Abstract
Inflammation and subsequent cyclooxygenase-2 (COX-2) activity has long been linked with the development of cancer, although little is known about any epigenetic effects of COX-2. A product of COX-2 activation, levuglandin (LG) quickly forms covalent bonds with nearby primary amines, such as those in lysine, which leads to LG-protein adducts. Here, we demonstrate that COX-2 activity causes LG-histone adducts in cultured cells and liver tissue, detectable through LC–MS, with the highest incidence in histone H4. Adduction is blocked by a γ-ketoaldehyde scavenger, which has no effect on COX-2 activity as measured by PGE2 production. Formation of the LG-histone adduct is associated with an increased histone solubility in NaCl, indicating destabilization of the nucleosome structure; this is also reversed with scavenger treatment. These data demonstrate that COX-2 activity can cause histone adduction and loosening of the nucleosome complex, which could lead to altered transcription and contribute to carcinogenesis.
Cyclooxygenase-2 (COX-2) expression is associated with the development of many cancers,1,2 and the enzyme plays a key role in the progression of chronic gastrointestinal inflammation to cancer.3 Predictably, treatment with COX inhibitors decreases a person’s total risk of cancer.4 Prevention studies as well as animal models suggest that increased COX-2 activity is both an early event in carcinogenesis, which contributes to the cellular transformation to malignancy, as well as a sustained event in some colorectal and lung cancers that can be associated with metastasis and poorer clinical prognosis.5,6 As predicted by these data, inhibiting COX-2 activity with non-steroidal anti-inflammatory drugs (NSAIDS) or COX-2-specific inhibitors over time reduces a person’s total risk of colon, breast, lung, and prostate cancers.4
Despite the promise of these drugs in cancer prevention, the gastrointestinal toxicity associated with long-term NSAID treatment and increased cardiovascular events associated with COX-2-specific inhibitors7 limit their clinical use. A better understanding of the specific downstream contributions of COX-2 to carcinogenesis could lead to new treatments that bypass these undesirable effects.
The product of COX-2, prostaglandin H2 (PGH2), is converted enzymatically into other prostaglandins, and indeed PGE2 is a well-described promoter of carcinogenesis.8 However, depending on the animal model, deletion of microsomal PGE2 synthase-1 can either prevent9 or accelerate10 tumorigenesis, indicating that the contribution of COX-2 to cancer, particularly to cellular transformation, is probably multifaceted.
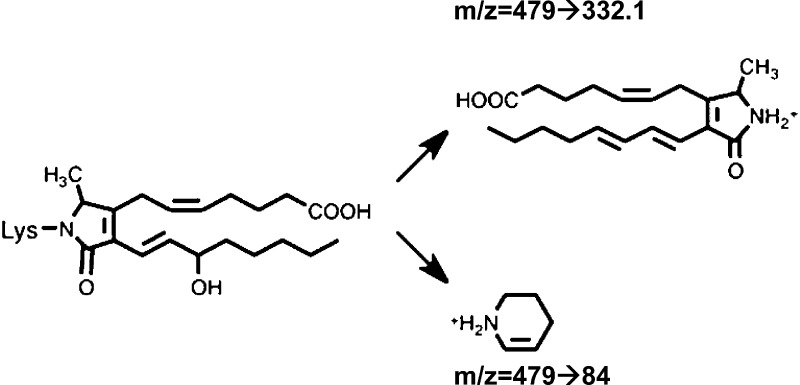
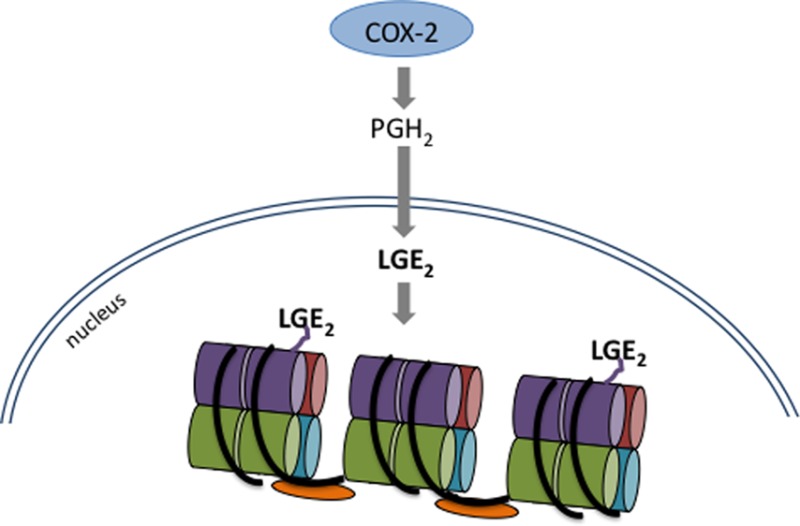
Besides enzymatic conversion, PGH2 also spontaneously rearranges in aqueous solution to form the highly reactive levuglandins LGE2 and LGD2. The γ-ketoaldehyde levuglandins (LGs) constitute about 20% of total PGH2 rearrangement products.11,12 Newly formed LGs react almost immediately with free amino groups, such as those in lysine,13,14 which leads to stable covalent LG-protein adducts measurable by mass spectrometry (Figure 1) or protein–protein cross-links.14,15 Following COX-2 activity, LG adducts of protein form in cells16 and in tissues.17 Proteins rich in lysine are thus especially susceptible to adduction, and due to the perinuclear localization of COX-2 and a PGH2 half-life measured in minutes,12 we speculated that it would be possible for PGH2 to cross the nuclear envelope before rearranging to LGE2, allowing formation of the LG adduct on the lysine-rich histones.
Figure 1.
Structure of the LG-lysyl adduct and the fragment ions monitored in positive ion mode (+H).
We have identified LG-histone adducts in multiple cancer cell lines as well as rat liver, with highest measurable amounts of adduct on the H4 histone. Adduct formation is dependent on COX-2 expression and activity, and we have found a small-molecule salicylamine derivative that scavenges LG to reduce adduct formation on histones without affecting COX-2 activity. Lysines, with their short side chain and positive charge, are critical for histone ionic interaction with DNA, and we find that this interaction is decreased with the introduction of a bulky negative charge from LG. Covalent histone modifications are a major method of controlling gene expression.18 Changes to lysyl modifications of histones19,20 are associated with human cancer. These findings link COX-2 induction with perturbation of normal DNA–histone interactions and provide a novel role for the enzyme in carcinogenesis.
Materials and Methods
Materials
All reagents and chemicals were purchased from Sigma-Aldrich (St Louis, MO) unless otherwise noted. Methanol and acetonitrile were from Fisher Scientific (Pittsburgh, PA) and were HPLC grade or higher. 14C-Arachidonic acid (AA) was obtained from PerkinElmer Life Sciences (Boston, MA). The following γ-ketoaldehyde scavenger molecules were synthesized by V. Amarnath as previously described:21 pentylpyridoxamine (PPM), 3-methoxysalicylamine (3-MoSA), and 5-ethylsalicylamine (EtSA).
Treatment of Cells
To stimulate COX-2 expression, A549 or RAW264.7 cells were treated overnight with 5 ng/mL IL-1β (A549) or 10 μg/mL LPS and 10 U/mL IFNγ (RAW264.7) in serum-free medium. When indicated, cells were pretreated with indomethacin, aldehyde scavengers (glucosamine, 3-MoSA, PPM, or EtSA), or vehicle (ethanol for indomethacin, H2O for scavengers) for 45 min and then given 20 μM arachidonic acid (AA) or DMSO vehicle for 1 h before lysing.
Histone Extraction
Cultured cells were lysed in hypotonic buffer (10 mM Tris/10 mM NaCl/3 mM MgCl2) containing 1 mM pyridoxamine and 100 μM indomethacin to prevent the artifactual formation of LGE2 during the lytic process.22 After allowing the cells to swell on ice, membranes were disrupted by addition of Triton X-100 (0.5% final concentration) and vortexing. Nuclei were isolated by centrifugation for 10 min at 1000 × g, and the resulting pellet was washed with PBS. Histones were extracted in 0.4 N H2SO4, precipitated with trichloracetic acid, and washed with acetone, following ref (23). With this method, histones are the predominant proteins and contaminating nuclear proteins are reduced23 (Figure 3A). Histones were resolubilized in dilute NaOH, and pH was neutralized with HCl. Protein concentration was determined using the method of Bradford. For tissue, a portion of frozen liver was homogenized in buffer containing 40 mM sodium citrate and 1% Triton X-100 with 3 mM Trolox and 100 μM indomethacin. The supernatant was separated from settled debris, and nuclei were pelleted at 500 × g and washed. Nuclear pellets were frequently transferred to fresh tubes to avoid contamination by floating lipid debris; any remaining was removed with a cotton swab. Histones were extracted as above. All centrifugation steps were carried out at 4 °C.
Figure 3.
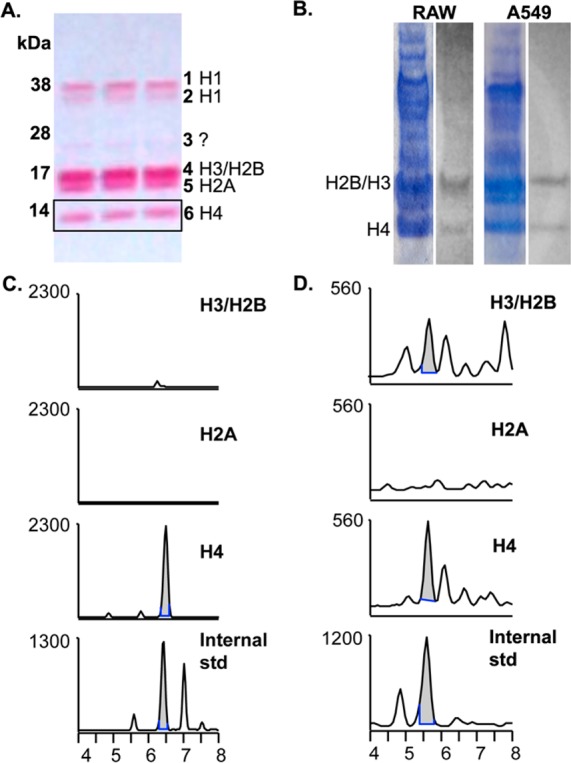
LG-lysyl adducts are predominantly detected on histone H4. (A). A Ponceau stain of a sample A549 histone extraction is shown, along with band identities.23 Histones were extracted from nuclei in 0.4 N H2SO4, resolved on 4–12% SDS-PAGE gradient gel, and transferred to nitrocellulose. H3 and H2B tend to run together as one band. (B). RAW264.7 or A549 cells were stimulated to express COX-2 and given 20 μM 14C-AA for 1 h. Cells were lysed, nuclei were isolated, and histones were extracted, concentrated, and resolved on SDS-PAGE prior to transfer to nitrocellulose and exposure to film. Shown is the Coomassie stain of the SDS-PAGE gel (left) and the result of autoradiography (right). We have observed that Ponceau, Coomassie R-250, Coomassie G-250, and silver stains each preferentially detect different histone or acid-soluble proteins. (C, D). RAW264.7 (C) or A549 (D) cells were stimulated to express COX-2 and treated with 20 μM AA for 1 h prior to histone extraction. Then 350–400 μg of total histone was loaded onto 4–12% SDS-PAGE gel and transferred to nitrocellulose. Individual bands were excised horizontally, and proteins were digested directly off the nitrocellulose by serial incubations with Pronase and aminopeptidase. The results were analyzed by LC–ESI/MS/MS, and the chromatographs of the H3/H2B and H4 bands shown against the LG-lysyl internal standard. The H2A chromatograph is shown as a representative negative result; no co-migrating peaks were seen in any other bands.
Sample Preparation and Mass Spectrometry
Histone samples were prepared for mass spectrometry by addition of ammonium bicarbonate to 5 mM final concentration before digestion to single amino acids by protease step-digestion as previously described.22 In the case of immunoblots, proteins were digested directly off the nitrocellulose through incubation of the nitrocellulose strip in 30 μg/mL Pronase in ammonium bicarbonate buffer and later addition of aminopeptidase. All samples were centrifuged at 2000 × g for 10 min after final digestion to remove precipitate, spiked with 0.2 ng of 13C-lysyl-lactam internal standard, and purified on prepared tC18 cartridges (Waters Corp., Milford, MA). Samples on tC18 cartridges were washed with water and then 30% methanol before being eluted in 80% methanol and concentrated by evaporation. Samples were evaluated by electrospray ionization (ESI) LC–MS/MS on a ThermoFisher TSQ Quantum triple quadrapole mass spectrometer in positive ion mode and quantitated by isotopic dilution as previously described,14 with the exception of a reduced flow rate of 0.1 mL/min.
Measurement of PGE2
A sample of cellular media was taken just prior to lysis and centrifuged to remove any cellular debris. For PGE2 analysis, samples were spiked with 2 ng of [2H7] PGE2 as an internal standard. Prostaglandins were isolated and derivatized for analysis by GC–MS, operating in negative ion chemical ionization (NICI) mode and monitoring selected ions as previously described.11 For the [2H4] PGE2 internal standard, m/z = 528. To account for the deuterium-protium exchange at the position C12 of [2H7] PGE2, the summation of the signals obtained at m/z = 530, m/z = 531, and m/z = 532 was performed.
Autoradiography
A549 or RAW264.7 cells were treated overnight to stimulate COX-2 expression and given 20 μM 14C-AA (116 μCi) for 1 h. Histones were isolated as described above and separated on 4–12% SDS-PAGE gels (Life Technologies), which were stained with Coomassie and exposed to film for 2 weeks.
Salt Extraction
A549 cells, 60–80% confluent, were stimulated and treated with AA ± 500 μM EtSA before cells were scraped in lysis media for nuclear isolation as above. After addition of Tritox X-100 and vortexing, 1.5 mL of each sample was aliquoted into an eppendorf and centrifuged to separate nuclei. Pellets were washed with PBS, and all buffer was removed. Nuclear pellets were resuspended in extraction buffer containing 0.6, 0.9, 1.2, or 1.5 M NaCl (with 10 mM Tris, pH 7.5, 3 mM MgCl2, 0.5% NP-40, and protease inhibitor cocktail) and incubated for 10 min on ice as in ref (24). Following this extraction period, the nuclei were centrifuged at 16,000 × g to obtain the soluble fraction. This was sonicated, denatured at 95 °C, and analyzed by SDS-PAGE and Western blotting, using Ponceau stain to visualize proteins and confirm equal loading and anti-H4 antibody (Abcam, Cambridge, MA).
Statistical Analyses
All data were analyzed using Prism software (GraphPad, La Jolla, CA). Data are expressed as means ± SE, and statistical significance was determined using one-way ANOVA followed by Tukey’s post-test or Dunnett’s multiple comparisons post-test, when appropriate. A p value of <0.05 was considered significant.
Results
Levuglandins Form Adducts on Histones in Cultured Cells and Whole Tissue
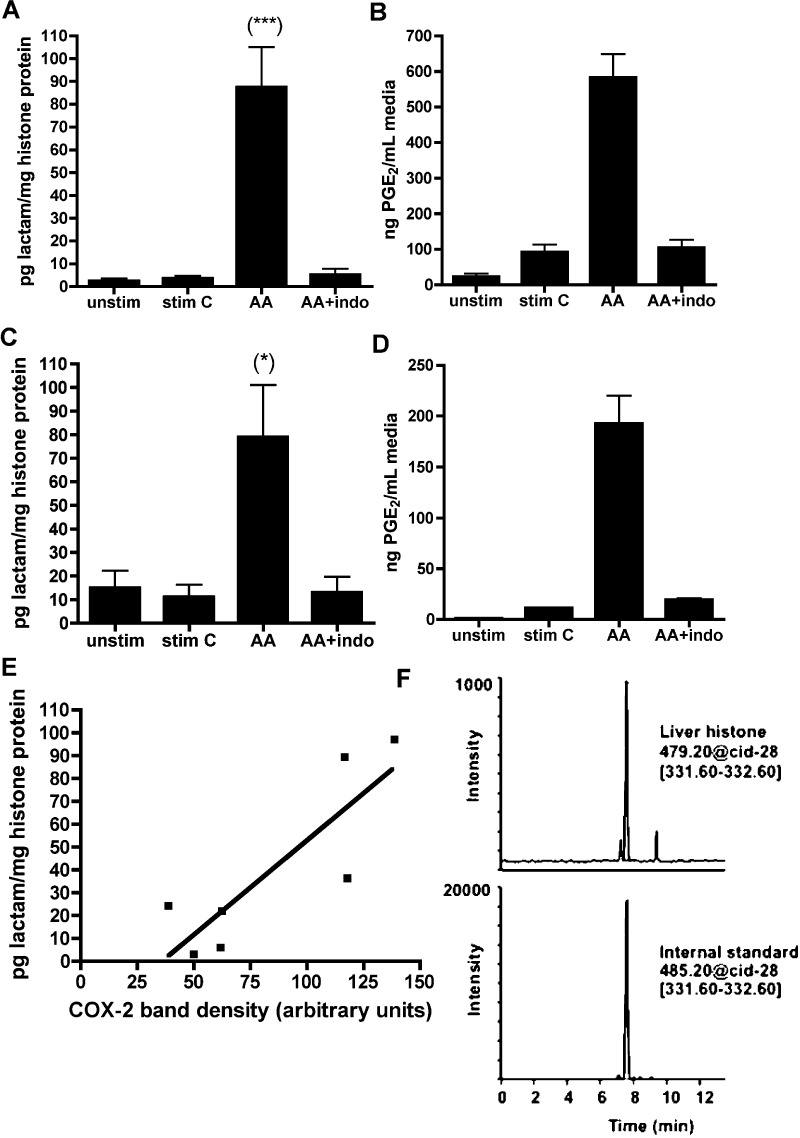
With mass spectrometry, we have identified LG-lysyl adducts on histones in RAW264.7 macrophages (Figure 2A) as well as A549 cultured lung epithelial cells (Figure 2C). COX-2 is upregulated in these cells upon cytokine stimulation, and addition of exogenous AA leads to formation of LG-histone adducts. Formation of these adducts is blocked with indomethacin, further indicating a COX-dependent mechanism (Figure 2A and C). Very few LG-histone adducts are formed in these cell lines without addition of exogenous AA, and PGE2 analysis of cell media from each group indicates there is comparatively little endogenous AA mobilized following induction of COX-2 (Figure 2B and D). Although few adducts are formed at basal levels in our cell lines, we find the LG-lysyl adduct in rat liver histones (Figure 2E), where levels correlate with COX-2 expression, demonstrating COX-2-dependent adduct formation under physiological conditions.
Figure 2.
LG-lysine adducts are found in cells and tissue, dependent on COX-2 activity. RAW264.7 mouse macrophage (A) and A549 human lung carcinoma (C) cells were stimulated to express COX-2 and then given 20 μM arachidonic acid (AA) or vehicle. A subgroup of cells was preincubated 45 min with 50 μM indomethacin. As a measure of COX activity, PGE2 was determined by GC–MS from cell media prior to lysis (B and D). Nuclei were isolated, and histones were extracted and digested to individual amino acids prior to LC–ESI/MS/MS analysis. *p < 0.05; ***p < 0.001 by ANOVA followed by Tukey’s post-test (n ≥ 5). (E). Histones were extracted from nuclei of rat liver and analyzed as above for LG-lactam adduct. COX-2 protein was analyzed by Western blotting and plotted against lactam adduct levels. Each point corresponds to one liver, and shown is the line of regression (r2 = 0.7237). Pearson r = 0.8507; two-tailed p = 0.0152. (F) LC–MS chromatograph of histones isolated from a rat liver with relatively high COX-2 expression (COX-2 band intensity of 117 arbitrary units).
LG-Histone Adducts Are Restricted to Specific Histone Isoforms
Using 0.4 N H2SO4 to extract histones results in a relatively pure preparation, with histones corresponding to known molecular weights (Figure 3A; ref (23)). Incubation of 14C-AA with stimulated A549 or RAW264.7 cells led to a 14C-containing band in the histone preparation that corresponded with H4 and H3/H2B, despite the fact that other histones are represented in equal or greater quantity (Figure 3B). We treated stimulated RAW264.7 macrophages or A549 lung carcinoma cells with 20 μM AA for 1 h and directly digested and analyzed the SDS-PAGE bands as labeled in Figure 3A. The H4 band yielded a predominant peak corresponding with the internal LG-lysyl standard, while lower or no signal was seen in the H3/H2B band and no corresponding peaks were seen in other bands (Figure 3C and D, data not shown). Thus, there is consistent evidence for formation of an LG-lysine lactam adduct on H4. The radiolabeled AA product adducted to H3/H2B probably includes structures in addition to the LG-lysine lactam. These results, from two separate cell lines, suggest that there is specificity in the reaction of LG with histones.
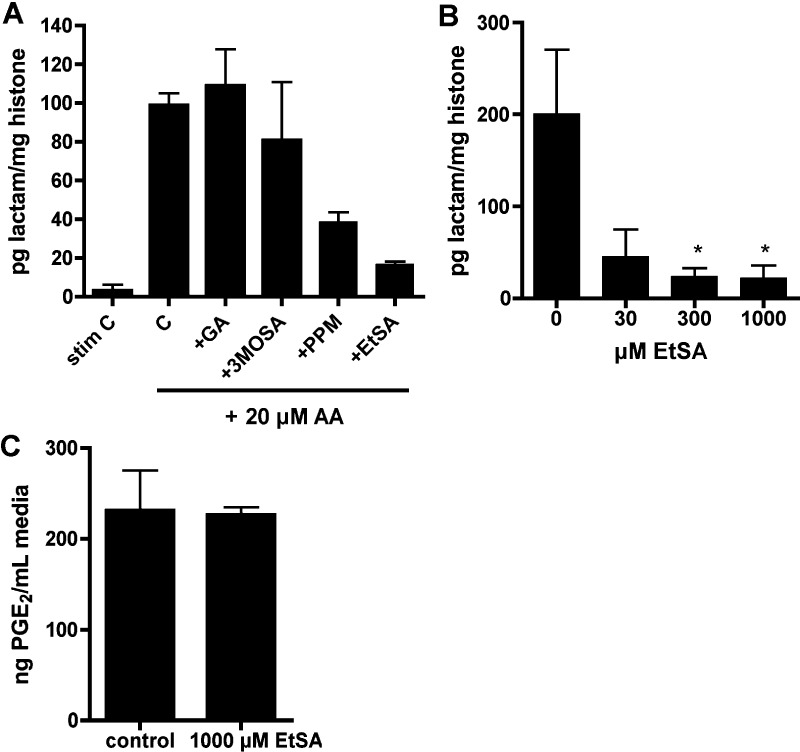
Scavenger 5-Ethylsalicylamine (EtSA) Reduces LG-Histone Adduct Formation without Affecting COX-2 Activity
We have previously characterized a number of small-molecule γ-ketoaldehyde scavengers that did not inhibit COX-2 and then further determined a subset that was free of cytotoxicity.21 From this subset, we then screened a number of these scavengers for the ability to reduce formation of LG-histone adducts. In RAW264.7 cells, only the scavenger 5-ethylsalicylamine (EtSA) was able to block adduct formation (Figure 4A). EtSA also inhibited LG-histone adduct formation in stimulated, AA-treated A549 cells, without affecting PGE2 production at the highest concentration tested (Figure 4B and C).
Figure 4.
Scavenger EtSA blocks LG-lysyl adduct formation in RAW264.7 and A549 histones, without affecting COX-2 activity. (A) Scavengers were screened in RAW264.7 cells for the ability to decrease LG adduct formation on histones. Scavengers used were glucosamine (GA), 3-methoxysalicylamine (3-MoSA), pentylpyridoxamine (PPM), and 5-ethylsalicylamine (EtSA). Cells were stimulated to express COX-2, pretreated 45 min with 500 μM scavenger or vehicle (H2O), and given 20 μM AA for 1 h before lysing and extraction of histones. Histone proteins were analyzed by LC–ESI/MS/MS for LG-lysyl lactam adduct, n = 2. (B) Stimulated A549 cells were pretreated with 30, 300, or 1000 μM EtSA prior to 1 h with 20 μM AA, and histones were analyzed for LG-lysyl adduct. *p < 0.05 by one-way ANOVA followed by Dunnett’s multiple comparisons post-test (n = 3–5). (C). A549 cells were stimulated, pretreated for 45 min with 1000 μM EtSA or H2O vehicle, and given 20 μM AA for 1 h. Media was analyzed by GC–MS for PGE2 (n = 3). There was no effect on PGE2 production at lower doses of EtSA (data not shown).
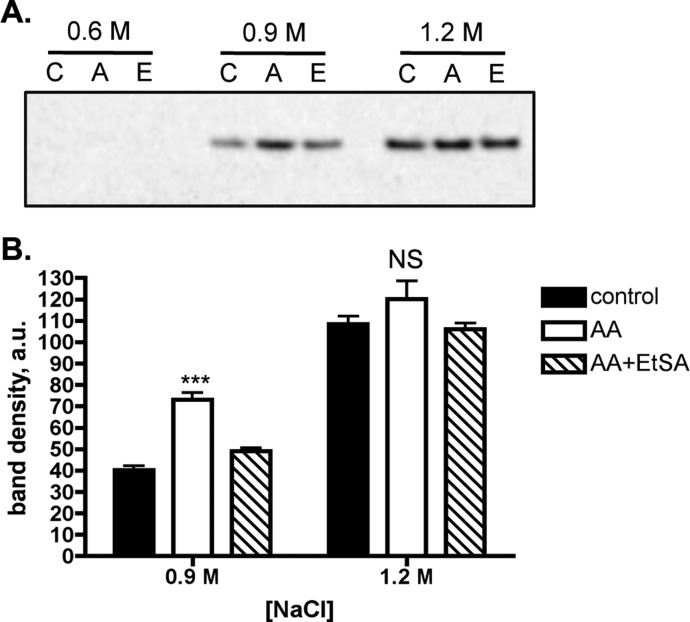
Formation of LG-Lysyl Adduct on Histone H4 Decreases DNA–Histone Interaction
To examine the functional effect of LG-histone adduction, we performed salt fractionation of A549 nuclei to determine histone solubility. In this assay, loosely bound histone is released at lower salt concentrations than tightly bound proteins.25 We found that in stimulated, AA-treated A549 cells, histone H4 was eluted at lower salt concentrations than in stimulated control cells; this was reversed after treatment with the scavenger EtSA (Figure 5).
Figure 5.
LG-lysyl adduct formation on histone H4 decreases DNA–histone interaction. A549 cells were stimulated and given DMSO vehicle (C lanes) or 20 μM AA for 1 h (A lanes). A subgroup of cells was treated with 500 μM EtSA 45 min prior to adding AA (E lanes). Nuclei were extracted with 0.6, 0.9, or 1.2 M NaCl buffer, and the supernatant was evaluated by Western blotting for histone H4. Shown is a representative Western blot (A) as well as the pooled results of 4 experiments (B). Different exposure times may have been used for the 0.9 and 1.2 M bands. ***p < 0.001 by one-way ANOVA followed by Tukey’s multiple comparisons post-test. NS, not significant.
Discussion
This study, establishing that COX-2 catalysis can cause changes in DNA–histone interactions through formation of LG-histone adducts, suggests a new hypothesis for the contribution of COX-2 to the etiology of cancer. Oxidative damage is known to cause N6-formylation of H1 histone,26 and epigenetic modification affecting COX-2 transcription is well-described,27 but the LG-lysyl histone adduct we describe here is an entirely novel finding that links inflammation and COX-2 activation with histone modification.
Here we demonstrate COX-2-dependent formation of LG-histone adducts in cells and tissues. Whereas COX-2 blockade by treatment with indomethacin decreases LG-histone adduct formation in A549 or RAW264.7 cells, this method of antagonism cannot separate the myriad effects of other COX-2 products from the effects of the LG-histone adducts. Therefore, we screened a number of small molecule levuglandin scavenger molecules for their ability to decrease LG-histone adduction. LG will react with these molecules 3 orders of magnitude faster than lysine,28 and we have previously shown that scavenger treatment decreases total cellular levels of LG-lysine adducts without affecting PGE2 production.21 These scavengers are orally bioavailable29,30 and able to decrease total LG-protein adduct levels when given to mice in drinking water.17 In future studies, aside from allowing investigation of LG-protein modification independent of COX activity, use of these scavengers may bypass the cardiovascular and gastric side effects seen with COX inhibitors.
Interestingly, not all histones are targeted, but cellular LG adducts seem to preferentially form on the H4 and, to a lesser extent, on H3/H2B. Whether this specificity is a reflection of histone availability in the nucleosome, accessibility of lysine residues, or a more favorable microenvironment for adduct formation remains to be shown. Incubation with 14C-AA in cells led to a stronger autoradiographic band at H3/H2B compared to H4, while LC–MS analysis indicated that H4 was the major form adducted by LGs. This discrepancy can be explained by several mechanisms. Protein-associated radioactivity would come from any product derived from 14C-AA, including LGs but also PGJ2/PGA2 cyclopentenone31 or arachidonate ester32 adducts. In addition, the occurrence of LG-lysyl adducts is almost certainly underreported with our current approach. Our internal standard and mass spectroscopy method are specific for detection of a single LG-lysyl adduct with an m/z equivalent to that of the lactam structure, but the initial Schiff base intermediate of LG-lysyl adducts can oxidize to form other structure such as hydroxylactam14 or intraprotein or protein–DNA cross-links,15,33 which would go undetected in our method. For these reasons, the autoradiograms cannot be quantitatively compared with the LC–ESI/MS/MS results as they do not measure the same molecular structures.
H4, along with H2A, H2B, and H3 histones, comprise the histone octamer around which DNA is “packaged” into nucleosomes. The interaction of histone N-terminal tails with DNA is critical to DNA compaction and organization and is dependent on the numerous positively charged lysine and arginine residues present; a mesoscopic model demonstrates that H4 tails are the most important in mediating internucleosomal interactions.34 A single lysyl acetylation on K16 of H4 modulates chromatin compaction and interaction of numerous chromatin-associated proteins; constitutive acetylation of this residue confers a folding defect comparable to deletion of the entire H4 tail.35 After LG-histone adduct formation we do find disruption of histone-DNA binding, resulting in increased DNA extraction in a salt solution. This decreased histone–DNA interaction may increase DNA transcriptional access to previously silent oncogenes and contribute to the development of cancer.
The complex patterns of lysyl acetylation and methylation comprise a “histone code” that regulates chromatin access and transcription;18 it is plausible that irreversible adduction of lysyl residues could disrupt this code or directly alter the access of DNA-interacting proteins. Changes in histone modifications are known to result in altered DNA methylation, deregulation of oncogenes, genomic instability, impaired DNA repair, and defects in cell cycle checkpoints.36,37 Changes in lysyl modifications of H4 in particular are a common hallmark of human cancers and are associated with a global loss of DNA methylation.20 Further elucidation of the effects of LG-histone adduction on histone modification, DNA-histone interactions, and transcription should increase our understanding of the molecular mechanisms whereby COX-2 contributes to cancer development and progression.
Acknowledgments
The authors are grateful for the valuable advice of Scott Hiebert.
Glossary
Abbreviations
- LG
levuglandin
- COX-2
cyclooxygenase-2
- NSAID
non-steroidal anti-inflammatory drug
- PGH2
prostaglandin H2
- PPM
pentylpyridoxamine
- 3-MOSA
3-methoxysalicylamine
- EtSA
5-ethylsalicylamine
- AA
arachidonic acid
- ESI
electrospray ionization
- NICI
negative ion chemical ionization
Author Contributions
∥ These authors contributed equally to this work.
Supported by the Center for Molecular Toxicology, NIH P30 ES000267, American Cancer Society PF-09-127-01-DMC (E.J.C.), GM15431, and the Thomas F. Frist, Sr. Chair in Medicine (J.A.O.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Agrawal A.; Fentiman I. S. (2008) NSAIDs and breast cancer: a possible prevention and treatment strategy. Int. J. Clin. Pract. 62, 444–449. [DOI] [PubMed] [Google Scholar]
- Harris R. E.; Beebe-Donk J.; Alshafie G. A. (2007) Reduced risk of human lung cancer by selective cyclooxygenase 2 blockade: results of a case control study. Int. J. Biol. Sci. 3, 328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konturek P. C.; Kania J.; Burnat G.; Hahn E. G.; Konturek S. J. (2005) Prostaglandins as mediators of COX-2 derived carcinogenesis in gastrointestinal tract. J. Physiol. Pharmacol. 56(Suppl 5), 57–73. [PubMed] [Google Scholar]
- Harris R. E. (2009) Cyclooxygenase-2 (cox-2) blockade in the chemoprevention of cancers of the colon, breast, prostate, and lung. Inflammopharmacology 17, 55–67. [DOI] [PubMed] [Google Scholar]
- Lim B. J.; Jung S. S.; Choi S. Y.; Lee C. S. (2010) Expression of metastasis-associated molecules in non-small cell lung cancer and their prognostic significance. Mol. Med. Rep. 3, 43–49. [DOI] [PubMed] [Google Scholar]
- Sheehan K. M.; Sheahan K.; O’Donoghue D. P.; MacSweeney F.; Conroy R. M.; Fitzgerald D. J.; Murray F. E. (1999) The relationship between cyclooxygenase-2 expression and colorectal cancer. J. Am. Med. Assoc. 282, 1254–1257. [DOI] [PubMed] [Google Scholar]
- Bertagnolli M. M. (2007) Chemoprevention of colorectal cancer with cyclooxygenase-2 inhibitors: two steps forward, one step back. Lancet Oncol. 8, 439–443. [DOI] [PubMed] [Google Scholar]
- Castellone M. D.; Teramoto H.; Williams B. O.; Druey K. M.; Gutkind J. S. (2005) Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science 310, 1504–1510. [DOI] [PubMed] [Google Scholar]
- Nakanishi M.; Montrose D. C.; Clark P.; Nambiar P. R.; Belinsky G. S.; Claffey K. P.; Xu D.; Rosenberg D. W. (2008) Genetic deletion of mPGES-1 suppresses intestinal tumorigenesis. Cancer Res. 68, 3251–3259. [DOI] [PubMed] [Google Scholar]
- Elander N.; Ungerbäck J.; Olsson H.; Uematsu S.; Akira S.; Söderkvist P. (2008) Genetic deletion of mPGES-1 accelerates intestinal tumorigenesis in APC Min/+ mice. Biochem. Biophys. Res. Commun. 372, 249–253. [DOI] [PubMed] [Google Scholar]
- Boutaud O.; Brame C. J.; Salomon R. G.; Roberts L. J. 2nd; Oates J. A. (1999) Characterization of the lysyl adducts formed from prostaglandin H2 via the levuglandin pathway. Biochemistry 38, 9389–9396. [DOI] [PubMed] [Google Scholar]
- Salomon R. G.; Miller D. B.; Zagorski M. G.; Coughlin D. J. (1984) Solvent-induced fragmentation of prostaglandin endoperoxides. New aldehyde products from PGH2 and a novel intramolecular 1,2-hydride shift during endoperoxide fragmentation in aqueous solution. J. Am. Chem. Soc. 106, 6049–6060. [Google Scholar]
- Brame C. J.; Salomon R. G.; Morrow J. D.; Roberts L. J. 2nd (1999) Identification of extremely reactive gamma-ketoaldehydes (isolevuglandins) as products of the isoprostane pathway and characterization of their lysyl protein adducts. J. Biol. Chem. 274, 13139–13146. [DOI] [PubMed] [Google Scholar]
- Boutaud O.; Brame C. J.; Chaurand P.; Li J.; Rowlinson S. W.; Crews B. C.; Ji C.; Marnett L. J.; Caprioli R. M.; Roberts L. J. 2nd; Oates J. A. (2001) Characterization of the lysyl adducts of prostaglandin H-synthases that are derived from oxygenation of arachidonic acid. Biochemistry 40, 6948–6955. [DOI] [PubMed] [Google Scholar]
- Iyer R. S.; Ghosh S.; Salomon R. G. (1989) Levuglandin E2 crosslinks proteins. Prostaglandins 37, 471–480. [DOI] [PubMed] [Google Scholar]
- Boutaud O.; Li J.; Zagol I.; Shipp E. A.; Davies S. S.; Roberts L. J. 2nd; Oates J. A. (2003) Levuglandinyl adducts of proteins are formed via a prostaglandin H2 synthase-dependent pathway after platelet activation. J. Biol. Chem. 278, 16926–16928. [DOI] [PubMed] [Google Scholar]
- Boutaud O.; Andreasson K. I.; Zagol-Ikapitte I.; Oates J. A. (2005) Cyclooxygenase-dependent lipid-modification of brain proteins. Brain Pathol. 15, 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T.; Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Esteller M. (2007) Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 8, 286–298. [DOI] [PubMed] [Google Scholar]
- Fraga M. F.; Ballestar E.; Villar-Garea A.; Boix-Chornet M.; Espada J.; Schotta G.; Bonaldi T.; Haydon C.; Ropero S.; Petrie K.; Iyer N. G.; Perez-Rosado A.; Calvo E.; Lopez J. A.; Cano A.; Calasanz M. J.; Colomer D.; Piris M. A.; Ahn N.; Imhof A.; Caldas C.; Jenuwein T.; Esteller M. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37, 391–400. [DOI] [PubMed] [Google Scholar]
- Zagol-Ikapitte I.; Amarnath V.; Bala M.; Roberts L. J. 2nd; Oates J. A.; Boutaud O. (2010) Characterization of scavengers of gamma-ketoaldehydes that do not inhibit prostaglandin biosynthesis. Chem. Res. Toxicol. 23, 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagol-Ikapitte I.; Masterson T. S.; Amarnath V.; Montine T. J.; Andreasson K. I.; Boutaud O.; Oates J. A. (2005) Prostaglandin H(2)-derived adducts of proteins correlate with Alzheimer’s disease severity. J. Neurochem 94, 1140–1145. [DOI] [PubMed] [Google Scholar]
- Shechter D.; Dormann H. L.; Allis C. D.; Hake S. B. (2007) Extraction, purification and analysis of histones. Nat. Protoc. 2, 1445–1457. [DOI] [PubMed] [Google Scholar]
- Bhaskara S.; Knutson S. K.; Jiang G.; Chandrasekharan M. B.; Wilson A. J.; Zheng S.; Yenamandra A.; Locke K.; Yuan J.-L.; Bonine-Summers A. R.; Wells C. E.; Kaiser J. F.; Washington M. K.; Zhao Z.; Wagner F. F.; Sun Z.-W.; Xia F.; Holson E. B.; Khabele D.; Hiebert S. W. (2010) Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 18, 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. M. (1978) Fractionation of nucleosomes by salt elution from micrococcal nuclease-digested nuclei. J. Cell Biol. 79, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T.; Zhou X.; Taghizadeh K.; Dong M.; Dedon P. C. (2007) N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc. Natl. Acad. Sci. U.S.A. 104, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Alvarez A.; Llorente-Izquierdo C.; Mayoral R.; Agra N.; Bosca L.; Casado M.; Martin-Sanz P. (2012) Evaluation of epigenetic modulation of cyclooxygenase-2 as a prognostic marker for hepatocellular carcinoma. Oncogenesis 1, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarnath V.; Amarnath K.; Davies S.; Roberts L. J. n. (2004) Pyridoxamine: an extremely potent scavenger of 1,4-dicarbonyls. Chem. Res. Toxicol. 17, 410–415. [DOI] [PubMed] [Google Scholar]
- Zagol-Ikapitte I.; Amarnath V.; Jadhav S.; Oates J. A.; Boutaud O. (2011) Determination of 3-methoxysalicylamine levels in mouse plasma and tissue by liquid chromatography-tandem mass spectrometry: application to in vivo pharmacokinetics studies. J. Chromatogr. 879, 1098–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagol-Ikapitte I. A.; Matafonova E.; Amarnath V.; Bodine C. L.; Boutaud O.; Tirona R. G.; Oates J. A.; Roberts L. J. 2nd; Davies S. S. (2010) Determination of the pharmacokinetics and oral bioavailability of salicylamine, a potent gamma-ketoaldehyde scavenger, by LC/MS/MS. Pharmaceutics 2, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne G. L.; Musiek E. S.; Morrow J. D. (2005) The cyclopentenone (A2/J2) isoprostanes--unique, highly reactive products of arachidonate peroxidation. Antioxid. Redox Signaling 7, 210–220. [DOI] [PubMed] [Google Scholar]
- Muszbek L.; Laposata M. (1993) Covalent modification of proteins by arachidonate and eicosapentaenoate in platelets. J. Biol. Chem. 268, 18243–18248. [PubMed] [Google Scholar]
- Murthi K. K.; Friedman L. R.; Oleinick N. L.; Salomon R. G. (1993) Formation of DNA-protein cross-links in mammalian cells by levuglandin E2. Biochemistry 32, 4090–4097. [DOI] [PubMed] [Google Scholar]
- Arya G.; Schlick T. (2006) Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. U.S.A. 103, 16236–16241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M.; Ishii H.; Sun J.-M.; Pazin M. J.; Davie J. R.; Peterson C. L. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847. [DOI] [PubMed] [Google Scholar]
- Fuks F. (2005) DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15, 490–495. [DOI] [PubMed] [Google Scholar]
- Füllgrabe J.; Kavanagh E.; Joseph B. (2011) Histone onco-modifications. Oncogene 30, 3391–3403. [DOI] [PubMed] [Google Scholar]