Abstract
In previous work, we identified two trimeric peptide ligands (designated WRW and KYY) which bound specifically to porcine parvovirus (PPV) and demonstrated their ability to capture and remove the virus from solutions containing 7.5% human blood plasma. This paper examines the influences of peptide density and the presence of an ethylene oxide spacer arm on the efficiency of virus capture using these two ligands. The WRW peptide bound the most virus from plasma solutions at the lowest peptide density tested (0.008 mmol/g dry resin) and binding was enhanced by the presence of the spacer arm. On the other hand, the KYY peptide bound the most virus at the same low peptide density but it performed better in the absence of the spacer arm. Of the two, the binding efficiency of the WRW peptide was more sensitive to peptide density and spacer arm presence. These results indicate that low peptide densities enhance binding selectivity, facilitating specific peptide-virus binding even in the presence of plasma proteins which can theoretically bind non-specifically.
1. Introduction
Processes designed to produce protein therapeutic products from animal or mammalian cell culture, or human plasma fractionation, are required to have a minimum of two virus clearance steps, each of which must remove 99.99% or 4 logs of contaminating virus (1). Inactivation methods that disrupt the lipid membrane surrounding enveloped viruses are successful (2) in this regard, but the nonenveloped viruses can be more difficult to remove. Currently, nonenveloped viruses are most commonly removed by either nanofiltration (3) or anion exchange chromatography (4, 5) at the end of the purification process. Recently, we identified small peptides ligands that could remove porcine parvovirus (PPV) from 7.5% human plasma solutions using a chromatographic column (6). These small peptides could find use for the selective affinity capture of PPV using column or membrane-based processes designed to achieve required virus clearance, at relatively low cost and high stability.
Affinity adsorption might also be of value in the removal of viruses from blood before transfusion, in a manner that is similar to recent findings demonstrating the removal of prion infectivity from leukoreduced whole blood using small ligand column chromatography (7). Also, chromatography is often used to purify viruses for use as vaccines or gene therapy vectors (8). Adeno-associated virus, a human parvovirus, is often purified by chromatography (9–11), and is currently being considered as a potential gene therapy vector. Therefore, the discovery and application of small peptide ligands with high binding specificity for nonenveloped viruses may result in the production of novel affinity-based separation and purification methods which could increase the yield and purity of virus vaccines or gene therapy vectors using a smaller number of process steps.
In prior work, small peptide ligands at a peptide density of 0.10 mmol/g dry resin and containing a polyethylene oxide (PEO) spacer arm in addition to the manufacturer’s spacer of unknown length were tested for their ability to remove PPV from solution (6). All of the trimer peptides examined were able to clear the PPV with which they were challenged when the virus was diluted in saline. However, when the virus was spiked into 7.5% human blood plasma and subjected to column chromatography, significant breakthrough occurred for all of the peptides tested. The prevailing lead peptide, WRW, was able to clear all challenged infectious virus in the first three column volumes only, which was equivalent of 1.5 ml of challenge for a 0.5 ml column. It was hypothesized that plasma proteins were non-specifically binding to the peptide ligands, reducing the peptides available for PPV binding.
In an effort to improve the selectivity of the peptides by reducing the opportunity for non-specific binding to plasma proteins, we investigated changes in peptide density and the addition of an extended PEO spacer arm. Specifically, resins manufactured with the WRW or KYY peptides at low (0.008 mmoles/g), medium (0.10 mmoles/g) and high (0.4 mmoles/g) density, and with or without an additional PEO spacer arm, were evaluated for their ability to remove PPV from 7.5% human blood plasma (6). The other peptide resins evaluated earlier for their ability to remove PPV from plasma (RAA and KRK) were also tested, but their ability to remove PPV was inferior to WRW and KYY.
2. Materials|Methods
2.1. Materials
Phosphate buffered saline (PBS) containing 0.01 M phosphate, 0.138 M NaCl and 0.0027 M KCl, pH 7.4 was purchased from Sigma (St. Louis, MO) and human blood plasma was donated by the American Red Cross (Rockville, MD). Eagle’s Minimum Essential Media (EMEM) was purchased from Quality Biologicals (Gaithersburg, MD). The sterile PBS, trypsin, penicillin/streptomycin and glutamine used for cell culture were purchased from Invitrogen (Carlsbad, CA).
2.2. Virus Propagation, Purification, and Titration
The porcine parvovirus (PPV) NADL-2 strain was titrated and propagated on porcine kidney (PK-13) cells, which were a gift from the American Red Cross (Rockville, MD). The PK-13 cells were maintained and the PPV propagated as described in Heldt et al. (12) using complete media, which consisted of EMEM supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, and 10% non-heat inactivated fetal calf serum (Hyclone, Logan, UT). After propagation of the virus, the cell culture supernatant was clarified (6) and stored at −80°C until further use. This crude cell culture supernatant was used in all column chromatography experiments.
Radioactive PPV was prepared by metabolically incorporating a radiolabel during virus propagation by addition of 35S methionine and cysteine to the cell culture media, as described by Heldt el al. (6). The radioactive PPV cell culture lysate was subjected to a freeze/thaw cycle at − 20°C and stored (as crude radiolabelled virus stock) at 4°C for up to four weeks prior to use in experiments.
Further purification of crude, radiolabelled PPV stocks was done by gel filtration chromatography and this virus preparation was used for the equilibrium isotherm experiments. Sephacryl 300 HR gel (Sigma, St. Louis, MO) was packed into an Omni column of 70 mm length. A Pharmacia Biotechnology FPLC (fast protein liquid chromatography) equipped with two P-50 pumps, an LC-501 Plus controller, Frac-100 fraction collector and associated equipment, was used to perform the separation. A 20-µl aliquot of crude labeled virus stock was injected onto the column at a flow rate of 0.5 ml/min. Fractions 1-ml in volume were collected and analyzed for radioactivity and virus infectivity. The first radioactive peak, usually corresponding to fractions 4 and 5, was collected and designated as the purified labeled virus stock.
All infectivity measurements were made using the MTT assay, which uses the tetrazolium salt MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) to detect cell proliferation. This assay has been previously correlated to the TCID50 (50% tissue culture infectious dose), a common method for the titration of infectious viruses (12).
2.3. Chromatography Using Peptide Resins
Peptide resins were synthesized on Toyopearl Amino 650M resin (Tosoh Biosciences, Montgomeryville, PA) by Peptides International (Louisville, KY). The peptides were attached to the resin at a peptide density of 0.40, 0.10, and 0.008 mmol/g of dry resin. For some of the resins, a 15–20 Å ethylene oxide spacer arm was added between the peptide and the resin; it was omitted for others. The purpose of the spacer arm was to aid in the flexibility of the peptide and also to allow the peptide to access canyons on the virus surface (6, 13). All amines on the resin surface that were not linked to the peptide ligands were capped with acetic anhydride (acetylated) to reduce non-specific electrostatic interactions with the virus and contaminating plasma proteins. The peptide resins were packed into disposable PIKSI columns (ProMetic Biosciences Ltd, Cambridge, England) with a total of 0.5 ml of settled resin in PBS per column. Starting titers between 5.2 – 6.2 log10 (MTT/ml) were used for all column experiments. The starting titers were different due to different stocks of virus that were used for some experiments. The starting titers were normalized in the log clearance calculation shown in equation 1. An acetylated control resin was synthesized by directly acetylating the Amino 650M resin (6).
| (1) |
2.4. Equilibrium Isotherms
The purified, radiolabelled PPV stock was serially diluted at a ratio of 1:2 to achieve six different concentrations ranging from 373 to 6323 counts per minute per milliliter (CPM/ml). A 50-µl volume of settled resin, which corresponded to 10 mg of dry resin, was placed into twelve microcentrifuge tubes, and as much buffer as possible was removed with a pipette without disturbing the resin. Five-hundred microliters of each virus dilution was added to each resin, in duplicate. The resins and virus were placed on an end-over-end rotator (Barnstead-Thermolyne Labquake Shaker-Rotisserie, 8 rpm, Waltham, MA) at room temperature for 1.5 hours. The tubes were briefly centrifuged to settle the resin, and the radioactive reading of the supernatant was determined using a Packard 1500 Tri-Carb Liquid Scintillation Analyzer (Wellesley, MA). Radioactivity counts before and after contact with the resin was used to determine the amount of virus that bound to the resin by determining a total material balance. Infectivity determination using the MTT assay was also done for confirmation (12).
3. Results and Discussion
In earlier work, trimer peptides that bound to PPV were identified using solid-phase combinatorial peptide libraries, and their ability to remove PPV from buffer solutions and solutions containing 7.5% human plasma was evaluated (6). The peptides used for chromatographic evaluation were synthesized on Amino 650M resin, which has a particle diameter of about 65 µm, a pore size of about 100 nm, and a total surface area of 31 m2/g. The amino resin had primary amine groups on the surface at a density of 0.4 mmol/g dry resin, which corresponds to the maximum ligand capacity for this resin. In the work reported here, the peptide density was varied from 0.008 to 0.4 mmol/g for peptides synthesized on the Amino 650M resin; the value of an additional spacer arm was also evaluated.
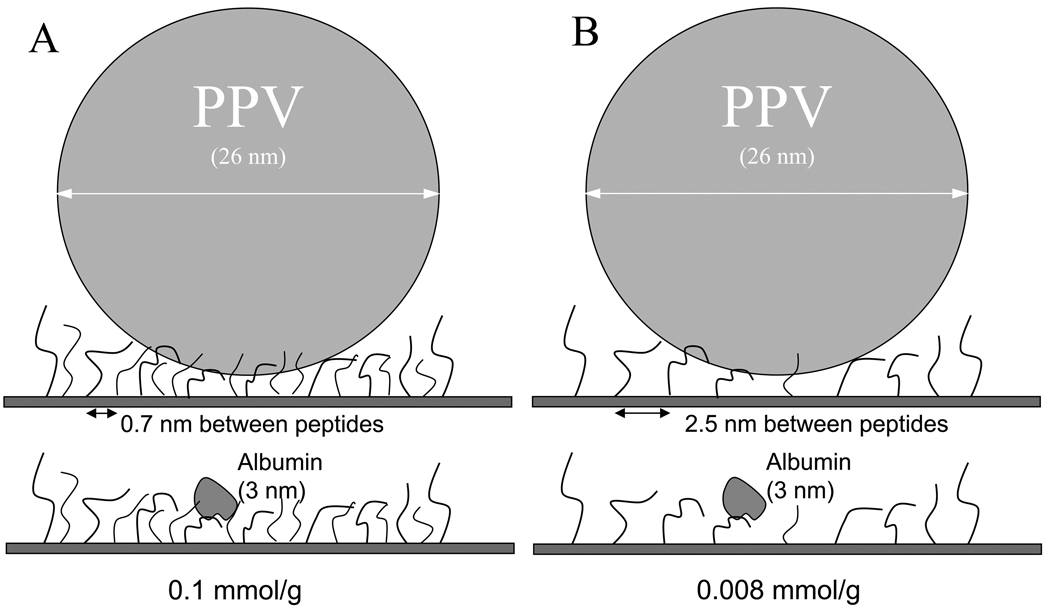
The purpose of varying the peptide density was to improve the selectivity of the peptides for PPV in the presence of competing proteins such as those found in plasma. Most plasma proteins are smaller in diameter when compared to infectious parvovirus particles (Table 1). Our hypothesis was that reduction of peptide density would prevent the binding of smaller blood proteins, which would not be able to span across multiple peptides, while still allowing multimeric binding of the virus to the ligands (14), as illustrated in Figure 1 for one sample protein. Due to the multitude of proteins present in plasma, the non-specific binding of the individual proteins was not assessed. This would need to be evaluated before commercialization of this virus removal system.
Table 1.
Size of PPV relative to select plasma proteins
Figure 1.
Hypothesized effect of high versus low density peptides. (A) At a concentration of 0.1 mmol/g, multiple peptides bind to both the virus and competing proteins (e.g., albumin). (B) At a density of 0.008 mmol/g, multiple peptide-virus contact points promotes virus binding, but limited (monomeric) peptide binding to smaller completing proteins reduces the likelihood and strength of non-specific binding, hence improving the performance of the peptides for virus capture.
The peptide density was controlled by adding a known ratio of alanine that was either protected with an Fmoc (9-fluorenylmethoxycarbonyl) or tBoc (tert-butyl-dicarbonate) group and this mixture was coupled to the resin (22, 23). The tBoc group was removed and the free amine of the alanine was acetylated. This was followed by removal of the Fmoc group and subsequent peptide synthesis directly on this alanine. By changing the ratio of Fmoc and tBoc alanine, different peptide densities could be achieved. The final peptide density was measured based on the concentration of the Fmoc group that was released after the final amino acid was added to the synthesized peptide chain. The exact spacing of the peptides on the resin is not controlled with this method, since the peptide density is controlled purely by the ratio of Fmoc and tBoc alanine added to the coupling reaction. There is a point at which the low peptide densities can no longer be accurately detected by the amount of Fmoc released in the final amino acid synthesis, and this results in the lower limit of peptide density. Due to these synthesis constraints, the lowest peptide density that could be tested was 0.008 mmol/g dry resin.
The self-assembly of the surface proteins of PPV results in the presence of “spikes” and “canyons” on the surface of the virion, with an estimated distance between the highest and the lowest points of about 35 Å (13). It is suspected that the so-called canyons contain the virus receptor binding site(s) (16), which consists of highly conserved regions of amino acid sequence. Theoretically, a resin which contained a spacer arm of about 35 Å in length and a low peptide density should be capable of binding the greatest amount of virus because it would facilitate increased contact between the ligand(s) and the virus surface binding sites. This, in turn, might increase binding selectivity in the presence of competing proteins. The amine group on the Toyopearl Amino 650M resin is separated from the base resin by a PEO spacer arm of undisclosed length. Making the assumption that the PEO spacer arm length approximated 10–15 Å, we chose to expand this length by 15–20 Å by the addition of four EO groups (6). Taken together, this should result in an approximate total spacer arm length of 35 Å. This entire spacer arm of the resin, which including the spacer added by Toyopearl and the additional EO groups, is depicted in Figure 2.
Figure 2.
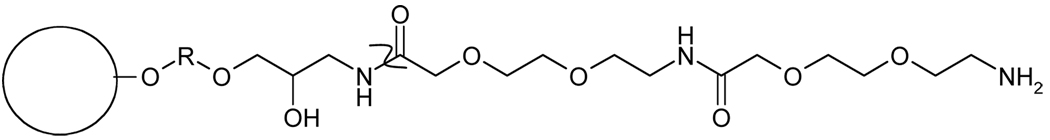
Surface chemistry of the Amino 650M resin containing the ethylene oxide spacer arm. The curved line (“Z”) denotes the junction between the spacer arm provided by the resin manufacturer and the ethylene oxide spacer arm added to the resin. The R denotes (CH2CH2O)x, a hydrophilic spacer arm of undisclosed length. The N-terminus of the spacer arm is attached to the carboxylic acid on the peptide through a peptide bond.
3.1. Removal of PPV in 7.5% Human Blood Plasma
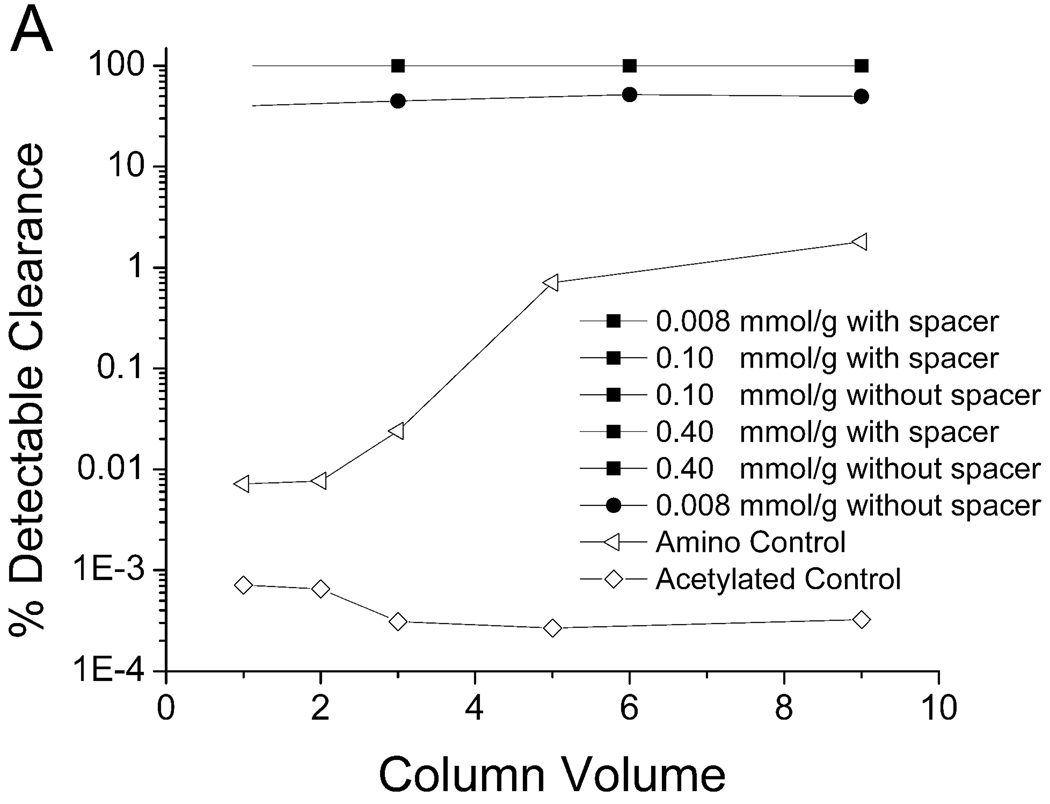
The degree of PPV clearance, when the virus was suspended in saline, achieved using the WRW and KYY resins, is shown in Figure 3. This is expressed as the percent of virus removed as a function of column volume. With a few exceptions, all of the peptide densities were able to clear all detectable PPV, regardless of the presence or absence of a spacer arm. The peptide resins were compared to two control resins, i.e., an amino control and an acetylated control. The latter was chosen because the peptides were attached to the amine groups on the resin, and all amine groups that did not contain peptide were acetylated. Consequently, the acetylated resin was a control for the resin surface. The acetylated resin bound none of the PPV in solution, as shown in Figure 3. The amino control resin, which has amine groups on the surface and is a weak ion exchange resin, was used because ion exchange resins may have inherent virus removal properties (24, 25); this allowed for comparison of peptide ligands to an anion exchange resin. The amino resin removed more virus from solution in the later column volumes than in the earlier column volumes (Figure 3). This was most likely due to the presence of cellular debris in the cell culture supernatant spike that was used in the experiments, which initially bound to the amino control resin and created additional binding sites for the virus as the experiment progressed (6).
Figure 3.
Clearance of PPV in PBS. (A) Peptide WRW; all peptide-spacer combinations shown by the filled squares were able to clear all detectable PPV; the 0.008 mmol/g resin without a spacer arm (filled circles) was less efficient. (B) Peptide KYY; all peptide-spacer combinations shown by the filled squares were able to clear all detectable PPV; the 0.40 mmol/g resin without a spacer arm (filled circles) was less efficient. A 0.5 ml settled resin column was used at a flow rate of 0.1 ml/min, thus each column volume consisted of 0.5 ml of flow through. The starting titer ranged from 6.1–7.2 log10(MTT/ml). A 4.5–5.6 log clearance represents close to 100% clearance.
Since most of the peptide resins were able to clear virtually all detectable virus in the first 10 column volumes, the maximum capacity of the resin was never reached. The actual virus titers added to these columns differed slightly but all were in the range of 105 to 106 infectious particles per milligram of dry resin. PPV produces many non-infectious particles for every infectious particle, with an estimated infectious to non-infectious particle ratio as high as 1:100,000 (16). Assuming a ratio this high, the resin may actually be binding as many as 1010 particles per milligram of dry resin.
The theoretical amount of virus that the resin could bind was determined by assuming that none of the virus penetrates into the pores of the resin. The resin pore size is 100 nm and, in principle, is large enough for the penetration of a virus particle that is 18–26 nm in diameter. However, penetration was considered unlikely because the virus is large enough to exhibit hindered diffusion through the pores, resulting in plugging of the pores as soon as the viruses adsorb to the ligands on the resin. Viruses diffuse slowly due to their size; therefore, it is likely that the virus will bind to a peptide that is near the pore opening and once bound, the pore size would be reduced, preventing additional viruses from penetrating the pores. This phenomenon was observed previously using a PL-SAX resin of 400 nm pore size with adenovirus (80–110 nm in diameter) (26), in which case the internal surface area of the resin was not fully utilized (8), most likely due to pore plugging. Based on this assumption and given a 26 nm spherical virus, the theoretical amount of viruses that could pack on the outside surface of a smooth 65 µm diameter bead (the average diameter of the Toyopearl 650M resin) is 1011 virus particles per milligram.
3.2. Removal of PPV in 7.5% Human Blood Plasma
To challenge the peptide resins with a more complex mixture, PPV was spiked into a solution containing 7.5% human blood plasma. This corresponds to approximately 5 mg/ml of protein, which represents both the approximate amount of protein in the final formulation of a protein product, and the amount of plasma that is present in red blood cell concentrate. By observing the trends in virus breakthrough, the PPV removal efficiency of the different peptide resins could be compared. Negative controls (amino and acetylated controls) were included in all experiments.
3.2.1. Peptide WRW
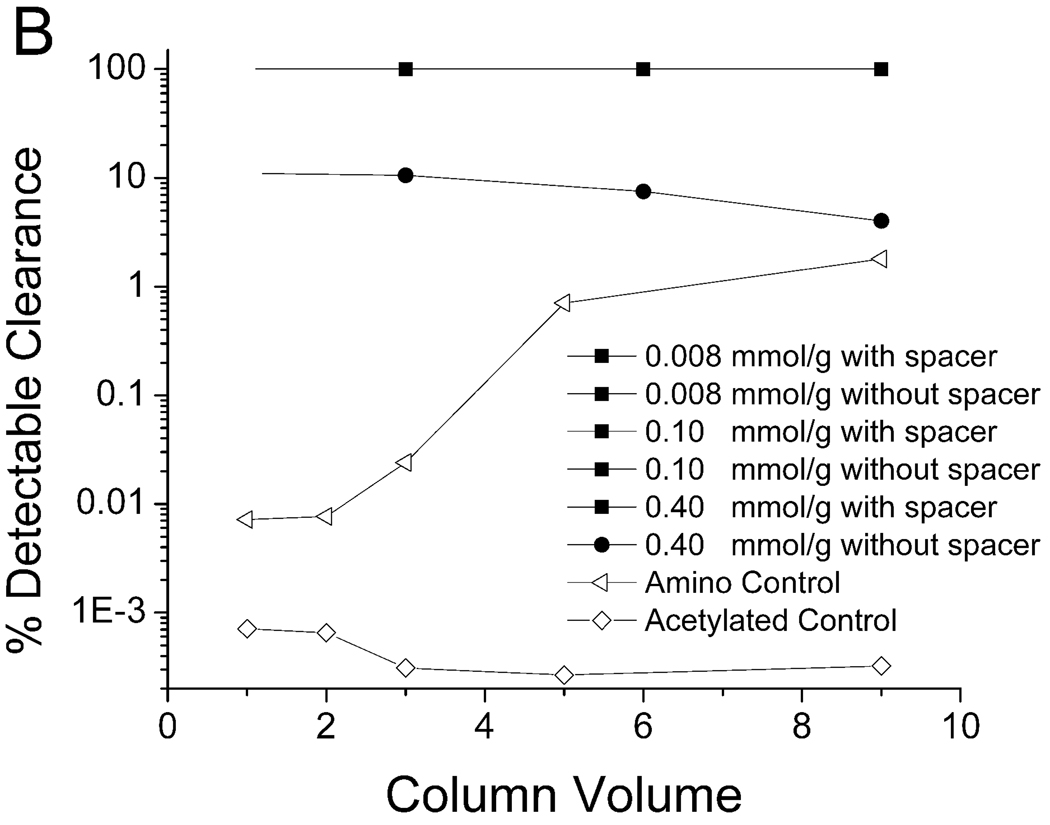
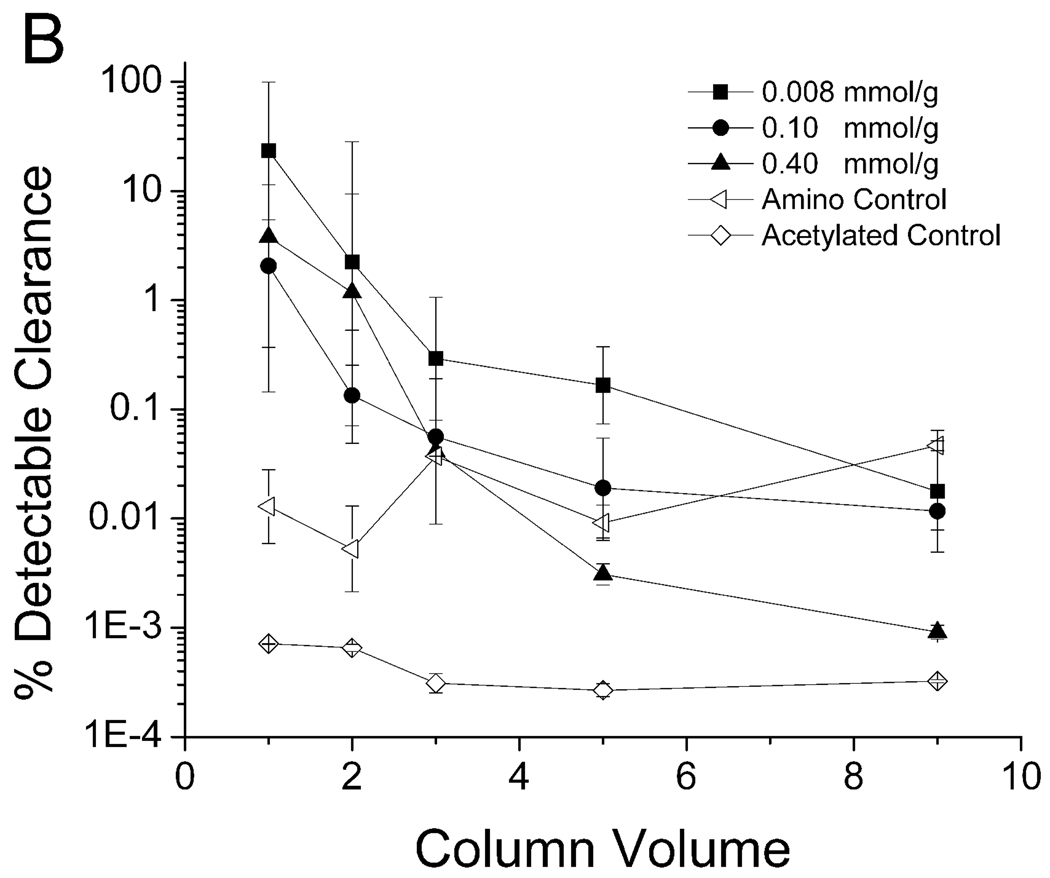
The ability of the WRW resin to remove PPV at different resin peptide densities with and without the ethylene oxide spacer arm is shown in Figure 4. The binding of virus to the WRW resin was highly influenced by the presence of a spacer arm. When the spacer arm was present (Figure 4A), the resin bound all of the detectable PPV in the first three column volumes for both the 0.008 mmol/g resin and the 0.10 mmol/g resin concentrations. None of the resins without the spacer (Figure 4B), nor the resins with higher peptide density (0.4 mmol/g) with the spacer arm (Figure 4A), were able to achieve this degree of removal.
Figure 4.
Clearance of PPV by peptide WRW in 7.5% human blood plasma with (A) and without (B) the spacer arm. A 4.6–5.6 log clearance, depending on starting titer, represents close to 100% clearance. Experimental conditions described in Figure 3.
The WRW resin also required the positively-charged amine group on the end of the peptide in order to facilitate PPV binding (Figure 4A). When the terminal amine on the WRW resin was acetylated, the ability of the resin to bind the virus was greatly decreased. The acetylation of the terminal amine did not completely eliminate all binding of PPV to WRW, as the acetylated peptide still bound more virus than the acetylated control.
The dynamic binding of PPV to the WRW resin in column adsorption studies is quantified in Table 2. This again highlights the importance of the low density and the spacer arm in the binding of PPV to WRW. The log clearance of PPV for the first five column volumes, defined in Equation 1 in Materials and Methods, was used to rank the resins for their ability to remove PPV. It should be noted that the WRW resin at the lowest peptide density (0.008 mmol/g) was able to achieve a 3.08 log removal of PPV from 7.5% human plasma. This log removal is significantly larger than what was achieved with the weak ion exchange control resin or the high peptide density resin (0.4 mmol/g).
Table 2.
Total log clearance in the first five column volumes for peptide WRW
| Peptide Density (mmol/g) |
Spacer Arm | Log clearance in first five column volumes |
|---|---|---|
| 0.008 | + | 3.08 |
| 0.10 | + | 2.45 |
| 0.008 | − | 2.21 |
| 0.10 - N-acetylated | + | 1.73 |
| 0.40 | + | 1.48 |
| 0.40 | − | 1.19 |
| 0.10 | − | 1.16 |
| Amino | 0.68 | |
| Acetylated | 0.24 |
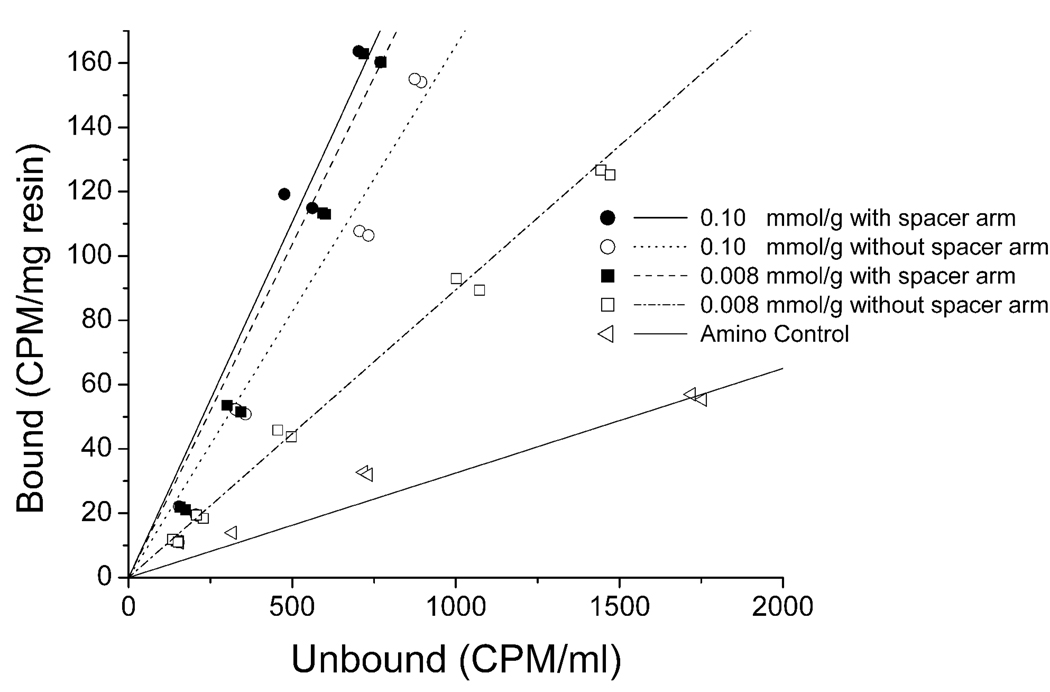
The WRW resin was also characterized for binding efficiency through measurement of equilibrium binding isotherms in PBS. Equilibrium isotherms can be used to illustrate the differences in affinity dissociation constant of the resin, as well as the total capacity. In the case of virus binding, it was not possible to reach the full resin capacities at increased virus concentrations, so only the linear portion of the isotherms could be constructed (Figure 5). The isotherms were fit to the linear form of the Langmuir adsorption isotherm,
| (2) |
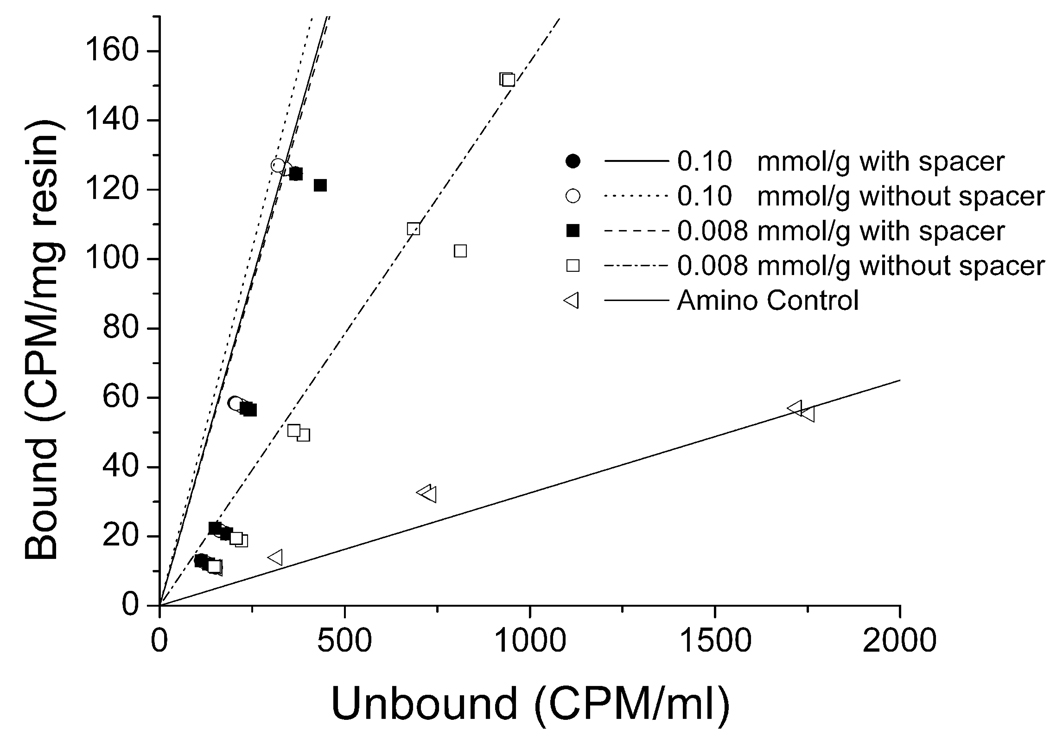
where q is the adsorption of virus bound to the resin, qmax is the maximum capacity of the resin, C is the unbound concentration of virus in solution, and Kd is the affinity dissociation constant. The slope of the isotherm represents qmax divided by Kd. Even though maximum capacity of the resin could not be determined, it is reasonable to assume that this is the same for all resins, since the capacity will be limited by the external area of the resin particles and is not likely to be strongly dependent on the ligand itself or its density. Because of this, the slopes of the linear form of the isotherm are a reflection of the Kd values of the different resins, with the largest slopes corresponding to the lower Kd values, indicating stronger binding. The equilibrium isotherms for WRW resins are shown in Figure 5. Two of the top three WRW resins, i.e., 0.10 mmol/g and the 0.008 mmol/g resin with a spacer arm, had virtually the same slope, while the resin at a concentration of 0.008 mmol/g without a spacer arm had a less steep slope, albeit more steep than that of the amino control.
Figure 5.
Equilibrium isotherm of PPV binding to peptide WRW. Filled symbols correspond to resins having the spacer arm, while open symbols correspond to resins without the spacer arm. The amino control is the bare Toyopearl Amino 650M resin without the peptide attached and represents an anion exchange resin. CPM (counts per minute).
It should be noted that the isotherms were measured in PBS buffer (Figure 5), while the dynamic binding experiments (Figure 4) were done in columns challenged with virus-inoculated 7.5% human plasma. The isotherms indicate resins and peptides constructed at low peptide density (0.008 mmol/g) had increased binding strength by approximately a factor of 3 when the spacer arm was present, while at a density of 0.10 mmol/g the spacer arm did not seem to increase the binding strength. It is interesting to note that all four peptide resins whose isotherms are shown in Figure 5 bound almost all of the PPV in PBS buffer (Figure 3), including the resin at low concentration (0.008 mmol/g) which lacked the spacer and showed the least efficient binding when challenged in the column format. This is consistent with the isotherm results. However, in competition with plasma proteins, the performance of the WRW resins with the spacer arm at 0.008 mmol/g and 0.10 mmol/g concentrations was clearly superior to that of resins without the spacer. The competition between virus and proteins is a complex phenomenon influenced by the ligand, its density and the presence or absence of a spacer. These discrepancies between experiments illustrate the need to evaluate binding in the relevant matrix, as it is difficult to predict such interactions without supporting experimental data.
3.2.2. Peptide KYY
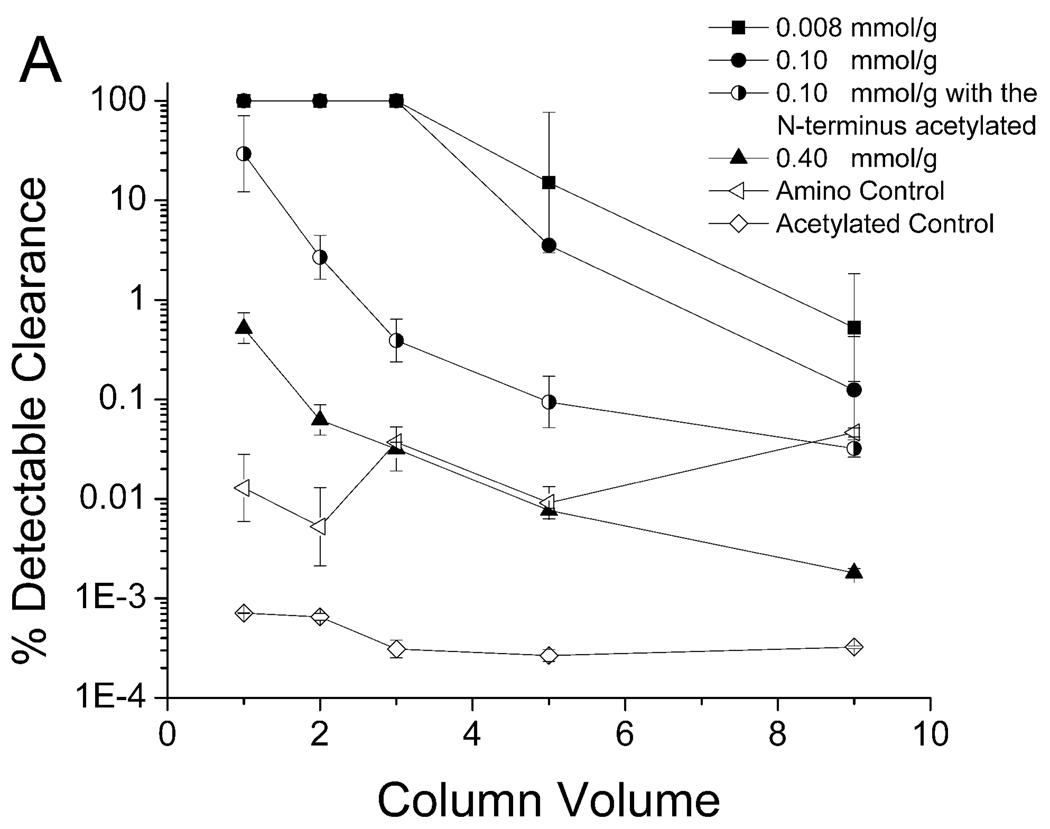
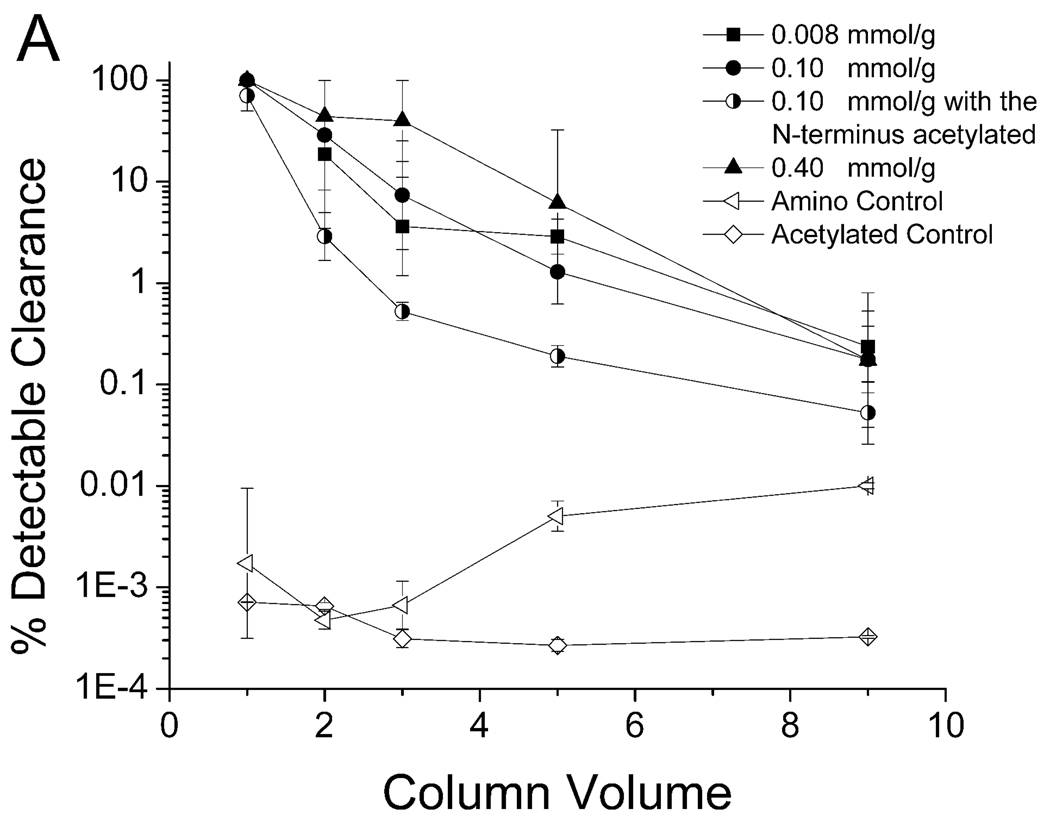
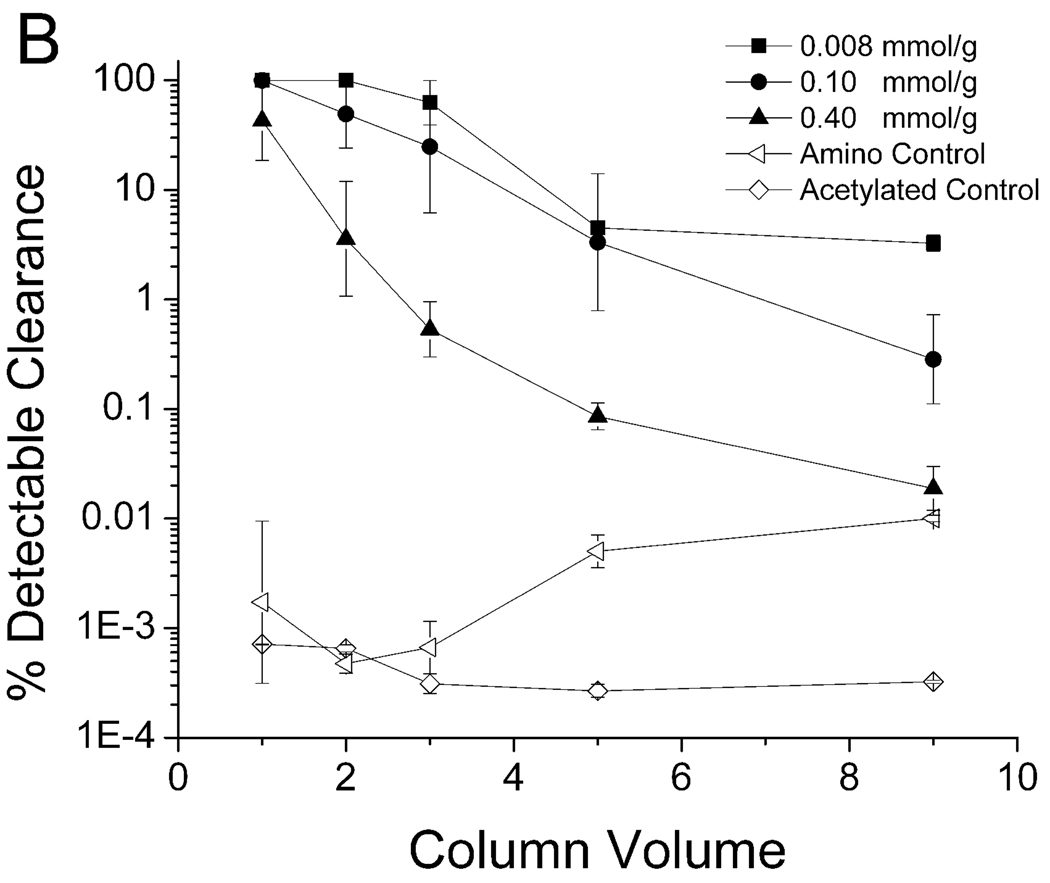
The binding between the KYY resin and PPV suspended in 7.5% human plasma is shown in Figure 6. The performance of the KYY resin was less sensitive to peptide density and spacer arm length when compared to peptide WRW. The best configuration for KYY was a concentration of 0.008 mmol/g without a spacer arm, which was able to clear all detectable PPV in the first two column volumes (Figure 6B). As was the case for the WRW peptide, the KYY resin also required the terminal amine group for PPV binding, as evidenced by a significant drop in binding efficiency when acetylated (Figure 6A).
Figure 6.
Clearance of PPV by peptide KYY with spacer arm (A) and without spacer arm (B) in 7.5% Human Blood Plasma. A 5.0–5.6 log clearance, depending on starting titer, represents close to 100% clearance. Experimental conditions described in Figure 3.
There were no clear trends on the effects of the spacer arm length and peptide density with respect to binding and clearance of PPV by peptide KYY (Table 3). The best configuration tested was at a peptide density of 0.008 mmol/g without a spacer arm and this configuration was able to clear 3.7 log10 of PPV in the first five column volumes, which was a much greater degree of removal than observed for any of the WRW resins (Table 2). This may be due, at least in part, by the relatively higher PPV titer used in the KYY challenges relative to the virus concentration used in the WRW challenge experiments. There were also differences in the shapes of the curves corresponding to the best performance of WRW columns (at 0.008 mmol/g with a spacer arm, Figure 4A) and KYY columns (at 0.008 mmol/g without a spacer arm, Figure 6B). Specifically, the WRW resin cleared all detectable PPV in the first three column volumes with a subsequent steep and rapid PPV breakthrough, whereas the KYY resin broke through by the third column volume, although the breakthrough was much more gradual.
Table 3.
Total log clearance in the first five column volumes for peptide KYY
| Peptide Density (mmol/g) |
Spacer Arm |
Log clearance in first five column volumes |
|---|---|---|
| 0.008 | − | 3.70 |
| 0.40 | + | 3.27 |
| 0.10 | − | 2.87 |
| 0.008 | + | 2.67 |
| 0.10 | + | 2.63 |
| 0.40 | − | 2.22 |
| 0.10 - N-acetylated | + | 2.03 |
| Amino | 0.02 | |
| Acetylated | 0 |
There was also a significant discrepancy in binding efficiency and removal when comparing KYY resins based on equilibrium isotherm measurements in buffer (Figure 7) and the dynamic column performance for virus suspended in 7.5% human blood plasma (Figure 6). The best performing resin in the column removal experiments (the 0.008 mmol/g resin without a spacer arm, Table 3) performed least well in the isotherm experiments (Figure 7). All of the KYY peptide densities (0.008 and 0.10 mmol/g resin) were able to clear all detectable PPV from buffer (Figure 3B) when tested in the column format. This indicates that the decrease in peptide density resulted in an increase in the Kd of that particular resin as applied to the binding of PPV in buffer, but it appeared to greatly improve the selectivity of the resin when applied to PPV seeded in 7.5% human blood plasma.
Figure 7.
Equilibrium isotherm of PPV binding to peptide KYY. Filled symbols correspond to resins having the spacer arm, while open symbols correspond to resins without the spacer arm. The amino control resin data is the same as shown in Figure 5. CPM (counts per minute).
4. Conclusions
The influence of peptide density and spacer arm length on the ability of two trimer peptides (WRW and KYY) previously reported to selectively bind (and remove) PPV from solutions containing 7.5% human blood plasma, was examined. The effects of peptide density and spacer arm length differed for the two peptides. Specifically, the WRW peptide resin performed better in the presence of the spacer arm when applied to PPV-seeded 7.5% human blood plasma, providing removal of all the detectable virus in the first three column volumes at the lowest peptide density tested (0.008 mmol/g). On the other hand, the KYY resin was much less sensitive to differences in spacer arm length and peptide density, demonstrating the best PPV binding efficiency at the lowest peptide density without a spacer arm. However, even this configuration only consistently bound PPV in the first two column volumes. It may be that the two peptides (WRW and KYY) are binding to different locations on the virus surface, which in turn may influence the stoichiometry necessary to promote optimal ligand-target interactions. The capture of viruses on solid surfaces is an interesting area that deserves further investigation. Although the example presented here focuses on the potential for removal of viruses from process streams, these ligands could also be applied to virus capture, purification, and/or detection. Clearly, small peptide ligands or other inexpensive, synthetic ligands could play an important role in any of these applications.
Supplementary Material
Acknowledgments
The authors would like to thank Pathogen Removal and Diagnostic Technologies (PRDT), a joint venture between the American Red Cross and ProMetic BioSciences Inc. and the NIH/NCSU Molecular Biotechnology Training Program for funding.
References
- 1.FDA. Points to consider in the manufacture and testing of monoclonal antibody products for human use. US Department of Health and Human Services: Rockville, MD; 1997. [Google Scholar]
- 2.Bryant BJ, Klein HG. Pathogen inactivation - The definitive safeguard for the blood supply. Arch Pathol Lab Med. 2007;131(5):719–733. doi: 10.5858/2007-131-719-PITDSF. [DOI] [PubMed] [Google Scholar]
- 3.Burnouf T, Radosevich M, Goubran HA, Willkommen H. Place of nanofiltration for assuring viral safety of biologicals. Curr Nanosci. 2005;1(3):189–201. [Google Scholar]
- 4.Lebing W, Remington KM, Schreiner C, Paul HI. Properties of a new intravenous immunoglobulin (IGIV-C, 10%) produced by virus inactivation with caprylate and column chromatography. Vox Sang. 2003;84(3):193–201. doi: 10.1046/j.1423-0410.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 5.Curtis S, Lee K, Blank GS, Brorson K, Xu Y. Generic/matrix evaluation of SV40 clearance by anion exchange chromatography in flow-through mode. Biotechnol Bioeng. 2003;84(2):179–186. doi: 10.1002/bit.10746. [DOI] [PubMed] [Google Scholar]
- 6.Heldt CL, Gurgel PV, Jaykus LA, Carbonell RG. Identification of Trimeric Peptides That Bind Porcine Parvovirus from Mixtures Containing Human Blood Plasma. Biotechnol Progr. 2008;24:554–560. doi: 10.1021/bp070412c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregori L, Gurgel PV, Lathrop JT, Edwardson P, Lambert BC, Carbonell RG, Burton SJ, Hammond DJ, Rohwer RG. Reduction in infectivity of endogenous transmissible spongiform encephalopathies present in blood by adsorption to selective affinity resins. Lancet. 2006;368(9554):2226–2230. doi: 10.1016/S0140-6736(06)69897-8. [DOI] [PubMed] [Google Scholar]
- 8.Trilisky EI, Lenhoff AM. Sorption processes in ion-exchange chromatography of viruses. J Chromatogr A. 2007;1142(1):2. doi: 10.1016/j.chroma.2006.12.094. [DOI] [PubMed] [Google Scholar]
- 9.Chahal PS, Aucoin MG, Kamen A. Primary recovery and chromatographic purification of adeno-associated virus type 2 produced by baculovirus/insect cell system. J Virol Methods. 2007;139(1):61. doi: 10.1016/j.jviromet.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 10.O'Riordan CR, Lachapelle AL, Vincent KA, Wadsworth SC. Scaleable chromatographic purification process for recombinant adeno-associated virus (rAAV) J Gene Med. 2000;2(6):444–454. doi: 10.1002/1521-2254(200011/12)2:6<444::AID-JGM132>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 11.Smith RH, Ding C, Kotin RM. Serum-free production and column purification of adeno-associated virus type 5. J Virol Methods. 2003;114(2):115. doi: 10.1016/j.jviromet.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Heldt CL, Hernandez R, Mudiganti U, Gurgel PV, Brown DT, Carbonell RG. A colorimetric assay for viral agents that produce cytopathic effects. J Virol Methods. 2006;135(1):56. doi: 10.1016/j.jviromet.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 13.Simpson AA, Herbert B, Sullivan GM, Parrish CR, Zadori Z, Tijssen P, Rossmann MG. The Structure of Porcine Parvovirus: Comparison with Related Viruses. J Mol Biol. 2002;315:1189–1198. doi: 10.1006/jmbi.2001.5319. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, Salm JR, Gurgel PV, Carbonell RG. Small Peptide Ligands for Affinity Separations of Biological Molecules. In: Galan M, Valle EMd, editors. Chemical Engineering: Trends and Developments. 2005. pp. 63–83. [Google Scholar]
- 15.Mengling WL. Diseases of Swine. 1999 [Google Scholar]
- 16.Muzyczka N, Berns KI. Parvoviridae: The Viruses and Their Replication. In: Knipe DM, Howely PM, editors. Field's Virology. 4th ed. Vol. 2. Lippincott-Raven; 2001. pp. 2327–2359. [Google Scholar]
- 17.Benhabbour SR, Liu L, Sheardown H, Adronov A. Protein Resistance of Surfaces Prepared by Chemisorption of Monothiolated Poly(ethylene glycol) to Gold and Dendronization with Aliphatic Polyester Dendrons: Effect of Hydrophilic Dendrons. Macromolecules. 2008;41(7):2567–2576. [Google Scholar]
- 18.Daniel F, Bradley L, David SM, Jiali L. Electrical characterization of protein molecules by a solid-state nanopore. Appl Phys Lett. 2007;91(5):053901. doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee K-B, Park S-J, Mirkin CA, Smith JC, Mrksich M. Protein Nanoarrays Generated By Dip-Pen nanolithography. Science. 2002;295(5560):1702. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Wang YJ, Kelner DN. Coagulation factor VIII: structure and stability. Int J Pharm. 2003;259(1–2):1. doi: 10.1016/s0378-5173(03)00227-8. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Rodriguez M, Attwood D, Ruso JM, Taboada P, Varela LM, Mosquera V. Adsorption of a cationic amphiphilic drug on human serum albumin: characterization of the complex. Phys Chem Chem Phys. 2001;3:1655–1660. doi: 10.1016/s0301-4622(01)00196-x. [DOI] [PubMed] [Google Scholar]
- 22.Merrifield B. Solid Phase Synthesis. Science. 1986;232(4748):341–347. doi: 10.1126/science.3961484. [DOI] [PubMed] [Google Scholar]
- 23.Daxecker H, Raab M, Bernard E, Devocelle M, Treumann A, Moran N. A peptide affinity column for the identification of integrin alpha(IIb)-binding proteins. Anal Biochem. 2008;374(1):203–212. doi: 10.1016/j.ab.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 24.Shukla AA, Hubbard B, Tressel T, Guhan S, Low D. Downstream processing of monoclonal antibodies--Application of platform approaches. J Chromatogr B. 2007;848(1):28. doi: 10.1016/j.jchromb.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 25.Kim IS, Choi YW, Lee SR, Woo HS, Lee S. Removal and inactivation of viruses during manufacture of a high purity antihemophilic factor VIII concentrate from human plasma. J Microbiol Biotechn. 2001;11(3):497–503. [Google Scholar]
- 26.The Universal Database of the International Committee on Taxonomy of Viruses (ICTVdb) http://www.ncbi.nlm.nih.gov/ICTVdb/index.htm.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.