Abstract
Amyloids are filamentous protein aggregates that can be formed by many different proteins and are associated with both disease and biological functions. The pathogenicities or biological functions of amyloids are determined by their particular molecular structures, making accurate structural models a requirement for understanding their biological effects. One potential factor that can affect amyloid structures is hydration. Previous studies of simple stacked β-sheet amyloids have suggested that dehydration does not impact structure, but other studies indicated dehydration-related structural changes of a putative water-filled nanotube. Our results show that dehydration significantly affects the molecular structure of the fungal prion-forming domain HET-s(218–289), which forms a β-solenoid with no internal solvent-accessible regions. The dehydration-related structural deformation of HET-s(218–289) indicates that water can play a significant role in complex amyloid structures, even when no obvious water-accessible cavities are present.
Amyloid fibrils are filamentous protein aggregates arranged in cross-β structures, in which the individual subunits contain β-strands that run approximately perpendicular to the fibril axis, forming β-sheets that run parallel to the fibril axis.1,2 A large number of proteins can form amyloids, exhibiting biological effects that can be pathological or functional.3 Pathological amyloids are implicated in diseases including Alzheimer’s disease, Parkinson’s disease, and type 2 diabetes.3 Functional amyloids are used in normal biological functions such as bacterial biofilm formation,4 human melanin synthesis,5 or as storage units for peptide hormones.6 Prions, proteinaceous infectious agents, tend to form amyloid fibrils that can also be pathological, as in Creutzfeldt-Jakob disease,7 or functional, such as HET-s,8 which is used in fungal mating-type determination. The biological activity of an amyloid is uniquely determined by its molecular structure, so accurate structural characterization is key to understanding the pathogenicity or function of the amyloid.
Previous studies have found that some amyloids appear to maintain their structures after dehydration,9−11 though these studies have been limited to observations of amyloids with stacked β-sheet architectures,9,11,12 in which the amyloid cores consist of approximately parallel β-sheets 8–10 Å apart.1,2 Amyloid fibrils formed from the yeast prion-forming domain Sup35NM have been shown to have in-register parallel β-sheet structures in hydrated and dehydrated states13 but different fiber diffraction patterns,14 indicating structural changes within the subunit while essentially maintaining the same cross-β hydrogen bonding arrangement. It has been suggested that the Huntington’s disease-related poly glutamine amyloid has a water-filled nanotube structure,15 and high resolution models of Alzheimer’s disease-related Aβ 1–40 amyloids have shown water-accessible cores,16,17 indicating that certain complex amyloid structures could be significantly impacted by dehydration. The fungal prion-forming domain HET-s(218–289) forms a two-rung β-solenoid structure18 with a hydrophobic core that is not solvent accessible.19 Previous studies on HET-s(218–289) hydration have shown that fibrils rehydrated after lyophilization showed identical spectra to fibrils that had not been lyophilized.20 In this study, we show that despite a lack of water-accessible structures, the complex molecular architecture of HET-s(218–289) is substantially affected by dehydration. The effects of dehydration on fibril structure are reversible but not completely so.
Experimental Procedures
Preparation of Fiber Diffraction Specimens
HET-s(218–289) was recombinantly expressed, purified, and fibrillized into the β-solenoid form at pH 7.5 as previously described.21 After fibrillization, fibrils were moved into pH 4.0, dilute HCl by three rounds of ultracentrifugation and resuspension. Oriented sols were made as described elsewhere.22 A solution of HET-s(218–289) was centrifuged at 264000g into a soft pellet and freeze–thawed five times. The pellet was sheared through a 0.5 mm glass capillary to induce orientation and sealed with an oxygen torch. Dried fiber specimens were prepared by hanging 12 μL droplets between sealed, silanized, and sanded capillaries and drying at nominal 100% relative humidity (RH). After drying, specimens were stored at 0%, 33%, 75%, 83%, 98%, and 100% RH in specimen chambers23 containing silica gel and saturated solutions of MgCl2, NaCl, KCl, K2SO4, and water, respectively. Zero percent relative humidity specimens were rehydrated by inundating the silica gel in the specimen chambers with water. Lyophilized specimens were prepared by lyophilizing HET-s(218–289) fibril solutions followed by resuspension in water before forming dried fibers.
Data Collection and Processing
X-ray fiber diffraction data were collected at the BioCAT beamline at the Advanced Photon Source at Argonne National Laboratory, and at the Biological Small-Angle X-ray Scattering beamline 4–2 at the Stanford Synchrotron Radiation Lightsource. Diffraction data were transformed into reciprocal space24 and background subtracted using WCEN.25 Meridional and equatorial plots were produced from processed patterns using WCEN by integrating over a 30° angle centered on each axis. Plots from each diffraction pattern were linearly scaled using a coefficient determined by least-squares fitting of the continuous equatorial diffraction data between 0.03 and 0.20 Å–1 (∼33–4 Å).
Results
Comparison of Hydrated Forms of HET-s(218–289)
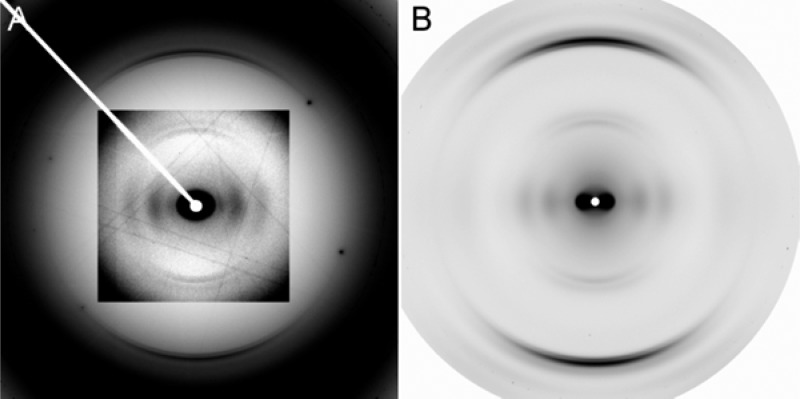
To compare the structures of fully hydrated and dried specimens under high humidity conditions, X-ray fiber diffraction was performed on oriented sols and dried fiber specimens (Figure 1). Oriented sols are concentrated HET-s(218–289) solutions sealed in glass capillaries, thus ensuring full hydration. Diffraction patterns from sols showed a large amount of water diffraction, seen around the edges of the patterns as a broad ring at ∼3.5 Å resolution (∼0.3 Å–1) (Figure 1A). The water diffraction is much stronger than the very weak amyloid diffraction, though the sample still shows orientation. The strong water and weak amyloid diffraction show that even after ultracentrifugation, the HET-s(218–289) pellets retain a large amount of water relative to protein. Diffraction from dried fibers maintained at nominal 100% relative humidity (RH) showed relatively stronger amyloid diffraction and better orientation than oriented sol patterns (Figure 1B). Qualitative comparisons of layer line spacings and relative equatorial intensities in these dried fiber patterns and oriented sol diffraction patterns indicated that the structures were similar. Quantitative comparisons could not be performed owing to the weak diffraction and low orientation of the oriented sol patterns. HET-s(218–289) β-solenoid diffraction has been discussed in detail elsewhere.21,26 Briefly, the two-rung repeating unit produces the characteristic 4.7 Å cross-β meridional reflection, reflecting the distance between β-strands. An additional 9.4 Å meridional reflection is caused by the two-strand thick repeating unit, while off-meridional diffraction maxima at 8.5 Å arise from the helical symmetry of the fibril. Equatorial diffraction consists of a series of diffraction maxima of comparable intensities, indicative of a roughly cylindrical, packed structure, distinct from a stacked β-sheet architecture.26,27
Figure 1.
X-ray fiber diffraction from HET-s(218–289) for different specimen types. (A) Oriented sol. (B) Dried fiber at nominal 100% RH. Inset in A has adjusted intensities to emphasize low-angle diffraction; for full patterns, see Figure S1, Supporting Information.
Fiber Diffraction of HET-s(218–289) under Different Relative Humidities
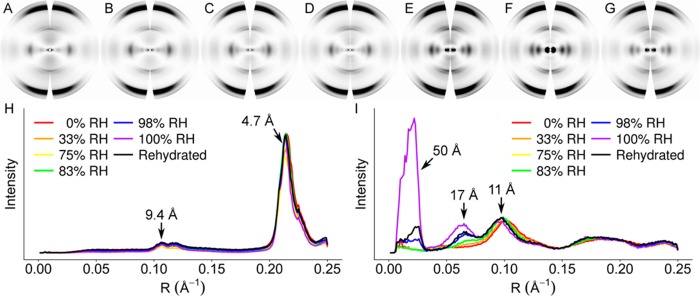
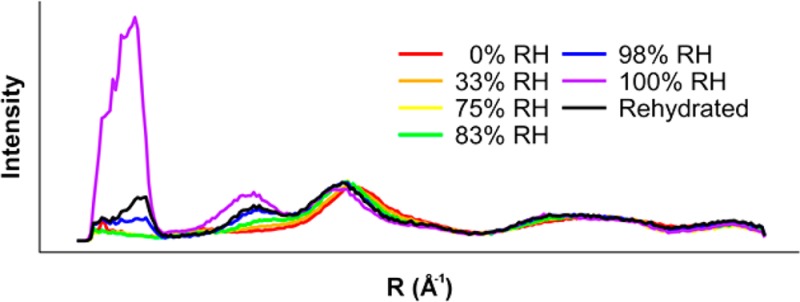
To determine the impact of dehydration on HET-s(218–289) structure, we collected diffraction data from specimens first dried at nominal 100% RH and further dried to 0%, 33%, 75%, 83%, and 98% RH (Figure 2A–F). To assess the reversibility of dehydration, the 0% RH specimen was rehydrated at nominal 100% RH, and diffraction data were collected (Figure 2G). For comparison, meridional and equatorial intensities were integrated, plotted, and scaled by least-squares fitting of the equatorial data between ∼30 and 4 Å (∼0.03–0.25 Å–1) resolution (Figure 2H,I). Dehydration did not affect the projection of the structure on the fiber axis; meridional relative intensities did not change (Figure 2H). Meridional diffraction spacings were also not perturbed, indicating that the helical parameters of the fibrils were unchanged. Diffraction along the equator, however, showed substantial changes on dehydration. By 83% RH, there was virtually complete loss of ∼50 Å and ∼17 Å maxima. While very low resolution diffraction such as the ∼50 Å maximum can sometimes be related to interfibril packing within a specimen, this is unlikely to be the case in HET-s(218–289), as we have consistently observed this intensity maximum in diffraction from many different preparations and specimens.21,26 The relative strengthening of the ∼11 Å (0.09 Å–1) maximum suggested a collapse of the triangular hydrophobic core of HET-s(218–289) into a more parallel arrangement, resembling a stacked β-sheet structure. Rehydration of the desiccated specimen resulted in incomplete recovery of low resolution diffraction maxima, with an equatorial plot closely resembling the 98% RH plot.
Figure 2.
Fiber diffraction from HET-s(218–289) at different relative humidities. Background-subtracted diffraction patterns of (A) 0% RH, (B) 33% RH, (C) 75% RH, (D) 83% RH, (E) 98% RH, (F) 100% RH, and (G) 0% RH rehydrated to nominal 100% RH. Plots of (H) meridians and (I) equators. Relative intensities are 5× stronger in (H) than in (I).
To quantitate the level of structural changes caused by dehydration, correlation coefficients (CC) were calculated between the equatorial data from each pair of specimens in two different resolution ranges. In the intermediate resolution range (∼30–4 Å, ∼0.03–0.25 Å–1) meaningful correlation (CC > 0.8)26,28 with the 100% RH specimen was lost by 83% RH (Table 1). When the full available resolution range was used (100–4 Å, 0.01–0.25 Å–1), meaningful correlation was lost by 98% RH (Table 2), although comparisons using CCs across the full resolution range may not be as useful as comparisons using intermediate resolutions, owing to the high sensitivity of the ∼50 Å intensity maximum to dehydration. CCs also showed that the rehydrated specimen correlated more closely with the 98% RH specimen than with the nominal 100% RH specimen.
Table 1. Correlation Coefficients from ∼30–4 Å Resolution (∼0.03–0.25 Å–1).
| sample | 0% RH | 33% RH | 75% RH | 83% RH | 98% RH | 100% RH | rehydrated |
|---|---|---|---|---|---|---|---|
| 0% RH | 1.00 | 0.98 | 0.90 | 0.92 | 0.80 | 0.52 | 0.76 |
| 33% RH | 1.00 | 0.97 | 0.98 | 0.89 | 0.65 | 0.86 | |
| 75% RH | 1.00 | 1.00 | 0.97 | 0.78 | 0.95 | ||
| 83% RH | 1.00 | 0.96 | 0.76 | 0.94 | |||
| 98% RH | 1.00 | 0.87 | 0.99 | ||||
| 100% RH | 1.00 | 0.90 | |||||
| rehydrated | 1.00 |
Table 2. Correlation Coefficients from 100–4 Å Resolution (0.01–0.25 Å–1).
| sample | 0% RH | 33% RH | 75% RH | 83% RH | 98% RH | 100% RH | rehydrated |
|---|---|---|---|---|---|---|---|
| 0% RH | 1.00 | 0.98 | 0.91 | 0.92 | 0.77 | –0.09 | 0.64 |
| 33% RH | 1.00 | 0.97 | 0.98 | 0.86 | –0.07 | 0.72 | |
| 75% RH | 1.00 | 1.00 | 0.94 | –0.01 | 0.81 | ||
| 83% RH | 1.00 | 0.93 | –0.04 | 0.80 | |||
| 98% RH | 1.00 | 0.24 | 0.95 | ||||
| 100% RH | 1.00 | 0.46 | |||||
| rehydrated | 1.00 |
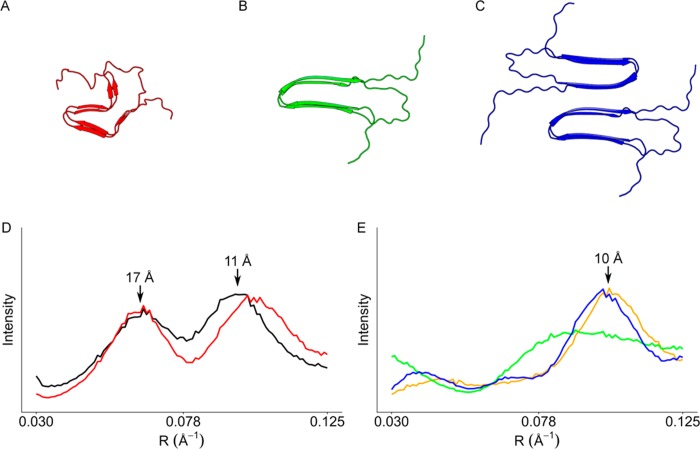
In order to be more comparable to fibrils studied by ssNMR,20 HET-s(218–289) fibrils in solution were lyophilized and rehydrated prior to fiber diffraction specimen preparation. The resultant fibrils provided fiber diffraction patterns consistent with two-rung β-solenoid diffraction (Figure S2, Supporting Information), but poor orientation prevented quantitative comparisons owing to the inability to perform accurate background subtraction. Qualitative comparisons of relative intensities of the ∼17 and ∼11 Å equatorial maxima (Figure 3) are more similar to the 98% RH and rehydrated specimens (Figure 2E,G), which show a more intense ∼11 Å relative to the ∼17 Å, while the 100% RH specimens show the opposite (Figure 2F).
Figure 3.
Molecular modeling of HET-s(218–289). (A) ssNMR two-rung β-solenoid model (PDB: 2KJ3). (B) One- and (C) two-protofilament two-rung stacked β-sheet models. Experimental and calculated equatorial diffraction for (D) β-solenoids and (E) stacked β-sheets. Black line: experimental data at 100% RH. Red line: calculated data from the model in A. Orange line: experimental data at 0% RH. Green line: calculated data from the model in B. Blue line: calculated data from the model in C.
Modeling the Effect of Dehydration
Molecular models were constructed by modifying the ssNMR structure of HET-s(218–289) in efforts to reproduce the types of structural change that can give rise to our observed fiber diffraction patterns. Our modifications of the structure take into account interatomic contacts but are not otherwise optimized. While our modifications are hypothetical, the modeling emphasizes the overall changes in architecture necessary to produce diffraction patterns consistent with the experimental results. Comparisons between simulated and experimental data were performed at resolutions between 0.030 and 0.125 Å–1 (∼33–8 Å), as agreement between the ssNMR model and experimental data was only reliable in this range, probably owing to disorder within the fibril that we were unable to simulate and problems with comparing diffraction patterns that represent static averaged data and ssNMR models, which represent ensemble structures. We have previously shown that the fibrils that provide the characteristic β-solenoid diffraction patterns also produce ssNMR spectra identical to those used for ssNMR structure determination.26 Simulated diffraction from the ssNMR structure (Figure 3A) fit the 100% RH experimental data well (Figure 3D), with a CC of 0.85.
The ssNMR model of HET-s(218–289) was modified to fit the 0% RH experimental data by straightening the β-strands into a stacked β-sheet structure (Figure 3B). The simulated equator from this structure showed the loss of the ∼17 Å maximum but did not produce an ∼10 Å maximum with the same sharpness and intensity as the experimental pattern (Figure 3E), resulting in a CC with the experimental data of 0.73. However, by using the same stacked β-sheet model (Figure 3B) in a two-protofilament arrangement (Figure 3C), the calculated equator showed increased intensity and sharpness in the ∼10 Å maximum, the result of increased ∼10 Å sampling caused by increasing the number of β-sheets stacked together. The CC between the two-protofilament model and the experimental data was 0.97.
Discussion
We have shown that dehydration of amyloid fibrils with complex structures can significantly perturb the molecular structure. Similar dehydration-related changes to amyloid diffraction patterns have been observed elsewhere,29 but knowledge of the hydrated structure of HET-s(218–289)18 provides us with further insights into the nature of the structural changes. By molecular modeling and comparisons of calculated and experimental diffraction patterns, we have shown that the observed dehydration-related structural change is consistent with a transition to a stacked β-sheet structure. Reproduction of the medium resolution diffraction from our dehydrated specimens required a two-protofilament model, suggesting that dehydration may induce the close association of fibrils. Previous ssNMR observations of HET-s(218–289) had shown that fibrils that had never been dried or had been lyophilized and rehydrated had identical spectra, indicating that dehydration of HET-s(218–289) fibrils was fully reversible.20 Our observations suggest that drying of our fiber diffraction specimens is reversible, but not completely. Taken together with the ssNMR results, these observations suggest that structural changes in the individual subunits are reversible, but long-range fibril interactions, which cannot be seen with ssNMR, are not fully reversible. We might speculate that this irreversibility could be caused by the formation of stable close interactions of the fibrils during dehydration, as suggested by our molecular modeling.
Our results with HET-s(218–289) show that dehydration effects can occur even in the absence of water-accessible channels. This observation indicates that water on the surface of the fibril can play a substantial role in its molecular structure. This does not appear to be the case for stacked β-sheet amyloids,9−11 possibly because the crystalline nature of the β-sheet packing provides structural stability against deformation caused by dehydration. Our observations show that differences in hydration can affect amyloid structure, suggesting that hydration must be controlled during studies in order to obtain reproducible results. Our results show that water can play an important role in amyloid structure even when no obvious water-accessible features are present, emphasizing the importance of the surface solvent.
Acknowledgments
We thank the staff of beamline 4-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) and BioCAT at the Advanced Photon Source (APS). The SSRL is a national user facility operated by Stanford University on behalf of the DOE. The SSRL Structural Molecular Biology Program is supported by DOE and NIH. The APS is supported by DOE. BioCAT is supported by NIH grant 9 P41 GM103622.
Glossary
Abbrevations
- RH
relative humidity
- CC
correlation coefficient
Supporting Information Available
Fiber diffraction from a HET-s(218–289) oriented sol and X-ray fiber diffraction from lyophilized and rehydrated HET-s(218–289). This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by U.S. National Institutes of Health (NIH) grants AG002132 (PI GS; Program Director Stanley Prusiner) and F31-AG040947 to W.W.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Eanes E. D.; Glenner G. G. (1968) X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 16, 673–677. [DOI] [PubMed] [Google Scholar]
- Astbury W. T.; Dickinson S.; Bailey K. (1935) The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J. 29, 2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F.; Dobson C. M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366. [DOI] [PubMed] [Google Scholar]
- Barnhart M. M.; Chapman M. R. (2006) Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey R. P.; Shewmaker F.; McPhie P.; Monterroso B.; Thurber K.; Wickner R. B. (2009) The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis. Proc. Natl. Acad. Sci. U.S.A. 106, 13731–13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji S. K.; Perrin M. H.; Sawaya M. R.; Jessberger S.; Vadodaria K.; Rissman R. A.; Singru P. S.; Nilsson K. P. R.; Simon R.; Schubert D.; Eisenberg D.; Rivier J.; Sawchenko P.; Vale W.; Riek R. (2009) Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B. (2007) Prions, in Fields Virology (Knipe D., and Howley P. M., Eds) 5th ed., pp 3059–3091, Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- Saupe S. J. (2011) The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin. Cell Dev. Biol. 22, 460–468. [DOI] [PubMed] [Google Scholar]
- Squires A. M.; Devlin G. L.; Gras S. L.; Tickler A. K.; MacPhee C. E.; Dobson C. M. (2006) X-ray scattering study of the effect of hydration on the cross-β structure of amyloid fibrils. J. Am. Chem. Soc. 128, 11738–11739. [DOI] [PubMed] [Google Scholar]
- Maurstad G.; Prass M.; Serpell L. C.; Sikorski P. (2009) Dehydration stability of amyloid fibrils studied by AFM. Eur. Biophys. J. 38, 1135–1140. [DOI] [PubMed] [Google Scholar]
- Morris K. L.; Rodger A.; Hicks M. R.; Debulpaep M.; Schymkowitz J.; Rousseau F.; Serpell L. C. (2013) Exploring the sequence–structure relationship for amyloid peptides. Biochem. J. 450, 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makin O. S.; Serpell L. C. (2004) Structural characterisation of islet amyloid polypeptide fibrils. J. Mol. Biol. 335, 1279–1288. [DOI] [PubMed] [Google Scholar]
- Shewmaker F.; Kryndushkin D.; Chen B.; Tycko R.; Wickner R. B. (2009) Two prion variants of Sup35p have in-register parallel β-sheet structures, independent of hydration. Biochemistry 48, 5074–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto A.; Hasegawa K.; Suzuki H.; Taguchi H.; Namba K.; Yoshida M. (2004) β-Helix is a likely core structure of yeast prion Sup35 amyloid fibers. Biochem. Biophys. Res. Commun. 315, 739–745. [DOI] [PubMed] [Google Scholar]
- Perutz M. F.; Finch J. T.; Berriman J.; Lesk A. (2002) Amyloid fibers are water-filled nanotubes. Proc. Natl. Acad. Sci. U.S.A. 99, 5591–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M.; Box H.; Bian W.; Kendall A.; Tycko R.; Stubbs G. (2012) Fiber diffraction data indicate a hollow core for the Alzheimer’s Aβ 3-fold symmetric fibril. J. Mol. Biol. 423, 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.-X.; Qiang W.; Yau W.-M.; Schwieters C. D.; Meredith S. C.; Tycko R. (2013) Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 154, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melckebeke H.; Wasmer C.; Lange A.; AB E.; Loquet A.; Böckmann A.; Meier B. H. (2010) Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid-state NMR spectroscopy. J. Am. Chem. Soc. 132, 13765–13775. [DOI] [PubMed] [Google Scholar]
- Van Melckebeke H.; Schanda P.; Gath J.; Wasmer C.; Verel R.; Lange A.; Meier B. H.; Böckmann A. (2011) Probing water accessibility in HET-s(218–289) amyloid fibrils by solid-state NMR. J. Mol. Biol. 405, 765–772. [DOI] [PubMed] [Google Scholar]
- Baxa U.; Wickner R. B.; Steven A. C.; Anderson D. E.; Marekov L. N.; Yau W.-M.; Tycko R. (2007) Characterization of β-sheet structure in Ure2p1–89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry 46, 13149–13162. [DOI] [PubMed] [Google Scholar]
- Wan W.; Wille H.; Stöhr J.; Baxa U.; Prusiner S. B.; Stubbs G. (2012) Degradation of fungal prion HET-s(218–289) induces formation of a generic amyloid fold. Biophys. J. 102, 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory J.; Holmes K. C. (1965) Methods of preparing orientated tobacco mosaic virus sols for X-ray diffraction. J. Mol. Biol. 13, 796–801. [Google Scholar]
- McDonald M.; Kendall A.; Tanaka M.; Weissman J. S.; Stubbs G. (2008) Enclosed chambers for humidity control and sample containment in fiber diffraction. J. Appl. Crystallogr. 41, 206–209. [Google Scholar]
- Fraser R. D. B.; Macrae T. P.; Miller A.; Rowlands R. J. (1976) Digital processing of fibre diffraction patterns. J. Appl. Crystallogr. 9, 81–94. [Google Scholar]
- Bian W.; Wang H.; McCullough I.; Stubbs G. (2006) WCEN: a computer program for initial processing of fiber diffraction patterns. J. Appl. Crystallogr. 39, 752–756. [Google Scholar]
- Wan W.; Bian W.; McDonald M.; Kijac A.; Wemmer D. E.; Stubbs G. (2013) Heterogeneous seeding of a prion structure by a generic amyloid form of the fungal prion-forming domain HET-s(218–289). J. Biol. Chem. 288, 29604–29612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H.; Bian W.; McDonald M.; Kendall A.; Colby D. W.; Bloch L.; Ollesch J.; Borovinskiy A. L.; Cohen F. E.; Prusiner S. B.; Stubbs G. (2009) Natural and synthetic prion structure from X-ray fiber diffraction. Proc. Natl. Acad. Sci. U.S.A. 106, 16990–16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A.; Nave C.; Marvin D. A. (1995) Pf1 filamentous bacteriophage: refinement of a molecular model by simulated annealing using 3.3 Å resolution X-ray fibre diffraction data. Acta Crystallogr., Sect. D 51, 792–804. [DOI] [PubMed] [Google Scholar]
- Russi S.; Juers D. H.; Sanchez-Weatherby J.; Pellegrini E.; Mossou E.; Forsyth V. T.; Huet J.; Gobbo A.; Felisaz F.; Moya R.; McSweeney S. M.; Cusack S.; Cipriani F.; Bowler M. W. (2011) Inducing phase changes in crystals of macromolecules: Status and perspectives for controlled crystal dehydration. J. Struct. Biol. 175, 236–243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.