Abstract
Circulating tumor cells (CTCs) are low frequency cells found in the bloodstream after having been shed from a primary tumor. These cells are research targets because of the information they may potentially provide about both an individual cancer as well as the mechanisms through which cancer spreads in the process of metastasis. Established technologies exist for CTC isolation, but the recent progress and future of this field lie in nanomaterials. In this review, we provide perspective into historical CTC capture as well as current research being conducted, emphasizing the significance of the materials being used to fabricate these devices. The modern investigation into CTCs initially featured techniques that have since been commercialized. A major innovation in the field was the development of a microfluidic capture device, first fabricated in silicon and followed up with glass and thermopolymer devices. We then specifically highlight the technologies incorporating magnetic nanoparticles, carbon nanotubes, nanowires, nanopillars, nanofibers, and nanoroughened surfaces, graphene oxide and their fabrication methods. The nanoscale provides a new set of tools that has the potential to overcome current limitations associated with CTC capture and analysis. We believe the current trajectory of the field is in the direction of nanomaterials, allowing the improvements necessary to further CTC research.
Keywords: nanomaterial, circulating tumor cell (CTC), magnetic nanoparticle (MNP), carbon nanotube, nanopillar, nanowire, nanofiber, nanoroughened surface, graphene oxide
Vocabulary
- CTC
circulating tumor cell, a cancer cell shed from the primary tumor that has intravasated into the bloodstream
- Metastasis
the spread of cancer as a result of tumor cells from the primary tumor disseminating to and proliferating in a secondary location
- Nanomaterial
structures that exist in the nanolength scale and afford the advantages of increased surface area and interaction with extracellular features
- DAPI
4′,6-diamidino-2-phenylindole, a nuclear stain
- CK
cytokeratin, an intermediate filament expressed by epithelial cells
- CD45
cluster of differentiation 45, an extracellular protein expressed by leucocytes
- EpCAM
epithelial cellular adhesion molecule, an extracellular marker expressed by epithelial cells commonly used in immunoseparation
- PDMS
polydimethylsiloxane, an inexpensive polymer commonly used in microfluidic prototyping
- MNP
magnetic nanoparticle, a nanomoiety used in surface modification, immunocapture, and capture by filtration
- CNT
carbon nanotube, an allotrope of carbon that has been used in circulating tumor cell capture structures and as the conducting layer on an electrode
- Nanopillar, nanowire, nanofiber
nanoconstructions present on a capture surface to increase capture agent presentation and cell–surface interactions
- Graphene oxide
a planar carbon nanomaterial that has been used in conducting electrodes and in cell capture
- CVD
chemical vapor deposition
- RIE
reactive ion etching
An intimidating aspect of cancer is its ability to spread, with metastasis causing 90% of cancer-related deaths.1,2 Metastasis is a multistep process in which tumor cells escape from the primary tumor site, enter the bloodstream, arrest at a secondary site, extravasate, and proliferate to form secondary tumor colonies (Figure 1).3,4 To migrate through the primary tissue and intravasate into the blood, the cell experiences several changes. Increased mobility and the loss of adhesive proteins allow the cell to traverse the extracellular matrix components and the basement membrane in its initial location.2,5−7 Many of these transformations are characteristic of the epithelial-mesenchymal transition (EMT), a process normally seen in embryogenesis that allows cancer cells to gain the motile phenotype coupled with the loss of distinguishing epithelial markers.8−10 The cell persists through phenotypic refinement, but without interaction with its environment, it would be unable to progress. An aggressive tumor cell is able to fight impediments to intravasation posed by the microenvironment such as hypoxia and an immune response using the tools at hand: stromal cells and their secreted factors are hijacked into both helping the tumor cell enter the bloodstream as well as prepare the secondary environment for colonization.2 Similarly, once the tumor cell has left the primary environment and entered the blood, it can aggregate with platelets to avert the immune response and gain protection from the shear stress caused by fluid flow. But arrival at the secondary microenvironment does not guarantee proliferation as the cell may remain dormant or even die.3 Only a small percentage of these tumor cells will ultimately grow into micrometastases, and of those micrometastases few still will proceed into full blown macrometastatic lesions. This process occurs in parallel to the development of the primary tumor, and often before that tumor is initially detected.11 Less than 0.00004% of initially disseminated cells complete the metastatic process,12 and yet, it is these few cells which lead that charge of cancer mortality. To be able to isolate and identify these cells is a clear direction of interest in cancer research.
Figure 1.
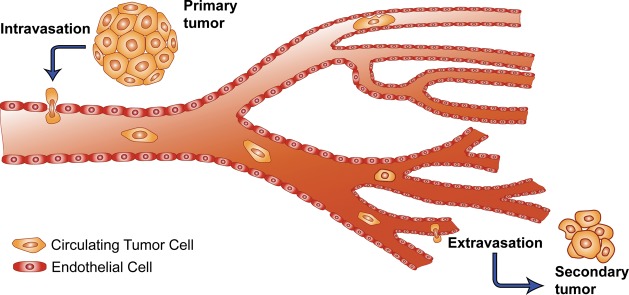
Schematic view of the metastatic process showing CTC transit: the CTCs exit the primary tumor, intravasate into the bloodstream, circulate, and extravasate into a secondary site where they may ultimately achieve different fates including dormancy and full-blown metastasis.3 Adapted with permission from ref (3). Copyright 2002. Nature Publishing Group.
The ease of drawing blood coupled with the wealth of potential information about metastatic mechanisms make circulating tumor cells (CTCs) a tantalizing target for study, particularly given their clinical relevance. Several studies correlate CTC counts with various clinical time points including overall survival and progression free survival. Most of these studies set discrete cut-offs and have evaluated the associations within cohorts in cancers such as breast,13−15 colorectal,16−18 prostate,19 and melanoma.20 In some cases, these cut-offs have been shown to better predict overall survival than traditional biomarkers such as PSA.19 However, while CTCs have been used in American Society of Clinical Oncology (ASCO) recommendations,21 more widespread use of CTCs as prognostic indicators is hindered by the lack of Stage III clinical data.22 Difficulties in including CTCs as potential biomarkers in clinical trials are a result of several factors, including the low number of CTCs recovered (hindering downstream analysis), lack of biological and molecular characterization of CTCs, and questions regarding the usefulness of CTC enumeration.23
To facilitate enumeration and isolation, various devices and technologies have been developed to capture CTCs. However, their operating principles are often at odds with heterogeneous and metaphoric nature of the cells of interest. While each of these detection and capture methods has their advantages, the drawbacks associated with the inherent properties of CTCs, as well as the challenging goal of capturing and culturing viable cells, mean current engineering must turn to new disciplines such as nanomaterials in order to build upon the wealth of technology already established (Figure 2).
Figure 2.
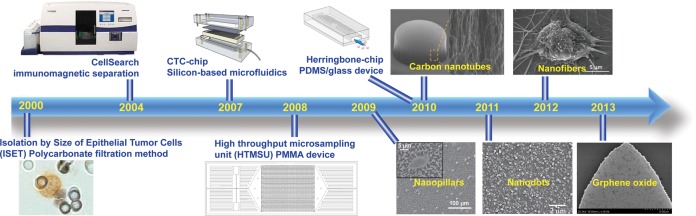
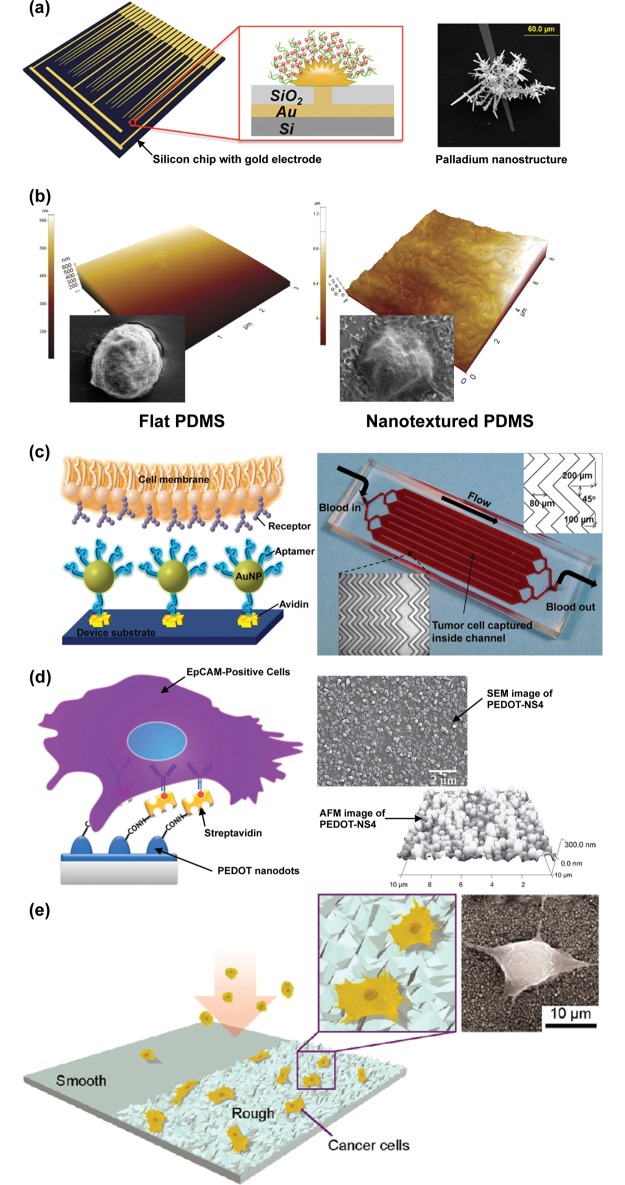
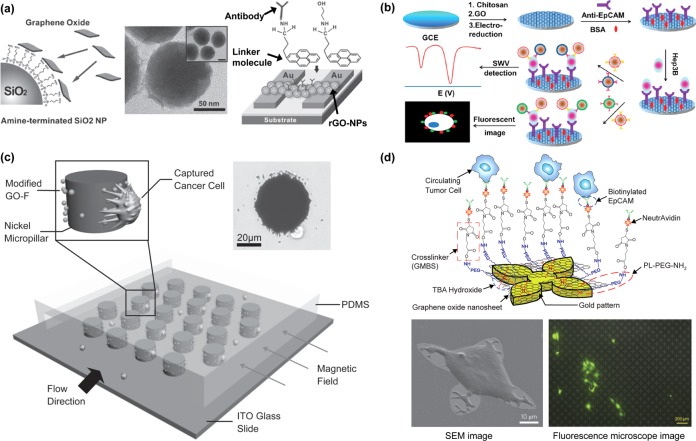
Recently developed CTC technologies. Isolation by Size of Epithelial Tumor Cells (ISET).33 Adapted with permission from ref (33). Copyright 2000 Elsevier. CellSearch. Adapted with permission. Copyright 2014. Janssen Diagnostics LLC. CTC-chip.46 Adapted with permission from ref (46). Copyright 2007 Nature Publishing Group. High-throughput microsampling unit (HTMSU).66 Adapted with permission from ref (66). Copyright 2008 American Chemical Society. Herringbone-chip.54 Nanopillars.90 Adapted with permission from ref (90). Copyright 2009 John Wiley & Sons, Inc. Carbon nanotubes.86 Adapted with permission from ref (86). Copyright 2011 John Wiley & Sons, Inc. Nanodots.104 Adapted with permission from ref (104). Copyright 2011 John Wiley & Sons, Inc. Nanofibers.98 Adapted with permission from ref (98). Copyright 2012 John Wiley & Sons, Inc. Graphene oxide.124 Adapted with permission from ref (124). Copyright 2013 Nature Publishing Group.
To provide an exhaustive litany of CTC isolation, detection, and analysis technologies would result in an unwieldy catalog; indeed, there have been over 12 000 publications on the subject since the year 2000.24 While other articles cover more clinical subject matter23 or emphasize different subsets of strategies,25 our goal is summarize the recent history of the field and the technologies in use and development today to provide a context for the future directions of this research and the tools available to solve challenges going forward.
Commercially Available CTC Isolation Technologies
Although in the past two decades tremendous progress has been made in the field of CTC isolation techniques, there are very few technologies that are commercially available for clinical and research use. Here we describe a selection of methods which take advantage of magnetic particles and size based filtration to separate CTCs.
FDA Approved CTC Technology
The CellSearch system (Veridex LLC) was the first US Food and Drug Administration (FDA) approved system for the detection and enumeration of CTCs in metastatic breast,26 prostate,27 and colon cancer patients.17 Using magnetic beads coated with antibodies against the epithelial cellular adhesion molecule (EpCAM), CellSearch isolated CTCs from the peripheral blood, after which they were fixed with 4% paraformaldehyde (PFA), immunostained with fluorescently labeled anti-cytokeratin (CK, an epithelial intermediate filament), anti-CD45 (a membrane antigen expressed by leucocytes), and DAPI (4′,6-diamidino-2-phenylindole, a nuclear stain), and enumerated by automated cell image capture and analysis. Cells designated as CTCs were characterized by low eccentricity, size greater than 5 μm, a visible nucleus, positive staining for CK, and negative staining for CD45. To verify the accuracy, precision, and linearity of the CellSearch system, Allard et al. evaluated the number of CTCs per 7.5 mL of blood using spiked samples as well as in 145 healthy donors, 199 patients with nonmalignant diseases, and 964 patients with various types of metastatic carcinomas.28 The average recovery of SKBR-3 tumor cells spiked into 7.5 mL of blood was 85%. In blood samples from cancer patients, between 0 and 23 618 CTCs were recovered per 7.5 mL, with 36% of specimens yielding at least 2 CTCs. Cristofanilli et al. first demonstrated convincing data for the prognostic relevance of CTCs in breast cancer patients though use of CellSearch technology.26 In a multi-institutional study of 177 patients with measurable metastatic breast cancer, 61% of pretreatment patient samples had greater than or equal to 2 CTCs. Increased progression-free survival and overall survival were correlated with falling below a cutoff of 5 CTCs in 7.5 mL of peripheral blood drawn at the time points of before a new line of therapy was administered and of 3 to 4 weeks after initiation of therapy. These results exemplify the value of CTCs for delineating treatment groups and auditing the therapeutic response of metastatic disease.
While the CellSearch system represented a breakthrough in CTC separation technology both in principles and in clinical applications, it is not without room for improvement. Given the rarity of CTCs, higher recovery and sensitivity would be desirable for most applications. Additionally, increased purity and the isolation of viable cells would allow more downstream analysis that could be informative for the study of cancer biology and for use in personalized medicine. The system itself requires expensive equipment. This technology represents an innovative milestone in CTC research, but it is a platform upon which the body of literature can build.
Size-Based Filtration Techniques
On the basis of his observation that tumor cells in the blood were often larger than other blood components such as erythrocytes, leukocytes, and platelets, Seal first used a simple sieve as a filter to separate what are now known as CTCs from the blood in 1964.29 The sieve material was a perforated Markrofol tape with 4.5 μm pore size. The filter was able to separate nearly 100% of HeLa cells spiked into whole blood, and retained cancer cells from 19 out of 50 cancer patient samples. Concurrently, irradiated and etched plastic filters with precisely controlled hole size and density were first described for potential use in cell separation.30 In 1992 these principles were combined using microporous polycarbonate membranes with a described application of separating and staining CD4+ and CD8+ lymphocytes following a preliminary immunoseparation.31 Rye et al. used immunomagnetic beads and three antibodies, MOC31, BM2, and LU-BCRU-G7, to enlarge and isolate cancer cells from single cell suspensions as well as bone marrow, blood, ascites, and tissue biopsies.32 Following a magnetic separation and wash, the sample solution was filtered with a 20 μm nylon monofilament filter (Figure 3a). The isolated cells were grown on the filters for 10 days (Figure 3b). These filters were then used either for visualization via immunohistochemistry using cytospin or for nude mouse xenografts. Filters with cultured melanoma cells were directly implanted into the mice, causing tumors in 4–6 weeks. Tumors presented even given a low number of cells, possibly a result of the increased viability afforded from the lack of trypsinization given the direct implantation or the preservation of a 3D environment throughout the change in culture conditions.
Figure 3.
Size-based filtration techniques. (a) Melanoma cells preincubated with immunobeads captured on a nylon monofilament filter.32 Adapted with permission from ref (32). Copyright 1997 Elsevier. (b) Culture of captured melanoma cells on nylon monofilament filter.32 Adapted with permission from ref (32). Copyright 1997 Elsevier. (c) Stained cells as separated using the polycarbonate filter found in ISET (Isolation by Size of Epithelial Tumor Cells) technology [1, spiked tumor cells; 2, membrane pores; 3, leukocytes].33 Adapted with permission from ref (33). Copyright 2000 Elsevier. (d) Parylene C microfilter released from silicon mold.36 Adapted with permission from ref (36). Copyright 2007 Elsevier. (e) Two-layer microfilter to protect cells from damaging forces.38 Adapted with permission from ref (38). Copyright 2010 Springer Science+Business Media, LLC. (f) CTC clusters isolated by RIA (reversible bead attachment for cell isolation and analysis) and different levels of HER2 expression in CTCs isolated from metastatic breast cancer patients.39 Adapted with permission from ref (39). Copyright 2007 John Wiley & Sons, Inc.
Isolation by Size of Epithelial Tumor Cells (ISET) was an improvement over previous filtration methods as it did not require a preliminary separation through techniques such as immunomagnetic and flow cytometric cell separation.33 Following dilution of the blood sample, the CTCs could be separated from the solution using a Track-Etch polycarbonate membrane with 8 μm diameter cylindrical pores. To characterize ISET, cells from liver, breast, cervical, and prostate cancer cell lines (HepG2, Hep3B, MCF-7, HeLa, and LNCaP) were spiked into 1 mL of peripheral blood (Figure 3c). Captured cells could then undergo genetic analyses such as fluorescence in situ hybridization (FISH) and the polymerase chain reaction (PCR). Efficacy of the system was further assessed using samples from primary liver cancer patients, with CTCs found in 23 of the 44 patients and none of the healthy control subjects.34 Cells were classified as CTCs based on size, nucleus to cytoplasm area ratio, and nuclear irregularities. Primer extension preamplification (PEP) and PCR were performed on the separated tumor cells as proof of ISET’s applicability in cancer study. Zabaglo et al. modified the cell filtration technique with laser scanning cytometry (LSC), allowing for the relocation of cells expressing cytokeratin for further analysis.35
While track etching allows for uniform pore diameter, the distribution of pores in the membrane can be irregular. Microfabrication techniques can be used to create a more uniform pore density. Zheng et al. used microfabrication with parylene-C, a polymer affording many advantageous qualities such as excellent mechanical properties, optical transparency, and low biofouling.36 Parylene-C was deposited on a silicon wafer and patterned with oxygen plasma in reactive ion etching (RIE) to create a regularly porous membrane filter which could then be released from the silicon wafer using deionized water (Figure 3d). Chrome/gold electrodes were integrated, allowing on-membrane lysis for further PCR analysis. This membrane was then tested with patient samples and the results were compared with those obtained from CellSearch: the parylene filter microdevice detected CTCs in 51 of the 57 patients tested, whereas CellSearch only detected CTCs in 26 of the 57 samples.37 The membrane design was modified for inclusion in a 3D hexagonal patch which featured a second membrane layer to reduce the stress caused by interaction of captured cells with the pores (Figure 3e).38
The strategy of enlarging cells of interest prior to filtration has been used with microfilters as well. Following incubation with microbeads conjugated with anti-EpCAM via a linker containing a photocleavable o-nitrobenzyl group, diluted blood samples were flowed through a microfilter chip with 8 μm pores.39 White blood cells continued through the membrane while enlarged cancer cells were retained. The microbeads were removed through i-line light irradiation, leaving viable cells that could then be analyzed with immunocytochemistry and quantitative fluorescence to determine in situ Human Epidermal Growth Factor Receptor (HER2) expression. Breast cancer cell line cells spiked into blood were separated with an average efficiency of 89%. Twelve metastatic breast cancer patient samples were assayed with all containing CTCs in the range of 1–31 CTCs/mL (Figure 3f).
Modifications to the filtration process are an area of continuing research, with advances being made in both the materials used and their fabrication. This further optimizes a technique that allows viable cell separation for staining and morphological observation as well as an immediate platform for cell culture. However, most filtration methods are still plagued by inconvenient preprocessing steps such as dilutions, flow cytometry, and immunomagnetic separation, which also affect overall throughput. Membrane clogging can result in issues with purity, while variability in CTC size can lead to the loss of these rare cells.
Microfluidic-Based CTC Devices
Microfluidic devices provide innovative solutions to logistical problems, affording the advantages of high sensitivity, low cost, low reagent usage, small size, and several established fabrication techniques.40 Operating on this length scale allows for laminar flow, yielding parallel streamlines with minimal mixing resulting only from diffusion.41 In general, these laboratories-on-a-chip or micro total analysis systems (μTAS) consist of several elements from the microfluidic tool box including pumps, valves, reservoirs, and mixers, in addition to other thermal and electrical components.42 Devices are often constructed from glass or silicon substrates; however, the low expense and plethora of simple fabrication methods have led to the development of a number of polymer-based devices.41 PDMS (polydimethylsiloxane) in particular has emerged as a fundamental material in microfluidics, facilitating inexpensive prototyping.43 Arising from an array of small scale analytical techniques,40 an early biological application of microfluidics was a device that performed PCR, improving upon large-scale methods by decreasing the time of each step.44 Other applications include on-chip molecular separation, protein analysis, immunosensing, and electronics cooling.45 Given the many benefits of microfluidics, including a length scale amenable to cellular analysis, it is unsurprising that microfluidic devices are a staple in CTC isolation and analysis.
Silicon-Based CTC Chips
Nagrath et al. pioneered a silicon-based microfluidic device for the capture of CTCs from cancer patient blood samples.46 This device featured viable cell isolation in addition to lacking sample preprocessing steps. The CTC-chip had an array of 78 000 silicon microposts which were subsequently coated with anti-EpCAM capture antibodies (Figure 4a,b). The optimal flow rate was determined to be 1–2 mL/h in order to maximize the capture efficiency, which was greater than 60% for the NCI-H1650 cells spiked into whole blood. As the normalized number of CTCs recovered from the blood has been correlated with efficacy of treatment,47 demonstrating an emerging clinical significance of the enumeration of CTCs, CTCs were enumerated in samples from 68 patients with non-small-cell lung (n = 55), prostate (n = 26), pancreatic (n = 15), breast (n = 10), and colon (n = 10) cancers. When fluorescent staining was used, CTCs were identified as those stained cells with DAPI+/CK+/CD45–, and were detected in 115 of 116 (99%) samples. While the number of CTCs in a patient sample did not reflect the size of the primary tumor, it did correspond to the patient’s response to treatment. Because of its high yield, sensitivity, specificity, and clinical relevance, this chip represented great strides in the field of CTC isolation. The research team went on to show further clinical applications using the Scorpion Amplification Refractory Mutation System (SARMS) technology to identify the T790M mutation in the epidermal growth factor receptor (EGFR).48 This mutation is associated with tumor resistance to tyrosine kinase inhibitors, and the data gained from this study could potentially be used to determine the appropriate course of treatment. With the use of the CTC-chip, CTCs were isolated from non-small-cell lung cancer patient samples for analysis and compared with results from the primary tumor as well as free plasma DNA. The mutation was found in the CTCs of 11 of 12 patients whose primary tumors featured this mutation and the free plasma DNA of 4 out of these 12 patients. Increased CTC counts were correlated with tumor progression and further EGFR mutations. This demonstrated the utility of an immunocapture micropost device in a noninvasive genotyping procedure.
Figure 4.
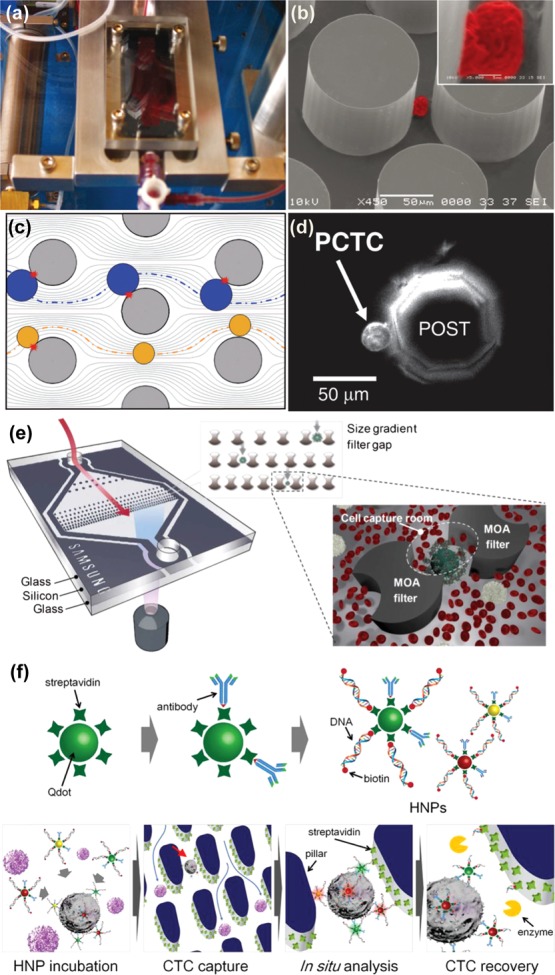
Silicon-based microfluidic CTC separation technologies. (a and b) Setup of post-based CTC-chip developed by Nagrath et al. Captured NCI-H1650 cell imaged with scanning electron microscopy (color added) with high magnification inset.46 Adapted with permission from ref (46). Copyright 2007 Nature Publishing Group. (c and d) Computer simulated particle paths for larger cells (blue) compared with smaller cells (yellow) for geometrically enhanced differential immunocapture (GEDI). Prostate cancer tumor cell captured on octagonal post of GEDI microdevice.49 Adapted with permission from ref (49). Copyright 2010 The Royal Society of Chemistry. (e) Filtration unit with two different filter gaps for the capture of CTCs that have undergone size amplification with magnetic particles.52 Adapted with permission from ref (52). Copyright 2012 The Royal Society of Chemistry. (f) Hybrid nanoparticles consisting of antibodies, quantum dots, and specific DNA sequences for the labeling, capture, and release of CTCs in a capture and recovery chip.53 Adapted with permission from ref (53). Copyright 2013 Elsevier.
In addition to the circular posts used by Nagrath et al., microposts on silicon chips have been designed to optimize capture through the manipulation of streamlines using alternative post geometries. Termed geometrically enhanced differential immunocapture (GEDI),49 octagonal posts were staggered to maximize collisions with cells (Figure 4c). The immunocapture aspect was achieved with a monoclonal antibody against the prostate-specific membrane antigen (PSMA), J591, and as such, the device was characterized with the LNCaP prostate cancer cell line. Applications of this device were demonstrated by performing capture with prostate cancer patient samples (Figure 4d), as well as proof-of-principle ex vivo drug testing using the taxanes docetaxel and paclitaxel.50 The device has been additionally functionalized with antibodies against HER2 to capture cancer cells known to have this membrane receptor upregulated.51 Following the selection of the most optimal anti-HER2 antibody, the device was characterized with high and low HER2-expressing breast cancer cell lines, yielding capture efficiencies of 78% and 26%, respectively. Clinical efficacy was verified through the analysis of nine blood samples from five breast cancer patients and two gastric cancer patients with an average of 74 CTCs/mL identified in breast cancer patients and 120 CTCs/mL in gastric cancer patients.
Silicon can also be incorporated into a glass device, combining the advantages of a transparent substrate with the ability of silicon to be finely patterned. Using anodic bonding and chemical mechanical polishing, Kim et al. were able to fabricate a cell capture device with a multiobstacle architecture (MOA) filter to trap size-enhanced cells (Figure 4e).52 Incubation of a cell solution with melamine microbeads conjugated with anti-EpCAM effectively increased the size of EpCAM-expressing cells, causing them to become trapped in the “cell capture rooms,” made from silicon, while the smaller white blood cells were able to escape. The device was characterized by spiking MCF-7 cells/mL in whole blood, yielding an 89.7% recovery. Effective bright field imaging was achieved as both the top and bottom of the device are glass.
In another example of silicon on glass technology, pillars were deep reactive ion etched into a silicon wafer to form a capture surface which was then anodically bonded to glass wafer patterned with microfluidic chambers, a capture and recovery chip (CRC) (Figure 4f).53 This CRC was functionalized with streptavidin to capture cells pretreated with hybrid nanoparticles (HNPs) consisting of an antibody, a quantum dot, and biotinylated DNA, which bound to the avidin on the surface. Three different HNPs were prepared, each with a different antibody (anti-EpCAM, anti-EGFR, or HER2), quantum dot (Qdot(525 nm), Qdot(565 nm), or Qdot(625 nm)), and DNA sequence, allowing the specific marking and release of different breast cancer cell lines. Respective quantum dot fluorescence ratios were reflective of established cell line expression patterns, and cell spike experiments using whole blood yielded capture efficiencies of 81.3%, 91.2%, and 90.0% for the cell lines MCF-7, SKBR-3, and MDA-MB-231, with identification accuracies of 94.7%, 99.3%, and 83.3% based off said fluorescence patterns. Using restriction enzymes, cells were released into 96-well plates with efficiencies of 78.6%, 93.7%, and 86.0%, where they were able to adhere and proliferate.
Glass/PDMS-Based CTC Chips
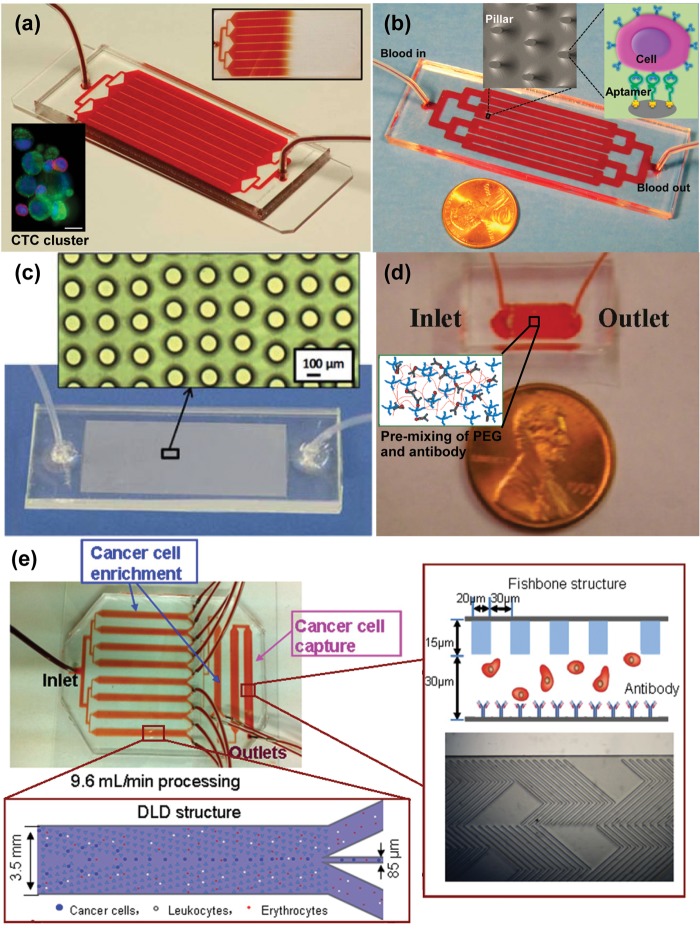
Glass is an attractive substrate for CTC capture chips due to its transparency for clear imaging, allowing for a wide variety of light microscopy based techniques yielding both bright field and fluorescent images. Shortly after the advent of the CTC-chip, the Toner group released a herringbone CTC capture chip (Figure 5a) that made use of passive mixing to increase encounters between flowing cells and anti-EpCAM-functionalized PDMS microchannels.54 The creation of microvortices was induced in this “Herringbone-chip” through chevron patterns on a PDMS ceiling, disrupting streamlines and increasing the capture efficiency. Once tethered to the chip through the antibody–antigen interaction, cells could be stained for DAPI, CK, and CD45. The capture efficiency for prostate cancer cells (PC-3) spiked into whole blood was 91.8%, with 14% purity and 95% viability. The device was further tested with metastatic prostate cancer patient samples, with 93% detection rate. The optical transparency of the glass allowed for further analysis in the form of fluorescence in situ hybridization (FISH) to determine androgen receptor copy number in LNCaP cells. Off-chip analysis included RNA isolation for real-time PCR (RT-PCR), allowing the identification of a specific TMPRSS2-ERG translocation. The utility of the Herringbone-chip was demonstrated through its use in a study of the epithelial–mesenchymal transition (EMT) in breast cancer CTCs.55 Using multiple capture antibodies, anti-EpCAM, anti-EGFR, and anti-HER2, CTCs were isolated and subjected to RNA in situ hybridization, showing the presence of mesenchymal marker-displaying CTC clusters as well as the epithelial to mesenchymal spectrum of CTCs.
Figure 5.
Glass/PDMS-based microfluidic CTC separation technologies. (a) Multiple flow channels shown with blood flowing through the device. The Herringbone-chip was able to isolate clusters of CTCs in addition to single cells (shown in lower inset).54 (b) Microfluidic device consisting of over 59 000 micropillars functionalized with DNA aptamers.58 Adapted with permission from ref (58). Copyright 2012 American Chemical Society. (c) PDMS post structure device fabricated using a 2-step AGEpp-PEI inking process.61 Adapted with permission from ref (61). Copyright 2011 The Royal Society of Chemistry. (d) Alginate hydrogel containing PEG conjugated to capture antibodies (inset).62 Adapted with permission from ref (62). Copyright 2011 American Chemical Society. (e) Deterministic lateral displacement is used to sort cells that are ultimately captured on an antibody-functionalized substrate.64 Adapted with permission from ref (64). Copyright 2013 Elsevier.
Herringbone micromixers have been optimized in a geometrically enhanced mixing (GEM) chip to increase throughput and purity in anti-EpCAM immunocapture.56 When the width of the grooves within the micromixer was increased, purity was increased to 84% for pancreatic cell line (L3.6pl) cells spiked into whole blood. However, a high number of white blood cells were present when patient samples were analyzed, leaving room for additional optimization. As CTCs were detected in 17 of 18 patients studied, and CTC number corresponded to tumor size in three stage IV metastatic pancreatic cancer patients analyzed over the course of treatment, this technology appears to hold promise for use in the clinical setting.
Purity can also be improved through the chemical patterning of the capture surface. Anti-EpCAM capture can be strategically combined with E-selectin such that the specific interaction of EpCAM expressing cells (the cancer cells in the blood) are preserved during a wash with a calcium-chelating buffer that disrupts the white blood cells bound in the E-selectin regions of the device.57 In cell spike experiments with CTCs and white blood cells represented, respectively, by MCF-7 and HL-60 cells, patterning with alternating regions of anti-EpCAM alone and anti-EpCAM combined with E-selectin resulted in an 84.1% capture ratio for the model CTCs, while only 5.2% of spiked model white blood cells remained on the device. This patterning was achieved through the use of two different conjugation chemistries. Photopolymerized poly(acrylic acid) served both as the basis for the combination anti-EpCAM/E-selectin regions subsequently functionalized through N-hydroxysuccinimide (NHS)/1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) chemistry and to block the silanization that ultimately presents anti-EpCAM alone. The device represents a proof of principle but will require testing with more clinically relevant models.
In contrast to the commonly used antibodies, aptamers are also used as capture agents. Easily generated using systematic evolution of ligands by exponential enrichment (SELEX), they bind their antigens with high specificity. Sheng et al. demonstrated an aptamer-mediated microfluidic device with elliptical glass micropillars in the glass substrate (Figure 5b).58 The glass substrate was isotropically wet-etched using a mixture of HF/HNO3/H2O to fabricate micropillars. Biotinylated aptamers59,60 have been used coupled with avidin on the surface of the glass microchannels/micropillars to capture tumor cells spiked into whole blood containing other nontarget cells leading to a high throughput (∼93% capture efficiency, processing 1 mL of blood in 28 min) and high viability (94%) device.
PDMS microposts were used for heightened capture in a glass/PDMS device sealed using a two-step inking process (Figure 5c).61 Cross-linking occurred when polyethyleneimine (PEI) was inked on the glass substrate and brought into contact with PDMS modified with polymerized allyl glycidyl ether (AGE). This device was functionalized with anti-EpCAM, capturing 80–90% of cells spiked into buffer and 70% of cells spiked into whole blood.
Alginate hydrogels were introduced to the glass/PDMS setup for improved rare cell capture and release.62 While alginate hydrogels have advantages such as simple formation and dissolution, they are also prone to nonspecific binding. To improve the specificity of capture, modified four-armed, amine-terminated PEG molecules capable of binding antibodies were added to the hydrogel and examined by extracting endothelial progenitor cells (EPCs) from whole blood (physiological concentration of 10 000 cells/mL). This hydrogel was injected into a device fabricated in PDMS irreversibly bonded to glass (Figure 5d). One-third of the EPCs were captured with a purity of 74% and a viability of 90% under optimal conditions, showing the potential for use of this method in other types of rare cell capture. Release of captured cells was performed using the calcium chelator ethylenediaminetetraacetic acid (EDTA) as the alginate hydrogels were cross-linked with divalent cations; however, this limits practical sample processing as blood is often stored in the presence of EDTA. Shah et al. modified this hydrogel via photo-cross-linking to demonstrate capture and release of cancer cells including PC-3 and SKBR-363 even in the presence of calcium chelators such as EDTA. The hydrogels were generally stable but could be degraded using the enzyme alginate lyase, allowing for viable cell release.
Liu et al. integrated size-based and affinity-based techniques in a glass/PDMS device for enrichment followed by capture.64 CTC enrichment was performed with deterministic lateral displacement (DLD) using a triangular micropost array (Figure 5e). The larger (cancer) cells were directed centrally due to collisions with the microposts, while the smaller (white blood) cells followed the paths of the laminar streamlines to designated wasted chambers. This resulted in 1500× enrichment. Cancer cells then flowed through a fishbone-patterned PDMS chamber which directed capture on an antibody-functionalized glass substrate. This sequence allowed for over 90% capture efficiency of 100/mL spiked cancer cells in 10× diluted blood at a high throughput of 9.6 mL/min.
Thermopolymer-Based CTC Chips
Poly(methyl methacrylate) (PMMA) can serve as an alternative to silicon as a substrate for microfluidic CTC capture and analysis devices due to its low cost and excellent optical transparency. Polymers such as PMMA are advantageous due to their amenability to convenient fabrication techniques such as hot embossing and injection molding. Carboxylic acid groups can be selectively generated on the surfaces of PMMA by UV exposure to explore electroless deposition, protein concentration, and cancer cell capture (Figure 6a).65 Increased roughness can be induced using high intensity light, providing increased surface area for functionalization. Additionally, thermal bonding occurs at low enough temperatures to preserve these microfeatures.66 With the use of these properties of PMMA, a high-throughput microsampling unit (HTMSU) was developed. The HTMSU was functionalized with anti-EpCAM monoclonal antibodies for capture and included a conductivity sensor which enabled enumeration (Figure 6b). The device was initially characterized with MCF-7, SW620, and HT29 cells (breast and two colorectal cell lines, respectively), showing the potential for cell release via trypsin and cell analysis using the polymerase chain reaction (PCR) and ligase detection reaction (LDR) (Figure 6c).66,67 The benefits of the high aspect ratio features of PMMA are not limited to antibody capture. The anti-EpCAM monoclonal antibodies were exchanged for RNA aptamers against the prostate-specific membrane antigen (PSMA) for tissue-specific recognition.68
Figure 6.
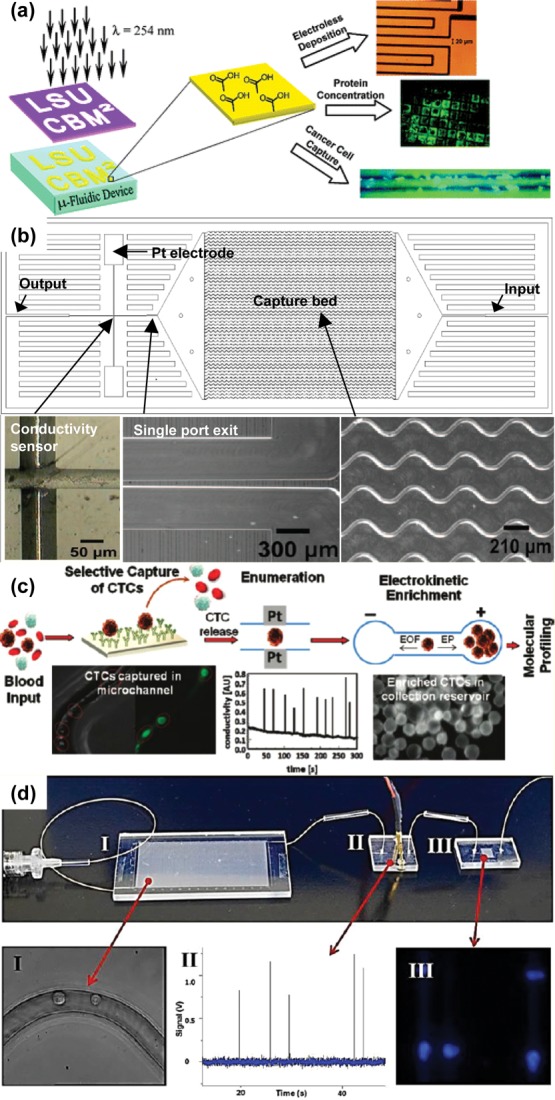
Thermopolymer-based microfluidic CTC separation technologies. (a) Surface modification of PMMA by UV exposure.65 Adapted with permission from ref (65). Copyright 2005 American Chemical Society. (b) HTMSU facilitates capture using serpentine channels and integrates enumeration using a conductivity sensor near the outlet.66 Adapted with permission from ref (66). Copyright 2008 American Chemical Society. (c) Overall schematic of CTC immunocapture and release for downstream analysis.67 Adapted with permission from ref (67). Copyright 2011 American Chemical Society. (d) Picture of the assembled microfluidic modules for CTC isolation and analysis: (I) HT-CTC selection module; (II) impedance sensing module; (III) staining and imaging module.70 Adapted with permission from ref (70). Copyright 2013 American Chemical Society.
Cyclic olefin copolymer (COC) has the advantages of PMMA in addition to having increased optical transmissivity; the implication of this is increased UV activation-produced carboxyl groups for increased capture antibody presentation, yielding higher capture efficiencies and purities.69 This polymer was used as the substrate for a high-throughput (HT) CTC selection module that comprises a component of a CTC isolation and analysis system (Figure 6d).70 Following capture in high aspect ratio sinusoidal anti-EpCAM functionalized microchannels, CTCs were released using trypsin. The cells flowed across an impedance sensor embedded in PMMA for enumeration into a 2D array fabricated in PMMA for staining and confined imaging. The capture efficiency for the selection module was dependent on channel length, with an overall average of 83.1% and a maximum of greater than 90%. Collection efficiency of the staining and imaging module was determined by comparing stained cells with numbers generated by the impedance sensor. Fixed cells were collected with higher efficiency than unfixed cells (96% vs 85%). A lower collection efficiency of 72% was exhibited by analyzed pancreatic ductal adenocarcinoma (PDAC) patient samples, possibly a result of the 15% misclassification rate inherent in the impedance sensor due to the enumeration of white blood cells. Samples analyzed from metastatic and local PDAC patients averaged 53 and 11 CTCs/mL, respectively, with a high purity of 86%, although cell viability was negatively affected by trypsinization.
The integration of microfluidics represents a dramatic change in the CTC separation paradigm, allowing for increased purity, yield, and sensitivity when compared to the CellSearch system. However, for CTC technology to progress, new techniques will be required to further increase these metrics, providing a greater number of a purer population of viable cells available for additional downstream analysis. These improvements will allow not only for greater study but also for room to adjust throughput for easier sample processing. Through interaction on the scale of extracellular structures, increased capture agent presentations, and the ability to transition to capture agent independent methods, nanomaterials provide the advantages necessary to take these next steps in CTC research.
CTC Devices Incorporating Nanomaterials
Magnetic Nanoparticles
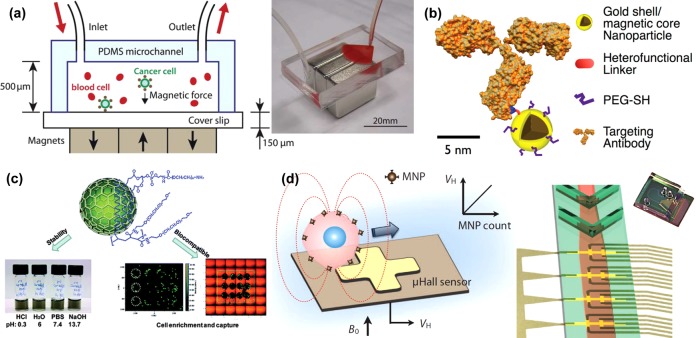
Magnetic nanoparticles (MNPs) can be used to take advantage of surface expression as well as innate physical properties of the cell. Following the exchange of blood plasma for dilution buffer, MNPs functionalized with anti-EpCAM were used to bind selected cells in the presence of a magnetic field in a reversibly bonded PDMS chamber using NdFeB block magnets (Figure 7a).71 The PDMS was then removed, facilitating microscopy. Cells were stained for CK, CD45, and DAPI and evaluated using computer automation with 90% capture ratios observed for spiked cell lines. An additional example of immunomagnetic separation included a dynamic setup to minimize cell aggregation and settling while maximizing capture.72 With the use of a magnet which was positioned in direct contact with the microchannel (with the exception of the inlet, preventing accumulation), cells labeled with Fe3O4 MNPs were separated in up to six devices that were rocked and repositioned during flow. Staining was performed with DAPI, anti-CK, and anti-CD45. Characterization with Colo205, PC-3, and SKBR-3 cell lines yielded 97%, 107%, and 94% capture ratios. Devices tested with patient samples showed comparable results to portions of those same samples exported to CellSearch for comparison. Like the Herringbone-chip, cell clusters were found. This strategy was adapted to account for variable antibody expression through the use of hybrid magnetic/plasmonic nanocarriers modified with several different antibodies (Figure 7b).73 Modified MNPs were conjugated with one of several biomarkers: anti-EpCAM, anti-HER2, anti-EGFR, anti-CK, and anti-MUC1. CK was detected through spontaneous internalization of the labeled MNP. Various combinations of nanoparticles were tested with cell lines with differing expression patterns. For example, a multiplex assay of anti-EpCAM and anti-MUC1 modified MNPs increased the capture of the low EpCAM expressing cell line BT-20 from the 45% yield achieved with anti-EpCAM alone to 78%.
Figure 7.
Magnetic nanoparticles (MNPs) in CTC capture and analysis. (a) Anti-EpCAM functionalized MNPs bind to CTCs and remove them from solution through the use of block magnets.71 Adapted with permission from ref (71). Copyright 2011 The Royal Society of Chemistry. (b) Schematic of an antibody modified using a heterofunctional linker and an immunomagnetic nanocarrier.73 Adapted with permission from ref (73). Copyright 2013 American Chemical Society. (c) DSPE-PEG-NH2 and C18-PMH-mPEG functionalized graphite-coated magnetic nanocrystals.74 Adapted with permission from ref (74). Copyright 2013 American Chemical Society. (d) The magnetic moment of a MNP conjugated with a specific antibody is sensed as the MNP-covered cell flows over a series of micro-Hall sensors.75 Adapted with permission from ref (75). Copyright 2012 American Association for the Advancement of Science.
Internalized MNPs were used for cancer cell detection using a magnetic microarray.74 FeCo MNPs were coated in graphite using chemical vapor deposition after which they were modified with two types of PEG, DSPE-PEG-NH2 and C18-PMH-mPEG, allowing for biocompatibility and aqueous solubility (Figure 7c). The MNP conjugates were spontaneously internalized by the ovarian cancer cell line SKOV3, allowing for cells to be attracted by strong magnetic fields. Cells were isolated from whole blood at a yield of at least 75% using a specifically designed microarray.
The inherent properties of the MNPs themselves have been exploited to characterize CTCs. Through the immunolabeling of three different sizes (10, 12, and 16 nm) of MNP with various biomarkers, a cell expressing those biomarkers can, in turn, be labeled with these MNP complexes.75 Levels of expression can be detected due to the directly proportional relationship between the magnetic moment of the targeted cell and the number of bound MNPs. Biomarker expression was determined by the magnetization curve representative of the different MNP sizes. Using a micro-Hall detector to detect the magnetic moments of cells in a solution, Issadore et al. counted CTCs from patient blood. MNP-complex-labeled cell solutions were processed through a microchannel array featuring a chevron ceiling, focusing the cells toward the bottom center of the channel over the implanted micro-Hall detectors (Figure 7d). The four biomarkers (HER2/neu, EGFR, EpCAM, and mucin-1) detected CTCs in all 20 ovarian cancer patients, whereas CellSearch performed on samples from those same patients only found CTCs in five. This same four-biomarker panel was used with a novel bioorthogonal nanoparticle detection (BOND) strategy to amplify cell labeling by immunolabeled MNP, to allow detection by a micronuclear magnetic resonance (μNMR) platform, as described in a recent review.76
Separation of CTCs from the blood through use of immunomagnetic nanobeads was achieved through lateral magnetophoresis, allowing for high throughput and purity.77 Microchannels were molded in SU-8 photoresist above ferromagnetic wires. Preimmunomagnetic nanobead labeled blood and buffer were introduced in two separate inlets and converged in the main channel, flowing above the wire-containing substrate. The application of a magnetic field drew the labeled cells into a separate smaller outlet from the larger outlet into which the blood cells naturally flowed due to the laminar nature of the channels. This setup led to 90% recovery in cell spike solutions and an average of 85% and 83% purities for 3 breast and 3 lung cancer patient samples, respectively. Downstream analysis was demonstrated through the RT-PCR detection of thyroid transcription factor-1 (TTF-1) expression levels in the isolated cells.
MNPs were also strategically self-assembled for use in CTC capture, detection, and downstream analysis.78 Layer-by-layer assembly was used to coat nanospheres with alternating layers of poly(ethylene imine) and MNPs. A series of reactions ultimately presented carboxylic acid groups on the surface of the magnetic nanospheres, allowing for NHS/EDC chemistry to be used for functionalization with anti-EpCAM. Characterization with 100 cells/mL spiked into whole blood of EpCAM-expressing cell lines showed capture efficiencies of greater that 92%. A total of 90.5% of captured cells were viable and could be cultured in vitro. CK19 and EGFR mutations could be detected through RT-PCR. Varied volumes of blood samples from patients with colon, liver, lung, and breast cancer patients were analyzed, and CTCs were detected in all.
Another property of MNPs used in CTC research is their ability to heat upon excitation. When conjugated with the S6 aptamer, gold-coated MNPs were able to recognize HER2 positive breast cancer cells, SKBR-6, from HER2 negative cells, LNCaP, MDA-MB, or HaCaT, following a 2 h incubation.79 Visualization occurred through the use of Cy-3 conjugated S6 aptamers, and photothermal destruction was achieved through the application of a 670 nm OEM laser. However, this killed up to 12% of unlabeled cells in solution, leaving room for optimization.
Immunomagnetic labeling was combined with a magnetic sifter for high-throughput (10 mL/h) cell capture and subsequent release.80 In the presence of a magnet, samples prelabeled with anti-EpCAM-conjugated MNPs were drawn to the edge of the 40 μm pores in a silicon nitride membrane that was coated with an 80% nickel and 20% iron permalloy; unlabeled cells continued through. Release occurred following the removal of the magnet and a buffer wash, releasing 92.7% of captured cells. Between 31 and 96 CTCs/mL were detected in each of six non-small-cell lung cancer patients. With antibodies specific to wild-type and mutated EGFR, additional analysis could be performed by applying extracted membrane proteins to a magneto-nanosensor biochip.
Using genetic engineering techniques, Maeda et al. were able to harness the cellular machinery of the bacterium Magnetospirillum magneticumAMB-1 to modify its naturally produced nanoscale bacterial magnetic particles (BacMPs).81 The BacMPs were enclosed in a membrane, which has a number of integral proteins that can be used in gene fusion to effectively conjugate a desired protein to the BacMP. Maeda et al. fused the biotin carboxyl carrier protein (BCCP) as well as Protein G to the bacterial protein Mms13. Streptavidin conjugated with a quantum dot bound the expressed biotin, functionalizing the particle for imaging. To perform cell capture, the solution of interest was incubated with an anti-EpCAM antibody followed by mixing with the nanocomposite, which bound the constant fragment of the antibody with the fused Protein G. A magnetic separation was performed, yielding 92% recovery in experiments using cell lines.
As shown by these examples, the nanoscale affords a breadth of advantages not available to traditional magnetic methods. Cellular internalization, signature size-based characteristic magnetic curves, and natural production and scale-up are several features that can be exploited by operating in the nano regime.
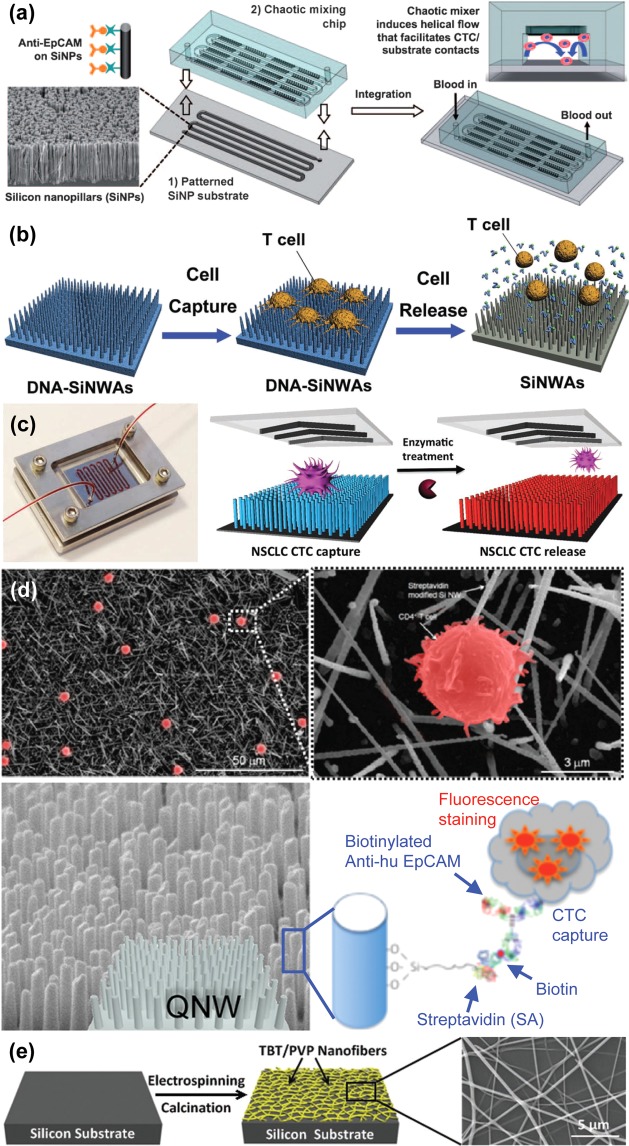
Vertically Aligned Carbon Nanotubes
The robust mechanical and electrical properties of carbon nanotubes (CNTs) make them another nanoscale tool being explored for cellular separation and characterization with potential applications in CTC research. With the use of well-established procedures,82,83 vertically aligned carbon nanotubes (VACNTs) can be grown using chemical vapor deposition (CVD), producing nanostructures for use capture and analysis.
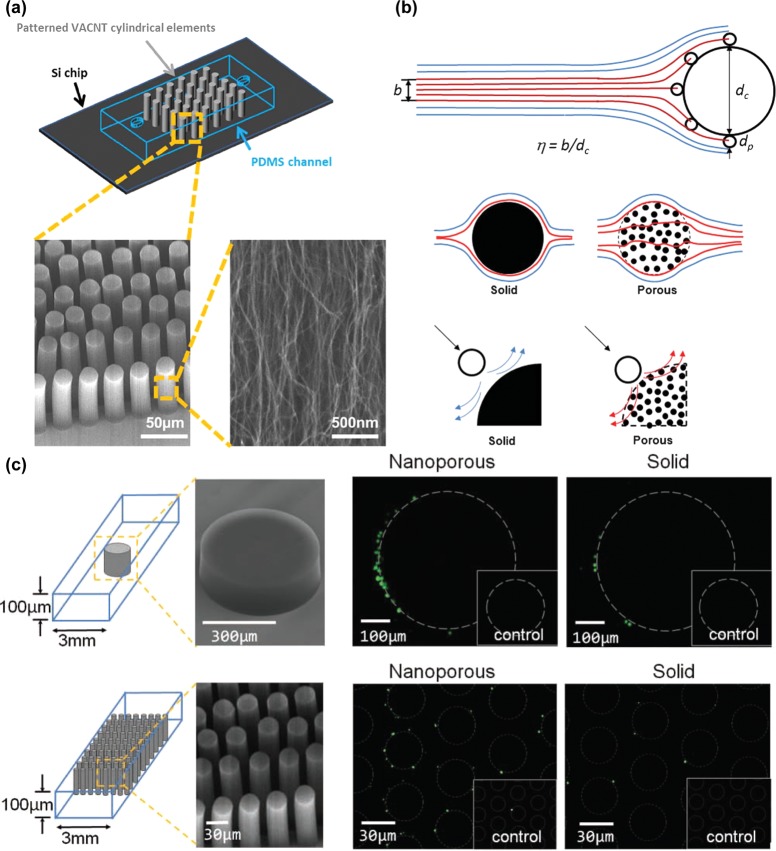
The deposition of CNTs was patterned to give two-tiered structures. Fachin et al. demonstrated the fabrication of VACNT “forests” consisting of nanotube posts (Figure 8a). This allowed capture on two scales: collisions with the post and adhesion within the porous nanotube interior.84 The smaller scale exploited the high surface area of the interior, while on the larger scale, the porosity allowed flow through the post, decreasing the width of the boundary layer and increasing the frequency of collisions of particles with the post (Figure 8b). With the use of the interior nonporous CNT elements, a higher level of control and flexibility was achieved, allowing for an increased ability to manipulate particle flow, fluid flow, geometric control, and molecular diffusion. A porous post reduced the near-surface hydrodynamic resistance that occurs in a solid post, which decreases the interception efficiency; higher capture was then achieved due to flow through and around functional surfaces. When VACNT designs were utilized, it was possible to capture multiple species on a single chip.85 A potential drawback to this design was the decreased density of capture antibodies extent on the post to bind cells, a disadvantage inherent in a highly porous structure. However, the streamline manipulation allowed improvement in capture relative to a control. Using these porous structures, Chen et al. demonstrated 500-μm-diameter posts for CD4+ T-cell capture and 30-μm-diameter post arrays for Streptococcus pneumonia or Escherichia coli capture. Experimental results showed a 5.5-fold increase in capture efficiency of the porous single post configuration when compared with the solid post control and a 6.3-fold increase in porous post array capture efficiency when compared with the solid analog (Figure 8c).86
Figure 8.
Vertically aligned carbon nanotubes (VACNTs). (a) Scanning electron microscopy images of VACNTs grown by chemical vapor deposition and enclosed in a PDMS chamber.84 Adapted with permission from ref (84). Copyright 2010 IEEE. (b) Increased capture via porous structures is a result of streamline manipulation and hydrodynamic resistance reduction, allowing for more collisions occurring at closer distances.85 Adapted with permission from ref (85). Copyright 2012 The Royal Society of Chemistry. (c) Porous elements of varying diameters improve cell capture and allow more of the post to be active in capture when compared with a comparably size solid control pillar.86 Adapted with permission from ref (86). Copyright 2011 John Wiley & Sons, Inc.
Detection upon capture of cancer cells was achieved through the unique electrical properties of carbon nanotubes: VACNTs can serve as a conductive layer on an electrode. Abdolahad et al. then harnessed the ability of cells to bind and spread on nanostructures to create a biosensor with cells as a dielectric material. This electrical cell impedance sensing biosensor (ECIS) could sense changes in impedance levels, revealing the presence of a cell. The mechanism of attachment has not yet been elucidated, but the failure of prefixed cells to bind the nanotubes suggests that the natural deformability of the cell may play a role.87
In the case of CNTs, material attributes in addition to scale-based features can be used to further investigate the presence and properties of cells, making these technologies well-suited for application in CTC research. Capture and study is facilitated both through the interactions between the cells and the CNTs and the interactions between the surrounding fluid and the CNTs, presenting many potential applications. Halloysite nanotubes have also been used to provide similar advantages in rare cell capture with selectins, as reviewed recently.88
Nanopillars, Nanowires, and Nanofibers
The ability of cell surface components to interact with nanofeatures due to their mutual scale is a fundamental asset in the biological application of nanotechnology, as described in a recent review article.89 The fabrication of such elements for cell capture has been performed using multiple materials in several structures.
Silicon was etched or deposited into nanopillars or nanowires to increase surface contact with extracellular features. Wang et al. used a silver and hydrofluoric acid etching process to create a surface of silicon nanopillars (SiNP). After attachment of streptavidin through NHS/maleimide chemistry, biotinylated anti-EpCAM was coupled, joining the capture antibody to the SiNP.90 The capture efficiency of SiNP modified substrates (45–65%) was up to 10 times higher than that of the flat silicon substrates (4–14%). The effect of SiNP length on capture was tested, resulting in speculation that the optimized lengths corresponded to lengths amenable to interaction with extracellular structures. These researchers having optimized the substrate with MCF-7 cells spiked into whole blood, a SiNP surface was used in conjunction with a chaotic micromixer for both cell line and patient sample CTC capture (Figure 9a).91 The results from patient sampling were compared with CellSearch analysis, and showed a marked improvement over this established method, detecting CTCs in 20 out of 26 patients, while CellSearch found CTCs in only 8 of those patients. Using a chemically etched silicon nanostructured surface with similar dimensions under the name silicon nanowires (SiNW), Hou et al. were able to conjugate a temperature sensitive polymer, poly(N-isopropylacrylamide) (PIPAAm), to the high surface area substrate, designated SiNWS, allowing for controlled release of captured CTCs.92 Further conjugated with anti-EpCAM antibodies, the functionalized SiNWS-bound PIPAAm facilitated cell capture at a ratio of over 70% of labeled MCF-7 cells spiked into blood and 90% cell release for 1000 MCF-7 cells/mL with 90% viability.
Figure 9.
Nanopillar, nanowire, and nanofiber structures. (a) Chaotic micromixer induces increased contact between flowing cells and anti-EpCAM functionalized silicon nanopillars (SiNPs) substrates.91 Adapted with permission from ref (91). Copyright 2011 John Wiley & Sons, Inc. (b) T lymphocyte cell capture on DNA-silicon nanowire arrays (SiNWAs) and cell release using exonuclease I to break down aptamers.93 Adapted with permission from ref (93). Copyright 2011 John Wiley & Sons, Inc. (c) Aptamer-coated NanoVelcro Chip for capturing and releasing NSCLC CTCs94 Adapted with permission from ref (94). Copyright 2013 John Wiley & Sons, Inc. (d) CD4+ T lymphocytes (shown by SEM images) may be selectively captured by quartz nanowires (QNWs) functionalized with anti-EpCAM.95,96 Adapted with permission from refs (95) and (96). Copyright 2010, 2012, American Chemical Society. (e) Titanium nanofibers are fabricated through electrospinning and calcination prior to functionalization for ultimate use in cell capture.98 Adapted with permission from ref (98). Copyright 2012 John Wiley & Sons, Inc.
The role of the antibody as a capture moiety may be assumed by DNA aptamers. These aptamers were conjugated to the elements of a silicon nanowire array (SiNWA) to isolate CD4+ T lymphocytes (Figure 9b).93 In addition to showing increased specificity as a result of both the aptamers and the nanostructured surface, the application of exonuclease I allowed for the release of 97% of captured cells with 90% viability. A similar method of rare cell capture and release via DNA aptamers of cancer cells was put forth as an update to the NanoVelcro Chip (Figure 9c).94 This device was validated using the non-small-cell lung cancer (NSCLC) cell line A549, and release was conducted with the enzyme Benzonase Nuclease. Increased purity was achieved through a second capture/release pass through the device, resulting in greater than 95% purity. The clinical utility of this method was demonstrated through genetic analysis of the released cells, using PCR and Sanger sequencing to reveal a mutation characteristic of A549 cells, KRASG12S.
SiNWs can also be fabricated using chemical vapor deposition. Kim et al. then treated these structures with oxygen plasma to yield hydroxyl groups capable of reacting with 3-aminoprophyltiethoxysilane (APTES), which, in turn, was conjugated with the cross-linking agent glutaraldehyde (GA), which ultimately reacted with streptavidin.95 Samples were mixed with biotinylated anti-CD4 antibodies, which could bind CD4+ cells and be bound by the streptavidin-functionalized surface (Figure 9d) in a capture technique that could by translated from T lymphocyte separation to separation of CTCs using a different biomarker. Using the same APTES/GA chemistry, Lee et al. performed CTC capture using anti-EpCAM conjugated with quartz nanowire (QNW) of comparable dimensions, additionally integrating laser scanning cytometry to conduct automated analysis.96 This group also used this chemistry to conjugate streptavidin to SiNWs fabricated using a silver-assisted chemical etching procedure.97 Pretreated solutions of A549 cells spiked into sheep blood underwent separation with an average capture efficiency of 92.6%. Cellular affinity for the structured surface was apparent given the extracellular protrusions visualized through SEM imaging.
Nanofabrication can also be conducted with titanium oxide, which can be electrospun into nanofibers of 100–300 nm diameter (Figure 9e).98 Horizontally oriented as opposed to the vertical orientation of the nanoposts and nanowires described above, titanium nanofibers (TiNFs) provide a scaffold for CTC capture. Zhang et al. fabricated TiNFs from a spun composite of titanium n-butoxide (TBT) and polyvinyl pyrrolidone (PVP). Through 3-mercaptopropyl trimethoxysliane (MPTMS) and N-maleimidobutyryloxy succinimide ester (GMBS), streptavidin was joined to the fiber, and biotin/avidin chemistry was exploited to join a biotinylated anti-EpCAM to the surface to enable cell capture. Performance was verified using samples from gastric and colorectal cancer patients. SEM imaging displayed cell spreading and the interaction of cellular structures with the nanostructures on the substrate surface.
Electrospinning was used to coat a laser microdissection slide with poly(lactic-co-glycolic acid) (PLGA) nanofibers, yielding a transparent capture surface with the advantages of a nanostructured surface.99 For melanoma-specific capture, biotinylated anti-CD146 was presented by streptavidin linked to the PLGA nanofibers by NHS chemistry. The device was characterized using the melanoma cell line M229, with an 87% capture efficiency when cell suspensions were flowed at 1 mL/h, with a slightly higher capture ratio when operated at 0.5 mL/h. Two patient samples demonstrated the clinical application of this device as well as potential for downstream analysis. Following four biomarker immunocytochemical analysis, which detected 43 and 36 CTCs, individual cells could be extracted by laser microdissection for subsequent whole genome analysis and Sanger sequencing. This allowed for the detection of the BRAFV600E mutation, which is highly relevant in the use of therapeutic BRAF inhibitors.
Through various chemical fabrication methods and coupling chemistries, a wide array of substrate surface structures has become available for use in CTC research. Nanopillar, wire, and fiber geometries exploit both size scale and high surface area to increase the interface between capture substrates and cellular structures, improving upon capture with specific antibodies alone.
Nanoroughened Structures
The adhesion preference of CTCs differs from that of blood cells, making nanoroughened surfaces an alternative technique for CTC capture.100 Nanoroughened surfaces increase the surface area available for adhesion, binding, and reactions. Through deposition, molding, and etching, nanoroughened surfaces have been fabricated to facilitate capture and postprocessing of rare cells, including CTCs.
The ability of RNA to reveal the tissue of origin or mutations associated with cancer progression makes it an important resource in CTC investigation. As such, its isolation and analysis is an area of interest that can be aided by the use of nanoroughened surfaces. Following the extraction of RNA from magnetic bead-captured CTCs, Ivanov et al. were able to recognize the cells as originating from the prostate by using electrodes covered in peptide nucleic acid probes for prostate specific antigen (PSA) RNA (Figure 10a).101 The cells were further characterized by including probes for TMPRSS/ERG Type III, a gene fusion commonly found in prostate cancer. To increase the presentation of probes on the surface of the electrodes, a nanostructured surface of palladium was deposited by electroplating. The increase in binding of RNA due to the increased accessibility allowed for a lower detection limit, which is especially desirable given the small population size of CTCs. RNA binding was sensed by the electrodes when the local increase in negative charge created an electrochemical signal through interaction with an electrocatalytic solution of Ru(NH3)63+ and Fe(CN)63-. The efficacy of this technology was shown using prostate cancer patient samples in an attempt to correlate CTC levels with Gleason score.
Figure 10.
Nanotextured surfaces. (a) Nanostructured microelectrodes with two different redox-active probes, Ru(NH3)63+, which accumulates on the sensor based on the amount of target mRNA, and Fe(CN)63-, which can regenerate the Ru(II) species.101 Adapted with permission from ref (101). Copyright 2012 American Chemical Society. (b) PDMS surfaces characterized by increased roughness as shown by atomic force microscopy show increased cell spreading and attachment (inset: scanning electron microscope images).102 Adapted with permission from ref (102). Copyright 2011 American Cancer Society. (c) Enhanced cell capture using gold nanoparticles with multiple aptamers for multivalent interaction.103 Adapted with permission from ref (103). Copyright 2013 American Chemical Society. (d) Poly(3, 4-ethylenedioxy)thiophene (PEDOT) dots functionalized with anti-EpCAM antibodies can capture EpCAM-positive cells.104 Adapted with permission from ref (104). Copyright 2011 John Wiley & Sons, Inc. (e) Cancer cells preferentially adhere to reactive ion etched glass surfaces, shown as a schematic and with scanning electron microscopy.100 Adapted with permission from ref (100). Copyright 2012 American Chemical Society.
RNA in and of itself is used in CTC technology through capture via RNA aptamers. Wan et al. utilized RNA aptamers against the epidermal growth factor receptor (EGFR) to isolate human glioblastoma cells, which are known to overexpress that receptor.102 To render this capture device more efficient, a nanoroughened PDMS surface was fabricated using a poly(d,l-lactide-co-glycolide) (PLGA) mold. The resulting PDMS exhibited increased surface area available for functionalization beginning with its treatment with ozone plasma and piranha solution. The resulting substrates were then soaked in APTES, creating amino groups. Once the amino groups were converted to isothiocyanate, DNA probes with added amino groups could be conjugated, and the RNA aptamers could ultimately hybridize with those probes. The initial increased area as a result of the nanoroughened PDMS allowed for more reactive groups at each step, carrying through to yield an increased density of surface aptamers for capture in the final step. Following capture, SEM imaging revealed that the cells spread over the nanotextured surface, while cells captured using a flat control device maintained a spherical morphology, highlighting the amenability of these modified substrates as cellular interfaces (Figure 10b).
DNA aptamers were also used on increased surface area substrates to improve rare cell capture.103 Sheng et al. used an avidin-coated glass substrate which bound biotinylated gold nanoparticles (AuNPs) which had been previously conjugated with DNA aptamers to capture the cells of interest, in this case the leukemia cell line CCRF-CEM. The AuNPs created a surface capable of displaying a higher concentration of aptamers resulting in multivalent binding for cell capture (Figure 10c), giving greater than 90% capture efficiencies for as few as 100 CEM cells spiked into whole blood. Initial challenges posed by red blood cells blocking aptamer binding of target cells were overcome by incorporating a herringbone pattern into the PDMS capture chamber, taking advantage of the versatility of PDMS molding to introduce micromixing.
Another polymer in addition to PDMS used as a substrate for biological capture is poly(3,4-ethylenedioxy)thiophene (PEDOT). Sekine et al. deposited a carboxylated PEDOT in nanodots onto the surface of indium tin oxide (ITO)-coated glass using an electrolyte solution and various voltages (Figure 10d).104 Increased voltage gave larger particle sizes and lower particle densities, and while the root-mean-square roughness initially increased with voltage, it eventually reached a maximum as the particles began to fuse. Capture anti-EpCAM was then appended to the surface through NHS/EDC and biotin/avidin chemistry. The anti-EpCAM nanostructured surface showed increased capture of EpCAM-expressing cells relative to flat films conjugated with anti-EpCAM when the surface was characterized and optimized using cell lines, suggesting possible applications in the field of CTC study.
Surfaces were modified to present P-selectin molecules in contrast to the antibodies and aptamers often used in cell capture.105 Negatively charged colloidal silica nanoparticles were attached to the inside of a tube through either poly-l-lysine or titanium(IV) butoxide, increasing the surface area for P-selectin adsorption. The capture system was evaluated with the acute myeloid leukemia KG1a cell line, showing a 50% capture yield for cells spiked into blood.106
The established interactions between cell surface features and nanoscale structures suggest the possibility of capture without the use of a specific capture moiety. Chen et al. exploited the selective adhesion of cancer cells to nanotextured surfaces when compared with blood cells by reactive ion etching a glass surface for use in CTC capture (Figure 10e).100 Increased roughness correlated with increased capture, while the conjugation of anti-EpCAM to the nanoroughened surface was not shown to make a significant difference in capture for increasingly roughened surfaces. The use of selection by EpCAM expression causes a significant loss of information within CTCs due to the inherent selection of a specific EpCAM-expressing population of cells that may not give a complete picture of the cancer. When nanoroughened surfaces are utilized, the use of capture antibodies is not necessary, showing the potential of nanoscale features to capture a metamorphic population of cells.
When every additional cell captured represents a significant increase in the amount of information that can be obtained, the ability of nanoroughened surfaces to increase capture yield through increased surface area for aptamer and antibody binding and display or through direct capture of the cells via extracellular structure interaction shows their promise in the expanding field of CTC research.
Graphene Oxide
Graphene oxide is a promising material as a component in applications such as delivery of water-insoluble cancer drugs,107 biosensors for bacterium assays and DNA detection,108,109 energy-storage materials,110,111 paper-like materials,112,113 and polymer composites.114,115 It is a derivative of graphene with oxygen functional groups on its basal planes and edges. Converting graphene oxide to graphene is one method used to manufacture graphene.114,116 Ruoff’s group demonstrated a solution-based approach to obtain individual graphene oxide sheets involving chemical oxidation of graphite to hydrophilic graphite oxide, followed by exfoliation through ultrasonication in water.117 Graphene oxide has certain advantages for biological applications. It is easy to functionalize graphene oxide through polyethylene glycol (PEG)-based chemistry.118 Additionally, graphene oxide particle size can be controlled by sonication time and filtration.119 Furthermore, the optical transparency of graphene oxide is one of its promising characteristics for biological and medical research, allowing for improved imaging.120
Myung et al. demonstrated a novel biosensor with graphene oxide nanoparticles for detection of breast cancer biomarkers.121 The negatively charged graphene oxide was coated on amine-terminated silicon oxide nanoparticles (NPs), as shown through transmission electron microscopy (TEM) (Figure 11a). The graphene oxide coated NPs were self-assembled on the surface functionalized oxide substrate with a metal electrode. To enable electrical conductivity to access sensing functionality, the graphene oxide was reduced to graphene by exposure to hydrazine vapor. Through the measurement of the relative conductance change of the devices, HER2 was detected.
Figure 11.
Graphene oxide. (a) Functionalized reduced graphene oxide nanoparticles for use in HER2 detection.121 Adapted with permission from ref (121). Copyright 2011 John Wiley & Sons, Inc. (b) Ultrasensitive graphene-enhanced fluorescent and electrochemical CTC detection procedures.122 Adapted with permission from ref (122). Copyright 2012 American Chemical Society. (c) Schematic of the micropillar device with modified GO-Fe3O4 MNPs (GO-F) and images of a single nickel micropillar capturing a cancer cell.123 Adapted with permission from ref (123). Copyright 2011 John Wiley & Sons, Inc. (d) Functionalization chemistry stemming from the self-assembly of PEG-functionalized graphene oxide dispersed with tetrabutylammonium, resulting in anti-EpCAM end groups that capture target cells; SEM images and fluorescence microscope image of captured and cultured MCF-7 cells.124 Adapted with permission from ref (124). Copyright 2013 Nature Publishing Group.
Graphene oxide was also used as a starting point for the electrical detection, capture, and fluorescent imaging of CTCs.122 A glassy carbon electrode was pretreated with chitosan, after which graphene oxide was deposited and reduced. Anti-EpCAM was conjugated to the surface using the cross-linker glutaraldehyde, allowing for cell capture. Anti-EpCAM and anti-GPC3, a marker for hepatocellular carcinoma, conjugated to SiO2 nanoparticles modified with ZnSe, CdTe1, or CdTe2 were incubated with the captured cells, allowing for enumeration via square-wave voltammetric (SWV) measurements as well as fluorescent imaging (Figure 11b). Cells were detected at concentrations as low as 5 cells/mL using spiked Hep3B cells in PBS, representing proof of principle. Spiked cells in blood and clinical samples will be necessary for further device characterization.
Graphite oxide was used as an intermediate for conjugation chemistry in a magnetic nanoparticle–micropost system (Figure 11c).123 Magnetic nanoparticles were embedded in graphite oxide sheets, which subsequently underwent NHS/EDC chemistry to bind streptavidin. Biotinylated anti-EpCAM was then used for functionalization. Nickel micropillars under a magnetic field attracted magnetic nanoparticles, creating both a textured surface for increased interaction with cells and increased antibody presentation. Capture of spiked cells from blood was achieved with greater than 40% efficiency. By removing the magnetic field and flowing a wash buffer, 92.9% of captured cells were released, 78% of which were viable.
Recently, Yoon et al. demonstrated the graphene oxide chip for sensitive capture of CTCs.124 Graphene oxide nanosheets functionalized with polyethylene glycol (PEG) were able to self-assemble on a gold-patterned silicon surface through use of a positively charged intercalating agent.125,126 A series of linker chemistries including cross-linker and biotin–avidin chemistry were then used to ultimately functionalize the substrate with an anti-EpCAM antibody (Figure 11d). To characterize the graphene oxide chip, MCF-7, Hs-578T, and PC-3 cell lines were spiked into buffer or blood and flowed through the chip. The captured cells were cultured on the patterned gold surface with graphene oxide sheets, making use of the advantageous virtually two-dimensional capture surface. Graphene oxide sheets have been shown to enhance cell proliferation because of their biocompatibility with cells.127,128 The culture of captured CTCs would allow for increased downstream analysis previously rendered difficult due to the low number of CTCs, such as RT-PCR or drug testing both for the development of new therapeutics and of individualized treatment.129 Blood samples from patients with breast, pancreatic, and early lung cancer were processed on the graphene oxide devices and 2–23 CTCs/mL were captured.
Nanomaterials such as graphene oxide present the many advantages of operating at that length scale: increased available surface area, convenient established chemistries, and a breadth of biological applications (Table 1). As shown by the graphene oxide CTC capture device, 2D carbon can serve as a foundation upon which a new generation of rare cell isolation and characterization devices may be built, enabling the scale-up and downstream analysis necessary to address the problems currently facing cancer researchers. Graphene oxide is representative of nanomaterials that can be integrated into CTC research because of its enabling of successful high sensitivity capture, suggesting its continued use in CTC devices.
Table 1. Summary of Nanomaterial Advantages.
| nanomaterial | advantages | examples (references) |
|---|---|---|
| Magnetic nanoparticles | Cellular internalization, signature size-based characteristic magnetic curves, potential natural production, easy scale-up | (71−75, 77−81) |
| Vertically aligned carbon nanotubes | Increased internal and external surface area, cell-nanotube and fluid-nanotube interactions, conductivity | (84−87) |
| Nanopillars, nanowires, nanofibers | Interactions with extracellular features, increased surface area, potential thermosensitivity | (90−99) |
| Nanoroughened surfaces | Interactions with extracellular features, increased surface area, potential for antigen independent capture | (101−106) |
| Graphene oxide | Interactions with extracellular features, increased surface area, conductivity, 2-dimensional | (121−124) |
Summary and Future Perspectives
The isolation and study of circulating tumors cells has been a research aim for over a century, and an FDA-approved separation device has been available for nearly a decade. Improving upon this established but imperfect technology, microfluidics allows the precise flow control and parameter manipulation to achieve the purity, specificity, and yield necessary for thorough investigation into the full potential of this small population of cells. Microfluidic devices for CTC capture were initiated using the building blocks of silicon and PDMS, but further increases in CTC capture metrics will, in turn, require further innovation.
Nanomaterials are able to address the problems of insufficient capture efficiency and low purity through their high surface-area-to-volume ratio.89 Carbon nanotubes, graphene oxide, nanopillars, nanowires, magnetic nanoparticles, and nanorough-featured surfaces can be manufactured and tailored to fit the desired application through microfabrication technologies, such as reactive ion etching, chemical vapor deposition, and photolithography, and customizable chemistries, such as biotin–avidin affinity. This field is filling its toolbox in an analogous fashion to the introduction of microfluidics to CTC isolation and is the future of rare cell separation as the field progresses.
The ideal CTC isolation technology must be sufficiently versatile to capture a heterogeneous population of cells, gentle to ensure their viability, and able to release the cells for further culture and study. This device would have high yield, sensitivity, and purity while simultaneously operating at high throughput. Fabrication should be simple, repeatable, and inexpensive. As such, materials for use in CTC isolation should have the desirable properties afforded by nanomaterials. Nanomaterials make available a wide range of established conjugation chemistries and provide the many benefits of increased surface area. They can be incorporated into microfluidic platforms, facilitating prototyping. Current CTC isolation technologies have been optimized in many of these respects, but a truly comprehensive device has yet to be developed.
To make full use of the availability of CTCs through a simple liquid biopsy and the information they may provide, CTC culture and additional genomic and proteomic analysis is necessary to conduct the translational research necessary to put a stop to metastatic cancer. However, these are likely attainable goals given the recent progress made with the help of nanomaterials and their promise for future study.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Director’s New Innovator Award (1DP2OD006672-01), a Career Development Program of the Gastrointestinal Specialized Program of Research Excellence (GI SPORE) award (CA 130810), and a Department of Defence (DoD) Office of the Congressionally Directed Medical Research Programs (CDMRP) Career Development Award to S.N.
Author Contributions
† These authors contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Weigelt B.; Peterse J. L.; van’t Veer L. J. Breast Cancer Metastasis: Markers and Models. Nat. Rev. Cancer 2005, 5, 591–602. [DOI] [PubMed] [Google Scholar]
- Gupta G. P.; Massagué J. Cancer Metastasis: Building a Framework. Cell 2006, 127, 679–695. [DOI] [PubMed] [Google Scholar]
- Chambers A. F.; Groom A. C.; MacDonald I. C. Metastasis: Dissemination and Growth of Cancer Cells in Metastatic Sites. Nat. Rev. Cancer 2002, 2, 563–572. [DOI] [PubMed] [Google Scholar]
- Fidler I. J. The Pathogenesis of Cancer Metastasis: The ‘Seed and Soil’ Hypothesis Revisited. Nat. Rev. Cancer 2003, 3, 453–458. [DOI] [PubMed] [Google Scholar]
- Thiery J. P. Epithelial-Mesenchymal Transitions in Tumour Progression. Nat. Rev. Cancer 2002, 2, 442–454. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H.; Wyckoff J.; Condeelis J. Cell Migration in Tumors. Curr. Opin. Cell Biol. 2005, 17, 559–564. [DOI] [PubMed] [Google Scholar]
- Christiansen J. J.; Rajasekaran A. K. Reassessing Epithelial to Mesenchymal Transition as a Prerequisite for Carcinoma Invasion and Metastasis. Cancer Res. 2006, 66, 8319–8326. [DOI] [PubMed] [Google Scholar]
- van Denderen B. J. W.; Thompson E. W. Cancer: The to and fro of Tumour Spread. Nature 2013, 493, 487–488. [DOI] [PubMed] [Google Scholar]
- Tsai J. H.; Donaher J. L.; Murphy D. A.; Chau S.; Yang J. Spatiotemporal Regulation of Epithelial-Mesenchymal Transition Is Essential for Squamous Cell Carcinoma Metastasis. Cancer Cell 2012, 22, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocaña O. H.; Córcoles R.; Fabra Á.; Moreno-Bueno G.; Acloque H.; Vega S.; Barrallo-Gimeno A.; Cano A.; Nieto M. A. Metastatic Colonization Requires the Repression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell 2012, 22, 709–724. [DOI] [PubMed] [Google Scholar]
- Klein C. A. Parallel Progression of Primary Tumours and Metastases. Nat. Rev. Cancer 2009, 9, 302–312. [DOI] [PubMed] [Google Scholar]
- Paterlini-Brechot P.; Benali N. L. Circulating Tumor Cells (CTC) Detection: Clinical Impact and Future Directions. Cancer Lett. 2007, 253, 180–204. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M.; Hayes D. F.; Budd G. T.; Ellis M. J.; Stopeck A.; Reuben J. M.; Doyle G. V.; Matera J.; Allard W. J.; Miller M. C.; et al. Circulating Tumor Cells: A Novel Prognostic Factor for Newly Diagnosed Metastatic Breast Cancer. J. of Clin. Oncol. 2005, 23, 1420–1430. [DOI] [PubMed] [Google Scholar]
- Budd G. T.; Cristofanilli M.; Ellis M. J.; Stopeck A.; Borden E.; Miller M. C.; Matera J.; Repollet M.; Doyle G. V.; Terstappen L. W. M. M.; et al. Circulating Tumor Cells versus Imaging—Predicting Overall Survival in Metastatic Breast Cancer. Clin. Cancer Res. 2006, 12, 6403–6409. [DOI] [PubMed] [Google Scholar]
- Hayes D. F.; Cristofanilli M.; Budd G. T.; Ellis M. J.; Stopeck A.; Miller M. C.; Matera J.; Allard W. J.; Doyle G. V.; Terstappen L. W. W. M. Circulating Tumor Cells at Each Follow-up Time Point during Therapy of Metastatic Breast Cancer Patients Predict Progression-Free and Overall Survival. Clin. Cancer Res. 2006, 12, 4218–4224. [DOI] [PubMed] [Google Scholar]
- Sastre J.; Maestro M. L.; Puente J.; Veganzones S.; Alfonso R.; Rafael S.; García-Saenz J. A.; Vidaurreta M.; Martín M.; Arroyo M.; Sanz-Casla M. T.; et al. Circulating Tumor Cells in Colorectal Cancer: Correlation with Clinical and Pathological Variables. Ann. Oncol. 2008, 19, 935–938. [DOI] [PubMed] [Google Scholar]
- Cohen S. J.; Punt C. J. A.; Iannotti N.; Saidman B. H.; Sabbath K. D.; Gabrail N. Y.; Picus J.; Morse M. A.; Mitchell E.; Miller M. C.; et al. Prognostic Significance of Circulating Tumor Cells in Patients with Metastatic Colorectal Cancer. Ann. Oncol. 2009, 20, 1223–1229. [DOI] [PubMed] [Google Scholar]
- Cohen S. J.; Punt C. J. A.; Iannotti N.; Saidman B. H.; Sabbath K. D.; Gabrail N. Y.; Picus J.; Morse M.; Mitchell E.; Miller M. C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients with Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [DOI] [PubMed] [Google Scholar]
- de Bono J. S.; Scher H. I.; Montgomery R. B.; Parker C.; Miller M. C.; Tissing H.; Doyle G. V.; Terstappen L. W. W. M.; Pienta K. J.; Raghavan D. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [DOI] [PubMed] [Google Scholar]
- Mocellin S.; Hoon D.; Ambrosi A.; Nitti D.; Rossi C. R. The Prognostic Value of Circulating Tumor Cells in Patients with Melanoma: A Systematic Review and Meta-Analysis. Clin. Cancer Res. 2006, 12, 4605–4613. [DOI] [PubMed] [Google Scholar]
- Pantel K.; Brakenhoff R. H.; Brandt B. Detection, Clinical Relevance and Specific Biological Properties of Disseminating Tumour Cells. Nat. Rev. Cancer 2008, 8, 329–340. [DOI] [PubMed] [Google Scholar]
- Danila D. C.; Fleisher M.; Scher H. I. Circulating Tumor Cells as Biomarkers in Prostate Cancer. Clin. Cancer Res. 2011, 17, 3903–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. D.; Casavant B. P.; Lang J. M. Rapid Translation of Circulating Tumor Cell Biomarkers into Clinical Practice: Technology Development, Clinical Needs and Regulatory Requirements. Lab Chip 2014, 14, 24–31. [DOI] [PubMed] [Google Scholar]
- Hyun K.-A.; Jung H.-I. Advances and Critical Concerns with the Microfluidic Enrichments of Circulating Tumor Cells. Lab Chip 2014, 14, 45–56. [DOI] [PubMed] [Google Scholar]
- Jin C.; McFaul S. M.; Duffy S. P.; Deng X.; Tavassoli P.; Black P. C.; Ma H. Technologies for Label-Free Separation of Circulating Tumor Cells: from Historical Foundations to Recent Developments. Lab Chip 2013, 14, 32–44. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M.; Budd G. T.; Ellis M. J.; Stopeck A.; Matera J.; Miller M. C.; Reuben J. M.; Doyle G. V.; Allard W. J.; Terstappen L. W. M. M.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [DOI] [PubMed] [Google Scholar]
- Olmos D.; Arkenau H.-T.; Ang J. E.; Ledaki I.; Attard G.; Carden C. P.; Reid A. H. M.; A’Hern R.; Fong P. C.; Oomen N. B.; et al. Circulating Tumour Cell (CTC) Counts as Intermediate End Points in Castration-Resistant Prostate Cancer (CRPC): A Single-Centre Experience. Ann. Oncol. 2009, 20, 27–33. [DOI] [PubMed] [Google Scholar]
- Allard W. J.; Matera J.; Miller M. C.; Repollet M.; Connelly M. C.; Rao C.; Tibbe A. G. J.; Uhr J. W.; Terstappen L. W. M. M. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but Not in Healthy Subjects or Patients with Nonmalignant Diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [DOI] [PubMed] [Google Scholar]
- Seal S. H. A Sieve for the Isolation of Cancer Cells and Other Large Cells from the Blood. Cancer 1964, 17, 637–642. [DOI] [PubMed] [Google Scholar]
- Fleischer R. L.; Price P. B.; Symes E. M. Novel Filter for Biological Materials. Science 1964, 143, 249–250. [DOI] [PubMed] [Google Scholar]
- Truchaud A.; Caldani C.; Bisconte J.-C. Filtration Cytometry: A New Concept for Parallel Real Time Analysis of Bacteria, Cells, and Particles. Biol. Cell 1992, 76, 245. [Google Scholar]
- Rye P. D.; Høifødt H.; Overli G.; Fodstad O. Immunobead Filtration: A Novel Approach for the Isolation and Propagation of Tumor Cells. Am. J. Pathol. 1997, 150, 99–106. [PMC free article] [PubMed] [Google Scholar]
- Vona G.; Sabile A.; Louha M.; Sitruk V.; Romana S.; Schütze K.; Capron F.; Franco D.; Pazzagli M.; Vekemans M.; et al. Isolation by Size of Epithelial Tumor Cells: A New Method for the Immunomorphological and Molecular Characterization of Circulating Tumor Cells. Am. J. Pathol. 2000, 156, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona G.; Estepa L.; Béroud C.; Damotte D.; Capron F.; Nalpas B.; Mineur A.; Franco D.; Lacour B.; Pol S.; et al. Impact of Cytomorphological Detection of Circulating Tumor Cells in Patients with Liver Cancer. Hepatology 2004, 39, 792–797. [DOI] [PubMed] [Google Scholar]
- Zabaglo L.; Ormerod M. G.; Parton M.; Ring A.; Smith I. E.; Dowsett M. Cell Filtration-Laser Scanning Cytometry for the Characterisation of Circulating Breast Cancer Cells. Cytometry, Part A 2003, 55A, 102–108. [DOI] [PubMed] [Google Scholar]
- Zheng S.; Lin H.; Liu J.-Q.; Balic M.; Datar R.; Cote R. J.; Tai Y.-C. Membrane Microfilter Device for Selective Capture, Electrolysis and Genomic Analysis of Human Circulating Tumor Cells. J. Chromatogr., A 2007, 1162, 154–161. [DOI] [PubMed] [Google Scholar]
- Lin H. K.; Zheng S.; Williams A. J.; Balic M.; Groshen S.; Scher H. I.; Fleisher M.; Stadler W.; Datar R. H.; Tai Y.-C.; et al. Portable Filter-Based Microdevice for Detection and Characterization of Circulating Tumor Cells. Clin. Cancer Res. 2010, 16, 5011–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.; Lin H.; Lu B.; Williams A.; Datar R.; Cote R.; Tai Y.-C. 3D Microfilter Device for Viable Circulating Tumor Cell (CTC) Enrichment from Blood. Biomed. Microdevices 2011, 13, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J.; Oh J. H.; Oh J. M.; Park J.-M.; Lee J.-G.; Kim M. S.; Kim Y. J.; Kang H. J.; Jeong J.; Kim S. I.; et al. Efficient Isolation and Accurate In Situ Analysis of Circulating Tumor Cells Using Detachable Beads and a High-Pore-Density Filter. Angew. Chem., Int. Ed. 2013, 52, 8337–8340. [DOI] [PubMed] [Google Scholar]
- Whitesides G. M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [DOI] [PubMed] [Google Scholar]
- Becker H.; Locascio L. E. Polymer Microfluidic Devices. Talanta 2002, 56, 267–287. [DOI] [PubMed] [Google Scholar]
- Haeberle S.; Zengerle R. Microfluidic Platforms for Lab-on-a-Chip Applications. Lab Chip 2007, 7, 1094–1110. [DOI] [PubMed] [Google Scholar]
- McDonald J. C.; Whitesides G. M. Poly(dimethylsiloxane) as a Material for Fabricating Microfluidic Devices. Acc. Chem. Res. 2002, 35, 491–499. [DOI] [PubMed] [Google Scholar]
- Khandurina J.; McKnight T. E.; Jacobson S. C.; Waters L. C.; Foote R. S.; Ramsey J. M. Integrated System for Rapid PCR-Based DNA Analysis in Microfluidic Devices. Anal. Chem. 2000, 72, 2995–3000. [DOI] [PubMed] [Google Scholar]
- Erickson D.; Li D. Integrated Microfluidic Devices. Anal. Chim. Acta 2004, 507, 11–26. [Google Scholar]
- Nagrath S.; Sequist L. V.; Maheswaran S.; Bell D. W.; Irimia D.; Ulkus L.; Smith M. R.; Kwak E. L.; Digumarthy S.; Muzikansky A.; et al. M., Isolation of Rare Circulating Tumour Cells in Cancer Patients by Microchip Technology. Nature 2007, 450, 1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethdorf S.; Wikman H.; Pantel K. Review: Biological Relevance of Disseminated Tumor Cells in Cancer Patients. Int. J. Cancer 2008, 123, 1991–2006. [DOI] [PubMed] [Google Scholar]
- Maheswaran S.; Sequist L. V.; Nagrath S.; Ulkus L.; Brannigan B.; Collura C. V.; Inserra E.; et al. Detection of Mutations in EGFR in Circulating Lung-Cancer Cells. N. Engl. J. Med. 2008, 359, 366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn J. P.; Pratt E. D.; Denning D.; Liu H.; Bander N. H.; Tagawa S. T.; Nanus D. M.; Giannakakou P. A.; Kirby B. J. Capture of Circulating Tumor Cells from Whole Blood of Prostate Cancer Patients Using Geometrically Enhanced Differential Immunocapture (GEDI) and a Prostate-Specific Antibody. Lab Chip 2010, 10, 27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby B. J.; Jodari M.; Loftus M. S.; Gakhar G.; Pratt E. D.; Chanel-Vos C.; Gleghorn J. P.; Santana S. M.; Liu H.; Smith J. P.; et al. Functional Characterization of Circulating Tumor Cells with a Prostate-Cancer-Specific Microfluidic Device. PLoS One 2012, 7, e35976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti G.; Sung M. S.; Vahdat L. T.; Shah M. A.; Santana S. M.; Altavilla G.; Kirby B. J.; Giannakakou P. Isolation of Breast Cancer and Gastric Cancer Circulating Tumor Cells by Use of an Anti HER2-Based Microfluidic Device. Lab Chip 2014, 14, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S.; Sim T. S.; Kim Y. J.; Kim S. S.; Jeong H.; Park J.-M.; Moon H.-S.; Kim S. I.; Gurel O.; Lee S. S.; et al. SSA-MOA: A Novel CTC Isolation Platform Using Selective Size Amplification (SSA) and a Multi-Obstacle Architecture (MOA) Filter. Lab Chip 2012, 12, 2874–2880. [DOI] [PubMed] [Google Scholar]
- Lee H. J.; Cho H.-Y.; Oh J. H.; Namkoong K.; Lee J. G.; Park J.-M.; Lee S. S.; Huh N.; Choi J.-W. Simultaneous Capture and In Situ Analysis of Circulating Tumor Cells Using Multiple Hybrid Nanoparticles. Biosens. Bioelectron. 2013, 47, 508–514. [DOI] [PubMed] [Google Scholar]
- Stott S. L.; Hsu C.-H.; Tsukrov D. I.; Yu M.; Miyamoto D. T.; Waltman B. A.; Rothenberg S. M.; Shah A. M.; Smas M. E.; Korir G. K.; et al. Isolation of Circulating Tumor Cells Using a Microvortex-Generating Herringbone-chip. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 18392–18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M.; Bardia A.; Wittner B. S.; Stott S. L.; Smas M. E.; Ting D. T.; Isakoff S. J.; Ciciliano J. C.; Wells M. N.; Shah A. M.; et al. Circulating Breast Tumor Cells Exhibit Dynamic Changes in Epithelial and Mesenchymal Composition. Science 2013, 339, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng W.; Ogunwobi O. O.; Chen T.; Zhang J.; George T. J.; Liu C.; Fan Z. H. Capture, Release and Culture of Circulating Tumor Cells from Pancreatic Cancer Patients Using an Enhanced Mixing Chip. Lab Chip 2014, 14, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launiere C.; Gaskill M.; Czaplewski G.; Myung J. H.; Hong S.; Eddington D. T. Channel Surface Patterning of Alternating Biomimetic Protein Combinations for Enhanced Microfluidic Tumor Cell Isolation. Anal. Chem. 2012, 84, 4022–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng W.; Chen T.; Kamath R.; Xiong X.; Tan W.; Fan Z. H. Aptamer-Enabled Efficient Isolation of Cancer Cells from Whole Blood Using a Microfluidic Device. Anal. Chem. 2012, 84, 4199–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. A.; Xu Y.; Xia Z.; Fan Z. H.; Tan W. Enrichment of Cancer Cells Using Aptamers Immobilized on a Microfluidic Channel. Anal. Chem. 2008, 81, 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Phillips J. A.; Yan J.; Li Q.; Fan Z. H.; Tan W. Aptamer-Based Microfluidic Device for Enrichment, Sorting, and Detection of Multiple Cancer Cells. Anal. Chem. 2009, 81, 7436–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkuri M. D.; Al-Ejeh F.; Shi J. Y.; Palms D.; Prestidge C.; Griesser H. J.; Brown M. P.; Thierry B. Plasma Functionalized PDMS Microfluidic Chips: Towards Point-of-Care Capture of Circulating Tumor Cells. J. Mater. Chem. 2011, 21, 8841–8848. [Google Scholar]
- Hatch A.; Hansmann G.; Murthy S. K. Engineered Alginate Hydrogels for Effective Microfluidic Capture and Release of Endothelial Progenitor Cells from Whole Blood. Langmuir 2011, 27, 4257–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A. M.; Yu M.; Nakamura Z.; Ciciliano J.; Ulman M.; Kotz K.; Stott S. L.; Maheswaran S.; Haber D. A.; Toner M. Biopolymer System for Cell Recovery from Microfluidic Cell Capture Devices. Anal. Chem. 2012, 84, 3682–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Zhang W.; Huang F.; Feng H.; Shu W.; Xu X.; Chen Y. High Throughput Capture of Circulating Tumor Cells Using an Integrated Microfluidic System. Biosens. Bioelectron. 2013, 47, 113–119. [DOI] [PubMed] [Google Scholar]
- McCarley R. L.; Vaidya B.; Wei S.; Smith A. F.; Patel A. B.; Feng J.; Murphy M. C.; Soper S. A. Resist-Free Patterning of Surface Architectures in Polymer-Based Microanalytical Devices. J. Am. Chem. Soc. 2005, 127, 842–843. [DOI] [PubMed] [Google Scholar]
- Adams A. A.; Okagbare P. I.; Feng J.; Hupert M. L.; Patterson D.; Göttert J.; McCarley R. L.; Nikitopoulos D.; Murphy M. C.; Soper S. A. Highly Efficient Circulating Tumor Cell Isolation from Whole Blood and Label-Free Enumeration Using Polymer-Based Microfluidics with an Integrated Conductivity Sensor. J. Am. Chem. Soc. 2008, 130, 8633–8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri U.; Njoroge S. K.; Witek M. A.; Adebiyi M. G.; Kamande J. W.; Hupert M. L.; Barany F.; Soper S. A. High-Throughput Selection, Enumeration, Electrokinetic Manipulation, and Molecular Profiling of Low-Abundance Circulating Tumor Cells Using a Microfluidic System. Anal. Chem. 2011, 83, 2301–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri U.; Balamurugan S.; Adams A. A.; Okagbare P. I.; Obubuafo A.; Soper S. A. Highly Efficient Capture and Enumeration of Low Abundance Prostate Cancer Cells Using Prostate-Specific Membrane Antigen Aptamers Immobilized to a Polymeric Microfluidic Device. Electrophoresis 2009, 30, 3289–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. M.; Witek M. A.; Hupert M. L.; Brady C.; Pullagurla S.; Kamande J.; Aufforth R. D.; Tignanelli C. J.; Torphy R. J.; Yeh J. J.; et al. UV Activation of Polymeric High Aspect Ratio Microstructures: Ramifications in Antibody Surface Loading for Circulating Tumor Cell Selection. Lab Chip 2014, 14, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamande J. W.; Hupert M. L.; Witek M. A.; Wang H.; Torphy R. J.; Dharmasiri U.; Njoroge S. K.; Jackson J. M.; Aufforth R. D.; et al. Modular Microsystem for the Isolation, Enumeration, and Phenotyping of Circulating Tumor Cells in Patients with Pancreatic Cancer. Anal. Chem. 2013, 85, 9092–9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino K.; Huang Y.; Lane N.; Huebschman M.; Uhr J. W.; Frenkel E. P.; Zhang X. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip 2011, 11, 3449–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-y.; Hoshino K.; Chen P.; Wu C.-h.; Lane N.; Huebschman M.; Liu H.; Sokolov K.; Uhr J.; Frenkel E.; Zhang J. J. Immunomagnetic Nanoscreening of Circulating Tumor Cells with a Motion Controlled Microfluidic System. Biomed. Microdevices 2013, 15, 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-H.; Huang Y.; Chen P.; Hoshino K.; Liu H.; Frenkel E. P.; Zhang J. X. J.; Sokolov K. V. Versatile Immunomagnetic Nanocarrier Platform for Capturing Cancer Cells. ACS Nano 2013, 7, 8816–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Hong G.; Wang H.; Welsher K.; Tabakman S. M.; Sherlock S. P.; Robinson J. T.; Liang Y.; Dai H. Graphite-Coated Magnetic Nanoparticle Microarray for Few-Cells Enrichment and Detection. ACS Nano 2012, 6, 1094–1101. [DOI] [PubMed] [Google Scholar]
- Issadore D.; Chung J.; Shao H.; Liong M.; Ghazani A. A.; Castro C. M.; Weissleder R.; Lee H. Ultrasensitive Clinical Enumeration of Rare Cells ex Vivo Using a Micro-Hall Detector. Sci. Transl. Med. 2012, 4, 141ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C. M.; Ghazani A. A.; Chung J.; Shao H.; Issadore D.; Yoon T.-J.; Weissleder R.; Lee H. Miniaturized Nuclear Magnetic Resonance Platform for Detection and Profiling of Circulating Tumor Cells. Lab Chip 2014, 14, 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Han S.-I.; Park M.-J.; Jeon C.-W.; Joo Y.-D.; Choi I.-H.; Han K.-H. Circulating Tumor Cell Microseparator Based on Lateral Magnetophoresis and Immunomagnetic Nanobeads. Anal. Chem. 2013, 85, 2779–2786. [DOI] [PubMed] [Google Scholar]
- Wen C.-Y.; Wu L.-L.; Zhang Z.-L.; Liu Y.-L.; Wei S.-Z.; Hu J.; Tang M.; Sun E.-Z.; Gong Y.-P.; Yu J.; et al. Quick-Response Magnetic Nanospheres for Rapid, Efficient Capture and Sensitive Detection of Circulating Tumor Cells. ACS Nano 2014, 8, 941–949. [DOI] [PubMed] [Google Scholar]
- Fan Z.; Shelton M.; Singh A. K.; Senapati D.; Khan S. A.; Ray P. C. Multifunctional Plasmonic Shell–Magnetic Core Nanoparticles for Targeted Diagnostics, Isolation, and Photothermal Destruction of Tumor Cells. ACS Nano 2012, 6, 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earhart C. M.; Hughes C. E.; Gaster R. S.; Ooi C. C.; Wilson R. J.; Zhou L. Y.; Humke E. W.; Xu L.; Wong D. J.; Willingham S. B.; et al. Isolation and Mutational Analysis of Circulating Tumor Cells from Lung Cancer Patients with Magnetic Sifters and Biochips. Lab Chip 2014, 14, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y.; Yoshino T.; Matsunaga T. Novel Nanocomposites Consisting of In Vivo-Biotinylated Bacterial Magnetic Particles and Quantum Dots for Magnetic Separation and Fluorescent Labeling of Cancer Cells. J. Mater. Chem. 2009, 19, 6361–6366. [Google Scholar]
- Wardle B. L.; Saito D. S.; García E. J.; Hart A. J.; de Villoria R. G.; Verploegen E. A. Fabrication and Characterization of Ultrahigh-Volume-Fraction Aligned Carbon Nanotube–Polymer Composites. Adv. Mater. 2008, 20, 2707–2714. [DOI] [PubMed] [Google Scholar]
- Hart A. J.; Slocum A. H. Rapid Growth and Flow-Mediated Nucleation of Millimeter-Scale Aligned Carbon Nanotube Structures from a Thin-Film Catalyst. J. Phys. Chem. B 2006, 110, 8250–8257. [DOI] [PubMed] [Google Scholar]
- Fachin F.; Chen G. D.; Toner M.; Wardle B. L. Integration of Bulk Nanoporous Elements in Microfluidic Devices with Application to Biomedical Diagnostics. J. Microelectromech. Syst. 2011, 20, 1428–1438. [Google Scholar]
- Chen G. D.; Fachin F.; Colombini E.; Wardle B. L.; Toner M. Nanoporous Micro-Element Arrays for Particle Interception in Microfluidic Cell Separation. Lab Chip 2012, 12, 3159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. D.; Fachin F.; Fernandez-Suarez M.; Wardle B. L.; Toner M. Nanoporous Elements in Microfluidics for Multiscale Manipulation of Bioparticles. Small 2011, 7, 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolahad M.; Taghinejad M.; Taghinejad H.; Janmaleki M.; Mohajerzadeh S. A Vertically Aligned Carbon Nanotube-Based Impedance Sensing Biosensor for Rapid and High Sensitive Detection of Cancer Cells. Lab Chip 2012, 12, 1183–1190. [DOI] [PubMed] [Google Scholar]
- Hughes A. D.; King M. R. Nanobiotechnology for the Capture and Manipulation of Circulating Tumor Cells. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2012, 4, 291–309. [DOI] [PubMed] [Google Scholar]
- Wang L.; Asghar W.; Demirci U.; Wan Y. Nanostructured Substrates for Isolation of Circulating Tumor Cells. Nano Today 2013, 8, 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Wang H.; Jiao J.; Chen K.-J.; Owens G. E.; Kamei K.-i.; Sun J.; Sherman D. J.; Behrenbruch C. P.; Wu H.; et al. Three-Dimensional Nanostructured Substrates toward Efficient Capture of Circulating Tumor Cells. Angew. Chem., Int. Ed. 2009, 121, 9132–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Liu K.; Liu J.; Yu Z. T. F.; Xu X.; Zhao L.; Lee T.; Lee E. K.; Reiss J.; Lee Y.-K.; et al. Highly Efficient Capture of Circulating Tumor Cells by Using Nanostructured Silicon Substrates with Integrated Chaotic Micromixers. Angew. Chem., Int. Ed. 2011, 50, 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S.; Zhao H.; Zhao L.; Shen Q.; Wei K. S.; Suh D. Y.; Nakao A.; Garcia M. A.; Song M.; Lee T.; et al. Capture and Stimulated Release of Circulating Tumor Cells on Polymer-Grafted Silicon Nanostructures. Adv. Mater. 2013, 25, 1547–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Liu X.; Su B.; Li J.; Jiang L.; Han D.; Wang S. Aptamer-Mediated Efficient Capture and Release of T Lymphocytes on Nanostructured Surfaces. Adv. Mater. 2011, 23, 4376–4380. [DOI] [PubMed] [Google Scholar]
- Shen Q.; Xu L.; Zhao L.; Wu D.; Fan Y.; Zhou Y.; OuYang W.-H.; Xu X.; Zhang Z.; Song M.; et al. Specific Capture and Release of Circulating Tumor Cells Using Aptamer-Modified Nanosubstrates. Adv. Mater. 2013, 25, 2368–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. T.; Kim D.-J.; Kim T.-J.; Seo D.-W.; Kim T.-H.; Lee S.-Y.; Kim K.; Lee K.-M.; Lee S.-K. Novel Streptavidin-Functionalized Silicon Nanowire Arrays for CD4+ T Lymphocyte Separation. Nano Lett. 2010, 10, 2877–2883. [DOI] [PubMed] [Google Scholar]
- Lee S.-K.; Kim G.-S.; Wu Y.; Kim D.-J.; Lu Y.; Kwak M.; Han L.; Hyung J.-H.; Seol J.-K.; Sander C.; et al. Nanowire Substrate-Based Laser Scanning Cytometry for Quantitation of Circulating Tumor Cells. Nano Lett. 2012, 12, 2697–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-K.; Kim D.-J.; Lee G.; Kim G.-S.; Kwak M.; Fan R. Specific Rare Cell Capture Using Micro-Patterned Silicon Nanowire Platform. Biosens. Bioelectron. 2014, 54, 181–188. [DOI] [PubMed] [Google Scholar]
- Zhang N.; Deng Y.; Tai Q.; Cheng B.; Zhao L.; Shen Q.; He R.; Hong L.; Liu W.; Guo S.; et al. Electrospun TiO2 Nanofiber-Based Cell Capture Assay for Detecting Circulating Tumor Cells from Colorectal and Gastric Cancer Patients. Adv. Mater. 2012, 24, 2756–2760. [DOI] [PubMed] [Google Scholar]
- Hou S.; Zhao L.; Shen Q.; Yu J.; Ng C.; Kong X.; Wu D.; Song M.; Shi X.; Xu X.; et al. Polymer Nanofiber-Embedded Microchips for Detection, Isolation, and Molecular Analysis of Single Circulating Melanoma Cells. Angew. Chem., Int. Ed. 2013, 52, 3533–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.; Weng S.; Zhang F.; Allen S.; Li X.; Bao L.; Lam R. H. W.; Macoska J. A.; Merajver S. D.; Fu J. Nanoroughened Surfaces for Efficient Capture of Circulating Tumor Cells without Using Capture Antibodies. ACS Nano 2012, 7, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.; Stojcic J.; Stanimirovic A.; Sargent E.; Nam R. K.; Kelley S. O. Chip-Based Nanostructured Sensors Enable Accurate Identification and Classification of Circulating Tumor Cells in Prostate Cancer Patient Blood Samples. Anal. Chem. 2012, 85, 398–403. [DOI] [PubMed] [Google Scholar]
- Wan Y.; Mahmood M. A. I.; Li N.; Allen P. B.; Kim Y.-t.; Bachoo R.; Ellington A. D.; Iqbal S. M. Nanotextured Substrates with Immobilized Aptamers for Cancer Cell Isolation and Cytology. Cancer 2012, 118, 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng W.; Chen T.; Tan W.; Fan Z. H. Multivalent DNA Nanospheres for Enhanced Capture of Cancer Cells in Microfluidic Devices. ACS Nano 2013, 7, 7067–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine J.; Luo S.-C.; Wang S.; Zhu B.; Tseng H.-R.; Yu H.-H. Functionalized Conducting Polymer Nanodots for Enhanced Cell Capturing: The Synergistic Effect of Capture Agents and Nanostructures. Adv. Mater. 2011, 23, 4788–4792. [DOI] [PubMed] [Google Scholar]
- Han W.; Allio B. A.; Foster D. G.; King M. R. Nanoparticle Coatings for Enhanced Capture of Flowing Cells in Microtubes. ACS Nano 2009, 4, 174–180. [DOI] [PubMed] [Google Scholar]
- King M. R.; Western L. T.; Rana K.; Liesveld J. L. Biomolecular Surfaces for the Capture and Reprogramming of Circulating Tumor Cells. J. Bionic Eng. 2009, 6, 311–317. [Google Scholar]
- Liu Z.; Robinson J. T.; Sun X.; Dai H. PEGylated Nanographene Oxide for Delivery of Water-Insoluble Cancer Drugs. J. Am. Chem. Soc. 2008, 130, 10876–10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty N.; Berry V. Graphene-Based Single-Bacterium Resolution Biodevice and DNA Transistor: Interfacing Graphene Derivatives with Nanoscale and Microscale Biocomponents. Nano Lett 2008, 8, 4469–4476. [DOI] [PubMed] [Google Scholar]
- Lu C. H.; Yang H. H.; Zhu C. L.; Chen X.; Chen G. N.; Graphene A. Platform for Sensing Biomolecules. Angew. Chem., Int. Ed. 2009, 48, 4785–4787. [DOI] [PubMed] [Google Scholar]
- Stoller M. D.; Park S.; Zhu Y.; An J.; Ruoff R. S. Graphene-Based Ultracapacitors. Nano Lett. 2008, 8, 3498–3502. [DOI] [PubMed] [Google Scholar]
- Xu J.; Wang K.; Zu S.-Z.; Han B.-H.; Wei Z. Hierarchical Nanocomposites of Polyaniline Nanowire Arrays on Graphene Oxide Sheets with Synergistic Effect for Energy Storage. ACS Nano 2010, 4, 5019–5026. [DOI] [PubMed] [Google Scholar]
- Dikin D. A.; Stankovich S.; Zimney E. J.; Piner R. D.; Dommett G. H.; Evmenenko G.; Nguyen S. T.; Ruoff R. S. Preparation and Characterization of Graphene Oxide Paper. Nature 2007, 448, 457–460. [DOI] [PubMed] [Google Scholar]
- Park S.; Lee K. S.; Bozoklu G.; Cai W.; Nguyen S. T.; Ruoff R. S. Graphene Oxide Papers Modified by Divalent Ions-Enhancing Mechanical Properties via Chemical Cross-Linking. ACS Nano 2008, 2, 572–578. [DOI] [PubMed] [Google Scholar]
- Stankovich S.; Dikin D. A.; Dommett G. H.; Kohlhaas K. M.; Zimney E. J.; Stach E. A.; Piner R. D.; Nguyen S. T.; Ruoff R. S. Graphene-Based Composite Materials. Nature 2006, 442, 282–286. [DOI] [PubMed] [Google Scholar]
- Ramanathan T.; Abdala A. A.; Stankovich S.; Dikin D. A.; Herrera-Alonso M.; Piner R. D.; Adamson D. H.; Schniepp H. C.; Chen X.; Ruoff R. S.; et al. Functionalized Graphene Sheets for Polymer Nanocomposites. Nat. Nanotechnol. 2008, 3, 327–331. [DOI] [PubMed] [Google Scholar]
- Park S.; Ruoff R. S. Chemical Methods for the Production of Graphenes. Nat. Nanotechnol. 2009, 4, 217–224. [DOI] [PubMed] [Google Scholar]
- Stankovich S.; Dikin D. A.; Piner R. D.; Kohlhaas K. A.; Kleinhammes A.; Jia Y.; Wu Y.; Nguyen S. T.; Ruoff R. S. Synthesis of Graphene-Based Nanosheets via Chemical Reduction of Exfoliated Graphite Oxide. Carbon 2007, 45, 1558–1565. [Google Scholar]
- Dreyer D. R.; Park S.; Bielawski C. W.; Ruoff R. S. The Chemistry of Graphene Oxide. Chem. Soc. Rev. 2010, 39, 228–240. [DOI] [PubMed] [Google Scholar]
- Sun X.; Liu Z.; Welsher K.; Robinson J. T.; Goodwin A.; Zaric S.; Dai H. Nano-Graphene Oxide for Cellular Imaging and Drug Delivery. Nano Res. 2008, 1, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K. P.; Bao Q.; Eda G.; Chhowalla M. Graphene Oxide as a Chemically Tunable Platform for Optical Applications. Nat. Chem. 2010, 2, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Myung S.; Solanki A.; Kim C.; Park J.; Kim K. S.; Lee K.-B. Graphene-Encapsulated Nanoparticle-Based Biosensor for the Selective Detection of Cancer Biomarkers. Adv. Mater. 2011, 23, 2221–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Xue P.; Kang Y.; Hui K. M. Highly Specific and Ultrasensitive Graphene-Enhanced Electrochemical Detection of Low-Abundance Tumor Cells Using Silica Nanoparticles Coated with Antibody-Conjugated Quantum Dots. Anal. Chem. 2013, 85, 3166–3173. [DOI] [PubMed] [Google Scholar]
- Yu X.; He R.; Li S.; Cai B.; Zhao L.; Liao L.; Liu W.; Zeng Q.; Wang H.; Guo S.-S.; et al. Magneto-Controllable Capture and Release of Cancer Cells by Using a Micropillar Device Decorated with Graphite Oxide-Coated Magnetic Nanoparticles. Small 2013, 9, 3895–3901. [DOI] [PubMed] [Google Scholar]
- Yoon H. J.; Kim T. H.; Zhang Z.; Azizi E.; Pham T. M.; Paoletti C.; Lin J.; Ramnath N.; Wicha M. S.; Hayes D. F.; et al. Sensitive Capture of Circulating Tumour Cells by Functionalized Graphene Oxide Nanosheets. Nat. Nanotechnol. 2013, 8, 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z.; Barlow D. E.; Sheehan P. E. The Assembly of Single-Layer Graphene Oxide and Graphene Using Molecular Templates. Nano Lett. 2008, 8, 3141–3145. [DOI] [PubMed] [Google Scholar]
- Wang H.; Wang X.; Li X.; Dai H. Chemical Self-Assembly of Graphene Sheets. Nano Res. 2009, 2, 336–342. [Google Scholar]
- Chang Y.; Yang S.-T.; Liu J.-H.; Dong E.; Wang Y.; Cao A.; Liu Y.; Wang H. In Vitro Toxicity Evaluation of Graphene Oxide on A549 Cells. Toxicol. Lett. 2011, 200, 201–210. [DOI] [PubMed] [Google Scholar]
- Lee W. C.; Lim C. H. Y. X.; Shi H.; Tang L. A. L.; Wang Y.; Lim C. T.; Loh K. P. Origin of Enhanced Stem Cell Growth and Differentiation on Graphene and Graphene Oxide. ACS Nano 2011, 5, 7334–7341. [DOI] [PubMed] [Google Scholar]
- Kang J. H.; Krause S.; Tobin H.; Mammoto A.; Kanapathipillai M.; Ingber D. E. A Combined Micromagnetic-Microfluidic Device for Rapid Capture and Culture of Rare Circulating Tumor Cells. Lab Chip 2012, 12, 2175–2181. [DOI] [PubMed] [Google Scholar]