Abstract
RNA interference (RNAi) is the sequence-specific degradation of mRNA by short double-stranded RNA molecules. The technology, introduced only 5 years ago, has stimulated many fantasies regarding the future of functional gene analysis and gene therapy. Given its ease of application, its high efficiency and remarkable specificity, RNAi holds great promise for broad in vitro and in vivo application in all areas of biomedicine. Despite its potential, the major obstacle to the use of RNAi (as for all previous gene silencing approaches) is the need for efficient and sustained delivery of small interfering RNA into primary mammalian cells, and specific targeting of particular cell types in vivo.
Keywords: functional genomics, gene silencing, primary mammalian cell, small interfering RNA, transfection
Introduction
In the postgenomic era it has become a major challenge to develop efficient reverse genetic approaches (i.e. from genotype to phenotype) to evaluate the function of a vast number of newly identified genes. Furthermore, specific silencing of disease-relevant genes (e.g. from tumours, pathogens, or inflammatory mediators) is an interesting therapeutic strategy. In this respect RNA interference (RNAi) technology, which allows targeted 'knockdown' of individual genes by so-called small interfering RNAs (siRNAs) [1], has already opened up new avenues for functional analyses in vitro, and holds great promise for analytical as well as therapeutic applications in vivo.
Although other gene silencing approaches, using antisense oligonucleotides, ribozymes, or DNAzymes, have been introduced over the past 25 years, their application has been restricted to certain areas. Only one antisense-based pharmacological agent has thus far been approved. In contrast to those technologies, RNAi represents a physiological process that occurs naturally in many eukaryotes, where it has evolved probably as a mechanism to defend against invading nucleic acids such as viruses and transposons [2,3], and therefore it is easily applicable to a large variety of organisms, cell types and genes. The technology has remarkable target specificity and requires only low amounts of siRNA effector molecules per cell, which can even be expressed directly in situ, allowing long-term silencing of target genes. This makes RNAi an interesting tool for the analysis of loss-of-function phenotypes in vivo and it may also lead to the development of new gene therapeutic approaches.
As for all gene silencing approaches, the critical step toward application of RNAi in mammals is the delivery of effector molecules into the target cell. What has been accomplished rather easily in cell lines represents a much greater challenge in hard-to-transfect primary mammalian cells, which are of course the ultimate targets.
This review briefly summarizes our current knowledge of the mechanism of RNAi, the technical basis for its application to functional gene analysis in mammalian cells in vitro and in vivo, and potential therapeutic applications.
Mechanism of RNA interference
The phenomenon of RNAi, originally described in the nematode worm C. elegans by Fire and colleagues [4] in 1998, has been recognized as a general mechanism in many organisms (Fig. 1) [1,5]. Basically, RNAi is induced within the cytoplasm when long, double-stranded RNA (dsRNA) is recognized by Dicer, a multidomain RNase III enzyme. Dicer processes dsRNA into short (21–25 nucleotide [nt]) duplexes that are termed siRNAs [6-10]. Like products of other RNase III enzymes, siRNA duplexes contain 5' phosphate and 3' hydroxyl termini, and two single-stranded nucleotide overhangs on their 3' ends [10]. These structural features are important for the entry of siRNAs into the RNAi pathway because blunt-ended siRNAs or those that lack a 5' phosphate group are ineffective in triggering gene silencing [11,12]. The generated siRNA associates with a multiprotein complex, the RNA-induced silencing protein complex (RISC), which becomes activated on ATP-dependent unwinding of the siRNA duplex [6,12]. One of the two siRNA strands is retained within the complex and confers sequence specificity in targeting of the mRNA by Watson–Crick base-pairing [6,11,13,14]. A perfectly homologous mRNA is cleaved at a single site in the centre of the duplex region formed with the guide siRNA, 10 nt from the 5' end of the latter [10,12,13,15]. Finally, RISC is released and the cleaved mRNA is further degraded by cellular exonucleases [16]. The specific degradation of mRNA in turn leads to decreased synthesis of the respective protein and eventually to a loss of protein function.
Figure 1.
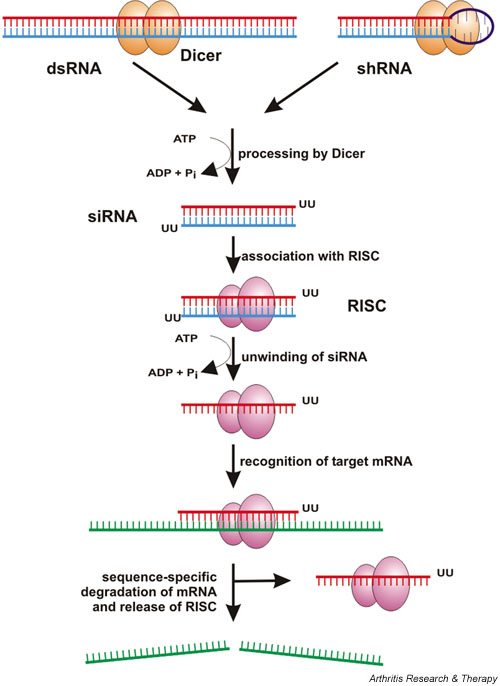
The RNA interference pathway. Long double-stranded RNA (dsRNA) or small hairpin RNA (shRNA) is processed by Dicer to form a small interfering RNA (siRNA), which associates with RNA-induced silencing protein complex (RISC) and mediates target sequence specificity for subsequent mRNA cleavage. (See text for further details.)
Concentrations of only a few siRNA molecules per cell can lead to a pronounced silencing effect, demonstrating the catalytic action of RISC [1,4]. Generally, although greatly diminished, residual mRNA levels can be detected. Hence, the RNAi-mediated silencing of a particular gene is generally referred to as a 'knockdown' rather than a 'knockout'.
RNA interference in mammalian cells
Originally, the RNAi pathway was thought to be nonfunctional in mammalian cells, where dsRNA longer than 30 base pairs induces a nonspecific antiviral response. This so-called interferon response is characterized by the activation of the RNA-dependent protein kinase [17], leading to phosphorylation of the translation initiation factor eIF-2α and thereby to a nonspecific arrest in translation and induction of apoptosis [18]. Moreover, the synthesis of 2'–5' polyadenylic acid results in the activation of the sequence nonspecific RNaseL [19].
The breakthrough for the use of RNAi in mammalian cells came when Elbashir and coworkers [20] and Caplen and colleagues [21] showed that siRNA, when directly introduced into mammalian cells, does not trigger the RNA-dependent protein kinase response but effectively elicits RNAi, presumably by directly associating with RISC. Targeted gene silencing in mammalian cells by the application of siRNA is well established. The high degree of sequence specificity inherent to the technology is emphasized by several reports showing that even a 1–2 nt mismatch in the siRNA sequence hampers targeted gene silencing [11,16,20,22,23].
Recently, evaluation of target gene specificity on a genome-wide level by applying gene expression profiling led to conflicting results. In two studies [24,25] no effects on nontarget genes were observed, although high concentrations (100 nmol/l) of siRNA were shown to induce stress-response and apoptosis-related genes. In contrast, Jackson and coworkers [26] challenged the idea of perfect sequence specificity of siRNA; they detected silencing of nontargeted genes with limited sequence similarity. As few as 11 contiguous nucleotides of identity to the siRNA were sufficient. Apparently, this off-target silencing was mediated not only by the antisense but also the sense strand of the siRNA. These findings highlight the need for careful selection of the siRNA sequences and appropriate specificity controls to verify functional effects.
Small interfering RNA selection
A synthetic siRNA consists of a 19 base-pair double-stranded region that is complementary to the gene of interest, contains 5' phosphate and 3' hydroxyl termini, and possesses two single-stranded nucleotides on the 3' ends [20].
Tuschl and coworkers [27] reported a number of guidelines for the design of siRNA molecules (Table 1). Several design tools are also available from the internet (Table 1). Although one can follow these guidelines it is still necessary to test several siRNAs, targeting distinct regions within the gene of interest, because there is great variability in the capacity of an individual siRNA to induce silencing [16,28]. One may have to test three or four siRNAs in order to find one that results in more than 90% reduction in target gene expression (unpublished data). The reason for this is not entirely understood but it may be related to one or more of the following factors: incorporation of siRNA into RISC and stability of RISC; base pairing with mRNA; cleavage of mRNA and turnover after mRNA cleavage; secondary and tertiary structures of mRNA; and binding of mRNA-associated proteins. Accordingly, Vickers and coworkers [28] found a significant correlation between mRNA sites that are RNase H sensitive (i.e. accessible) and sites that promote efficient siRNA-mediated mRNA degradation. Moreover, placing the recognition site of an efficient siRNA into a highly structured RNA region abrogated silencing.
Table 1.
Guidelines for siRNA design
| General guidelines for siRNA design | Select 23-nt long sequences from the mRNA conforming to the consensus 5'-AA [N19]UU-3' or 5'-NA [N19]NN-3' (where N is any nucleotide) |
| Avoid targeting of regions that are likely to bind regulatory proteins, such as 5'-UTR, 3'-UTR and regions close to the start site | |
| Choose sequences with GC content between 30% and 70% | |
| Avoid highly G-rich sequences | |
| Design sense and antisense N19 sequences, add two 2-deoxythymidine residues to the 3' ends | |
| Perform BLAST search to exclude potential homology to other genes | |
| Additional considerations for vector-based siRNA expression | Avoid more than three consecutive As or Ts in the targeting sequence |
| U6 promotor requires a guanine at position +1 | |
| H1 promotor prefers adenosine at position +1 | |
| Design oligonucleotides containing N19 targeting sequence, a loop forming spacer sequence (often 5'-TTCAAGAGA-3'), followed by the reverse complementary targeting sequence and five to six consecutive thymidine residues for termination of transcription | |
| Add respective restriction sites for cloning | |
| siRNA design tools on the internet | http://www.ambion.com |
| http://www.qiagen.com/siRNA | |
| http://jura.wi.mit.edu/bio/ | |
| http://www.dharmacon.com | |
| http://sinc.sunysb.edu/Stu/shilin/rnai.html |
Rules for the design of synthetic siRNAs according to Tuschl and coworkers [27] and some further considerations for vector-based small hairpin RNA (shRNA) expression are given. A collection of links to small interfering RNA (siRNA) design tools on the internet is provided. nt, nucleotide; UTR, untranslated region.
Two recent reports [29,30] found that the decision regarding which of the two strands of a siRNA molecule is incorporated into RISC was crucial in determining the efficiency of gene silencing. In order to target specifically a given mRNA for degradation, the antisense strand of the siRNA duplex, which is complementary to the mRNA, must be incorporated into the activated RISC. Schwarz and coworkers [29] and Khvorova and colleagues [30] found that the absolute and relative stabilities of the base pairs at the 5' ends of the two siRNA strands determine the degree to which each strand participates in the RNAi pathway. The strand with lower 5' end stability is preferred. As a consequence, a highly functional siRNA is characterized by lower internal stability at the 5' end of the antisense strand as compared with less effective duplexes. A further improved algorithm for the prediction of siRNA efficiency is highly desirable and will enable us to to improve quality and efficiency, and reduce the cost of the technology.
Modes of application and routes into the cell
To induce RNAi in mammalian cells, siRNA can either be directly transfected or produced endogenously within the target cell from expression plasmids [22,31-34]. Synthetic siRNA can be generated by chemical synthesis, by in vitro transcription using a T7 polymerase [34,35], or by Dicer digestion of long dsRNA [36]. Synthesized siRNA induces potent silencing at concentrations of 1–10 nmol/l [13].
siRNA expression vectors utilize mostly U6-snRNA or H1 (RNase P) promoters, both of which are members of the RNA polymerase III promoter family, which lack downstream transcriptional elements and produce a transcript without a cap or poly-A tail [37]. Transcription is terminated at a stretch of five to six thymidine residues, leading to the incorporation of two to three uracil residues at the 3' end, which is compatible with the two or three nt overhangs that are found to be indispensable for silencing activity in natural siRNAs.
Sense and antisense strands are either produced from two independent promoters and anneal within the cell [31], or more commonly the two strands are linked by a 9 base pair spacer leading to the expression of a stem-loop structure termed short hairpin RNA (shRNA). The hairpin is subsequently cleaved by Dicer to generate effective siRNA molecules [22,33,34,38] (Fig. 2). By incorporating a drug resistance gene or via episomally replicating plasmids, a long-lived knockdown effect can be achieved in cultured cells [31,39]. To facilitate the analysis of genes that are essential for cell survival and cell cycle regulation, two groups have generated inducible shRNA expression systems [40,41]. However, the specificity of gene knockdown must be tightly controlled, because Bridge and coworkers [42] recently reported the induction of an interferon response by a substantial number of shRNA expression vectors tested, perhaps caused by the accumulation of nonprocessed Pol III transcripts within the cell.
Figure 2.
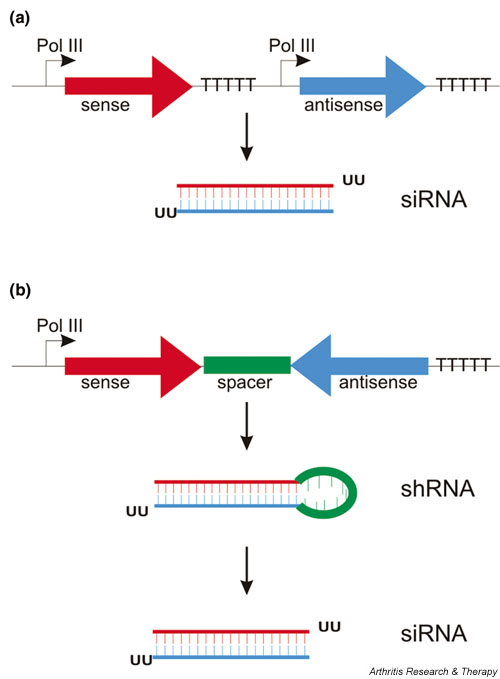
Approaches to endogenous expression of siRNAs in mammalian cells. (a) Sense and antisense strand of the siRNA duplex are expressed from separate promoters. (b) siRNA duplex is expressed as a stem-loop structure (small hairpin RNA [shRNA]) from a single promotor. Sense and antisense strands are separated by a loop-forming spacer. The construct is further processed by Dicer within the cell to form a functional siRNA. In both cases transcription is terminated by six consecutive thymidine residues.
Gene silencing occurs very rapidly after the transfection of an efficient siRNA. Although the kinetics may vary depending on the gene of interest, usually target mRNA levels will be diminished after 48 hours, reaching a minimum at 72 hours after transfection. A knockdown efficiency of 90–95% reduction in the amount of target mRNA can be achieved. However, the major drawback of the method is its transient gene silencing effect. The duration of the knockdown using synthetic siRNA is generally in the range of 3–5 days. Protein levels will return to normal 5–7 days after transfection [16,27]. The longevity of silencing depends on factors such as the abundance of target mRNA and protein, the stability of target protein, transcriptional feedback loops, and the number of cell divisions diluting the siRNA, rather than on the degradation of the siRNA itself.
Both chemically synthesized siRNA and shRNA expression plasmids can be delivered to cells using standard transfection methods. Thereby, the efficiency mainly depends on the type of cell that is targeted. Because of their small size, transfection of synthetic siRNAs is usually very efficient, even in primary mammalian cells. A number of cationic lipid-based or liposome-based transfection reagents optimized for the transfection of oligonucleotides are commercially available. In cells that are more resistant to chemical transfection methods (e.g. suspension cells), electroporation may achieve an efficient induction of RNAi. Transduction rates with siRNA of up to 80–90% have been reported for some haematopoietic cell lines and primary cells [43,44]. Optimized for the transfection of primary human cells with siRNA, Nucleofection™ technology (Amaxa biosystems, Cologne, Germany) appears to be a very efficient and convenient approach [45,46].
When using these conventional transfection strategies, the silencing effect is only transient. Exceptions are established cell lines that allow selection for integrated vectors. Viral gene delivery systems are perfectly suited to overcome these limitations; they are well established tools for efficient transduction of primary cells and some of them have the inherent ability to integrate into the host cell genome, thereby leading to stable transgene expression. Several adenoviral [47,48], onco-retroviral [49-51] and lentiviral [52-54] vectors have been utilized for the efficient delivery of shRNA expression cassettes. Adenoviral infection is transient whereas onco-retroviral vectors, based on the Moloney murine leukaemia virus or the murine stem cell virus, integrate into the host cell genome, leading to a prolonged silencing effect. Lentiviral vectors based on HIV-1 bear the additional advantage of efficiently transducing both dividing as well as nondividing cells, such as stem cells and terminally differentiated cells. Moreover, they are resistant to developmental silencing after integration of the provirus, and therefore they can be used to generate transgenic animals. Several groups have reported the use of lentiviral systems for the silencing of genes in a variety of cultured as well as primary cells, such as human and murine T cells [52,53], haematopoietic stem cells [53] and mouse dendritic cells [53,54]. Although onco-retroviruses and lentiviruses hold great promise as vehicles for gene therapy, two patients who were undergoing retroviral based therapy for X-linked severe combined immunodeficiency developed leukaemia [55,56]. This indicates that improved safety standards and ways to control the integration of the provirus are needed before retroviruses can be used to deliver siRNA for therapeutic purposes.
Towards in vivo application of small interfering RNA
RNAi has already been proven to be a powerful tool for dissecting and elucidating gene function, even on a genome-wide basis. The first example comes from C. elegans, in which Kamath and coworkers [57] reported the construction of a library of bacterial clones that express dsRNA, which corresponds to approximately 86% of the total gene products made by C. elegans. Also, the library has been used to screen for genes that are involved in body fat regulation, longevity and genome stability [58-60].
Thus far, in vivo gene silencing approaches are very limited in the mammalian system. Nonetheless, a number of potential candidate genes, especially in viral infections, cancers and inherited genetic disorders but also in chronic inflammatory diseases such as autoimmune arthritis, has been defined and successfully targeted in vitro. Consistent with its natural function as an antiviral defence mechanism, siRNA was found to inhibit in vitro replication of several viruses effectively, including HIV, hepatitis C virus and influenza virus, by interfering with various stages of the virus life cycles [38,52,61-67].
Similarly, several cancer-related genes have been targeted in proof-of-principle experiments, including cellular oncogenes and drug resistance genes. In these studies, RNAi was efficient and highly selective in targeting oncogenes resulting from chromosomal translocations [43,68] or carrying single point mutations, without affecting the wild-type allele [50,69].
Protocols must be established for efficient delivery of siRNA and selective targeting of specific cell types in order to allow future therapeutic applications and in vivo verification of results obtained from in vitro silencing experiments. Moreover, it must be determined whether transient gene silencing, as obtained by introduction of synthetic siRNA or expression plasmids, is sufficient for treatment, or whether the target gene must be silenced for an extended period of time by the use of viral expression systems.
Direct injection of siRNA into the blood would be ineffective because of rapid degradation of the RNA by serum ribonucleases. However, it was recently demonstrated that chemical modification can protect the siRNA molecule from degradation [70] and might even prolong the silencing effect due to slower depletion within the cell [71]. Thus far, synthetic siRNAs have been applied in animals via hydrodynamic transfection [72] (i.e. the intravenous injection of a substantial dose of siRNA within a large volume of liquid), resulting in a knockdown efficiency up to 70–80%, at least in some organs, including liver, kidney, spleen, lung and pancreas [73]. Using this method, the silencing of either Fas receptor [74] or caspase-8 [75] resulted in a clearly measurable protection from severe Fas-induced liver damage. In vivo application of siRNA against genes of the hepatitis B virus also led to an effective inhibition of virus replication [76].
This method is of course not applicable to humans. It is also limited by the fact that siRNA can only be delivered to a certain set of organs and it is not possible to target specific organs or cells. Development of cell-specific or organ-specific delivery systems for siRNA, as is required for broad in vivo application of this technique, is indeed a demanding task.
Prolonged gene silencing by stable integration of a siRNA expression vector is currently only possible in vitro. The subsequent in vivo adoptive transfer of these in vitro manipulated cells is an option in situations where a small number of cells can develop a dominant phenotype in vivo. This is the case for stem cells (e.g. embryonic stem [ES] cells) or haematopoietic stem cells, which either give rise to a complete new animal or at least generate defined organs.
An approach using siRNA-modified stem cells would be particularly useful for the analysis of gene function in vivo. So far this has mainly been done in knockout mice, which carry a nonfunctional mutation of the target gene, generated by homologous recombination in ES cells. The technique suffers from a number of limitations that could be overcome by RNAi technology, such as the need for cloning of the target gene, the time and effort required for generating a knockout mouse, and the potential embryonic lethality. In contrast to the all-or-nothing phenotype obtained from knockout animals, analysis of gene dosage effects may be possible by using siRNAs with variable silencing efficiency. Finally, the combination of multiple loss-of-function phenotypes in one generation would be possible. Lentiviral siRNA vectors have been used to generate stable transgenic 'knockdown' animals by infection of fertilized eggs [77]. In another study, Rubinson and coworkers [53] used lentiviral vectors expressing green fluorescent protein as a selection marker and an siRNA targeting CD8 for embryo infection. Between 25% and 50% of the resulting mice were transgenic and expressed both green fluorescent protein and siRNA in all tissues tested. Transgenic mice exhibited a reduction in CD8 expression of about 90%; however, the percentage of cells affected by gene silencing varied among individual mice and correlated with the number of integrated viruses per genome. Therefore, different siRNA expression levels may account for this variance. In an alternative approach, not involving the use of lentiviral vectors, transgenic 'knockdown' mice were generated by transfecting ES cells with a siRNA expression plasmid containing a drug resistance gene [78].
The adoptive transfer of in vitro modified cells may also be applicable to the modulation of an antigen-specific immune response (e.g. for the treatment of autoimmune diseases, allergies, or organ rejection). In these situations, a relatively small population of antigen-specific lymphocytes or antigen-presenting cells, previously modified by siRNA in vitro, may later dominate an antigen-specific immune response in vivo. This has recently been demonstrated by transfer of dendritic cells transfected with an siRNA against the immunomodulatory cytokine interleukin-12 [79]. However, for therapeutic use in humans, both the safety of stably transfected cells and the target specificity of the siRNA must be controlled more closely.
Conclusion
RNAi has rapidly evolved as a potent technology for the analysis of gene function in many organisms in vitro and in vivo. In mammals, at present RNAi is mainly restricted to the analysis of easily transfectable cell lines in vitro, but here it has already proven its efficiency in targeting a number of therapeutically relevant genes with high specificity. Recent work has set the scene for addressing gene function in primary cells both in vitro and in vivo, which is more pertinent to the definition of disease-related pathways and potential therapeutic targets. However, for therapeutic applications of siRNA in humans, new strategies must be developed that will allow the efficient and specific targeting of distinct organs or cell types.
Competing interests
None declared.
Abbreviations
dsRNA = double-stranded RNA; ES = embryonic stem (cell); nt = nucleotide; RISC = RNA-induced silencing protein complex; RNAi = RNA interference; shRNA = small hairpin RNA; siRNA = small interfering RNA.
Acknowledgments
Acknowledgements
We were unable to cite all relevant publications because of space constraints. Farah Hatam is gratefully acknowledged for critical reading of the manuscript. SR was supported by a grant from the Boehringer Ingelheim Fonds.
References
- Tijsterman M, Ketting RF, Plasterk RH. The genetics of RNA silencing. Annu Rev Genet. 2002;36:489–519. doi: 10.1146/annurev.genet.36.043002.091619. [DOI] [PubMed] [Google Scholar]
- Vance V, Vaucheret H. RNA silencing in plants: defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- Plasterk RH. RNA silencing: the genome's immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Gil J, Esteban M. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis. 2000;5:107–114. doi: 10.1023/A:1009664109241. [DOI] [PubMed] [Google Scholar]
- Player MR, Torrence PF. The 2–5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Chi JT, Chang HY, Wang NN, Chang DS, Dunphy N, Brown PO. Genomewide view of gene silencing by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci USA. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3' overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze O, Picard D. RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res. 2002;30:e46. doi: 10.1093/nar/30.10.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JW, Jones JT, Meyer T, Ferrell JE., Jr Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat Biotechnol. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar eB, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci USA. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de WM, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Schoer RA, Egan JE, Hannon GJ, Mittal V. From the cover: inducible, reversible, and stable RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2004;101:1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Scherr M, Battmer K, Winkler T, Heidenreich O, Ganser A, Eder M. Specific inhibition of bcr-abl gene expression by small interfering RNA. Blood. 2003;101:1566–1569. doi: 10.1182/blood-2002-06-1685. [DOI] [PubMed] [Google Scholar]
- McManus MT, Haines BB, Dillon CP, Whitehurst CE, Van Parijs L, Chen J, Sharp PA. Small interfering RNA-mediated gene silencing in T lymphocytes. J Immunol. 2002;169:5754–5760. doi: 10.4049/jimmunol.169.10.5754. [DOI] [PubMed] [Google Scholar]
- Bidere N, Lorenzo HK, Carmona S, Laforge M, Harper F, Dumont C, Senik A. Cathepsin D triggers Bax activation, resulting in selective apoptosis-inducing factor (AIF) relocation in T lymphocytes entering the early commitment phase to apoptosis. J Biol Chem. 2003;278:31401–31411. doi: 10.1074/jbc.M301911200. [DOI] [PubMed] [Google Scholar]
- Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, Dale JK, Puck J, Davis J, Hall CG, Skoda-Smith S, Atkinson TP, Straus SE, Lenardo MJ. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Shen C, Buck AK, Liu X, Winkler M, Reske SN. Gene silencing by adenovirus-delivered siRNA. FEBS Lett. 2003;539:111–114. doi: 10.1016/S0014-5793(03)00209-6. [DOI] [PubMed] [Google Scholar]
- Barton GM, Medzhitov R. Retroviral delivery of small interfering RNA into primary cells. Proc Natl Acad Sci USA. 2002;99:14943–14945. doi: 10.1073/pnas.242594499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/S1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Devroe E, Silver PA. Retrovirus-delivered siRNA. BMC Biotechnol. 2002;2:15. doi: 10.1186/1472-6750-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, Weinberg RA, Novina CD. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, Cavazzana-Calvo M, Fischer A. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- Marshall E. Gene therapy. Second child in French trial is found to have leukemia. Science. 2003;299:320. doi: 10.1126/science.299.5605.320. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, Tijsterman M. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci USA. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Surabhi RM, Gaynor RB. RNA interference directed against viral and cellular targets inhibits human immunodeficiency Virus Type 1 replication. J Virol. 2002;76:12963–12973. doi: 10.1128/JVI.76.24.12963-12973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Lee SK, Dykxhoorn DM, Novina C, Zhang D, Crawford K, Cerny J, Sharp PA, Lieberman J, Manjunath N, Shankar P. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J Virol. 2003;77:7174–7181. doi: 10.1128/JVI.77.13.7174-7181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, Chen J. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilda M, Fuchs U, Wossmann W, Borkhardt A. Killing of leukemic cells with a BCR/ABL fusion gene by RNA interference (RNAi) Oncogene. 2002;21:5716–5724. doi: 10.1038/sj.onc.1205653. [DOI] [PubMed] [Google Scholar]
- Ding H, Schwarz DS, Keene A, Affar el B, Fenton L, Xia X, Shi Y, Zamore PD, Xu Z. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell. 2003;2:209–217. doi: 10.1046/j.1474-9728.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, Chen J, Shankar P, Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Zender L, Hutker S, Liedtke C, Tillmann HL, Zender S, Mundt B, Waltemathe M, Gosling T, Flemming P, Malek NP, Trautwein C, Manns MP, Kuhnel F, Kubicka S. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci USA. 2003;100:7797–7802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Gish G, Lickert H, Jones N, Pawson T, Rossant J. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat Biotechnol. 2003;21:559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- Hill JA, Ichim TE, Kusznieruk KP, Li M, Huang X, Yan X, Zhong R, Cairns E, Bell DA, Min WP. Immune modulation by silencing IL-12 production in dendritic cells using small interfering RNA. J Immunol. 2003;171:691–696. doi: 10.4049/jimmunol.171.2.691. [DOI] [PubMed] [Google Scholar]


