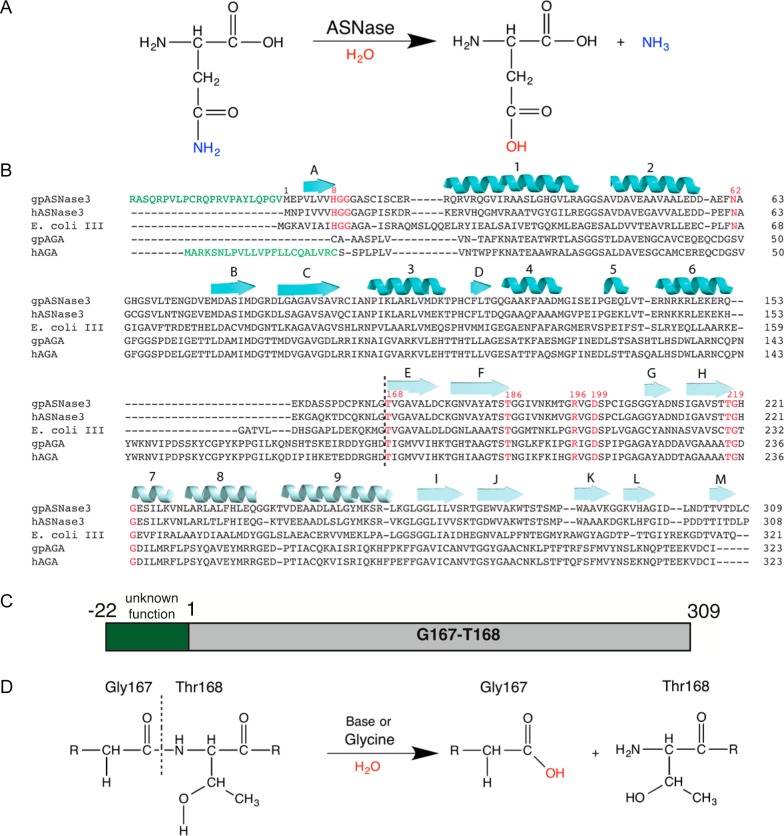
Figure 1.
Reaction schemes, sequence alignment, and constructs of guinea pig ASNase3. (A) Reaction scheme whereby asparaginase hydrolyzes asparagine into aspartate and ammonia. (B) Sequence alignment using Clustal Omega25 of gpASNase3 (H0VQC8_CAVPO), hASNase3 (ASGL1_HUMAN), E. coli type III (IAAA_ECOLI), gpAGA (H0UZ36_CAVPO), and hAGA (ASPG_HUMAN). The hAGA signal peptide and the uncharacterized peptide at the N-terminus of gpASNase3 are denoted in green. Conserved active site residues are in red. The black dashed line separates the α- and β-subunits after autoproteolytic cleavage. The cartoon β-strands and helices in cyan and light cyan represent the secondary structural elements of the α- and β-subunits, respectively, of gpASNase3. (C) Scheme of the uncharacterized H0VQC8_CAVPO UniProt entry. We refer to gpASNase3 as the catalytic domain (gray), lacking the preceding 23 residues (green). (D) Cleavage reaction scheme. GpASNase3 is cleaved between Gly167 and Thr168 through autoproteolysis (very slow) or accelerated by glycine (see text). The freed amino group of Thr168, the first residue of the β-subunit, is required for its l-asparaginase activity.