Abstract
Background
There is notable heterogeneity in the clinical presentation of patients with COPD. To characterize this heterogeneity, we sought to identify subgroups of smokers by applying cluster analysis to data from the COPDGene Study.
Methods
We applied a clustering method, k-means, to data from 10,192 smokers in the COPDGene Study. After splitting the sample into a training and validation set, we evaluated three sets of input features across a range of k (user-specified number of clusters). Stable solutions were tested for association with four COPD-related measures and five genetic variants previously associated with COPD at genome-wide significance. The results were confirmed in the validation set.
Findings
We identified four clusters that can be characterized as 1) relatively resistant smokers (i.e. no/mild obstruction and minimal emphysema despite heavy smoking), 2) mild upper zone emphysema predominant, 3) airway disease predominant, and 4) severe emphysema. All clusters are strongly associated with COPD-related clinical characteristics, including exacerbations and dyspnea (p<0.001). We found strong genetic associations between the mild upper zone emphysema group and rs1980057 near HHIP, and between the severe emphysema group and rs8034191 in the chromosome 15q region (p<0.001). All significant associations were replicated at p<0.05 in the validation sample (12/12 associations with clinical measures and 2/2 genetic associations).
Interpretation
Cluster analysis identifies four subgroups of smokers that show robust associations with clinical characteristics of COPD and known COPD-associated genetic variants.
Background
The clinical presentation of chronic obstructive pulmonary disease (COPD) is heterogeneous. Smoking-related damage manifests as airway wall thickening, loss of small airways, emphysematous lung destruction, and a range of extrapulmonary manifestations. However, these specific manifestations may vary in individual smokers. COPD heterogeneity has been broadly characterized as emphysema-predominant and airway predominant disease, 1;2 and the varying amounts of airway obstruction and emphysema present in an individual can be described with quantitative computed tomography (CT) measures. In addition to the emphysema-airway characterization, additional subtypes have been proposed in an effort to further refine our understanding of smoking-related lung damage. Some of these, such as upper lobe predominant emphysema and the “frequent-exacerbator” subtype, have important consequences for clinical management.3–5
The most widely accepted current definition of COPD is that of the Global Initiative for Chronic Obstructive Lung Disease (GOLD 2007).6 Based primarily on spirometry, GOLD 2007 confirms the diagnosis of COPD based on FEV1/FVC and classifies disease severity based on FEV1. This simplicity has arguably led to improved recognition, diagnosis and treatment of the disease.6;7 However, the GOLD 2007 criteria do not fully describe the heterogeneity of COPD,8;9 and the most recent GOLD 2011 criteria add clinical characteristics to define new classes.10 GOLD provides clear cutoffs to define presence/absence of COPD based on FEV1 and FEV1/FVC; however spirometric measures, as well as associated CT scan characteristics such as emphysema have a continuous distribution in the population, indicating that the smoking-related damage characteristic of COPD is likely a continuous process that can also be present in subjects who have not yet developed airflow obstruction meeting standard criteria.
One rationale for the simplicity of the GOLD 2007 criteria is that there is substantial overlap between different disease characteristics and among proposed subtypes. It is a challenge to synthesize the various smoking-related subtypes proposed in the literature, because subtypes may overlap or be defined in ways that are not complementary. In an effort to derive data-driven COPD classifications, investigators have recently employed unsupervised machine learning approaches.11–13 The benefit of such approaches is that they employ quantitative methods to define subtypes, but the challenge in applying these approaches for clinical subtype identification is that they are designed primarily for data exploration rather than specific hypothesis testing. As a result, the generalizability and reproducibility of machine-learned COPD subtype classifications in independent data samples has been largely unexplored.
We hypothesized that k-means, a widely used unsupervised clustering method, would identify novel, clinically relevant subtypes when applied to quantitative chest computed tomography (CT), spirometric, and clinical measures from the COPDGene study. The COPDGene study is a large epidemiologic and genetic study of over 10,000 current and former smokers with and without COPD that includes demographic and clinical information, spirometry, genome-wide SNP genotyping data, and inspiratory and expiratory CT scans. We specified a priori a set of clinically relevant clinical and genetic variables that would be used only to evaluate and interpret (but not to generate) clusters, and we split our data into a training and validation set to provide rigorous assessment of the reproducibility of our results.
Results
The characteristics of the training and validation samples are shown in Table 1, and the samples are comparable. The difference in sample size between the training and validation samples is due to differences in missing data (see Supplement).
Table 1.
Baseline Characteristics of the Training and Validation Data
| Training | Validation | |
|---|---|---|
| N | 4187 | 4101 |
| Age | 59.5 (9.0) | 59.7 (9.0) |
| Gender, % female | 46.7 | 45.9 |
| Race, % African-American | 32.0 | 31.4 |
| FEV1, % of predicted | 76.9 (25.2) | 77.1 (25.2) |
| FEV1/FVC | 0.67 (0.16) | 0.67 (0.16) |
| Pack-Years, median (IQR) | 39.3 (28.0) | 39.7 (27.0) |
| BMI | 28.9 (6.3) | 28.9 (6.1) |
| Emphysema at −950HU, median (IQR) | 1.8 (5.8) | 2.0 (6.1) |
| Upper/Lower Emphysema Ratio (IQR) | 0.8 (1.1) | 0.8(1.2) |
| Segmental Airway Wall Thickness | 61.4 (3.2) | 61.4 (3.3) |
| Upper/Lower Lobe Emphysema Difference (IQR) | −0.17 (2.0) | −0.14 (2.2) |
| Gas Trapping (IQR) | 14.5 (24.8) | 14.7 (25.3) |
| GOLD Unclassifiable*, % | 12.0 | 12.6 |
| Smoking controls, % | 43.8 | 43.8 |
| GOLD 1, % | 8.3 | 7.7 |
| GOLD 2, % | 19.2 | 19.4 |
| GOLD 3, % | 11.3 | 11.3 |
| GOLD 4, % | 5.4 | 5.3 |
Values are mean (SD) unless otherwise noted.
Smoking Intensity – average cigarettes smoked per day
GOLD unclassifiable refers to subjects with a FEV1% predicted <80 but FEV1/FVC > 0.7.
Defining Feature Subsets
Factor analysis on the comprehensive feature set identified four factors that individually accounted for at least 5% of the variance in the data. Features with the top loadings for these factors were functional residual capacity (FRC) % predicted, FEV1 % predicted, CT-quantified emphysema at −950 Hounsfield units (HU), and bronchodilator responsiveness as a % of FEV1. For the core feature set, correlation filtering yielded a set of four features - FEV1 % predicted, CT-quantified emphysema, segmental wall area %, and emphysema distribution (log ratio of upper third/lower third emphysema).
Prioritizing Clustering Solutions by Cluster Stability
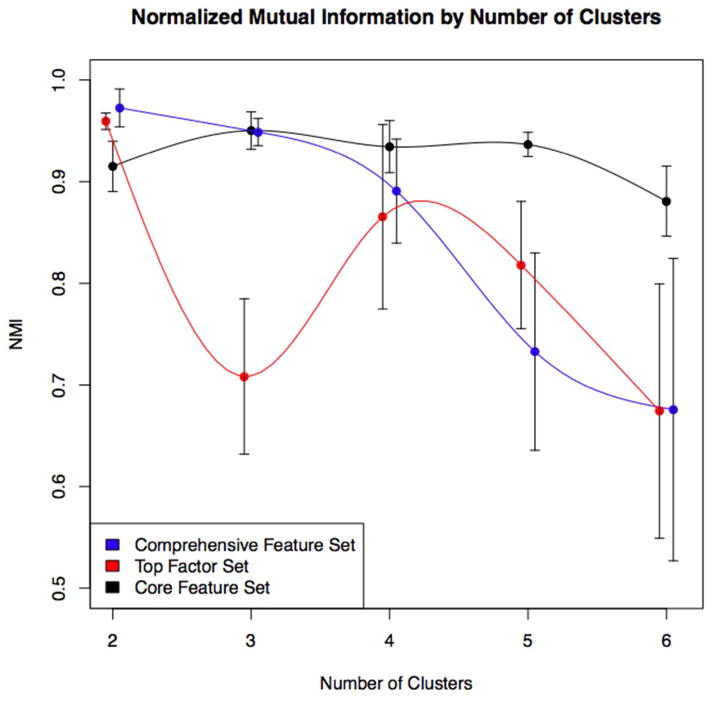
Cluster stability for the three feature sets is shown in Figure 1. Seven stable clustering solutions with NMI > 0.9 were prioritized for further evaluation. We examined the clinical and genetic associations of these seven solutions in the training sample. For the comprehensive and top factor feature sets, the highest stability results were for k=2. These solutions largely replicated the traditional COPD case-control distinction and were likely driven by the case-control design and recruitment strategy of COPDGene.
Figure 1.
Cluster stability as measured by average normalized mutual information (NMI) by number of clusters across the three input feature sets. High NMI values indicate high cluster stability. For the comprehensive and top factor feature sets, stability is greatest for the k=2 solution. For the core feature set very high stability is observed up to k=5. Dots and standard errors bars represent average NMI and standard errors over five-fold cross validation, respectively. Dots are slightly offset to improve visualization.
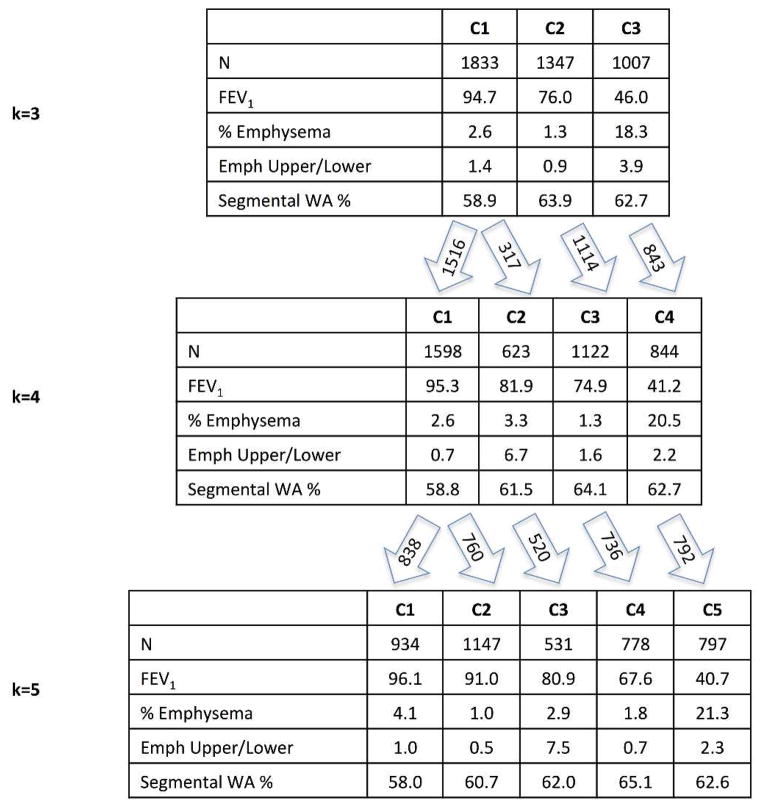
For the core feature set, highly stable clustering was observed for a range of k from 2 to 5. Figure 2 shows the characteristics of the clustering features for the k=3 to k=5 solutions and the pattern in which clusters emerge as k increases. Based on the strong pattern of cluster-specific clinical and genetic associations, the k=4 core feature (CF4) solution was selected for further validation.
Figure 2.
Average values of clustering features from core feature set solutions k=3 through 5. Arrows indicate relationships between these k-means derived clusters that share large numbers of individuals.
Cluster Characteristics
Cluster characteristics for the CF4 solution are shown in Table 2. The four clusters can be characterized as low susceptibility smokers, mild upper zone emphysema predominant, airway predominant, and severe emphysema.
Table 2.
Cluster Characteristics in Training and Validation Data for Core Feature Set Cluster Solution, k=4
| Training Sample | Validation Sample | |||||||
|---|---|---|---|---|---|---|---|---|
| C1:Mean | C2:Mean | C3:Mean | C4:Mean | C1:Mean | C2:Mean | C3:Mean | C4:Mean | |
| N | 1598 | 623 | 1122 | 844 | 1595 | 620 | 1060 | 826 |
| Age | 58.9 | 58.0* | 56.8 | 65.4 | 58.7 | 58.9* | 57.3 | 65.4 |
| Gender, % female | 0.44 | 0.53 | 0.52 | 0.40 | 0.43 | 0.51 | 0.53 | 0.40 |
| Race, % African-American | 0.30 | 0.46 | 0.37 | 0.19 | 0.29 | 0.45 | 0.37 | 0.17 |
| FEV1, percent of predicted | 95.3 | 81.9 | 74.9 | 41.2 | 95.7 | 81.6 | 73.8 | 42.0 |
| FEV1/FVC | 0.76 | 0.70 | 0.71 | 0.42 | 0.76 | 0.69 | 0.71 | 0.42 |
| BMI | 28.7 | 27.9 | 31.4* | 26.7 | 28.3 | 27.6 | 32.0* | 26.8 |
| Pack Years | 38.0 | 45.8 | 42.8 | 56.8 | 38.3 | 46.9 | 43.1 | 55.9 |
| Emphysema at −950HU | 2.6 | 3.3 | 1.3 | 20.5 | 2.7 | 3.6 | 1.4 | 20.7 |
| Segmental Airway Wall Thickness | 58.8 | 61.5 | 64.1 | 62.7 | 58.8 | 61.4 | 64.2 | 63.0 |
| Upper/Lower Emphysema Ratio | 0.7 | 6.7 | 0.6 | 2.2 | 0.7 | 8.3 | 0.6 | 2.3 |
| Upper/Lower Emphysema Difference | −0.3 | 1.4 | −0.3 | 2.6 | −0.3 | 1.7 | −0.3 | 2.9 |
| Gas Trapping+ | 12.9 | 16.5 | 13.4 | 52.1 | 13.1 | 17.3 | 13.3 | 52.7 |
Values represent the mean of each variable for each cluster unless otherwise specified.
Only the variables shown in bold were used as input variables for the primary clustering solution (CF4).
C1 = relatively resistant smokers, C2 = mild upper zone predominant emphysema, C3 = airway predominant, C4 = severe emphysema
p-value comparing mean in training to validation <0.05 for t-test
%LAA using −856 Hounsfield unit threshold on expiratory CT scan
Cluster 1 – Relatively Resistant Smokers
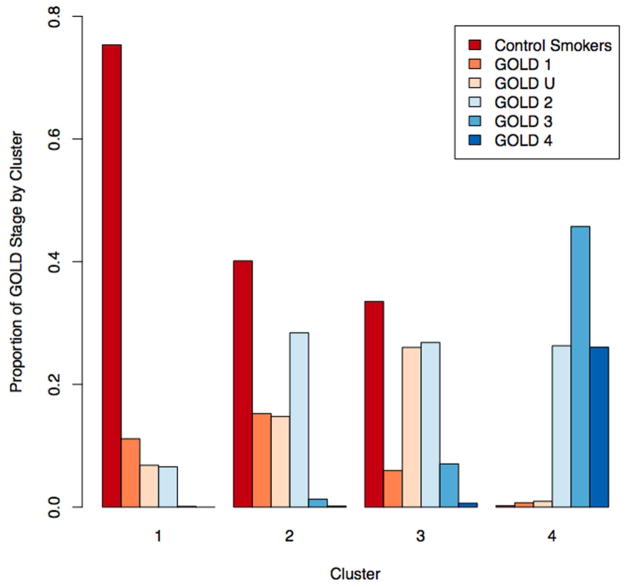
Cluster 1 represents 38% of the COPDGene training sample and is characterized by heavy smoking exposure with no or minimal airflow obstruction, as well as lower emphysema (p<0.001 for comparison with Clusters 2 and 4) and airway wall thickness (p<0.001 for all cluster comparisons) compared to the more severely affected clusters. The majority of individuals in the relatively resistant cluster are control smokers or GOLD Stage 1 (Figure 3).
Figure 3.
Proportion of individuals in each GOLD 2007 stage by Core Feature Set Clustering Solution (k=4). Cluster 1 (relatively smoking resistant individuals) consists largely of control smokers and GOLD 1–2 individuals. Cluster 4 (severe emphysema) consists largely of GOLD 2–4 individuals. Clusters 2 and 3 (upper zone emphysema and airway predominant) consist largely of control smokers, GOLD 1–2 and GOLD unclassifiable (GOLD U) individuals.
Cluster 2 – Mild Upper Zone Predominant Emphysema
Cluster 2 represents 15% of the training sample and is characterized by mild airflow obstruction and mild emphysema with marked upper zone predominance (p-values compared to other clusters <0.001). The average amount of emphysema in this group is modest (mean emphysema=3.31%), though the range is broad and nearly a quarter of this cluster has greater than 5% emphysema. As is shown in Figure 3, most of the individuals in the mild upper zone emphysema cluster are control smokers or GOLD Stages 1–2, with 15% unclassifiable by GOLD criteria.
Compared to the relatively resistant cluster, this cluster was more likely to experience an exacerbation, have a higher MMRC dyspnea score and BODE index, and more likely to have used the emergency room or been admitted to the hospital for a respiratory issue (Table 3). The NHW subjects in this group show a strong genetic association with rs1980057 near the HHIP gene (p=4.4×10−6). This cluster has a higher proportion of African-Americans than the airway predominant and severe emphysema clusters (p <0.001) and a higher proportion of women compared to the relatively smoking resistant and severe emphysema clusters (p <0.001).
Table 3.
Cluster Associations with COPD-Related Measures and COPD SNPs in Training and Validation Data for Core Feature Set Cluster Solution, k=4
| Training | Validation | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| C2:OR (CI) | C3:OR (CI) | C4:OR (CI) | C2:OR (CI) | C3:OR (CI) | C4:OR (CI) | |
|
| ||||||
| Exacerbations | 2.27 (1.97–2.61)*** | 3.16 (2.82–3.55)*** | 8.93 (7.97–10.01)*** | 2.17 (1.90–2.49)*** | 2.66 (2.38–2.98)*** | 7.80 (7.00–8.70)*** |
|
|
||||||
| MMRC | 2.81 (2.57–3.07)*** | 3.39 (3.14–3.66)*** | 10.88 (10.00–11.83)*** | 2.02 (2.01–2.40)*** | 3.00 (2.78–3.23)*** | 10.07 (9.26–10.94)*** |
|
|
||||||
| BODE | 3.37 (3.06–3.70)*** | 4.63 (4.27–5.02)*** | 66.52 (60.06–73.67)*** | 2.62 (2.38–2.88)*** | 4.23 (3.90–4.58)*** | 52.64 (47.62–58.19)*** |
|
|
||||||
| Hospitalizations/ER Visits | 4.07 (3.34–4.95)*** | 5.05 (4.24–6.01)*** | 11.82 (9.98–14.00)*** | 3.05 (2.53–3.68)*** | 4.13 (3.52–4.86)*** | 8.03 (6.86–9.39)*** |
|
| ||||||
| rs7671167 (FAM13A) | 0.95 (0.87–1.04)NS | 0.87 (0.81–0.93)* | 0.84 (0.78–0.91)* | 1.01 (0.92–1.10)NS | 0.89 (0.83–0.95)NS | 0.91 (0.85–0.98)NS |
|
|
||||||
| rs1980057 (HHIP) | 0.64 (0.58–0.70)*** | 0.92 (0.85–0.98)NS | 0.79 (0.73–0.85)*** | 0.80 (0.73–0.87)* | 1.09 (1.01–1.17)NS | 0.74 (0.69–0.80)*** |
|
|
||||||
| rs13180 (Chr15q25) | 0.82 (0.75–0.90)* | 1.04 (0.96–1.11)NS | 0.82 (0.76–0.88)** | 0.72 (0.66–0.79)*** | 0.99 (0.92–1.07)NS | 0.82 (0.76–0.88)** |
|
|
||||||
| rs8034191 (Chr15q25) | 1.33 (1.21–1.46)** | 1.03 (0.96–1.11)NS | 1.50 (1.39–1.61)*** | 1.30 (1.19–1.43)** | 0.89 (0.83–0.96)NS | 1.17 (1.09–1.26)* |
|
|
||||||
| rs7937 (Chr 19q13) | 1.30 (1.18–1.42)** | 1.16 (1.08–1.24)* | 1.20 (1.12–1.29)* | 1.08 (0.99–1.18)NS | 1.06 (0.99–1.14)NS | 1.46 (1.36–1.57)*** |
OR = odds ratio. Effect sizes represent odds ratio from logistic regression or proportional odds logistic regression in the case of Exacerbations, MMRC Score, and BODE index.
CI = 95% confidence interval.
In all instances, cluster 1 (i.e. the cluster with the highest mean FEV1 % of predicted) serves as the reference.
Effect Allele for rs7671167 = C, rs1980057 = T, rs13180 = C, rs8034191 = C, rs7937 = T.
0.01<p<=0.05,
0.001<p<=0.01,
p<=0.001, NS p>0.05
Cluster 3 – Airway Predominant Disease
Cluster 3 represents 27% of the training sample and is characterized by thicker airway walls, the lowest average emphysema of all clusters, and high BMI (p <0.001 for all measures). The overall distribution of GOLD 2007 stages in this group is similar to the mild upper zone emphysema cluster, with the exception of a higher proportion of GOLD Stage 3 and unclassifiable individuals (Figure 3).
This cluster is more likely than the relatively smoking resistant cluster to report COPD exacerbations and lung-related healthcare utilization, and they have higher MMRC score and BODE index (Table 3). It has a significantly higher proportion of women than the smoking resistant and severe emphysema clusters (p <0.001), and the overall strength of genetic associations between this cluster and COPD SNPs is weak.
Cluster 4 – Severe Emphysema
Cluster 4 represents 20% of the sample and is characterized by high emphysema, gas trapping and severe airflow obstruction (p <0.001 for all measures). This group consists primarily of GOLD 2–4 individuals. It has the lowest BMI, highest lifetime pack- years exposure, oldest average age (p <0.001 for all measures), and it is the most severely affected cluster in terms of COPD-related measures. The effect sizes of the associations between the severe emphysema cluster and the four COPD-related clinical variables are roughly twice as large as those observed for the upper zone emphysema and airway predominant clusters.
This cluster is strongly associated with rs1980057 (p=0.001) near HHIP and rs8034191 (p=5×10−8) in the Chromosome 15q locus that includes the nicotinic receptor genes CHRNA3 and CHRNA5 as well as IREB2 (Table 3). It has a significantly higher proportion of NHWs than all other clusters and a higher proportion of male subjects than the mild upper zone emphysema and airway predominant clusters (p values <0.001).
Validation of the CF4 Clustering Solution
To validate the CF4 clustering solution, we examined the characteristics and associations of CF4 clusters in the validation data sample. The characteristics of the CF4 clusters in the training and validation samples were similar (Table 2), demonstrating that the clusters can reliably be reproduced in a separate data sample.
The associations in the training and validation sample between CF4 clusters, COPD-related clinical measures and COPD SNPs are shown in Table 3. For the clinical variables, all 12 of the associations are highly significant in training and validation. For the genetic risk factors, the two associations in the training sample with p-values below the Bonferroni-determined threshold of p=0.0007 were both replicated at p<=0.05 in the validation sample. Furthermore, of the 11 genetic associations observed with p<=0.05 in the training sample, 7 were replicated at p<=0.05 in validation.
Robustness of CF4 Clusters After Adjustment for GOLD Stage
To determine whether the associations observed with these clusters and COPD-related clinical and genetic variables were driven by severity of airflow obstruction, we repeated the cluster association tests adjusting for GOLD 2007 stage and GOLD 2011 classes A–D (Supplemental Tables 2 and 3). All of the associations with clinical measures remained significant (p<=0.001). This suggests that the discovered clusters provide information independent from COPD severity as defined by GOLD.
In regard to genetic associations, the cluster associations showed divergent behavior in response to adjustment for GOLD 2007 stage and GOLD A–D classes. The genetic associations with cluster 4 were attenuated, whereas the strong association observed between cluster 2 (upper zone emphysema) and rs1980057 near HHIP was unaffected, suggesting that this association is due to properties of this cluster that are distinct from disease severity as assessed by severity of airflow obstruction.
Discussion
Using a large sample of smokers with a wide range of airflow obstruction and well-characterized with respect to COPD features, cluster analysis identified solutions demonstrating strong association with clinically relevant COPD-related measures and high repeatability in cross-validation. A filtered subset of input features yielded a four cluster result that is informative beyond the traditional COPD case-control distinction. These clusters can be described as: 1) relatively smoking resistant individuals, 2) individuals with mild upper zone predominant emphysema and airflow obstruction, 3) individuals with airway predominant disease, and 4) individuals with severe obstruction and emphysema. In addition to being relevant clinically, some of these clusters are strongly associated with known COPD-associated variants. These clusters and associations were validated in a second data sample from the same study population.
This analysis presents novel findings about smoking-related pulmonary subtypes. We describe a mild upper zone emphysema predominant cluster that has not been extensively described in previous studies, and we demonstrate that membership in this cluster is associated with a genetic variant in the HHIP gene. This cluster was identified in our study population for at least three reasons: first, our study population included CT scans from a range of smokers, including those with mild or no obstruction; second, we included emphysema distribution as an input feature for clustering; and third, our sample size is substantially larger than previously reported COPD cluster analysis studies. Our work also adds to the field by explicitly addressing the reproducibility of cluster analyses and by using intrinsic (i.e. cluster stability) and extrinsic (i.e. clinical and genetic associations) criteria for assessing multiple potential clustering solutions.
These results confirm some of the findings from previous subtyping efforts in COPD. First, most studies have identified a severely affected group, though the severity of emphysema and airway wall thickness in this group has been variable.12;21–23 Second, these findings affirm the concept of emphysema-predominant and airway-predominant COPD while providing additional insight regarding the role of emphysema distribution in COPD heterogeneity.2;5;13;21;22;24;25 The identification of emphysema and airway predominant groups, however, has not been universal. Garcia-Aymerich et al did not identify an airway predominant group, and instead identified a group with elevated BMI and increased comorbidities but with less prominent airway wall thickness on CT scan.12 In our study, the high average BMI and overrepresentation of women in the airway predominant group is of clinical and epidemiologic interest, and the female airway predominance recapitulates observations by Martinez et al in NETT.26
We examined the association of clusters with known COPD GWAS SNPs. While the directionality of associations varied between clusters for some SNPs, the analyzed SNPs did show a consistent direction of effect compared with the previous COPD susceptibility association literature in the comparison of the relatively smoking resistant cluster to the severe obstruction/emphysema cluster. The weak associations in our airway-predominant group are consistent with the findings in the ECLIPSE cohort, where no associations were identified with Pi10.27 In contrast, consistent associations with the HHIP and 15q loci were found for both the severe and mild upper lobe predominant emphysema group. This association in the latter group is particularly notable since the airway predominant group, with similar average lung function to the upper lobe predominant group, shows no strong genetic associations. These results are congruent with ECLIPSE where the associations of these loci with radiologist-scored emphysema were stronger than that for FAM13A.27 Together, these findings suggest that genetic associations in COPD may be subtype dependent.
This work has some limitations. It focuses primarily on continuous spirometric and quantitative CT measures; however, other aspects of COPD such as biomarker measurements and comorbidities were not included due either to their absence from our data or limitations of the k-means clustering method, which can yield spurious results when applied to a mixture of continuous and categorical variables. In the future, approaches that evaluate a range of clustering methods and a wider set of variables will be of interest. However, as this work demonstrates, the inclusion of more input features does not necessarily yield better clustering results. The optimal selection of features for clustering (i.e. feature selection), is a critical area for the application of unsupervised learning to disease subtyping that requires further exploration. This analysis is cross- sectional, and it is possible that these results may be confounded by differences in disease severity. This is an important limitation for all clustering efforts using cross-sectional data that could be addressed through analyses of longitudinal data or through the development of novel clustering methods. A number of subjects from the overall study were excluded from the clustering analysis due to missing data, primarily from CT scan-related variables, and there is some bias in the clustering subset compared to the excluded subjects. This limits the generalizability of the sample on which clustering was performed, though the included sample is large and consists of a broad spectrum of smoking-related disease.
In summary, k-means clustering in the COPDGene Study identifies four groups of smokers that are associated with important COPD-related measures even after adjustment for GOLD stage. Genetic association analysis with known COPD-associated variants shows strong, cluster-specific associations with these known genetic risk factors. This clustering approach is reproducible in independent data sets, facilitating the further study and characterization of these groups of smokers.
Methods
Data Collection
Quantitative measures of emphysema and airway wall thickness were generated with SLICER (http://www.slicer.org) and VIDA software (VIDA Diagnostics, Iowa City, IA; http://www.vidadiagnostics.com), respectively.(1) Dyspnea and lung disease-specific quality of life measures were obtained through the use of previously validated questionnaire items.(2;3)
Cross-Validation Estimates of Cluster Stability
To assess the stability of various cluster solutions, we used five-fold cross validation to derive estimates of cluster stability as quantified by the average normalized mutual information (NMI). Normalized mutual information quantifies the dependency between variables, and it ranges from 0 (no dependency) to 1 (high dependency). Unlike Pearson correlation, NMI captures nonlinear in addition to linear dependency between variables. This procedure was carried out entirely in the training portion of the data. Four-fifths of the training sample served as the cross-validation training set (CV Train) and the remaining one-fifth of the data served as cross-validation test set (CV Test). Using the learned centroids from the CV Train set, clusters were predicted in the CV Test set and then compared to the cluster results for that fold obtained by running k-means on the entire (original) training sample. NMI quantified the degree of agreement, and the average NMI results obtained from each of the five rounds of cross-validation were used to prioritize cluster solutions by stability.
Genetic Association Testing
Genetic associations were performed in non-Hispanic white (NHW) subjects only using additive genetic coding and adjusted for principal components of genetic ancestry. A Bonferonni-adjusted statistical significance of p=0.0007 for genetic associations in the training set was defined based on 70 genetic association tests performed. The threshold for validation in the independent sample was p=0.05.
Missing Data
We employed a complete cases approach and excluded individuals from analysis who were missing data in any of the variables used for clustering, cluster association testing or interpretation. There was no difference in age of pack-years between included and excluded subjects (Supplemental Table 8). There was statistically significant but relatively minor differences in FEV1 and FEV1/FVC, and there were significant differences in gender and racial composition. Subjects with missing data were more likely to be female and African-American. Of the 10,300 individuals enrolled in COPDGene, 108 non-smokers were excluded from analysis, as well as 63 individuals with inadequate spirometry data. Of the remaining 10,129 individuals, 511 did not receive an inspiratory or expiratory scan. An additional 953 subjects failed quality control for either the inspiratory or expiratory scan, and 64 subjects were excluded for an FRC/TLC ratio >1. Of the remaining 8,601 subjects, 143 had incomplete data for emphysema distribution. An additional 170 individuals were excluded due to missing data for the following variables: airway wall thickness (n=4), gas trapping (n=44), resting oxygen saturation (n=2), MMRC dyspnea score (n=11), and BODE (n=109).
Supplementary Material
Key Messages.
What is the key question?
Can distinct subtypes of pulmonary damage be identified in smokers?
What is the bottom line?
Cluster analysis in the COPDGene study identifies four clusters of smokers with distinct patterns of airway wall thickness, emphysema and emphysema distribution, and these subtypes show strong association with relevant clinical measures and known COPD-associated genetic variants.
Why read on?
This paper demonstrates robust, data-driven clustering results that identify clinically important subgroups of smokers in the largest COPD subtyping study to date.
Acknowledgments
Funding
This work was supported by U.S. National Institutes of Health (NIH) grants K08HL102265 (Castaldi), K08HL097029 (Cho), P01HL105339 (Silverman), and by Award Numbers R01HL089897 (Crapo) and R01HL089856 (Silverman) from the National Heart, Lung, And Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, And Blood Institute or the National Institutes of Health. The COPDGene® project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens and Sunovion.
COPDGene® Investigators – Core Units
Administrative Core: James Crapo, MD (PI), Edwin Silverman, MD, PhD (PI), Barry Make, MD, Elizabeth Regan, MD, PhD, Sarah Moyle, MS, Amy Willis, MA, Rochelle Lantz, Lori Stepp, Sandra Melanson, Douglas Stinson
Genetic Analysis Core: Terri Beaty, PhD, Barbara Klanderman, PhD, Nan Laird, PhD, Christoph Lange, PhD, Michael Cho, MD, Stephanie Santorico, PhD, John Hokanson, MPH, PhD, Dawn DeMeo, MD, MPH, Nadia Hansel, MD, MPH, Craig Hersh, MD, MPH, Peter Castaldi, MD, MSc, Jacqueline Hetmanski, MS, Margaret Parker, MS, Tanda Murray, MS
Imaging Core: David Lynch, MB, Joyce Schroeder, MD, John Newell, Jr., MD, John Reilly, MD, Harvey Coxson, PhD, Philip Judy, PhD, Eric Hoffman, PhD, George Washko, MD, Raul San Jose Estepar, PhD, James Ross, MSc, Ho Yun Lee, MD, Joon Beom Seo, MD, PhD, Atsushi Nambu, MD, PhD, Gongyoung Jin, MD, PhD, Song Soo Kim, MD, Mustafa Al Qaisi, MD, Rebecca Leek, Jordan Zach, Alex Kluiber, Jered Sieren, Heather Baumhauer, Verity McArthur, Demitry Kazlouski, Andrew Allen, Tanya Mann, Anastasia Rodionova, Deanna Richert, Joshua Jaramillo, Alexander McKenzie, Thomas Gethin-Jones, Jaleh Akhavan, Douglas Stinson
PFT QA Core, LDS Hospital, Salt Lake City, UT: Robert Jensen, PhD
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, PhD, Stacey Meyerer, Shivam Chandan, Samantha Bragan
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, USA: Douglas Everett, PhD, Andre Williams, PhD, Carla Wilson, MS, Anna Forssen, MS, Amber Powell, Joe Piccoli
Epidemiology Core, University of Colorado School of Public Health, Denver, USA: John Hokanson, MPH, PhD, Marci Sontag, PhD, Jennifer Black-Shinn, MPH, Gregory Kinney, MPH, PhDc, Sharon Lutz, MPH, PhD
COPDGene® Investigators – Clinical Centers
Ann Arbor VA: Jeffrey Curtis, MD, Ella Kazerooni, MD
Baylor College of Medicine, Houston, TX: Nicola Hanania, MD, MS, Philip Alapat, MD, Venkata Bandi, MD, Kalpalatha Guntupalli, MD, Elizabeth Guy, MD, Antara Mallampalli, MD, Charles Trinh, MD, Mustafa Atik, MD, Hasan Al-Azzawi, MD, Marc Willis, DO, Susan Pinero, MD, Linda Fahr, MD, Arun Nachiappan, MD, Collin Bray, MD, L. Alexander Frigini, MD, Carlos Farinas, MD, David Katz, MD, Jose Freytes, MD, Anne Marie Marciel, MD
Brigham and Women’s Hospital, Boston, MA: Dawn DeMeo, MD, MPH, Craig Hersh, MD, MPH, George Washko, MD, Francine Jacobson, MD, MPH, Hiroto Hatabu, MD, PhD, Peter Clarke, MD, Ritu Gill, MD, Andetta Hunsaker, MD, Beatrice Trotman-Dickenson, MBBS, Rachna Madan, MD
Columbia University, New York, NY: R. Graham Barr, MD, DrPH, Byron Thomashow, MD, John Austin, MD, Belinda D’Souza, MD
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., MD, Lacey Washington, MD, H Page McAdams, MD
Fallon Clinic, Worcester, MA: Richard Rosiello, MD, Timothy Bresnahan, MD, Joseph Bradley, MD, Sharon Kuong, MD, Steven Meller, MD, Suzanne Roland, MD
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, MD, MPH, Joseph Tashjian, MD
Johns Hopkins University, Baltimore, MD: Robert Wise, MD, Nadia Hansel, MD, MPH, Robert Brown, MD, Gregory Diette, MD, Karen Horton, MD
Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Los Angeles, CA: Richard Casaburi, MD, PhD, Janos Porszasz, MD, PhD, Hans Fischer, MD, PhD, Matt Budoff, MD, Mehdi Rambod, MD
Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, MD, Charles Trinh, MD, Hirani Kamal, MD, Roham Darvishi, MD, Marc Willis, DO, Susan Pinero, MD, Linda Fahr, MD, Arun Nachiappan, MD, Collin Bray, MD, L. Alexander Frigini, MD, Carlos Farinas, MD, David Katz, MD, Jose Freytes, MD, Anne Marie Marciel, MD
Minneapolis VA: Dennis Niewoehner, MD, Quentin Anderson, MD, Kathryn Rice, MD, Audrey Caine, MD
Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, MD, MS, Gloria Westney, MD, MS, Eugene Berkowitz, MD, PhD
National Jewish Health, Denver, USA: Russell Bowler, MD, PhD, Adam Friedlander, MD, David Lynch, MB, Joyce Schroeder, MD, John Newell, Jr., MD, Valerie Hale, MD, John Armstrong, II, MD, Debra Dyer, MD, Jonathan Chung, MD, Christian Cox, MD, Hakan Sahin, MD
Temple University, Philadelphia, PA: Gerard Criner, MD, Victor Kim, MD, Nathaniel Marchetti, DO, Aditi Satti, MD, A. James Mamary, MD, Robert Steiner, MD, Chandra Dass, MD, Libby Cone, MD
University of Alabama, Birmingham, AL: William Bailey, MD, Mark Dransfield, MD, Michael Wells, MD, Surya Bhatt, MD, Hrudaya Nath, MD, Satinder Singh, MD
University of California, San Diego, CA: Joe Ramsdell, MD, Paul Friedman, MD
University of Iowa, Iowa City, IA: Geoffrey McLennan, MD, PhD, Edwin JR van Beek, MD, PhD, Brad Thompson, MD, Dwight Look, MD, Alejandro Cornellas, MD
University of Michigan, Ann Arbor, MI: Fernando Martinez, MD, MeiLan Han, MD, Ella Kazerooni, MD
University of Minnesota, Minneapolis, MN: Christine Wendt, MD, Tadashi Allen, MD
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, MD, Joel Weissfeld, MD, MPH, Carl Fuhrman, MD, Jessica Bon, MD, Danielle Hooper, MD
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, MD, Sandra Adams, MD, Carlos Orozco, MD, Mario Ruiz, MD, Amy Mumbower, MD, Ariel Kruger, MD, Carlos Restrepo, MD, Michael Lane, MD
Reference List
- 1.BURROWS B, NIDEN AH, FLETCHER CM, JONES NL. CLINICAL TYPES OF CHRONIC OBSTRUCTIVE LUNG DISEASE IN LONDON AND IN CHICAGO. A STUDY OF ONE HUNDRED PATIENTS. Am Rev Respir Dis. 1964;90:14–27. doi: 10.1164/arrd.1964.90.1.14. [DOI] [PubMed] [Google Scholar]
- 2.BURROWS B, FLETCHER CM, Heard BE, JONES NL, Wootliff JS. The emphysematous and bronchial types of chronic airways obstruction. A clinicopathological study of patients in London and Chicago. Lancet. 1966;1(7442):830–835. doi: 10.1016/s0140-6736(66)90181-4. [DOI] [PubMed] [Google Scholar]
- 3.Hurst JR, Vestbo J, Anzueto A, Locantore N, Mullerova H, Tal-Singer R, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med. 2010;363(12):1128–1138. doi: 10.1056/NEJMoa0909883. [DOI] [PubMed] [Google Scholar]
- 4.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, et al. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348(21):2059–2073. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 5.Ziegler-Heitbrock L, Frankenberger M, Heimbeck I, Burggraf D, Wjst M, Haussinger K, et al. The EvA study: aims and strategy. Eur Respir J. 2012;40(4):823–829. doi: 10.1183/09031936.00142811. [DOI] [PubMed] [Google Scholar]
- 6.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176(6):532– 555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 7.Calverley PM. The GOLD classification has advanced understanding of COPD. Am J Respir Crit Care Med. 2004;170(3):211–212. doi: 10.1164/rccm.2405008. [DOI] [PubMed] [Google Scholar]
- 8.Agusti A, Calverley PM, Celli B, Coxson HO, Edwards LD, Lomas DA, et al. Characterisation of COPD heterogeneity in the ECLIPSE cohort. Respir Res. 2010;11:122. doi: 10.1186/1465-9921-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rennard SI, Vestbo J. The many “small COPDs”: COPD should be an orphan disease. Chest. 2008;134(3):623–627. doi: 10.1378/chest.07-3059. [DOI] [PubMed] [Google Scholar]
- 10.Vestbo J, Hurd SS, Agusti AG, Jones PW, Vogelmeier C, Anzueto A, et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease: GOLD Executive Summary. Am J Respir Crit Care Med. 2013;187(4):347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 11.Cho MH, Washko GR, Hoffmann TJ, Criner GJ, Hoffman EA, Martinez FJ, et al. Cluster analysis in severe emphysema subjects using phenotype and genotype data: an exploratory investigation. Respir Res. 2010;11:30. doi: 10.1186/1465-9921-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Aymerich J, Gomez FP, Benet M, Farrero E, Basagana X, Gayete A, et al. Identification and prospective validation of clinically relevant chronic obstructive pulmonary disease (COPD) subtypes. Thorax. 2011;66(5):430–437. doi: 10.1136/thx.2010.154484. [DOI] [PubMed] [Google Scholar]
- 13.Paoletti M, Camiciottoli G, Meoni E, Bigazzi F, Cestelli L, Pistolesi M, et al. Explorative data analysis techniques and unsupervised clustering methods to support clinical assessment of Chronic Obstructive Pulmonary Disease (COPD) phenotypes. J Biomed Inform. 2009;42(6):1013–1021. doi: 10.1016/j.jbi.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraiman R, Justel A, Svarc M. Selection of Variables for Cluster Analysis and Classification Rules. Journal of the American Statistical Association. 2008;103:1294– 1303. [Google Scholar]
- 16.R Development Core Team. R: A Language and Environment for Statistical Computing. 2011 http://www.R-project.org.
- 17.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42(3):200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, et al. A genomewide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5(3):e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5(3):e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21(4):947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimoto K, Kitaguchi Y, Kubo K, Honda T. Clinical analysis of chronic obstructive pulmonary disease phenotypes classified using high-resolution computed tomography. Respirology. 2006;11(6):731–740. doi: 10.1111/j.1440-1843.2006.00930.x. [DOI] [PubMed] [Google Scholar]
- 22.Pistolesi M, Camiciottoli G, Paoletti M, Marmai C, Lavorini F, Meoni E, et al. Identification of a predominant COPD phenotype in clinical practice. Respir Med. 2008;102(3):367–376. doi: 10.1016/j.rmed.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Burgel PR, Paillasseur JL, Caillaud D, Tillie-Leblond I, Chanez P, Escamilla R, et al. Clinical COPD phenotypes: a novel approach using principal component and cluster analyses. Eur Respir J. 2010;36(3):531–539. doi: 10.1183/09031936.00175109. [DOI] [PubMed] [Google Scholar]
- 24.Patel BD, Coxson HO, Pillai SG, Agusti AG, Calverley PM, Donner CF, et al. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;178(5):500–505. doi: 10.1164/rccm.200801-059OC. [DOI] [PubMed] [Google Scholar]
- 25.Hogg JC. A pathologist’s view of airway obstruction in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;186(5):v–vii. doi: 10.1164/rccm.201206-1130ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez FJ, Curtis JL, Sciurba F, Mumford J, Giardino ND, Weinmann G, et al. Sex differences in severe pulmonary emphysema. Am J Respir Crit Care Med. 2007;176(3):243–252. doi: 10.1164/rccm.200606-828OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillai SG, Kong X, Edwards LD, Cho MH, Anderson WH, Coxson HO, et al. Loci identified by genome-wide association studies influence different disease-related phenotypes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182(12):1498–1505. doi: 10.1164/rccm.201002-0151OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.