Abstract

siRNA nanomedicines can potentially treat many human diseases, but safe and effective delivery remains a challenge. DNA delivery polymers such as poly(β-amino ester)s (PBAEs) generally cannot effectively deliver siRNA and require chemical modification to enable siRNA encapsulation and delivery. An optimal siRNA delivery nanomaterial needs to be able to bind and self-assemble with siRNA molecules that are shorter and stiffer than plasmid DNA in order to form stable nanoparticles, and needs to promote efficient siRNA release upon entry to the cytoplasm. To address these concerns, we designed, synthesized, and characterized an array of bioreducible PBAEs that self-assemble with siRNA in aqueous conditions to form nanoparticles of approximately 100 nm and that exhibit environmentally triggered siRNA release upon entering the reducing environment of the cytosol. By tuning polymer properties, including bioreducibility and hydrophobicity, we were able to fabricate polymeric nanoparticles capable of efficient gene knockdown (91 ± 1%) in primary human glioblastoma cells without significant cytotoxicity (6 ± 12%). We were also able to achieve significantly higher knockdown using these polymers with a low dose of 5 nM siRNA (76 ± 14%) compared to commercially available reagent Lipofectamine 2000 with a 4-fold higher dose of 20 nM siRNA (40 ± 7%). These bioreducible PBAEs also enabled 63 ± 16% gene knockdown using an extremely low 1 nM siRNA dose and showed preferential transfection of glioblastoma cells versus noncancer neural progenitor cells, highlighting their potential as efficient and tumor-specific carriers for siRNA-based nanomedicine.
Keywords: nanoparticle, siRNA, drug delivery, polymer, cancer, nanomedicine
RNA interference (RNAi) is a naturally occurring cellular mechanism that ultimately results in sequence-specific gene knockdown and can be externally induced by intracellular delivery of short interfering RNA (siRNA).1 Targeted gene knockdown via siRNA delivery has exciting potential for the treatment of diseases caused by aberrant gene expression.2,3 However, safe and efficient intracellular siRNA delivery remains a challenging obstacle.
Promising siRNA delivery strategies have been suggested that employ lipid-based,4,5 inorganic,6−8 or polymeric materials9−11 similar to those designed for DNA delivery. Certain siRNA delivery material design parameters can be addressed using the same materials found to effectively deliver DNA. Cationic polymers with high buffering capacities, such as poly(ethylenimine) (PEI), promote nucleic acid compaction and protection, cellular internalization, and endosomal escape.12 Polymer degradability such as that afforded by hydrolytically cleavable poly(β-amino ester)s (PBAE)s results in cargo release far superior to nondegradable PEI.13
Two key delivery obstacles specific to siRNA are unstable particle formation and cytoplasmic targeting. The former concern results from the relatively small size and rigidity of siRNA, which is ∼200 times smaller than most plasmids used for DNA delivery and is stiffer than DNA.14,15 Shorter length results in reduced mutivalency of electrostatic interactions between a cationic polymer and anionic siRNA molecule, while rigidity may prevent siRNA from conforming into shapes favorable for binding and nanoparticle (NP) self-assembly. In addition, cytoplasmic targeting of siRNA is required for optimal gene knockdown, as the cytosol is the site of RNAi-induced mRNA degradation.16 Polymer bioreduction by glutathione (GSH) in the reducing cytoplasmic environment is a simple and specific method to engineer triggered cytoplasmic siRNA release.17 This can be achieved by the inclusion of bioreducible disulfide linkages as a cross-linking agent,18 adjacent to cationic groups on polymer end-caps19,20 or along the polymer backbone.21
The use of bioreducible moieties in other siRNA delivery vehicles has met with success in the past. Linear, low-molecular weight PEI segments linked with disulfide bonds were shown to be as effective as commercially available, branched 25 kDa PEI (bPEI) but less cytotoxic. In particular, this material was capable of roughly 50% knockdown of a fluorescent marker gene during in vitro, serum-free siRNA delivery to Chinese hamster ovary cells (CHO-K1) using 100 nM siRNA.9 Disulfide-containing poly(amido amine)s have shown successful in vitro siRNA delivery in human head and neck carcinoma cells (UM-SCC-14C),22 nonsmall cell lung carcinoma (H1299),11 and human prostate cancer cells in which ∼80% knockdown was achieved with 30 nM siRNA.23 The KALA peptide (a 30-residue peptide containing 3 Lys-Ala-Leu-Ala repeats)24 modified with cysteine residues and cross-linked to form a bioreducible polymer was electrostatically complexed with PEGylated siRNA.25 This delivery system achieved nearly 50% gene knockdown in vitro in 10% serum-containing media to MDA-MB-435 melanoma cells using ∼60 nM siRNA. PBAEs containing disulfides in the polymer end-caps achieved ∼70% knockdown in human umbilical vein cells in an in vitro study in which 60 nM siRNA was delivered in the presence of 2% serum.26
Recently, we have shown the synthesis and characterization of a single linear PBAE with disulfide bonds along the polymer backbone.21 In our current work, we sought to improve siRNA delivery with bioreducible PBAE-based nanoparticles by specifically addressing the instability of siRNA nanoparticles and the need for cytoplasmic targeting. We wanted to engineer a class of polymers for siRNA nanoparticle formation and efficient siRNA delivery by balancing polymer bioreducibility and hydrophobicity, as PBAE hydrophobicity may enhance particle stability and has been shown to promote enhanced delivery of both DNA and siRNA.19,27 Polymer bioreducibility was used to reduce potential cytotoxicity and to impart cytoplasmic targeting of siRNA release. To further elucidate ideal siRNA delivery criteria, we also examined the effects of changing nanoparticle formulation parameters and physical properties on gene knockdown and cytotoxicity. We also characterized the biological efficacy of these particles in both cancer and noncancer cells. The results presented herein show that bioreducible PBAE chemical properties and nanoparticle physical properties can be engineered for simple, safe, effective, and cancer-specific siRNA delivery.
Results and Discussion
We were able to successfully synthesize and characterize bioreducible and hydrophobic PBAEs. Bioreducible monomer 2,2′-disulfanediylbis(ethane-2,1-diyl) diacrylate BR6 was synthesized in a method similar to Chen et al.(28) Synthesis of bioreducible and hydrophobic polymers was achieved by mixing backbone monomers 2,2′-disulfanediylbis(ethane-2,1-diyl) (BR6) and hexane-1,6-diyl diacrylate (B6) at ratios of either 1:0, 3:1, 1:1, 1:3, or 0:1 prior to polymerization with side chain monomers 3-amino-1-propanol (S3), 4-amino-1-butanol (S4), or 5-amino-1-pentanol (S5). Polymers were then end-capped with small molecule 1-(3-aminopropyl)-4-methylpiperazine (E7) (Scheme 1). As an example, a polymer synthesized with a 3:1 BR6:B6 ratio, side chain S3, and end-capped with E7 will be referred to as “3:1 R637” in this manuscript, while the same polymer with a 0:1 BR6:B6 ratio will be referred to as “637.” BR6 has almost the same structure as B6, except that it contains a disulfide linkage. As the ratio of BR6:B6 increases, so does the bioreducibility of the combined polymer. Proton nuclear magnetic resonance (1H NMR) was used to confirm the identity and purity of the polymers (Supporting Information Figure S1), while gel permeation chromatography (GPC) was used to confirm the size and polydispersities of the polymers (Supporting Information Table S1).
Scheme 1. Polymer synthesis scheme. Diacrylate backbone monomers BR6 and B6 were randomly copolymerized at a ratio of X:Y with side chain S3, S4, or S5, (demonstrated with S4 above). The resulting acrylate-terminated base polymers were then end-capped with small molecules E6 or E7. The representative polymer shown above would be referred to as “X:Y R647”.

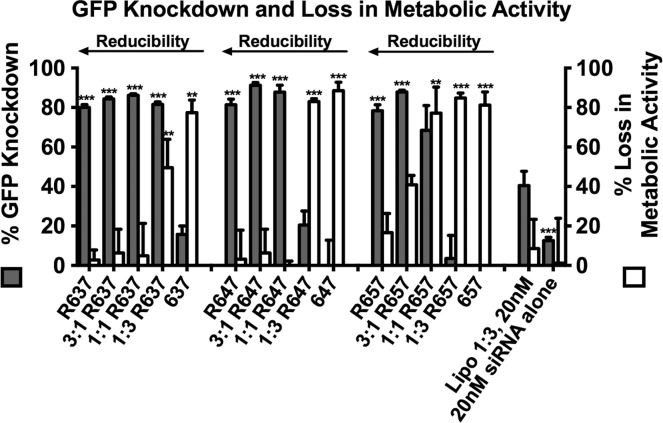
The in vitro siRNA delivery efficacy and cytotoxicity of each of these 15 polymers was evaluated in primary human glioblastoma (GBM 319) cells expressing constitutive GFP,29 using GFP-targeting or a scrambled control siRNA (scRNA). Lipofectamine 2000 and siRNA alone were used as controls with 20 nM siRNA. To evaluate the effect of polymer structure, nanoparticles were formed with all 15 polymers to yield final in vitro concentrations of 180 μg/mL polymer and 20 nM siRNA. These results, which are presented in Figure 1, show interesting trends with regard to polymer bioreducibility and hydrophobicity. First, the results show that as the polymer side chain becomes more hydrophobic, toxicity increases, a conclusion supported by the statistical results shown in Table 1. An example is polymer 1:1 R647 that formed nanoparticles that caused −9 ± 11% (essentially zero) loss in metabolic activity versus polymer 1:1 R657, which has a side chain longer by only one hydrocarbon but formed nanoparticles that caused 77 ± 13% loss in metabolic activity. Second, the results show that polymer bioreducibility significantly reduces cytotoxicity. For example, polymers based on 1:1 BR6:B6, in which ∼50% of repeat units are bioreducible, have dramatically less cytotoxicity than polymers based on 1:3 BR6:B6, in which only ∼25% of repeat units are bioreducible (Table 1). A particular example of this extreme toxicity change with a small change to polymer structure is 1:1 R647, which formed nanoparticles that caused no significant loss in metabolic activity, versus 1:3 R647-based nanoparticles, which caused a loss in metabolic activity of 83 ± 1%.
Figure 1.
Gene knockdown and loss in metabolic activity of polymers with varying bioreducibility and hydrophobicity. Results shown include day 1 loss in metabolic activity and day 9 gene knockdown of GFP+ GBM 319 cells transfected with all polymers using 180 μg/mL polymer and 20 nM siRNA targeting GFP, normalized to cells treated with the same NP formulation using scrambled control RNA. Lipofectamine 2000 is used as the control for statistical comparisons by one-way ANOVA with Dunnett’s post-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
Table 1. Two-Way ANOVA of Loss in Metabolic Activity of All Polymers.
| two-way ANOVA results of loss in metabolic activity | P-value | significance |
|---|---|---|
| Side Chains | ||
| S3 vs S4 | 0.5877 | ns |
| S3 vs S5 | <0.0001 | **** |
| S4 vs S5 | 0.0006 | *** |
| Base Monomers | ||
| R6 vs 3:1 | 0.721 | ns |
| R6 vs 1:1 | 0.2547 | ns |
| R6 vs 1:3 | <0.0001 | **** |
| R6 vs B6 | <0.0001 | **** |
| 3:1 vs 1:1 | 0.9266 | ns |
| 3:1 vs 1:3 | <0.0001 | **** |
| 3:1 vs B6 | <0.0001 | **** |
| 1:1 vs 1:3 | <0.0001 | **** |
| 1:1 vs B6 | <0.0001 | **** |
| 1:3 vs B6 | 0.7468 | ns |
The tunable toxicities of these polymers is interesting, as polymer hydrophobicity has been shown to promote enhanced nucleic acid delivery;19,27 therefore, hydrophobic polymers, such as 647, may be effective for siRNA delivery but are too toxic to have an effective therapeutic window. By combining hydrophobic monomers with bioreducible ones, we have been able to harness the useful properties of hydrophobicity while reducing cytotoxicity and promoting cytoplasmic cargo release. Polymer R647, for example, formed nanoparticles that achieved 81 ± 3% GFP knockdown versus 3:1 R647, which achieved 91 ± 1%, a significant increase (p < 0.05 by Student’s t test), resulting only from making 25% of repeat units more hydrophobic. Another interesting result from these studies was that eight of the polymeric nanoparticles tested achieved significantly higher GFP knockdown than Lipofectamine 2000 without causing significantly higher loss of metabolic activity. An example fluorescence image of 1:1 R647 nanoparticle treated cells demonstrating safety and efficacy is shown in Figure 2E.
Figure 2.
GFP knockdown and loss of metabolic activity in GFP+ GBM 319 cells transfected with various formulations of 1:1 R647 siRNA nanoparticles. All knockdown values are normalized to scrambled control RNA. (A) Transfection results using 180 μg/mL polymer with siRNA doses ranging from 1 to 160 nM. The Lipofectamine 2000 control shown used 20 nM siRNA. (B) Transfection results using 20 nM siRNA with polymer concentrations ranging from 11.25 to 360 μg/mL. (C) Correlation of knockdown efficiency and varying siRNA doses with polymer concentration fixed at 180 μg/mL fitted to a semilogarithmic line. (D) Correlation of knockdown efficiency and varying polymer concentrations with siRNA concentration fixed at 20 nM fitted to a linear regression. (E) Phase contrast (top) and fluorescence (bottom) images of GFP+ GBM cells treated with 1:1 R647 at 180 μg/mL and 20 nM of either siRNA targeting GFP (left) or scrambled control RNA (right) NPs. Lipofectamine 2000 is used as the control for statistical comparisons by one-way ANOVA with Dunnett’s post-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
To further elucidate the nanoparticle properties favorable for safe and effective siRNA delivery, we sought to examine the effects of changing nanoparticle formulation and the resulting physical properties associated with these changes. First, siRNA dose-dependency was examined by delivering siRNA at final in vitro doses ranging from 1 to 160 nM using polymer 1:1 R647 at a fixed concentration of 180 μg/mL (Figure 2A,C). An intriguing result from this experiment is that we were able to achieve significantly higher GFP knockdown using only 5 nM siRNA (76 ± 14%) compared to leading commercially available Lipofectamine 2000 with 20 nM siRNA (40 ± 7%). Importantly, none of the formulations tested were significantly more toxic to the cells than Lipofectamine 2000. Additionally, we achieved 63 ± 16% GFP knockdown with as little as 1 nM siRNA, demonstrating the efficiency of these bioreducible siRNA-containing nanoparticles. Interestingly, we did not see a particularly strong siRNA dose-dependent trend of GFP knockdown within the range of nanoparticle formulations tested, as almost all siRNA doses evaluated caused uniformly high knockdown. Of the nine polymer/siRNA doses tested, seven achieved more than 75% knockdown and were significantly more effective than Lipofectamine 2000. For these samples, knockdown correlated semilogarithmically with siRNA dose (R2 = 0.8649).
Polymer concentration dependency was examined by carrying out transfections with 20 nM siRNA and varying 1:1 R647 concentrations from 11.25 to 360 μg/mL (Figure 2B,D). Interestingly, GFP knockdown correlated linearly with polymer concentration with R2 = 0.9440. To elucidate the mechanisms behind these surprising results, we analyzed the nanoparticle physical properties associated with each delivery method to determine the size, zeta potential, nanoparticle concentration, and siRNA loading of each formulation. siRNA loading was calculated from the nanoparticle concentration, total siRNA dose, and siRNA molecular weight. Nanoparticle concentration measurements were quantified in a manner consistent with the protocol described by Bhise et al.(30)
To evaluate the ability of the polymers to complex with siRNA to form nanoparticles, we determined the polymer/siRNA weight ratios (wt/wts) at which siRNA became completely complexed. We performed a gel retention assay using 1:1 R647 with wt/wts ranging from 37.5 to 600 w/w (Figure 3). We found that siRNA is completely bound to 1:1 R647 at wt/wts as low as 150 w/w, but not at 75 or 37.5 w/w. This study also enabled us to validate that siRNA loading could not be accurately calculated for nanoparticle formulations at these lower wt/wts. To demonstrate the siRNA release efficacy of this polymer in a reducing environment comparable to the cytosol,17 particles were incubated in a solution of 5 mM glutathione for 15 min prior to electrophoresis. All formulations tested showed complete siRNA release, showing that siRNA unloading can occur within minutes of reaching the cytosol, in an environmentally triggered manner, due to the disulfide linkages in the polymer. While there are also ester linkages in these bioreducible PBAEs, the half-life for hydrolysis of the ester bonds in this class of materials is on the order of hours,31 while the half-life for bioreduction of the disulfide bonds is on the order of minutes in the presence of glutathione.21 While Figure 3 shows that intracellular siRNA release is likely driven by disulfide degradation, the hydrolytic degradability of the polymers may afford an additional reduction in potential toxicity.
Figure 3.

Gel retention assay of 1:1 R647 particles formed at varying w/w and incubated for 15 min at room temperature in the absence (top) or presence (bottom) of 5 mM GSH. Columns above each well indicate polymer to siRNA weight ratio (w/w).
Nanoparticle properties were measured using the same formulations shown in Figure 2A, in which siRNA dose was varied from 1 to 160 nM and 1:1 R647 concentration remained the same at 180 μg/mL. Nanoparticle diameter was shown to correlate with siRNA dose on a semilogarithmic scale (R2 = 0.9077), while zeta potential remained consistently between 18 and 22 mV (Figure 4A). Size measurements were also completed using 0 nM siRNA, which showed smaller polymeric particles, 78 ± 4 nm in size. Nanoparticle concentration remained nearly constant with changing siRNA dose, even with 0 nM siRNA, staying between 1.29 and 1.66 × 1011 particles/mL (R2 = 0.0013). siRNA loading was calculated and demonstrated a linear correlation with siRNA dose (R2 = 0.9980).
Figure 4.
Characterization of nanoparticle size, zeta potential, concentration, and loading of nanoparticles synthesized with 180 μg/mL 1:1 R647 and varying siRNA doses (A and C) or with 20 nM siRNA and varying polymer concentrations (B and D). Size and concentration were measured by NTA, zeta potential was measured using DLS, and siRNA loading was calculated from concentration. (A) Nanoparticle size positively correlates with siRNA dose on a semilogarthimic scale, while zeta potential does not change. (B) Nanoparticle size and zeta potential do not strongly correlate with polymer concentration. (C) Nanoparticle concentration remains consistent despite changing siRNA dose, while siRNA loading increases linearly. (D) Nanoparticle concentration linearly increases with polymer concentration resulting in exponential decay of siRNA loading with increasing polymer concentration.
We repeated the same experiments, this time using the nanoparticle formulations shown in Figure 2B, where siRNA dose remained constant at 20 nM and 1:1 R647 concentration was varied from 11.25 to 360 μg/mL. Polymer concentration did not correlate well with either nanoparticle diameter (R2 = 0.3012) or zeta potential (R2 = 0.4280) (Figure 4B). Nanoparticle concentration, however, fit a linear regression versus polymer concentration (R2 = 0.9496). This resulted in siRNA loading values that exponentially decayed with increasing polymer concentration (R2 = 0.9989), meaning that the most effective siRNA delivery formulations in this group consisted of the highest nanoparticle concentrations but with the lowest siRNA loading values.
When calculating siRNA loading, we were calculating average siRNA loading across a nanoparticle batch. We believe that siRNA loading within these batches is also roughly uniform based on the presence of a constant nanoparticle concentration across all siRNA doses as well as linearly increasing nanoparticle diameters with increasing siRNA dose. To further characterize nanoparticle size and siRNA loading, we performed transmission electron microscopy (TEM) on 1:1 R647 at 180 μg/mL with 20 nM siRNA and compared the size histogram results from NTA of this nanoparticle formulation to the corresponding nanoparticle formulation without siRNA (Figure 5). TEM shows a roughly uniform size distribution, with the presence of a few larger particles/aggregates (Figure 5A). This matches the NTA analysis as well (Figure 5B). The NTA histogram of the nanoparticles containing 20 nM siRNA (113 ± 2 nm) was distinct from that of the smaller nanoparticles containing 0 nM siRNA (78 ± 4 nm). Due to this greater size and monodisperse distribution, it is most likely that as siRNA dose is increased, each nanoparticle contains more siRNA per particle, rather than a significant fraction of nanoparticles remaining empty, which would have resulted in a bimodal particle distribution.
Figure 5.
Characterization of nanoparticle size distribution. (A) TEM image of nanoparticles made with 1:1 R647 at 180 μg/mL and 20 nM siRNA. (B) Nanoparticle size distribution of 1:1 R647 at 180 μg/mL and either 20 or 0 nM siRNA as measured by NTA.
We envision that these bioreducible siRNA nanoparticles could potentially be used for local intracranial delivery of siRNA for the treatment of glioblastoma. In this approach, they would applied in a manner analogous to the GLIADEL Wafer following surgical resection of glioblastoma. Although we intend for this potential therapeutic to be used for local delivery, rather than systemic delivery, we also looked at transfection in the presence of serum proteins, which are known to reduce the efficacy of many gene delivery systems. We found that bioreducible PBAE polymers can enable high (80 ± 4%) GFP knockdown in the presence of 10% serum-containing media and knockdown can persist for at least 2 weeks (Supporting Information Figure S2).
To evaluate the therapeutic potential of the bioreducible nanoparticles in vitro, primary human glioblastoma and primary human noncancer neural progenitor cells were both transfected with the same bioreducible nanoparticles containing siRNA that could cause cell death to successfully transfected cells (Figure 6). We evaluated the brain cancer killing potential of these nanoparticles, their potential off-target cytotoxicity to noncancer human primary cells, and the potential for the nanoparticles to effectively target siRNA delivery to brain cancer cells over healthy brain cells.
Figure 6.
Delivery of death siRNA to cancer and noncancer brain cells using polymer R646. (A and B) Delivery efficacy of death positive control siRNA to cancer (GBM 319) and noncancer (fNPC 34) cells using a range of bioreducible PBAE nanoparticle formulations. All percent siRNA-mediated death values are calculated versus cell counts of scrambled control RNA-treated cells. (C and D) Toxicity of nanoparticle treatments to GBM 319 and fNPC 34 cells. (E) Phase contrast images of fNPC 34 (top) and GBM 319 (bottom) cells following treatment with nanoparticles containing 120 nM of either scrambled RNA or death positive control siRNA.
To accomplish this goal, we optimized siRNA delivery to human fetal neural progenitor cells (fNPC 34s) and GBM 319 cells over a range of polymer and siRNA concentrations. Out of the polymers synthesized from the monomers in Scheme 1, we found R646 to be optimal for siRNA delivery to the fNPC 34 cells, which included minimizing cytotoxicity, and it was therefore utilized for subsequent functional siRNA delivery studies. Functional delivery efficacy was detected using a positive control cell death-inducing siRNA, with which successfully delivered siRNA would result in cell death. Cell death was measured using DAPI and propidium iodide (PI) to stain healthy and dying cells, respectively, and cell death was calculated using fluorescence microscopy followed by quantification with ImageJ software. Nanoparticle toxicity was calculated by normalizing scrambled RNA (scRNA)-treated cell counts to untreated cell counts, while siRNA delivery efficacy was calculated by normalizing death siRNA-treated cell counts to scRNA-treated cell counts.
For all R646 nanoparticle formulations tested, the GBM 319 cells showed near-complete siRNA-mediated cell death (79–97, while the fNPC 34 cells had 0–27% siRNA-mediated cell death under the same conditions. For certain nanoparticle formulations, high knockdown leading to specific cell death of human glioblastoma cells was achieved (>90%), while nonspecific cytotoxicity to both human glioblastoma and healthy human neural cells was kept low (<20%). As the siRNA itself was not specific to GBM 319 cells over fNPC 34 cells and the same particle and siRNA doses were utilized, this finding suggests that bioreducible PBAE-based nanoparticles themselves preferentially transfect cancer cells over noncancer cells. This presents an interesting opportunity in siRNA-based nanomedicine as future siRNA targets and siRNA combinations may not need to be limited to proteins that would specifically affect cancer cell viability without harming the surrounding tissue, but instead could enable effective targeting and knockdown of any protein necessary to tumor survival as the material itself could provide cancer specificity.
We have synthesized and characterized novel siRNA delivery nanoparticles capable of near complete gene knockdown in human primary glioblastoma (GBM 319) cells. These nanoparticles are safe and effective even at very low siRNA doses. We were able to show that combining polymer hydrophobicity, a property known to promote enhanced siRNA and DNA delivery,19,27 with bioreducibility decreased the cytotoxic effects typical of hydrophobic polymers while optimizing environmentally triggered cytoplasmic cargo release to enhance siRNA delivery. We examined the effects of changing nanoparticle formulation and were able to show that, with this class of materials, nanoparticle concentration is largely determined by polymer concentration and that higher polymer concentrations promote enhanced siRNA delivery. Gene knockdown was shown to be very effective (91 ± 1%) with moderate doses of siRNA (20 nM), and effective (63 ± 16%) even at very low doses of siRNA (1 nM). Functional siRNA delivery efficacy was shown to be near complete (up to 97 ± 4%) in human primary glioblastoma, but less effective (up to 27 ± 9%) in human primary noncancer brain cells, which suggests that these particles could exhibit cancer specificity. Bioreducible PBAEs with tunable hydrophobicities have exciting potential as safe and efficient siRNA delivery vehicles for nanomedicine applications.
Materials and Methods
Materials
All chemicals used for the synthesis of monomer BR6 were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO) and used without further purification. All other monomers were purchased from Alfa Aesar (Ward Hill, MA). Lipofectamine 2000 and Opti-MEM I were purchased from Invitrogen (Carlsbad, CA) and used according to manufacturer’s instructions. Ambion Silencer eGFP and Ambion Silencer Negative Control #1 siRNA were purchased from Life Technologies. CellTiter 96 AQueous One MTS assay was purchased from Promega (Fitchburg, WI) and used according to manufacturer’s instructions.
BR6 Synthesis
Bis(2-hydroxyethyl) disulfide (15.4 g, 10 mmol) and triethylamine (TEA, 37.5 mL, 300 mmol) were dissolved in 450 mL of tetrahydrofuran (THF) (previously dried with Na2SO4) in a 1 L round-bottom flask; the contents were then flushed with N2 for 10 min and maintained under a N2 environment for the remainder of the reaction time. Acryloyl chloride (24.4 mL, 300 mmol) was dissolved in 50 mL of dried THF and added to the flask dropwise over 2 h while stirring, and the reaction was allowed to continue at room temperature for 24 h. Following reaction, TEA HCl precipitate was removed by filtration, and THF was removed by rotary evaporation. The product was dissolved in 200 mL of dichloromethane (DCM) and washed five times with 200 mL of aqueous 0.2 M Na2CO3 and three times with distilled water. The solution was dried with Na2SO4, and DCM was removed by rotary evaporation. The product 2,2′-disulfanediylbis(ethane-2,1-diyl) (BR6) was confirmed via1H NMR: (CDCl3, 400 Hz), δ2.95 (2H, t, CH2CHCOOCH2CH2S), δ3.95 (2H, t, CH2CHCOOCH2CH2S), δ5.8–5.9 (1H, d, CH2CHCOOCH2CH2S), δ6.1–6.2 (1H, dd, CH2CHCOOCH2CH2S), δ6.4–6.5 (1H, d, CH2CHCOOCH2CH2S).
Polymer Synthesis
Polymer synthesis was carried out in a method similar to Bhise et al.(32) The diacrylate base monomers used for polymerization were BR6 (see above) or hexane-1,6-diyl diacrylate (B6). Backbone monomers BR6 and hexane-1,6-diyl diacrylate (B6) were mixed at a molar ratio of 1:0, 3:1, 1:1, 1:3, or 0:1 prior to polymerization. Side chain monomers used were 3-amino-1-propanol (S3), 4-amino-1-butanol (S4), or 5-amino-1-pentanol (S5). The end-caps used were 2-(3-(aminopropyl)amino)methanol (E6) and 1-(3-aminopropyl)-4-methylpiperazine (E7). For all polymers, polymerization was completed using a base monomer to side chain ratio of 1.01:1 at 500 mg/mL in dimethyl sulfoxide (DMSO) at 90 °C for 24 h while stirring. The polymers were end-capped in DMSO at 100 mg/mL with 0.2 M E7 for 1 h at room temperature while shaking. Excess E7 monomer was not removed from the polymer solution; however, examining E7 monomer cytotoxicity showed that free E7 was not significantly cytotoxic to GBM 319 cells (Supporting Information Figure S3). As we have found with our prior work with this class of polymers,32 the step growth polymerization of the bifunctional monomers leads to short linear polymers without any byproducts or side reactions. Purity of polymers and the identity of copolymers is confirmed by 1H NMR spectra. The 1H NMR spectra for representative polymers R647, 1:1 R647, and 647 are shown in Supporting Information Figure S1. The integration of the peaks of the copolymers validate that base monomers B6 and BR6 incorporate into a copolymer at the same molar ratio as is used during polymer synthesis.
siRNA Delivery to GBM 319 and fNPC 34 Cells and Cell Viability
GFP+ GBM 319 glioblastoma cells were plated at a cell density of 15 000 cells/well in 96-well tissue culture plates in 89% GIBCO DMEM-F12, 1% GIBCO Antibiotic-Antimycotic (Invitrogen), and 10% Corning Cellgro Heat-Inactivated FBS and allowed to adhere overnight. fNPC 34 cells were utilized to compare siRNA delivery between primary human glioblastoma cells and healthy human primary cells found in the brain. fNPC 34 cells are primary fetal neural progenitor cells obtained as described previously following procedures approved by the Johns Hopkins University Institutional Review Board.33 fNPC 34 cells were plated at a density of 15 000 cells/well in 96-well tissue culture plates in 97% GIBCO DMEM-F12, 1% GIBCO Antibiotic-Antimycotic (Invitrogen), 2% B-27 Serum-Free Supplement, and 20 μg/mL each of basic fibroblast growth factor (Roche Applied Science) and epidermal growth factor (Sigma) and allowed to adhere overnight prior to transfection. The siRNAs used were either siRNA targeting eGFP with sequence 5′-CAAGCUGACCCUGAAGUUCTT (sense) and 3′-GAACUUCAGGGUCAGCUUGCC (antisense), or a scrambled control siRNA (scRNA) with sequence 5′-AGUACUGCUUACGAUACGGTT (sense) and 3′-CCGUAUCGUAAGCAGUACUTT (antisense). AllStars Human Cell Death siRNA was purchased from Qiagen. For all transfections, siRNA and polymers were diluted in 25 mM NaAc at 12 times the final concentration listed for each group, and siRNA and polymers were combined in a 1:1 (v/v) ratio and allowed to form cells for 10 min at room temperature. As an example, nanoparticles listed at final concentrations of “180 μg/mL and 20 nM siRNA” were formed by mixing a 2.16 mg/mL solution of polymer with a 240 nM solution of siRNA. The cell culture media was removed and replaced with serum-free media prior to adding nanoparticles. Nanoparticle formulations were diluted in each well in quadruplicates in a 1:6 (v/v) ratio to yield the final siRNA and polymer concentrations listed for each group. Cells were incubated with nanoparticles for 4 h for siGFP experiments and 2 h for death siRNA experiments, after which the nanoparticle solutions were removed and fresh, serum-containing media was added. Cytoxicity for siGFP transfections was assessed 24 h after transfection CellTiter 96 AQueous One MTS assay following manufacturer’s instructions and read using a BioTek Synergy 2 Microplate Reader.
Cell death in death siRNA experments was assessed by staining cells with propidium iodide (PI) in DMEM-F12 at 1:200 (v/v) PI prior to fixation and 750 nM 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI) following fixation in 10% formalin. Cell images were taken at 5× magnification using a Zeiss Axio observer A1 microscope with a Zeiss Axiocam MRm camera using AxioVision Release 4.8.2 software. Fluorescence was provided by an Exfo X-Cite series 120Q. Live and dead cells were quantified using ImageJ v1.47 software, and dead cells were subtracted from the live cell count to yield the total cell count for each well.
Flow Cytometry
All flow cytometry was completed at 9 days post-transfection using an Intellicyt high-throughput loader attached to an BD Accuri C6 flow cytometer (emission filter: 530/30 nm). Hypercyt software was used to discriminate events between each well and FlowJo 7 software was used to analyze the flow cytometry results. Cells were prepared for flow cytometry by 5 min of trypsinization with 30 μL of 0.05% trypsin-EDTA, followed by the addition of 170 μL of a buffer of PBS containing 1:50 (v/v) FBS and 1:200 (v/v) PI. Cell suspensions were moved to round-bottom 96-well plates and centrifuged for 5 min at 1000 rpm. A total of 170 μL of supernatant was removed, and cells were resuspended in the remaining buffer. PI signal was used to distinguish dead or dying cells from live cells so that the unhealthy cells could be removed from analysis. GFP knockdown was determined by finding the geometric mean FL1 fluorescence signal for each sample. Percent knockdown was calculated by normalizing the GFP expression of siRNA-treated cells to scRNA-treated cells. All the transfections were carried out using the same cell line, siRNAs, controls, and data collection protocols, and all formulations that caused >60% loss in metabolic activity were considered nonviable and excluded from further analysis.
Gel Retention Assay
Nanoparticles were formed using 0.01 mg/mL scrambled control RNA (scRNA) in 25 mM sodium acetate (NaAc) and polymer 1:1 R647 at weight ratios to scRNA ranging from 600 to 0 w/w (siRNA) alone. Polymer to siRNA ratios are also described as N:P ratios (Supporting Information Table S2). These were incubated for 10 min at room temperature to allow for particle formation. To compare the effects of a nonreducing and reducing environment on the particles, either PBS or PBS containing l-glutathione (GSH) to yield a final GSH concentration of 0 or 5 mM, respectively, was added and allowed to incubate at room temperature for 15 min. A solution of 30% glycerol was added to the particles in a 1:5 (v/v) ratio. The particles were loaded into a 1% agarose gel containing 1 μg/mL ethidium bromide and electrophoresed at 100 mV for 20 min. Gels were visualized using UV light exposure.
Particle Size and Concentration Determination: Nanoparticle Tracking Analysis
All nanoparticles were made in the same manner that they were for transfection and then diluted so that their sizes and concentrations could be accurately determined using Nanoparticle Tracking Analysis (NTA). NTA was performed using a NanoSight NS500 and analyzed using NanoSight NTA 2.4 software. As an example, particles for transfection groups labeled “180 μg/mL polymer with 20 nM siRNA” were synthesized by forming particles at a polymer concentration of 1.08 mg/mL and scRNA at 120 nM in NaAc, as these particles would be diluted in a 1:6 (v/v) ratio in media during transfection. For NTA, however, these particles were diluted in PBS following the protocol recommended by Bhise et al.(30) All measurements were repeated with three separate formulations for each condition. The NTA analysis reported the number-average hydrodynamic radius of the particles. All particle concentrations were reported as the number of particles per volume that would be present in the transfection wells.
siRNA loading was calculated by dividing the total amount of siRNA per transfection well by the number of particles per well. This calculation was only completed for particle formulations with wt/wt ratios high enough to completely bind all siRNA as determined by the gel retention assay. For this reason, any particle formulations with wt/wt ratios at or below 75 w/w were excluded from siRNA loading calculations.
Particle Zeta-Potential Determination: Dynamic Light Scattering
Particles were formed at the same concentrations and in the same manner as described for particle sizing. Particles were diluted 1:650 (v/v) in PBS and loaded into a disposable cuvette cell. Particle surface charge was determined via dynamic light scattering (DLS) using a Malvern Zetasizer NanoZS.
Transmission Electron Microscopy
Nanoparticles formed using 1:1 R647 at 180 μg/mL and 20 nM siRNA were imaged using transmission electron microscopy (TEM). The 1:1 R647 was diluted to 2.16 μg/mL in 25 mM NaAc, scRNA was diluted to 240 nM in NaAc, and the two solutions were combined in a 1:1 (v/v) ratio and allowed to form particles for 10 min at room temperature. Following particle formation, 5 μL of the nanoparticle solution was placed onto a carbon-coated copper TEM grid and allowed to dry. Particles were imaged using a Philips/FEI BioTwin CM120 transmission electron microscope.
Gel Permeation Chromatography
GPC was performed using a Waters GPC system with three Waters Styragel columns in a series (HR 1, HR 3, and HR4) and a Waters 2414 refractive index detector, both maintained at 40 °C throughout all samples, which were loaded using a Waters 717plus autosampler (Waters Corp., Milford, MA). All samples were loaded at 5 mg/mL using 94% THF, 5% DMSO, and 1% piperidine (v/v) as the eluent at a flow rate of 1.0 mL/min for 40 min. Polymer molecular weights were calculated relative to polystyrene standards (Shodex, Japan).
Statistics
All results are presented as mean ± standard error of the mean. Statistical significance results for all % GFP knockdown and % loss in metabolic activity were determined using a one-way ANOVA with Dunnett’s post-tests using Lipofectamine 2000 as the control. All particle formulations that caused >60% toxicity were considered nonviable and excluded from statistical testing. A two-way ANOVA with Tukey’s multiple comparisons post-test was also used to compare changes in loss in metabolic activities of cells treated with different polymers using side chain and base monomer as the parameters. R squared correlation values were calculated compared to either linear or nonlinear regressions as labeled in each figure caption. All significance tests with p < 0.05 were considered significant.
Supporting Information Available
Polymer characterization via GPC, polymer characterization via1H NMR, and additional GFP knockdown data. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported in part by the NIH (1R01EB016721 and R21CA152473). K.L.K. thanks the NIH Cancer Nanotechnology Training Center (R25CA153952) at the JHU Institute for Nanobiotechnology for fellowship support and S.Y.T. thanks the National Science Foundation for fellowship support. The authors thank the Microscopy and Imaging Core Module of the Wilmer Core Grant, EY001765 for flow cytometry studies. The authors thank Alfredo Quiñones-Hinojosa for the use of GBM 319 and fNPC 34 cells for these experiments.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Fire A.; Xu S. Q.; Montgomery M. K.; Kostas S. A.; Driver S. E.; Mello C. C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Wu W.; Sun M.; Zou G. M.; Chen J. MicroRNA and Cancer: Current Status and Prospective. Int. J. Cancer 2007, 120, 953–960. [DOI] [PubMed] [Google Scholar]
- Yadav S.; van Vlerken L. E.; Little S. R.; Amiji M. M. Evaluations of Combination MDR-1 Gene Silencing and Paclitaxel Administration in Biodegradable Polymeric Nanoparticle Formulations to Overcome Multidrug Resistance in Cancer Cells. Cancer Chemother. Pharmacol. 2009, 63, 711–722. [DOI] [PubMed] [Google Scholar]
- Akinc A.; Zumbuehl A.; Goldberg M.; Leshchiner E. S.; Busini V.; Hossain N.; Bacallado S. A.; Nguyen D. N.; Fuller J.; Alvarez R.; et al. A Combinatorial Library of Lipid-Like Materials for Delivery of RNAi Therapeutics. Nat. Biotechnol. 2008, 26, 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple S. C.; Akinc A.; Chen J.; Sandhu A. P.; Mui B. L.; Cho C. K.; Sah D. W.; Stebbing D.; Crosley E. J.; Yaworski E.; et al. Rational Design of Cationic Lipids for siRNA Delivery. Nat. Biotechnol. 2010, 28, 172–176. [DOI] [PubMed] [Google Scholar]
- Derfus A. M.; Chen A. A.; Min D. H.; Ruoslahti E.; Bhatia S. N. Targeted Quantum Dot Conjugates for siRNA Delivery. Bioconjugate Chem. 2007, 18, 1391–1396. [DOI] [PubMed] [Google Scholar]
- Elbakry A.; Zaky A.; Liebl R.; Rachel R.; Goepferich A.; Breunig M. Layer-by-Layer Assembled Gold Nanoparticles for siRNA Delivery. Nano Lett. 2009, 9, 2059–2064. [DOI] [PubMed] [Google Scholar]
- Kakizawa Y.; Furukawa S.; Ishii A.; Kataoka K. Organic-Inorganic Hybrid-Nanocarrier of siRNA Constructing through the Self-Assembly of Calcium Phosphate and Peg-Based Block Aniomer. J. Controlled Release 2006, 111, 368–370. [DOI] [PubMed] [Google Scholar]
- Breunig M.; Hozsa C.; Lungwitz U.; Watanabe K.; Umeda I.; Kato H.; Goepferich A. Mechanistic Investigation of Poly (Ethylene Imine)-Based siRNA Delivery: Disulfide Bonds Boost Intracellular Release of the Cargo. J. Controlled Release 2008, 130, 57–63. [DOI] [PubMed] [Google Scholar]
- Matsumoto S.; Christie R. J.; Nishiyama N.; Miyata K.; Ishii A.; Oba M.; Koyama H.; Yamasaki Y.; Kataoka K. Environment-Responsive Block Copolymer Micelles with a Disulfide Cross-Linked Core for Enhanced siRNA Delivery. Biomacromolecules 2009, 10, 119–127. [DOI] [PubMed] [Google Scholar]
- van der Aa L. J.; Vader P.; Storm G.; Schiffelers R. M.; Engbersen J. F. J. Optimization of Poly(amido amine)S as Vectors for siRNA Delivery. J. Controlled Release 2011, 150, 177–186. [DOI] [PubMed] [Google Scholar]
- Boussif O.; Lezoualch F.; Zanta M. A.; Mergny M. D.; Scherman D.; Demeneix B.; Behr J. P. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in Vivo: Polyethylenimine. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn D. M.; Langer R. Degradable Poly (Beta-Amino Esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA. J. Am. Chem. Soc. 2000, 122, 10761–10768. [Google Scholar]
- Hagerman P. J. Flexibility of RNA. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 139–156. [DOI] [PubMed] [Google Scholar]
- Kebbekus P.; Draper D. E.; Hagerman P. Persistence Length of RNA. Biochemistry 1995, 34, 4354–4357. [DOI] [PubMed] [Google Scholar]
- Kawasaki H.; Taira K. Short Hairpin Type of dsRNAs That Are Controlled by Trnaval Promoter Significantly Induce RNAi-Mediated Gene Silencing in the Cytoplasm of Human Cells. Nucleic Acids Res. 2003, 31, 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith O. W. Biologic and Pharmacologic Regulation of Mammalian Glutathione Synthesis. Free Radical Bio. Med. 1999, 27, 922–935. [DOI] [PubMed] [Google Scholar]
- Miyata K.; Kakizawa Y.; Nishiyama N.; Harada A.; Yamasaki Y.; Koyama H.; Kataoka K. Block Catiomer Polyplexes with Regulated Densities of Charge and Disulfide Cross-Linking Directed to Enhance Gene Expression. J. Am. Chem. Soc. 2004, 126, 2355–2361. [DOI] [PubMed] [Google Scholar]
- Tzeng S. Y.; Green J. J. Subtle Changes to Polymer Structure and Degradation Mechanism Enable Highly Effective Nanoparticles for siRNA and DNA Delivery to Human Brain Cancer. Adv. Healthcare Mater. 2013, 2, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng S. Y.; Hung B. P.; Grayson W. L.; Green J. J. Cystamine-Terminated Poly(beta-amino ester)s for siRNA Delivery to Human Mesenchymal Stem Cells and Enhancement of Osteogenic Differentiation. Biomaterials 2012, 33, 8142–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozielski K. L.; Tzeng S. Y.; Green J. J. A Bioreducible Linear Poly(beta-amino ester) for siRNA Delivery. Chem. Commun. 2013, 49, 5319–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader P.; van der Aa L. J.; Engbersen J. F. J.; Storm G.; Schiffelers R. M. Disulfide-Based Poly(amido amine)s for siRNA Delivery: Effects of Structure on siRNA Complexation, Cellular Uptake, Gene Silencing and Toxicity. Pharm. Res. 2011, 28, 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. H.; Christensen L. V.; Yockman J. W.; Zhong Z. Y.; Engbersen J. F. J.; Kim W. J.; Feijen J.; Kim S. W. Reducible Poly(amido ethylenimine) Directed to Enhance RNA Interference. Biomaterials 2007, 28, 1912–1917. [DOI] [PubMed] [Google Scholar]
- Wyman T. B.; Nicol F.; Zelphati O.; Scaria P.; Plank C.; Szoka F. C. Jr. Design, Synthesis, and Characterization of a Cationic Peptide That Binds to Nucleic Acids and Permeabilizes Bilayers. Biochemistry 1997, 36, 3008–3017. [DOI] [PubMed] [Google Scholar]
- Mok H.; Park T. G. Self-Crosslinked and Reducible Fusogenic Peptides for Intracellular Delivery of siRNA. Biopolymers 2008, 89, 881–888. [DOI] [PubMed] [Google Scholar]
- Tzeng S. Y.; Yang P. H.; Grayson W. L.; Green J. J. Synthetic Poly(ester amine) and Poly(amido amine) Nanoparticles for Efficient DNA and siRNA Delivery to Human Endothelial Cells. Int. J. Nanomed. 2012, 6, 3309–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine J. C.; Akanda M. I.; Li D.; Kozielski K. L.; Green J. J. Effects of Base Polymer Hydrophobicity and End-Group Modification on Polymeric Gene Delivery. Biomacromolecules 2011, 12, 3592–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Qiu X.; Ouyang J.; Kong J.; Zhong W.; Xing M. M. pH and Reduction Dual-Sensitive Copolymeric Micelles for Intracellular Doxorubicin Delivery. Biomacromolecules 2011, 12, 3601–3611. [DOI] [PubMed] [Google Scholar]
- Tzeng S. Y.; Guerrero-Cazares H.; Martinez E. E.; Sunshine J. C.; Quiñones-Hinojosa A.; Green J. J. Non-Viral Gene Delivery Nanoparticles Based on Poly (beta-amino esters) for Treatment of Glioblastoma. Biomaterials 2011, 32, 5402–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhise N. S.; Shmueli R. B.; Gonzalez J.; Green J. J. A Novel Assay for Quantifying the Number of Plasmids Encapsulated by Polymer Nanoparticles. Small 2012, 8, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine J. C.; Peng D. Y.; Green J. J. Uptake and Transfection with Polymeric Nanoparticles Are Dependent on Polymer End-Group Structure, but Largely Independent of Nanoparticle Physical and Chemical Properties. Mol. Pharm. 2012, 9, 3375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhise N. S.; Gray R. S.; Sunshine J. C.; Htet S.; Ewald A. J.; Green J. J. The Relationship between Terminal Functionalization and Molecular Weight of a Gene Delivery Polymer and Transfection Efficacy in Mammary Epithelial 2-D Cultures and 3-D Organotypic Cultures. Biomaterials 2010, 31, 8088–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin R.; Blank P. S.; Steinkamp A.; Rappaport S. M.; Ravin N.; Bezrukov L.; Guerrero-Cazares H.; Quinones-Hinojosa A.; Bezrukov S. M.; Zimmerberg J. Shear Forces During Blast, Not Abrupt Changes in Pressure Alone, Generate Calcium Activity in Human Brain Cells. PLoS One 2012, 7, e39421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.