Abstract
Whether for fundamental biological research or for diagnostic and drug discovery applications, protein micro- and nanoarrays are attractive technologies because of their low sample consumption, high-throughput, and multiplexing capabilities. However, the arraying platforms developed so far are still not able to handle membrane proteins, and specific methods to selectively immobilize these hydrophobic and fragile molecules are needed to understand their function and structural complexity. Here we integrate two technologies, electropolymerization and amphipols, to demonstrate the electrically addressable functionalization of micro- and nanosurfaces with membrane proteins. Gold surfaces are selectively modified by electrogeneration of a polymeric film in the presence of biotin, where avidin conjugates can then be selectively immobilized. The method is successfully applied to the preparation of protein-multiplexed arrays by sequential electropolymerization and biomolecular functionalization steps. The surface density of the proteins bound to the electrodes can be easily tuned by adjusting the amount of biotin deposited during electropolymerization. Amphipols are specially designed amphipathic polymers that provide a straightforward method to stabilize and add functionalities to membrane proteins. Exploiting the strong affinity of biotin for streptavidin, we anchor distinct membrane proteins onto different electrodes via a biotin-tagged amphipol. Antibody-recognition events demonstrate that the proteins are stably immobilized and that the electrodeposition of polypyrrole films bearing biotin units is compatible with the protein-binding activity. Since polypyrrole films show good conductivity properties, the platform described here is particularly well suited to prepare electronically transduced bionanosensors.
Keywords: protein nanoarrays, membrane proteins, amphipols, conducting polymers, self-assembly
During the past decade, the development of protein arrays with micrometer features has attracted great attention because of their potential applications in life science and biomedical diagnostics.1,2 This technology could provide high-throughput screening of protein functions and biomolecular interactions requiring low amounts of analytes and short processing times.3 Further miniaturization of protein and small organic molecule arrays in the nanometer regime could also improve detection sensitivity and provide important tools for investigating specific biomolecular reactions that are not afforded by micrometer-scale structures.4 Arrays of proteins with micrometer and/or submicrometer features have been fabricated by using patterning techniques such as microcontact printing,4,5 scanning near-field optical,6 dip-pen,7 photo-, and ion beam lithography,8−10 atomic force microscopy (AFM),11 and electrochemical-based approaches.12 However, only a handful of examples of multiple soluble protein nanoarrays has been reported,1 and, to date, none of the techniques mentioned above have been used to nanopattern multiple purified membrane proteins (MPs).13 Although MPs represent more than 60% of the drug targets and there is a strong interest in the production of MP micro- and nanoarrays for both basic and applied research, their complex nature, insolubility in aqueous solutions, and instability in the presence of detergents strongly complicate the fabrication of MP biochips.14−17 In particular, techniques involving drying steps and/or harsh conditions such as high temperatures and mechanical shear may denature MPs.14 MP stability often depends on the presence of its naturally associated lipid molecules, which is difficult to ensure in the presence of detergents. This requirement sets further constraints in the use of the solvents that are normally used to prevent evaporation and keep the protein hydrated in scanning probe lithography techniques.18
Synthetic polymers named “amphipols” (APols) offer a solution to some of the challenges mentioned above. Thanks to the numerous lateral hydrophobic side groups, these short soluble polymers associate with the transmembrane surface of MPs,19 rendering them soluble in aqueous solutions in the absence of detergent while stabilizing them in native-like functional states.20 APol-trapped MPs have been investigated by a variety of methods.19,20 Because the association between MPs and APols is quasi-irreversible in the absence of other surfactants,21,22 various functionalities can be added to the MPs by linking affinity, fluorescent, or radioactive tags to the APols without either chemical or genetic modification of the protein.22,23 Thus, trapping a MP in a biotin-labeled APol (BAPol) makes it possible to tether it to streptavidin (SA)-modified surfaces and to study its interactions with other biomolecules.23 As demonstrated by Charvolin etal.,23 BApols create an environment that gives stability and allows flexibility to the MPs while preventing their direct exposure to the solid substrate. Because APols are freely miscible22 and several APol molecules associate with each individual MP,20 it is straightforward to endow MP/APol complexes with multiple functions; several functional groups can be added to a MP in a defined stoichiometry by the simple method of trapping it in a mixture of appropriately functionalized APols. As an example, BAPols and fluorescently labeled APols (FAPols) can be used in combination to trap a MP while conferring it both an affinity and a fluorescent tag.
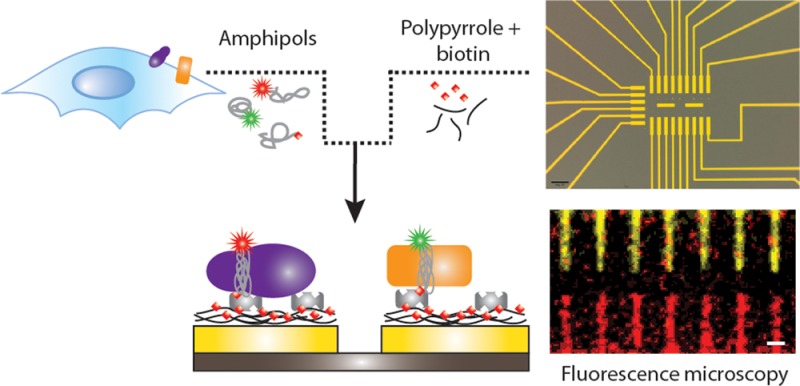
However, a suitable tool that mimics the native phospholipid membrane is not the only challenge that needs to be addressed in the production of MP nanoarrays. Here, we report a novel strategy to fabricate micro- and nanoarrays carrying multiple MPs, based on the combined use of BAPols and of the selective functionalization of gold electrodes by electropolymerization of a biotin-doped polypyrrole (PPy-biotin) film. The electrosynthesis of conducting polymers is an established method that allows the controlled and reproducible functionalization of electrodes at the micro- and nanoscale.24 Pyrrole functionalized with different biotin groups has been synthesized24−28 and polymeric films have been prepared and used as a support for the immobilization of biotin groups to electrode surfaces.24,25,29 After incubation with avidin, biotin-tagged biomolecules can be anchored to the remaining free sites of avidin to construct biosensors.24 However, as biotin is negatively charged under the conditions used for polymerization of PPy, it can also be incorporated in the polymeric film when used as a doping anion. This approach is particularly appealing, as it does not require any chemical synthesis and, as shown by George etal.,30 can be used as a platform to release biotin and attached drugs to cells. Here, we extend this method by demonstrating a straightforward approach for (membrane) proteins multiplexing on electrically contacted gold micro- and nanosurfaces. As schematically illustrated in Figure 1, the key to our method is the selective electropolymerization of conducting surfaces with a PPy film containing biotin sites, followed by the immobilization of (strept-)avidin. Subsequently, we take advantage of the strong interaction between (strept-)avidin and biotin31−33 to immobilize MPs onto selected structures with the protein forming a connecting bridge between the PPy-biotin film and the BAPol in which the MP is trapped. The platform is then decorated with two different MPs that retain their bioactivity on the surface, as evidenced by the binding of MP-specific antibodies. Finally, the versatility and the potentials of the approach are demonstrated by screening the interaction of BAPol-trapped MPs with various types of surfaces.
Figure 1.
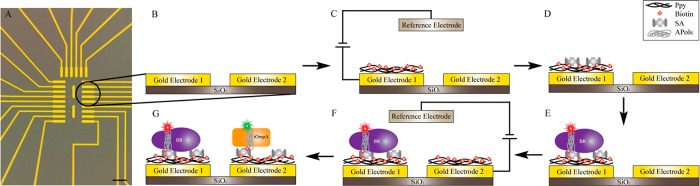
Schematic representation of the immobilization procedure. (A) Differential interference contrast image of the array of electrodes used in the experiments. Scale bar is 50 μm. The array consists of 24 electrically contacted gold surfaces deposited on top of an insulating SiO2 layer on a silicon substrate (as an example only two electrodes are drawn in B–G). (C) The array is immersed into a solution of 0.1 M pyrrole, 1 mM biotin, and 10 mM NaCl, and an oxidizing potential is applied to only one of the electrodes. (D) After functionalization of the surface with a PPy-biotin film, the entire array is incubated into a solution of SA. (E) The array is extensively washed with 10 mM potassium phosphate buffer pH 7.2 (PPB), before being incubated with a solution containing the first target MP (here bacteriorhodopsin shown in purple) trapped in BAPol/FAPol. (F) A second electrode is activated with the same electropolymerization procedure as in (C) and, after incubation of the array with SA, (G) a second MP can be immobilized on the electrode (tOmpA shown in orange). Biotin-binding molecules are colored in gray and biotin in red; the mesh of black curved lines represents the PPy film. The two MPs are shown trapped in APols (gray) with fluorophores represented as red and green stars.
Results and Discussion
Immobilization of Streptavidin Proteins on PPY-Biotin Film
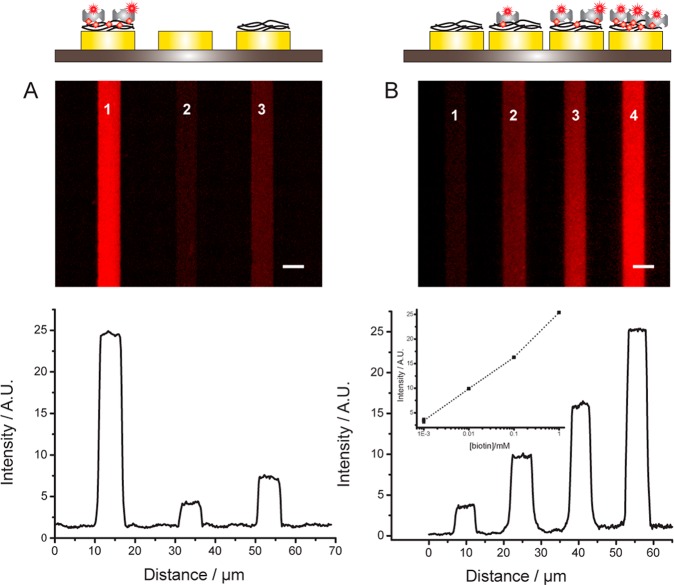
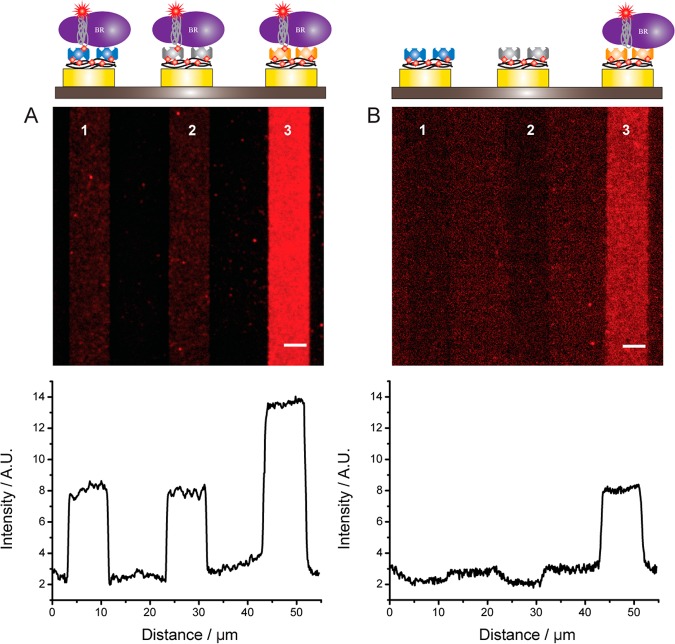
To demonstrate the feasibility of our immobilization strategy, we used an array of 24 gold electrodes 5 μm wide and spaced by 5–10 μm (Figure 1). We initially electrogenerated a PPy film doped with biotin on a gold surface. Subsequently, the entire chip was incubated with a solution of SA-Alexa Fluor 647 (SA-647). The presence of the fluorescent tag on SA enabled direct visualization of the protein binding to the electrodes by epifluorescence microscopy. The effect of doping the PPy film with biotin on protein binding can be evaluated by comparing the fluorescence intensity of three gold electrodes that were either (i) unmodified or functionalized with either (ii) a PPy-biotin film or (iii) a PPy film (Figure 2A and Supporting Figure 1). The image clearly demonstrates that SA binds only to the electrode functionalized with PPy-biotin. The fluorescence signal is homogeneously distributed (standard deviation of the mean = 0.07), and the normalized fluorescence intensity is at least 4× and 7× higher than that of the electrode functionalized only with PPy and of the bare gold electrode, respectively. Moreover the background fluorescence signal measured on the SiO2 surface is <3% of the specific signal, indicating that the washing procedure is effective and that SA does not bind to unmodified SiO2. To confirm that SA interacts specifically with the biotinylated film, fluorescently labeled SA was incubated with an excess of biotin prior to being applied to the PPy-biotin-modified electrodes. Under these conditions, the fluorescence intensity of the electrodes (Supporting Figure 2) is approximately the same as that measured on the electrode functionalized only with PPy, indicating that free biotin-binding sites are necessary for SA to bind to the PPy-biotin film and confirming the low level of nonspecific adsorption. The affinity of SA for the PPy-biotin film was evaluated by measuring the fluorescence intensity of the functionalized electrode after incubation with a range of concentrations of SA. For a given concentration of SA, the electrodes display a homogeneous fluorescence signal, confirming the quality and reproducibility of the immobilization strategy (Supporting Figure 3A). The signal intensity increases monotonically with increasing concentrations of SA, saturating at 1 μM (Supporting Figure 3B). Fitting the data to Hill’s equation yields a KD of 88 ± 12 nM, in good agreement with previously reported values for SA binding to biotin-modified surfaces.32
Figure 2.
Immobilization of SA on PPy-biotin films. (A) Fluorescence image and line profile of electrodes modified with PPy-biotin (1) and PPy films (3) after incubation with SA-647. Electrode 2 is bare gold. (B) Four electrodes functionalized with PPy-biotin films polymerized in the presence of variable concentrations of biotin (from left to right 0 mM, 0.01 mM, 0.1 mM, 1.0 mM). Each polymerization was followed by washing in PPB. After polymerization of all the electrodes, the chip was immersed for 30 min in a 1 μM SA-647 solution and washed with PPB + 0.001% Tween 20 (PPB-T20). Scale bar is 5 μm. (inset of B) The fluorescence signal intensity is plotted against the biotin concentration (squares are raw data and the dotted line is a guide to the eye).
The surface density of the proteins immobilized on the electrodes can be easily tuned by varying the concentration of biotin in the starting solution of pyrrole and doping ion (Figure 2B) or the charge deposited during electropolymerization (Supporting Figure 4). For example, by setting the biotin-doping levels between 0.01 and 1.0 mM, the density of anchoring SA can be modulated by a factor of ∼2.5 (as evaluated by fluorescence intensity; Figure 2B, inset). The final charge deposited during polymerization can also be used to tune the number of biotin moieties present at the surface of the PPy film, with a charge of 100 mC/cm2 showing the highest level of protein adsorption (Supporting Figure 4). Controlling the protein surface density is easier compared to other methods such as dip-pen nanolithography (DPN), where several parameters such as protein ink fluidity, tip–substrate contact time, and humidity need to be finely tuned to regulate the concentration of the deposited proteins.34
An important feature of our methodology is the ease with which biotin is incorporated into the PPy film. To demonstrate the stability of the doping, an electrode was functionalized with a PPy-biotin film and stored in buffer for 14 days at 4 °C. After incubation with SA-647, the surface showed binding properties similar to those of a freshly prepared electrode (as evaluated from the fluorescence intensity of the electrode), indicating that the biotin is strongly bound to the PPy film and it is not released in solution. Therefore, the PPy-biotin-based technique provides a method for specific, homogeneous, reproducible, easy-to-tune, and controlled functionalization of electrically contacted surfaces for stable immobilization of proteins via SA/biotin interaction.
Multiplexed Functionalization of Gold Surfaces with Soluble Proteins
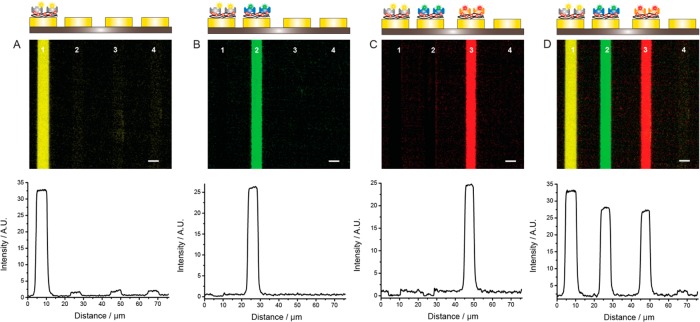
Figure 3 shows how the sequential functionalization of gold electrodes with PPy-biotin films can be used to selectively deposit three biotin-binding proteins with three different fluorescent labels onto the same chip. The electrodes were first incubated in a monomeric solution of pyrrole and biotin, and a PPy-biotin film was deposited on electrode 1 by applying a positive voltage, while the other electrodes were held at open circuit voltage (OCV). After rinsing with phosphate buffer, all electrodes were incubated in a solution of SA-Alexa Fluor 555, resulting in deposition of protein only on electrode 1 (Figure 3A). All electrodes were then rinsed and immersed in the monomeric pyrrole and biotin solution, and a potential was applied to electrode 2, resulting in a PPy-biotin film deposited only on this electrode. All electrodes were then washed, and the array was incubated with a solution of a different fluorescently labeled protein, neutravidin-Oregon Green. Only the electrode that had been freshly functionalized with a PPy-biotin film displayed distinct fluorescence intensity at the corresponding wavelength (Figure 3B), and no interaction with the previously functionalized electrode was observed. Functionalization of the second electrode on the same chip did not affect the surface previously biofunctionalized, as observed at the Alexa 555 wavelength. Subsequently, all electrodes were washed, a PPy-biotin film was electropolymerized on electrode 3, and all electrodes were incubated with an avidin-Alexa Fluor 633 solution (Figure 3C). Electrode 4 was kept at OCV throughout the experiment. In each of the three cycles, the whole chip was exposed to the pyrrole solution and to the proteins without the need of any specialized fluidic system and/or physical barrier. The surface functionalization is exclusively controlled by the ability to selectively modify one particular electrode at a time by electrochemistry. Figure 3C also demonstrates that labeled proteins adsorbed specifically only to the newly prepared PPy-biotin film. It was thus possible to saturate all active biotin sites on the functionalized electrodes, ensuring minimal cross-functionalization between electrically insulated surfaces. Finally, three fluorescence images were collected using appropriate excitation and emission filters and merged into a single image (Figure 3D). The merged image shows that this strategy can be used to selectively immobilize multiple proteins on adjacent electrodes. As proteins are not released from the electrodes during the different functionalization steps, the method presented here could be used to construct protein arrays containing very large protein libraries, where the only constraint is the number of electrodes.
Figure 3.
Sequential functionalization of gold electrodes with a PPy-biotin film allows for spatially and temporally controlled specific binding of several proteins with different fluorescent labels. (A) Electrode 1 was modified with a biotinylated PPy film. After exposing all the electrodes to SA-Alexa Fluor 555, only the functionalized surface showed a distinct fluorescence signal, while the rest of the chip remained dark. (B) Electrode 2 was functionalized with a PPy-biotin film, and the chip was incubated with another fluorescently labeled protein solution, neutravidin-Oregon Green. Only the second electrode displayed green fluorescence intensity. (C) A PPy-biotin film was electrogenerated on electrode 3, and the array was immersed in a solution of Alexa Fluor 633-labeled avidin. This resulted in a fluorescence signal recorded in the appropriate channel only on electrode 3. Electrode 4 was not functionalized at any time. (D) Merge of the three images obtained at the excitation and emission wavelengths specific for the three fluorophores after functionalization of the three electrodes. Scale bar is 5 μm.
Selective Functionalization of Surfaces with Membrane Proteins
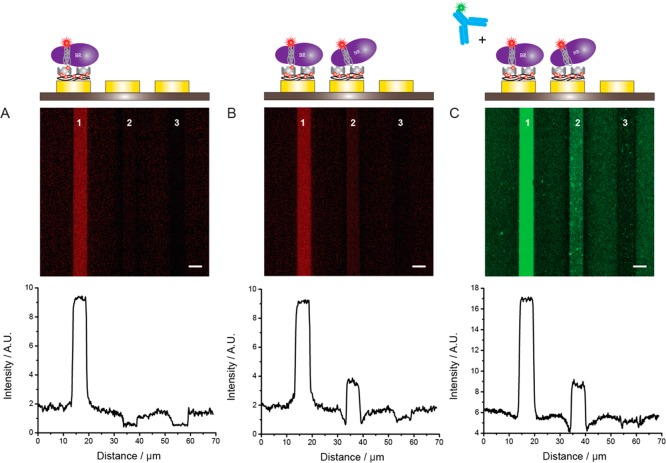
As the protein immobilization method described above does not involve the use of harsh solvents and/or surface drying steps and relies on self-assembly of proteins, it limits risks of protein denaturation and seems particularly well suited to the production of MP arrays. Once the PPy-biotin film is functionalized with biotin-binding proteins (e.g., (strept-)avidin), we can take advantage of the multiple binding sites of these molecules and further decorate the electrode surface. For our initial studies of MP immobilization, we took advantage of BAPols to immobilize MPs onto surfaces via biotin tags without altering their functions, as shown in a previous study in the case of bacteriorhodopsin (BR) and the nicotinic acetylcholine receptor.23,35 BR, a light-driven proton pump, was first trapped in a 1:1 mixture of BAPol and APol labeled with an Alexa Fluor 647 tag (FAPolA647)20,23 and then bound via the biotin tag to an electrode functionalized with a PPy-biotin film and SA. This approach is demonstrated in Figure 4A; the high and homogeneous fluorescence signal measured on the functionalized electrode confirms also that SA is still active after immobilization and can undergo a modular reaction with other biotin-tagged biomolecules such as proteins, oligonucleotides, and vesicles. These experiments demonstrate also how the miscibility of APols can be exploited for the creation of interesting complexes possessing both affinity and fluorescent tags.
Figure 4.
MPs anchoring to a PPy-biotin surface functionalized with SA. (A) Electrode 1 was modified with PPy-biotin; then the chip was incubated with 1 μM SA, washed with PPB-T20, and incubated with 1 μM BR trapped in BAPol/FAPolA647 dissolved in PBS buffer + 200 mM NaCl. After washing with PPB-T20 the electrodes were imaged. (B) Electrode 2 was modified with PPy-biotin; then the chip was incubated with 1 μM SA, washed with PPB-T20, and incubated with 1 μM BR trapped in APol/FAPolA647 dissolved in PBS buffer + 200 mM NaCl. Electrode 3 was not modified at any time. (C) The chip was incubated for 1 h with 5 nM anti-BR tagged with Alexa-488 dye, washed with PPB-T20 for 30 min, and imaged. Scale bar is 5 μm.
SA was also bound to a second PPy-biotin film, and the chip was then incubated with BR trapped in a mixture of unlabeled APol and FAPolA647 lacking the biotin moiety (Figure 4B). The fluorescence signal was at least 3 times higher with BAPols than with untagged APols. The strong difference in fluorescence intensity between the two electrodes indicates that the availability of biotin tags is fundamental for the attachment of BR onto the electrodes and that there is minimal nonspecific adsorption of APol-trapped BR to the SA surface. The specificity of the interaction is also supported by the fact that BR trapped in a BApol/FAPolA647 mixture can be immobilized neither onto bare gold electrodes nor onto PPy-biotin-coated surfaces. Previous studies have demonstrated the stability and functionality of BR after BAPol-mediated immobilization onto SA surfaces.23 Our images reveal that there is no significant decrease in fluorescence signal for the BR-functionalized electrode upon incubation with the pyrrole–biotin mixture or the SA solution, indicative of the stability of the immobilization.
In order to confirm the presence and the biological binding activity of BR in the array, we incubated the chip with a primary antibody raised against BR (anti-BR) labeled with an Alexa-488 dye (Figure 4C). Only the electrode corresponding to immobilized BR showed a strong fluorescence signal. A low level of fluorescence was observed on the electrode functionalized only with PPy-biotin and SA, indicative of some nonspecific adsorption of the antibody. These results not only confirm the presence of BR in the array but also indicate that the MP retains its ability to recognize specific ligands after immobilization onto the electrode, as already shown on other SA surfaces, where the function of the immobilized BR has been also confirmed.23
Micro- and Nanoarrays of MPS
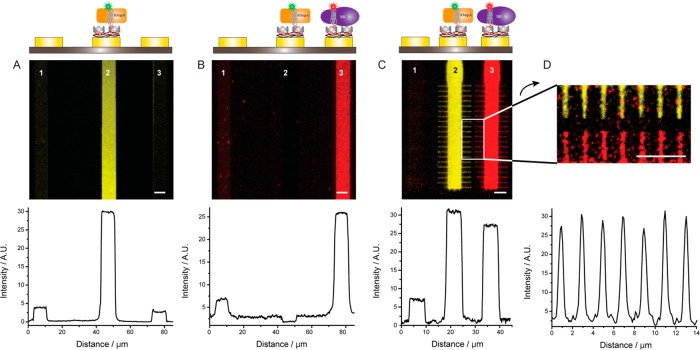
In Figure 5, the sequential functionalization of electrodes with the PPy-biotin layer and SA is used to decorate two adjacent surfaces with two distinct MPs. In addition to BR trapped in a mixture of BAPol and FAPolA647, we used the transmembrane domain of the outer membrane protein A (tOmpA), an eight-strand β-barrel MP from E. coli already characterized in APols.36 For our study tOmpA was trapped in a mixture of BAPol and APol fluorescently labeled with a 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) dye (FAPolNBD). First, SA was immobilized on a PPy-biotin layer electrogenerated on electrode 2. Then the chip was incubated in a solution of tOmpA, resulting in functionalization with the MP of electrode 2 only (Figure 5A). After rinsing with PPB, a PPy-biotin film was prepared on electrode 3, and the chip was exposed first to SA and then to a BR/BAPol/FAPolA647 solution. As shown in Figure 5B, the procedure resulted in decoration of only electrode 3 with minimal contamination of the electrode that had not been functionalized (electrode 1) or where MPs had already been immobilized and the biotin-binding sites blocked. These results further demonstrate that the BAPol-trapped MPs remain attached after incubation with the monomeric solution of pyrrole and are not detached by the application of the electric field needed for the activation of the adjacent electrode. Therefore, our procedure should be easily scalable to any number of different MPs and/or other biotin-labeled compounds by following a step-by-step incubation of the chip with (i) a pyrrole–biotin solution, with application of a potential only to the electrode(s) to be functionalized; (ii) SA; and (iii) the molecule of interest.
Figure 5.
Sequential and addressable functionalization of micro- and nanometer surfaces with MPs. (A) Electrode 2 was functionalized with a PPy-biotin film, and the chip incubated for 30 min with 1 μM SA and for 30 min with 1 μM tOmpA trapped in BAPol/FAPolNBD, washed with PPB-T20, and imaged. (B) Electrode 3 was functionalized with a PPy-biotin film. Then the chip was incubated for 30 min with 1 μM SA and for 30 min with 2 μM BR trapped in BAPol/FAPolA647, washed with PPB-T20, and imaged. Electrode 1 was not functionalized. (C) Two electrodes with 50 nm wide extensions were selectively functionalized with tOmpA trapped in BAPol/FAPolNBD (electrode 2) and BR trapped in BAPols/FAPolA647 (electrode 3). (D) Zoom of the nanosurfaces of (C); the line profiles show the fluorescence intensity measured across the nanometer-wide surfaces of both electrodes 2 and 3. Scale bar is 5 μm.
Due to the highly localized nature of the electric fields, this procedure can be implemented on submicrometer electrodes. This is shown in Figure 5C, where two electrodes with features as small as ∼50 nm were functionalized with BR and tOmpA. To demonstrate the scalability of our approach to the nanometer regime, we also selectively decorated 200 nm wide, 300 nm distant gold nanoelectrodes with tOmpA trapped in BAPol/FAPolNBD (Supporting Figure 5). Due to the resolution limit of the fluorescence microscope, the fine structure of the nanosurfaces cannot be resolved, but the images show a strong fluorescence signal only on the electrodes and not on the SiO2 insulating surfaces. Thus, we conclude that it is possible to biofunctionalize submicrometer surfaces, the only limit of this technique being the availability of an electrically contacted surface. The use of electrochemical polymerization to functionalize surfaces with MPs provides also an important advantage over lithography-based techniques that require challenging alignment procedures. As an example, nanowire and carbon nanotube arrays can be fabricated with diameters and pitches as small as 10 nm;37 selective functionalization of these nanostructures cannot be accomplished by stamping techniques,38 but it could be easily achieved by the electrochemical approach described here.
Protein arrays are optimal candidates for comparing the affinity of an analyte for different ligands. To test this application, we built a protein array containing three different biotin-binding proteins (avidin, SA, and neutravidin) and performed binding assays with BR trapped in different APol mixtures. The first array was incubated with BR trapped in a 1:1 mixture of BAPol and FAPolA647. Figure 6A shows a fluorescence image of the array after incubation. The electrode where avidin had been immobilized shows a fluorescence intensity that is almost double that recorded on the electrodes functionalized with SA and neutravidin. Compared with the latter surfaces, the avidin-electrode seems to have a higher affinity toward BR. However, the fluorescence image of a second protein array immersed in a solution of BR trapped in a 1:1 mixture of unfunctionalized APol and FAPolA647 also showed a signal on the avidin-decorated surface, whereas no fluorescence signal was detected on the electrodes bearing SA and neutravidin (Figure 6B). These experiments indicate a higher nonspecific binding of the APol-trapped BR to the avidin surface than to the SA and neutravidin surfaces, which can be likely attributed to electrostatic interactions taking place in the PPB solution. BR trapped in APols carries a high negative charge due to the many carboxylic side chains of the polymeric belt, all of which are ionized at pH 7.2.22,39 As a result, it adsorbs nonspecifically onto the positively charged avidin substrate (isoelectric point of ∼10.5). In contrast, SA- and neutravidin-modified surfaces have a slightly negative charge at pH 7.2 (isoelectric points 6.1 and 6.3, respectively40) and exhibit much less nonspecific binding of BR/APol complexes than avidin. This experiment shows that the net charge carried by the electrode surface plays an important role in the binding of APol-trapped MPs, and it indicates the surface of choice for APol-mediated MP immobilization. It also demonstrates the multiplexing potential of the developed protein array for diagnostics and drug screening applications, where the interaction between an analyte and several binding partners can be screened simultaneously.
Figure 6.
Effect of surface charge on MP binding. Three gold electrodes were functionalized with neutravidin (electrode 1), SA (electrode 2), and avidin (electrode 3), and the chip was incubated with (A) BR trapped in BAPol/FAPolA647 and (B) BR trapped in APol/FAPolA647 (no biotin tag) in PPB (no NaCl). Scale bar is 5 μm.
Conclusions
We describe here an electrochemical method to selectively and spatially functionalize gold micro- and nanoelectrodes with proteins using biotin-doped PPy films. The conditions used to electrodeposit the biotinylated surfaces are compatible with the stability and functionality of the avidin conjugates. The protein density on the surfaces can be easily controlled by varying a few electrochemical parameters, and different proteins can be immobilized onto separate electrodes by a step-by-step functionalization. Further, the studies presented here demonstrate the use of APols for constructing arrays of MPs, a class of biomolecules that require special handling techniques because of their hydrophobic nature. APols keep MPs soluble and can be used to endow them with one or more tags, allowing their incorporation into micro- and nanometer arrays and their visualization by fluorescence microscopy. On the basis of previous experience,20,41 trapping with APols can be expected to increase the shelf life of MPs and the storage temperature of chips as compared to detergent solutions. We also show that our platform can be used to detect the interaction of antibodies with immobilized MPs and to analyze the binding of ligands in a multiplexed assay.
The work presented here is a first step for fabricating functional arrays of MPs, providing that the method to isolate and stabilize the MP in solution allows keeping it functional. This is important for several reasons: (i) the method allows the selective immobilization of biomolecules on different surfaces on the same chip without the need of microfluidics; (ii) the addressable nature of electropolymerization avoids the alignment and registry limitations of other techniques such as dip-pen lithography; this method could be extended to functionalizing arrays of electrically contacted nanostructures (i.e., Bio-FET, nanowire arrays, etc.); (iii) the platform is suitable for the immobilization of other biotinylated proteins and (iv) has potential for high-throughput, parallel screening of multiple protein/ligand interactions for studies of MP functionality, proteomic analysis, and development of new diagnostics and pharmaceutical screening assays.
Experimental Section
Materials
Streptavidin-Alexa Fluor 647, streptavidin-Alexa Fluor 555, neutravidin-Oregon Green, and avidin-Alexa Fluor 633 were purchased from Life-Technologies (Invitrogen), dissolved in deionized water (18.2 MΩ·cm at 25 °C) to a concentration of 1 g/L, and stored at −20 °C before use. Pyrrole, NaCl, biotin, Tween-20, and potassium phosphate monobasic and dibasic were obtained from Sigma Aldrich and used as received. BioBeads SM2 were obtained from Bio-Rad. Buffers were freshly prepared in deionized water.
Membrane Protein Preparation and Trapping in APols
BR was isolated from the purple membrane of Halobacterium salinarium and solubilized in 40 mM octylglucoside in 25 mM sodium phosphate buffer pH 7.0.35 tOmpA was overexpressed as inclusion bodies in E. coli, folded, and purified in 20 mM C8E4 in 20 mM Tris-HCl buffer pH 8.0 as previously described.36 APol A8-35, BAPol, FAPolNBD, and FAPolA647 were synthesized and characterized according to previously described procedures.22,23,36 BAPol and A8-35 were mixed with FAPol in a 1:1 ratio according to the experiments and added at mass ratios of 1:5 protein to APols for BR and 1:4 protein to APols for tOmpA. The detergent was removed by incubating the MP/APols/detergent solutions overnight at 4 °C with BioBeads (mass ratio of 1:20 detergent to BioBeads) under constant stirring. After removing the BioBeads and centrifuging the sample for 20 min at 120000g at 4 °C to remove large aggregates, the sample was loaded onto a Superdex 200 10/300 GL column (GE Healthcare) and eluted with PPB. Fractions containing only MPs trapped in APols were stored. Protein concentrations were evaluated by measuring the sample absorbance at 280 nm and using extinction coefficients of 81 000 and 46 470 M–1·cm–1 for BR and tOmpA, respectively.23,36
Purification and Labeling of Antibodies
Antibodies against BR and tOmpA were obtained from AgroBio; the proteins were injected into rabbits and the sera collected after 49 days.23 Antibodies were purified from sera by affinity chromatography on a protein A high-performance spintrap column (GE Healthcare) and characterized by SDS-PAGE. Antibodies were fluorescently labeled with Alexa 488 dye via an amino coupling reaction. Briefly, the antibody solution was exchanged with a 0.1 M sodium bicarbonate buffer, pH 8.0, and then incubated with Alexa Fluor 488 succinimidyl ester (Life-Technologies) at a molar ratio of 10:1 dye to antibody for 1 h at room temperature (rt). Afterward, the free dye was removed using Micro Bio-Spin 30 columns (Bio-Rad) and the labeled antibodies were characterized by UV visible and fluorescence spectroscopy.
Array Design and Instrumentation
The metal electrodes were defined using standard electron beam lithography on p-doped Si(100) substrates covered with a 500 nm thick insulating thermal SiO2. Electron beam evaporation was used to evaporate a 100 nm thick Au layer on top of a 10 nm thick Ti adhesion layer. The array, composed of 24 electrodes 5 μm wide, was mounted on a chip carrier and cleaned with 2-propanol, ethanol, and deionized water before being fitted to a home-designed electrochemical cell. The assembly was tightly sealed with a nitrile NBR-70 O-ring (M Seals A/S) by applying a pressure between two plates using four screws and plugged into a multiplexing control unit. A three-electrode configuration was used for all the electrochemical experiments; a platinum wire mesh and a platinum wire were used as counter and reference electrodes, respectively. The distance between these two electrodes and the working electrode on the chip was kept as small as possible (<2 mm). The counter and reference electrodes were directly connected to a CH Instrument (CHI630B) electrochemical analyzer, whereas the multiplexing unit interfaced the potentiostat and the chip, allowing the multiple electrical control of the micro- and nanoelectrodes. The electrodes were imaged in PPB using a Leica DM5500 B upright optical microscope with epifluorescence optics. All fluorescence microscopy images presented here are false-color images acquired with the excitation and emission filter settings being Ex470/40 Em525/50 for the Alexa Fluor 488 and Oregon Green dye, Ex531/40 Em593/40 for the NBD and the Alexa Fluor 555 dye, and Ex620/60 Em700/75 for the Alexa 633 and Alexa 647 dye. Data were analyzed using ImageJ software. The fluorescence intensity was determined by measuring the average intensity over the electrode and subtracting the value of the background image. The figures are presented as follows: the top illustrations are schematic representations of the functionalized electrode and indicate the position of the electrodes; the central images are fluorescence microscopy images; and the bottom images are line profiles representing the average fluorescence intensity measured across the entire image.
Surface Functionalization
The PPy-biotin film was formed on selected electrodes by oxidative electropolymerization of a pyrrole (0.1 M), biotin (1 mM), and NaCl (10 mM) solution at 0.65 V vs Pt in deionized water (18.2 MΩ·cm at 25 °C) under an N2-saturated atmosphere. A low concentration of NaCl was used to facilitate the incorporation of biotin in the PPy film. The functionalized gold electrodes show a dark, homogeneous surface with a weak fluorescence at ∼600 nm (Supporting Figure 1). After modification of selected electrodes with the PPy-biotin film, all the electrodes of a chip were rinsed several times with PPB to remove monomers and oligomers from the surface of the array and then incubated for 30 min at rt with 1 μM biotin-binding proteins conjugated to a fluorescent dye. The electrodes were then washed thoroughly with PPB-T20 in order to remove nonspecifically adsorbed proteins and imaged. The detergent concentration was kept below its cmc in order to limit interactions with the immobilized proteins. The first control experiment was performed by electrogenerating a PPy film doped only with NaCl and incubating the chip with 1 μM SA-647 for 30 min. In the second control experiment an electrode was functionalized with a PPy-biotin film, and a solution of 1 μM SA-647 previously incubated with 1 mM biotin for 60 min at rt was applied to the chip for 30 min. In both control experiments, the chip was rinsed with PPB-T20 and imaged. MP immobilization was performed by incubating the chip with a 1 μM solution of the desired protein for 30 min at rt. The chip was washed thoroughly with PPB-T20 to remove any excess protein and imaged in PPB. For the antibody-binding experiments, the surface was first functionalized with the MPs as described above, and then the chip was exposed to 5 nm of fluorescently labeled anti-BR for 30 min. The chip was incubated and washed intensively with PPB-T20 for 10 min in order to remove nonspecifically adsorbed molecules.
Acknowledgments
E.D.P. is the recipient of a fellowship awarded by the Danish Research Council (FTP-12-132506). This work was supported by the UNIK Synthetic Biology, funded by the Danish Ministry for Science, Technology and Innovation, the Danish Agency for Science Technology and Innovation (The Danish Council for Strategic Research - ANaCell project), the French Centre National de la Recherche Scientifique, University Paris 7, the U.S. National Institutes of Health (grant R01AI092129 from the National Institute of Allergy and Infectious Diseases), and the “Initiative d’Excellence” program from the French State (grant “DYNAMO”, ANR-11-LABX-0011-01). We thank Fabrice Giusti for the gift of APol (A8-35), BAPol, and FAPolNBD, as well as for his guidance for the synthesis of FAPolA647.
Supporting Information Available
Supporting figures are available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Christman K. L.; Enriquez-Rios V. D.; Maynard H. D. Nanopatterning Proteins and Peptides. Soft Matter 2006, 2, 928–939. [DOI] [PubMed] [Google Scholar]
- Wingren C.; Borrebaeck C. A. K. Progress in Miniaturization of Protein Arrays - a Step Closer to High-Density Nanoarrays. Drug Discovery Today 2007, 12, 813–819. [DOI] [PubMed] [Google Scholar]
- Bano F.; Fruk L.; Sanavio B.; Glettenberg M.; Casalls L.; Niemeyer C. M.; Scoles G. Toward Multiprotein Nanoarrays Using Nanografting and DNA Directed Immobilization of Proteins. Nano Lett. 2009, 9, 2614–2618. [DOI] [PubMed] [Google Scholar]
- Coyer S. R.; Garcia A. J.; Delamarche E. Facile Preparation of Complex Protein Architectures with Sub-100-nm Resolution on Surfaces. Angew. Chem., Int. Ed. 2007, 46, 6837–6840. [DOI] [PubMed] [Google Scholar]
- Hoff J. D.; Cheng L. J.; Meyhofer E.; Guo L. J.; Hunt A. J. Nanoscale Protein Patterning by Imprint Lithography. Nano Lett. 2004, 4, 853–857. [Google Scholar]
- Leggett G. J. Bionanofabrication by near-Field Optical Methods. NanoBiotechnology 2007, 3, 223–240. [Google Scholar]
- Lee K. B.; Park S. J.; Mirkin C. A.; Smith J. C.; Mrksich M. Protein Nanoarrays Generated by Dip-Pen Nanolithography. Science 2002, 295, 1702–1705. [DOI] [PubMed] [Google Scholar]
- Christman K. L.; Requa M. V.; Enriquez-Rios V. D.; Ward S. C.; Bradley K. A.; Turner K. L.; Maynard H. D. Submicron Streptavidin Patterns for Protein Assembly. Langmuir 2006, 22, 7444–7450. [DOI] [PubMed] [Google Scholar]
- Zhang G. J.; Tanii T.; Kanari Y.; Ohdomari I. Production of Nanopatterns by a Combination of Electron Beam Lithography and a Self-Assembled Monolayer for an Antibody Nanoarray. J. Nanosci. Nanotechnol. 2007, 7, 410–417. [DOI] [PubMed] [Google Scholar]
- Kannan B.; Kulkarni R. P.; Majumdar A. DNA-Based Programmed Assembly of Gold Nanoparticles on Lithographic Patterns with Extraordinary Specificity. Nano Lett. 2004, 4, 1521–1524. [Google Scholar]
- Gu J. H.; Yam C. M.; Li S.; Cai C. Z. Nanometric Protein Arrays on Protein-Resistant Monolayers on Silicon Surfaces. J. Am. Chem. Soc. 2004, 126, 8098–8099. [DOI] [PubMed] [Google Scholar]
- Hoover D. K.; Lee E. J.; Chan E. W. L.; Yousaf M. N. Electroactive Nanoarrays for Biospecific Ligand Mediated Studies of Cell Adhesion. ChemBioChem 2007, 8, 1920–1923. [DOI] [PubMed] [Google Scholar]
- Valiokas R.; Vaitekonis Š.; Klenkar G.; Trinkunas G.; Liedberg B. Selective Recruitment of Membrane Protein Complexes onto Gold Substrates Patterned by Dip-Pen Nanolithography. Langmuir 2006, 22, 3456–3460. [DOI] [PubMed] [Google Scholar]
- Fang Y.; Frutos A. G.; Lahiri J. Membrane Protein Microarrays. J. Am. Chem. Soc. 2002, 124, 2394–2395. [DOI] [PubMed] [Google Scholar]
- Majd S.; Mayer M. Generating Arrays with High Content and Minimal Consumption of Functional Membrane Proteins. J. Am. Chem. Soc. 2008, 130, 16060–16064. [DOI] [PubMed] [Google Scholar]
- Bieri C.; Ernst O. P.; Heyse S.; Hofmann K. P.; Vogel H. Micropatterned Immobilization of a G Protein-Coupled Receptor and Direct Detection of G Protein Activation. Nat. Biotechnol. 1999, 17, 1105–1108. [DOI] [PubMed] [Google Scholar]
- Hong Y.; Webb B. L.; Su H.; Mozdy E. J.; Fang Y.; Wu Q.; Liu L.; Beck J.; Ferrie A. M.; Raghavan S. Functional GPCR Microarrays. J. Am. Chem. Soc. 2005, 127, 15350–15351. [DOI] [PubMed] [Google Scholar]
- Zheng Z. J.; Daniel W. L.; Giam L. R.; Huo F. W.; Senesi A. J.; Zheng G. F.; Mirkin C. A. Multiplexed Protein Arrays Enabled by Polymer Pen Lithography: Addressing the Inking Challenge. Angew. Chem., Int. Ed. 2009, 48, 7626–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popot J. L. Amphipols, Nanodiscs, and Fluorinated Surfactants: Three Nonconventional Approaches to Studying Membrane Proteins in Aqueous Solutions. Annu. Rev. Biochem. 2010, 79, 737–775. [DOI] [PubMed] [Google Scholar]
- Popot J. L.; Althoff T.; Bagnard D.; Baneres J. L.; Bazzacco P.; Billon-Denis E.; Catoire L. J.; Champeil P.; Charvolin D.; Cocco M. J.; et al. Amphipols from A to Z. Annu. Rev. Biophys. 2010, 40, 379–408. [DOI] [PubMed] [Google Scholar]
- Tribet C.; Diab C.; Dahmane T.; Zoonens M.; Popot J. L.; Winnik F. Thermodynamic Characterization of the Exchange of Detergents and Amphipols at the Surfaces of Integral Membrane Proteins. Langmuir 2009, 25, 12623–12634. [DOI] [PubMed] [Google Scholar]
- Zoonens M.; Giusti F.; Zito F.; Popot J. L. Dynamics of Membrane Protein/Amphipol Association Studied by Förster Resonance Energy Transfer: Implications for in Vitro Studies of Amphipol-Stabilized Membrane Proteins. Biochemistry 2007, 46, 10392–10404. [DOI] [PubMed] [Google Scholar]
- Charvolin D.; Perez J. B.; Rouviera F.; Giusti F.; Bazzacco P.; Abdine A.; Rappaport F.; Martinez K. L.; Popot J. L. The Use of Amphipols as Universal Molecular Adapters to Immobilize Membrane Proteins onto Solid Supports. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosnier S.; Galland B.; Gondran C.; Le Pellec A. Electrogeneration of Biotinylated Functionalized Polypyrroles for the Simple Immobilization of Enzymes. Electroanalysis 1998, 10, 808–813. [Google Scholar]
- Cosnier S.; Stoytcheva M.; Senillou A.; Perrot H.; Furriel R. P. M.; Leone F. A. A Biotinylated Conducting Polypyrrole for the Spatially Controlled Construction of an Amperometric Biosensor. Anal. Chem. 1999, 71, 3692–3697. [DOI] [PubMed] [Google Scholar]
- Darmanin T.; Bellanger H.; Guittard F.; Lisboa P.; Zurn M.; Colpo P.; Gilliland D.; Rossi F. Structured Biotinylated Poly(3,4-Ethylenedioxypyrrole) Electrodes for Biochemical Applications. RSC Adv. 2012, 2, 1033–1039. [Google Scholar]
- Torres-Rodriguez L. M.; Roget A.; Billon M.; Bidan G. Synthesis of a Biotin Functionalized Pyrrole and Its Electropolymerization: Toward a Versatile Avidin Biosensor. Chem. Commun. 1998, 1993–1994. [Google Scholar]
- Yang S. T.; Witkowski A.; Hutchins R. S.; Scott D. L.; Bachas L. G. Biotin-Modified Surfaces by Electrochemical Polymerization of Biotinyl-Tyramide. Electroanalysis 1998, 10, 58–60. [Google Scholar]
- Bidan G.; Billon M.; Galasso K.; Livache T.; Mathis C.; Roget A.; Torres-Rodriguez L. M.; Vieil E. Electropolymerization as a Versatile Route for Immobilizing Biological Species onto Surfaces - Application to DNA Biochips. Appl. Biochem. Biotechnol. 2000, 89, 183–193. [DOI] [PubMed] [Google Scholar]
- George P. M.; LaVan D. A.; Burdick J. A.; Chen C. Y.; Liang E.; Langer R. Electrically Controlled Drug Delivery from Biotin-Doped Conductive Polypyrrole. Adv. Mater. 2006, 18, 577–581. [Google Scholar]
- Iversen L.; Cherouati N.; Berthing T.; Stamou D.; Martinez K. L. Templated Protein Assembly on Micro-Contact-Printed Surface Patterns. Use of the Snap-Tag Protein Functionality. Langmuir 2008, 24, 6375–6381. [DOI] [PubMed] [Google Scholar]
- Rostgaard K. R.; Frederiksen R. S.; Liu Y.-C. C.; Berthing T.; Madsen M. H.; Holm J.; Nygård J.; Martinez K. L. Vertical Nanowire Arrays as a Versatile Platform for Protein Detection and Analysis. Nanoscale 2013, 5, 10226–10235. [DOI] [PubMed] [Google Scholar]
- Wilchek M.; Bayer E. A. Introduction to Avidin-Biotin Technology. Methods Enzymol. 1990, 184, 5–13. [DOI] [PubMed] [Google Scholar]
- Senesi A. J.; Rozkiewicz D. I.; Reinhoudt D. N.; Mirkin C. A. Agarose-Assisted Dip-Pen Nanolithography of Oligonucleotides and Proteins. ACS Nano 2009, 3, 2394–2402. [DOI] [PubMed] [Google Scholar]
- Gohon Y.; Dahmane T.; Ruigrok R. W. H.; Schuck P.; Charvolin D.; Rappaport F.; Timmins P.; Engelman D. M.; Tribet C.; Popot J. L.; et al. Bacteriorhodopsin/Amphipol Complexes: Structural and Functional Properties. Biophys. J. 2008, 94, 3523–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoonens M.; Catoire L. J.; Giusti F.; Popot J.-L. NMR Study of a Membrane Protein in Detergent-Free Aqueous Solution. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 8893–8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melosh N. A.; Boukai A.; Diana F.; Gerardot B.; Badolato A.; Petroff P. M.; Heath J. R. Ultrahigh-Density Nanowire Lattices and Circuits. Science 2003, 300, 112–115. [DOI] [PubMed] [Google Scholar]
- Wong I. Y.; Melosh N. A. Directed Hybridization and Melting of DNA Linkers Using Counterion-Screened Electric Fields. Nano Lett. 2009, 9, 3521–3526. [DOI] [PubMed] [Google Scholar]
- Gohon Y.; Pavlov G.; Timmins P.; Tribet C.; Popot J.-L.; Ebel C. Partial Specific Volume and Solvent Interactions of Amphipol A8-35. Anal. Biochem. 2004, 334, 318–334. [DOI] [PubMed] [Google Scholar]
- Petrlova J.; Masarik M.; Potesil D.; Adam V.; Trnkova L.; Kizek R. Zeptomole Detection of Streptavidin Using Carbon Paste Electrode and Square-Wave Voltammetry. Electroanalysis 2007, 19, 1177–1182. [Google Scholar]
- Pocanschi C. L.; Popot J.-L.; Kleinschmidt J. H. Folding and Stability of Outer Membrane Protein a (OmpA) from Escherichia coli in an Amphipathic Polymer, Amphipol A8-35. Eur. Biophys. J. 2013, 42, 103–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.