the 3WJ-MG and 2′-F A/C RNA-3WJ.
Further, we used real-time PCR to measure the Ta of pRNA-3WJ-MG and 3WJ-MG2′-F constructs (Figure 9C). The annealing curves show that the 2′-F A/C modified 3WJ resulted in the annealing temperature of 69.7 °C, higher than unmodified RNA variant with Ta value of 62.8 °C. This is consistent with the trend of increased RNA 3WJ stability using 2′-F modifications. Thus, the data demonstrate the effectiveness of the fluorine modifications regardless of the complexity of the structure, while retaining the functional conformation of the pRNA-3WJ.
Conclusions
In this paper, we obtained thermodynamic parameters for the pRNA-3WJ using real-time PCR and TGGE approaches. It was seen that the three fragments existed either in 3WJ complex or as monomers, with the intermediate of dimers almost undetectable. It seems that the three fragments can lead to the formation of 3WJ complex efficiently within a rapid time. It is also found that the formation of the three-component complex was governed by entropy, instead of enthalpy, as usually found in RNA complexes. By combining this beneficial assembly with the improved thermodynamic characteristics by 2′-fluoro modifications to the pRNA, 3WJ stable RNA nanoparticles can be constructed with high yield for the treatment of cancers and viral infections.
Acknowledgments
The authors would like to thank Jeannie Haak for help in preparing this manuscript and Zhengyi Zhao for help in preparing figures.
Glossary
Abbreviations
- pRNA
packaging RNA
- 2′-F
2′-fluoro chemical modification
- KD
dissociation constant
- 3WJ
three-way junction
- Tm
melting temperature
- Ta
annealing temperature
- rtPCR
real-time polymerase chain reaction
- TGGE
temperature gradient gel electrophoresis
- ΔG°
change of Gibbs free energy
- ΔH°
change in enthalpy
- ΔS°
change in entropy
- EB
ethidium bromide
- MG
Malachite green
The authors declare the following competing financial interest(s): P. Guo is a co-founder of Kylin Therapeutics, Inc., and Biomotor and RNA Nanotechnology Development Corp. Ltd.
This research was supported by NIH Grants EB003730 and CA151648 to P.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. Funding to Peixuan Guo’s Endowed Chair in Nanobiotechnology position is by the William Fairish Endowment Fund.
Funding Statement
National Institutes of Health, United States
References
- Guo P.; Zhang C.; Chen C.; Trottier M.; Garver K. (1998) Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell 2, 149–155. [DOI] [PubMed] [Google Scholar]
- Guo P. (2010) The emerging field of RNA nanotechnology. Nat. Nanotechnol. 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X.; Hemida M.; Zhang H. M.; Hanson P.; Ye Q.; Yang D. (2012) Current advances in Phi29 pRNA biology and its application in drug delivery. Wiley Interdiscip. Rev. RNA 3(4), 469–481. [DOI] [PubMed] [Google Scholar]
- Afonin K. A.; Danilov E. O.; Novikova I. V.; Leontis N. B. (2008) TokenRNA: A new type of sequence-specific, label-free fluorescent biosensor for folded RNA molecules. ChemBiochem 9, 1902–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D.; Moll W. D.; Deng Z.; Mao C.; Guo P. (2004) Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett. 4, 1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L.; Chworos A. (2006) The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct Biol. 16, 531–543. [DOI] [PubMed] [Google Scholar]
- Chworos A.; Severcan I.; Koyfman A. Y.; Weinkam P.; Oroudjev E.; Hansma H. G.; Jaeger L. (2004) Building programmable jigsaw puzzles with RNA. Science 306, 2068–2072. [DOI] [PubMed] [Google Scholar]
- Lescoute A.; Westhof E. (2006) Topology of three-way junctions in folded RNAs. RNA 12, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N. B.; Westhof E. (2003) Analysis of RNA motifs. Curr. Opin. Struct. Biol. 13, 300–308. [DOI] [PubMed] [Google Scholar]
- Bindewald E.; Afonin K.; Jaeger L.; Shapiro B. A. (2011) Multistrand RNA secondary structure prediction and nanostructure design including pseudoknots. ACS Nano 5, 9542–9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D.; Huang L.; Hoeprich S.; Guo P. (2003) Construction of phi29 DNA-packaging RNA (pRNA) monomers, dimers and trimers with variable sizes and shapes as potential parts for nano-devices. J. Nanosci. Nanotechnol. 3, 295–302. [DOI] [PubMed] [Google Scholar]
- Shu D.; Shu Y.; Haque F.; Abdelmawla S.; Guo P. (2011) Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 6, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F.; Shu D.; Shu Y.; Shlyakhtenko L.; Rychahou P.; Evers M.; Guo P. (2012) Ultrastable Synergistic Tetravalent RNA Nanoparticles For Targeting To Cancers. Nano Today 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y.; Haque F.; Shu D.; Li W.; Zhu Z.; Kotb M.; Lyubchenko Y.; Guo P. (2013) Fabrication of 14 Different RNA Nanoparticles for Specific Tumor Targeting without Accumulation in Normal Organs. RNA 19, 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin K. A.; Grabow W. W.; Walker F. M.; Bindewald E.; Dobrovolskaia M. A.; Shapiro B. A.; Jaeger L. (2011) Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat. Protoc. 6, 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severcan I, Geary C, Jaeger L, Bindewald E., Kasprzak W., and Shapiro B. A. (2009) Computational and Experimental RNA Nanoparticle Design, in Automation in Genomics and Proteomics: An Engineering Case-Based Approach (Alterovitz G. and Ramoni M., Eds.) pp 193–220, Wiley, Boston. [Google Scholar]
- Tabernero J.; Shapiro G. I.; Lorusso P. M.; Cervantes A.; Schwartz G. K.; Weiss G. J.; Paz-Ares L.; Cho D. C.; Infante J. R.; Alsina M.; Gounder M. M.; Falzone R.; Harrop J.; Seila White A. C.; Toudjarska I.; Bumcrot D.; Meyers R. E.; Hinkle G.; Svrzikapa N.; Hutabarat R. M.; Clausen V. A.; Cehelsky J.; Nochur S. V.; Gamba-Vitalo C.; Vaishnaw A. K.; Sah D. W.; Gollob J. A.; Burris H. A. III (2013) First-in-Man Trial of an RNA Interference Therapeutic Targeting VEGF and KSP in Cancer Patients with Liver Involvement. Cancer Discov. 3, 406–417. [DOI] [PubMed] [Google Scholar]
- Yingling Y. G.; Shapiro B. A. (2007) Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett. 7, 2328–2334. [DOI] [PubMed] [Google Scholar]
- Delebecque C. J.; Silver P. A.; Lindner A. B. (2012) Designing and using RNA scaffolds to assemble proteins in vivo. Nat. Protoc. 7, 1797–1807. [DOI] [PubMed] [Google Scholar]
- Lee J. B.; Hong J.; Bonner D. K.; Poon Z.; Hammond P. T. (2012) Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat. Mater. 11, 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. I.; Lee T. Y.; Kim S.; Sun X.; Hong S. W.; Yoo J. W.; Dua P.; Kang H. S.; Kim S.; Li C. J.; Lee D. K. (2012) Enhanced intracellular delivery and multi-target gene silencing triggered by tripodal RNA structures. J. Gene Med. 14, 138–146. [DOI] [PubMed] [Google Scholar]
- Guo P.; Haque F.; Hallahan B.; Reif R.; Li H. (2012) Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid Ther. 22, 226–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin K. A.; Bindewald E.; Yaghoubian A. J.; Voss N.; Jacovetty E.; Shapiro B. A.; Jaeger L. (2010) In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmawla S.; Guo S.; Zhang L.; Pulukuri S.; Patankar P.; Conley P.; Trebley J.; Guo P.; Li Q. X. (2011) Pharmacological characterization of chemically synthesized monomeric pRNA nanoparticles for systemic delivery. Mol. Ther. 19, 1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y.; Cinier M.; Shu D.; Guo P. (2011) Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 54, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchia L.; Giangrande P. H.; McNamara J. O.; de Franciscis V. (2009) Cell-specific aptamers for targeted therapies. Methods Mol. Biol. 535, 59–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H.; Shi W.; Freund L. B. (2005) Mechanics of receptor-mediated endocytosis. Proc. Natl. Acad. Sci. U. S. A. 102, 9469–9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K. K. (2005) The role of nanobiotechnology in drug discovery. Drug Discovery Today 10, 1435–1442. [DOI] [PubMed] [Google Scholar]
- Li W.; Szoka F. (2007) Lipid-based Nanoparticles for Nucleic Acid Delivery. Pharm. Res. 24, 438–449. [DOI] [PubMed] [Google Scholar]
- Maeda H. (2001) The enhanced permeability and retention (EPR) effect in tumor vasculature: the key role of tumor-selective macromolecular drug targeting. Adv. Enzyme Regul. 41, 189–207. [DOI] [PubMed] [Google Scholar]
- Maeda H.; Nakamura H.; Fang J. (2013) The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv. Drug Delivery Rev. 65, 71–79. [DOI] [PubMed] [Google Scholar]
- Guo P. (2005) RNA Nanotechnology: Engineering, Assembly and Applications in Detection, Gene Delivery and Therapy. J. Nanosci. Nanotechnol. 5(12), 1964–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S.; Tschammer N.; Mohammed S.; Guo P. (2005) Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum. Gene Ther. 16, 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima Y.; Abe H.; Abe N.; Aikawa K.; Ito Y. (2011) Branched RNA nanostructures for RNA interference. Chem. Commun. (Cambridge) 47, 8367–8369. [DOI] [PubMed] [Google Scholar]
- Chang C. I.; Lee T. Y.; Yoo J. W.; Shin D.; Kim M.; Kim S.; Lee D. K. (2012) Branched, Tripartite-Interfering RNAs Silence Multiple Target Genes with Long Guide Strands. Nucleic Acid Ther. 22, 30–39. [DOI] [PubMed] [Google Scholar]
- Khaled A.; Guo S.; Li F.; Guo P. (2005) Controllable Self-Assembly of Nanoparticles for Specific Delivery of Multiple Therapeutic Molecules to Cancer Cells Using RNA Nanotechnology. Nano Lett. 5, 1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov P. L.; Filiminov V. V. (1978) Thermodynamic analysis of transfer RNA unfolding. J. Mol. Biol. 122, 447–464. [DOI] [PubMed] [Google Scholar]
- Freier S. M.; Kierzek R.; Jaeger J. A.; Sugimoto N.; Caruthers M. H.; Neilson T.; Turner D. H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. U. S. A. 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A.; SantaLucia J. J.; Tinoco I. J. (1993) Determination of RNA structure and thermodynamics. Annu. Rev. Biochem. 62, 255–285. [DOI] [PubMed] [Google Scholar]
- Kawasaki A. M.; Casper M. D.; Freier S. M.; Lesnik E. A.; Zounes M. C.; Cummins L. L.; Gonzalez C.; Cook P. D. (1993) Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 36, 831–841. [DOI] [PubMed] [Google Scholar]
- Sugimoto N.; Nakano S.; Katoh M.; Matsumura A.; Nakamuta H.; Ohmichi T.; Yoneyama M.; Sasaki M. (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- Lesnik E. A.; Freier S. M. (1995) Relative Thermodynamic Stability of Dna, Rna, and Dna-Rna Hybrid Duplexes - Relationship with Base Composition and Structure. Biochemistry 34, 10807–10815. [DOI] [PubMed] [Google Scholar]
- Gyi J. I.; Conn G. L.; Lane A. N.; Brown T. (1996) Comparison of the thermodynamic stabilities and solution conformations of DNA center dot RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry 35, 12538–12548. [DOI] [PubMed] [Google Scholar]
- Diamond J. M.; Turner D. H.; Mathews D. H. (2001) Thermodynamics of three-way multibranch loops in RNA. Biochemistry 40, 6971–6981. [DOI] [PubMed] [Google Scholar]
- Brunel C.; Marquet R.; Romby P.; Ehresmann C. (2002) RNA loop-loop interactions as dynamic functional motifs. Biochimie 84, 925–944. [DOI] [PubMed] [Google Scholar]
- Liu J.; Guo S.; Cinier M.; Shlyakhtenko L.; Shu Y.; Chen C.; Shen G.; Guo P. (2010) Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging. ACS Nano 5, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A. E.; Turner D. H.; Kim J.; Lyttle M. H.; Muller P.; Mathews D. H.; Zuker M. (1994) Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. U. S. A. 91, 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A.; Breslauer K. J. (1987) Calculating Thermodynamic Data for Transitions of Any Molecularity from Equilibrium Melting Curves. Biopolymers 26, 1601–1620. [DOI] [PubMed] [Google Scholar]
- Zhang C. L.; Lee C.-S.; Guo P. (1994) The proximate 5′ and 3′ ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage ϕ29 DNA. Virology 201, 77–85. [DOI] [PubMed] [Google Scholar]
- Reid R. J. D.; Bodley J. W.; Anderson D. (1994) Characterization of the prohead-pRNA interaction of bacteriophage phi29. J. Biol. Chem. 269, 5157–5162. [PubMed] [Google Scholar]
- Ellington A. D.; Szostak J. W. (1992) Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355, 850–852. [DOI] [PubMed] [Google Scholar]
- Gold L. (1995) The SELEX process: a surprising source of therapeutic and diagnostic compounds. Harvey Lect. 91, 47–57. [PubMed] [Google Scholar]
- Guo S.; Huang F.; Guo P. (2006) Construction of folate-conjugated pRNA of bacteriophage phi29 DNA packaging motor for delivery of chimeric siRNA to nasopharyngeal carcinoma cells. Gene Ther. 13, 814–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeprich S.; ZHou Q.; Guo S.; Qi G.; Wang Y.; Guo P. (2003) Bacterial virus phi29 pRNA as a hammerhead ribozyme escort to destroy hepatitis B virus. Gene Ther. 10, 1258–1267. [DOI] [PubMed] [Google Scholar]
- Sarver N. A.; Cantin E. M.; Chang P. S.; Zaia J. A.; Ladne P. A.; Stephens D. A.; Rossi J. J. (1990) Ribozymes as Potential Anti-HIV-1 Therapeutic Agents. Science 24, 1222–1225. [DOI] [PubMed] [Google Scholar]
- Liu H.; Guo S.; Roll R.; Li J.; Diao Z.; Shao N.; Riley M. R.; Cole A. M.; Robinson J. P.; Snead N. M.; Shen G.; Guo P. (2007) Phi29 pRNA Vector for Efficient Escort of Hammerhead Ribozyme Targeting Survivin in Multiple Cancer Cells. Cancer Biol. Ther. 6, 697–704. [DOI] [PubMed] [Google Scholar]
- Pegtel D. M.; Cosmopoulos K.; Thorley-Lawson D. A.; van Eijndhoven M. A.; Hopmans E. S.; Lindenberg J. L.; de Gruijl T. D.; Wurdinger T.; Middeldorp J. M. (2010) Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U. S. A. 107, 6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Zhu X.; Zhang X.; Liu B.; Huang L. (2010) Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol. Ther. 18, 1650–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X.; Liu Z.; Hemida M. G.; Yang D. (2011) Targeted delivery of mutant tolerant anti-coxsackievirus artificial microRNAs using folate conjugated bacteriophage Phi29 pRNA. PLoS One 6, e21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W. C.; Nahvi A.; Roth A.; Collins J. A.; Breaker R. R. (2004) Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286. [DOI] [PubMed] [Google Scholar]
- Mulhbacher J.; St-Pierre P.; Lafontaine D. A. (2010) Therapeutic applications of ribozymes and riboswitches. Curr. Opin. Pharmacol. 10, 551–556. [DOI] [PubMed] [Google Scholar]
- de P. D.; Bentley M. V.; Mahato R. I. (2007) Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. RNA 13, 431–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke M. A. (2008) Chemical modification of siRNAs for in vivo use. Oligonucleotides 18, 305–319. [DOI] [PubMed] [Google Scholar]
- Conte M. R.; Conn G. L.; Brown T.; Lane A. N. (1997) Conformational properties and thermodynamics of the RNA duplex r(CGCAAAUUUGCG)(2): Comparison with the DNA analogue d(CGCAAATTTGCG)(2). Nucleic Acids Res. 25, 2627–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauzan B.; McMichael E.; Cave R.; Sevcik L. R.; Ostrosky K.; Whitman E.; Stegemann R.; Sinclair A. L.; Serra M. J.; Deckert A. A. (2013) Kinetics and Thermodynamics of DNA, RNA, and Hybrid Duplex Formation. Biochemistry 52, 765–772. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J.; Frank R.; Blocker H.; Marky L. A. (1986) Predicting Dna Duplex Stability from the Base Sequence. Proc. Natl. Acad. Sci. U. S. A. 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santalucia J.; Allawi H. T.; Seneviratne A. (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- Mathews D. H.; Turner D. H. (2002) Experimentally derived nearest-neighbor parameters for the stability of RNA three- and four-way multibranch loops. Biochemistry 41, 869–880. [DOI] [PubMed] [Google Scholar]
- Novikova I. V.; Hassan B. H.; Mirzoyan M. G.; Leontis N. B. (2010) Engineering cooperative tecto-RNA complexes having programmable stoichiometries. Nucleic Acids Res. 39(7), 2903–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdalet E.; Roldan C.; Olivar M. P.; Lysnes K. (2005) Quantifying RNA and DNA in planktonic organisms with SYBR Green II and nucleases. Part A. Optimisation of the assay. Sci. Mar. 69, 1–16. [Google Scholar]
- Reif R.; Haque F.; Guo P. (2013) Fluorogenic RNA Nanoparticles for Monitoring RNA Folding and Degradation in Real Time in Living Cells. Nucleic Acid Ther. 22(6), 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallan P. S.; Greene E. M.; Jicman P. A.; Pandey R. K.; Manoharan M.; Rozners E.; Egli M. (2011) Unexpected origins of the enhanced pairing affinity of 2 ′-fluoro-modified RNA. Nucleic Acids Res. 39, 3482–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadalavada D. M.; Bevilacqua P. C. (2009) Analyzing RNA and DNA folding using temperature gradient gel electrophoresis (TGGE) with application to in vitro selections. Methods Enzymol. 468, 389–408. [DOI] [PubMed] [Google Scholar]
- Hecker R.; Wang Z. M.; Steger G.; Riesner D. (1988) Analysis of RNA structures by temperature-gradient gel electrophoresis: viroid replication and processing. Gene 72, 59–74. [DOI] [PubMed] [Google Scholar]
- Petersheim M.; Turner D. H. (1983) Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry 22, 256–263. [DOI] [PubMed] [Google Scholar]
- Watts J. K.; Martin-Pintado N.; Gomez-Pinto I.; Schwartzentruber J.; Portella G.; Orozco M.; Gonzalez C.; Damha M. J. (2010) Differential stability of 2 ′ F-ANA.RNA and ANA.RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility. Nucleic Acids Res. 38, 2498–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh C.; Grate D.; Wilson C. (2000) 2.8 Å crystal structure of the malachite green aptamer. J. Mol. Biol. 301, 117–128. [DOI] [PubMed] [Google Scholar]
- Babendure J. R.; Adams S. R.; Tsien R. Y. (2003) Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc. 125, 14716–14717. [DOI] [PubMed] [Google Scholar]
- Guo P.; Schwartz C.; Haak J.; Zhao Z. (2013) Discovery of a new motion mechanism of biomotors similar to the earth revolving around the sun without rotation. Virology 446, 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Endrizzi J. A.; Shu Y.; Haque F.; Sauter C.; Shlyakhtenko L. S.; Lyubchenko Y.; Guo P.; Chi Y. I. (2013) Crystal Structure of 3WJ Core Revealing Divalent Ion-promoted Thermostability and Assembly of the Phi29 Hexameric Motor pRNA. RNA 19, 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]