Abstract
In this study the sterol and oxysterol profile of newborn brain from the Dhcr7Δ3-5/T93M mouse model of Smith–Lemli–Opitz syndrome (SLOS) has been investigated. This is a viable mouse model which is compound heterozygous containing one null allele and one T93M mutation on Dhcr7. We find the SLOS mouse has reduced levels of cholesterol and desmosterol and increased levels of 7- and 8-dehydrocholesterol and of 7- and 8-dehydrodesmosterol in brain compared to the wild type. The profile of enzymatically formed oxysterols in the SLOS mouse resembles that in the wild type but the level of 24S-hydroxycholesterol, the dominating cholesterol metabolite, is reduced in a similar proportion to that of cholesterol. A number of oxysterols abundant in the SLOS mouse are probably derived from 7-dehydrocholesterol, however, the mechanism of their formation is unclear.
Keywords: Cholesterol, 7-Dehydrocholesterol, 3β-Hydroxysterol-Δ7-reductase, Hydroxy-7-dehydrocholesterol, 7-Dehydrodesmosterol
1. Introduction
Smith–Lemli–Opitz syndrome (SLOS) is an autosomal recessive disorder characterised by a deficiency of 3β-hydroxysterol-Δ7-reductase (7-dehydrocholesterol reductase, DHCR7, EC 1.3.1.21), which catalyses conversion of 7-dehydrocholesterol (7-DHC) into cholesterol in the last step of cholesterol biosynthesis via the Kandustch–Russell pathway [1]. Reduced activity of DHCR7 results in elevated levels of the cholesterol precursor 7-DHC, and of 8-DHC, in tissues and in the circulation [2-4]. The disorder leads to dysmorphia and mental retardation [5]. SLOS has a relatively high incidence ranging from 1 in 10,000 to 1 in 60,000 in both Europe and America. For Caucasian populations carrier frequency for mutant alleles may be as high as 1 in 30, but it is probable that the condition often goes undiagnosed as patients with a mild disorder may have little evident phenotype, while early foetal loss may result in the severely affected [6]. Diagnosis is based on elevated plasma levels of DHC [4]. A number of mouse models for SLOS exist, these include those with a null mutation in Dhcr7, but homozygous animals die within 1 day of birth [7,8]. More recently a viable mouse model has been described which is compound heterozygous, containing one null allele (deletion of coding exons 3,4,5) and one T93M mutation on Dhcr7, i.e. Dhcr7Δ3-5/T93M [9,10]. These are the most severely affected viable SLOS animals and here we investigate the sterol/oxysterol content of their brain at birth.
In the adult mammal essentially all cholesterol in brain is derived through in situ biosynthesis from acetyl CoA [11], this is also true in the newborn mouse where more than 90% of cholesterol is synthesised in brain [12]. In the developing foetus after closure of the blood brain barrier (BBB), and in the very young animal, desmosterol, 7-DHC and also 7-dehydrodesmosterol (7-DHD) levels are elevated indicating rapid de novo synthesis of sterols through both the Kandustch–Russell and Bloch pathways [13]. The BBB is impervious to cholesterol excluding both its export and import. This defines the necessity for in situ de novo synthesis of cholesterol in brain and also its metabolism to sterols capable of passing the BBB and into the circulation [14]. What then is the situation in SLOS brain where 7-DHC levels are expected to be elevated and those of cholesterol to be reduced? In the current study we have attempted to answer this question by investigating the sterol and oxysterol profile of the newborn Dhcr7Δ3-5/T93M SLOS mouse.
2. Materials and methods
2.1. Dhcr7Δ3-5/T93M mice
Compound heterozygous Dhcr7Δ3-5/T93M mice were generated by crossing heterozygous phenotypically normal Dhcr7+/Δ3-5 females with homozygous Dhcr7T93M/T93M males as described in Marcos et al. [10]. Phenotypically normal + /T93M littermates were used as controls. All animal work conformed to NIH guidelines and was approved by Institutional Animal Care and Use Committee of the Children’s Hospital Oakland Research Institute.
2.2. Materials
Sterol/oxysterol standards were obtained from Avanti Polar Lipids (Alabaster, AL, USA), Steraloids, Inc. (Newport, RI, USA) or from previous studies in our laboratories [13]. (24E)26-Hydroxydesmosterol and (24Z)26-hydroxydesmosterol were kindly provided by Prof. Hans-Joachim Knölker from Technische Universität in Dresden. Cholesterol oxidase from Streptomyces sp. was from Sigma–Aldrich (St. Louis, MO, USA) and Girard P (GP) reagent [1-(carboxymethyl)pyridinium chloride hydrazide] from TCI Europe (Zwijndrecht, Belgium). Solid phase extraction (SPE) cartridges, Certified Sep-Pak C18, 200 mg, were from Waters, Inc. (Elstree, UK). Solvents were from Fisher-Scientific (Loughborough, Leicestershire, UK). Water, acetonitrile, methanol, ethanol and propan-2-ol were HPLC grade. Acetic acid 100% was from VWR International, Ltd. (Poole, Dorset, UK) and formic acid 98/100% from VWR International S.A.S. (Briare, France). Potassium phosphate buffer made from potassium dihydrogen phosphate (KH2PO4) was from Sigma–Aldrich (Japan).
Stock solutions of internal standards were made by dissolving 1 mg of 24R/S-[26,26,26,27,27,27-2H6]hydroxycholesterol and 10 mg of [25,26,26,26,27,27,27-2H7]cholesterol in 10 mL volumes of propan-2-ol. Ten microlitres of the oxysterol stock solution was diluted with 990 μL of ethanol to make a working solution of 1 ng/μL.
2.3. Isolation of sterols/oxysterols from newborn mouse brain
Mice were sacrificed and dissected brains immediately frozen in liquid nitrogen. Entire litters of newborn animals were sacrificed at one time. Whole brain (60–130 mg) was homogenised and sterols extracted in methanol:chloroform (1:1, v/v), and re-extracted in methanol. The dried extracts were transported from Oakland, CA to Swansea, UK in glass tubes refrigerated below −20 °C until analysis. The lipid extracts were re-constituted in 1.05 mL of ethanol containing 50 ng of 24R/S-[2H6]hydroxycholesterol and 50 μg of [2H7]cholesterol and ultrasonicated for 15 min at ambient temperature. The ethanolic extract was diluted with 0.45 mL of water and the resulting solution was sonicated for another 15 min. This mixture was centrifuged at 14,000 × g at 4 °C for 60 min and the supernatant was retained. This procedure was repeated on the lipid residue (with another 1.05 mL of ethanol containing internal standards followed by addition of 0.45 mL of water) and the supernatants pooled to give a final volume 3 mL of 70% ethanol containing 100 ng of 24R/S-[2H6]hydroxycholesterol and 100 μg of [2H7]cholesterol.
Oxysterols were separated from cholesterol and other sterols of similar hydrophobic nature (including desmosterol, 7- and 8-DHC and 7- and 8-DHD) by reversed-phase (RP) SPE on a 200 mg Certified Sep-Pak C18 cartridge as described by Meljon et al. [13]. The resultant oxysterol and cholesterol rich fractions (i.e. SPE1-FR1 and SPE1-FR3, respectively) were then split into two equal volumes, i.e. A and B, each of which was dried under reduced pressure and reconstituted in 100 μL of propan-2-ol.
2.4. Oxidation of 3β-hydroxy-5-ene sterols/oxysterols with cholesterol oxidase from Streptomyces sp. and derivatisation with Girard P reagent
Neutral sterols/oxysterols are neither strong proton donors nor strong proton acceptors, therefore to aid subsequent electrospray ionisation – mass spectrometry (ESI-MS) and tandem mass spectrometry (MSn) analysis we decided to use a derivatisation method to enhance ion formation. Steroids possessing a 3β-hydroxy-5-ene or 3β-hydroxy-5α-hydrogen structure were first transformed to 3-oxo-4-ene or 3-oxo analogues (Fig. 1A). This was achieved using the enzyme cholesterol oxidase [15]. Fractions A from above were incubated with cholesterol oxidase (0.26 u) in 1 mL of phosphate buffer while B fractions were incubated in buffer in the absence of enzyme. After 60 min at 37 °C the reaction was terminated by the addition of 2 mL of methanol. Then, 0.15 mL of glacial acetic acid and 0.15 g of GP hydrazine was added to each of the A and B fractions. The mixture was incubated at ambient temperature over night and protected from light (Fig. 1A). As GP hydrazine is added to the reaction mixture in great excess to provide a high yield of derivatised steroids it should subsequently be removed prior to ESI-MS analysis. Some workers have developed liquid chromatography (LC)–MS systems utilising a trap column and a switching system to wash away excess derivatisation reagent prior to sterol separation on an analytical LC-column and MS analysis [16,17], but we prefer to include a RP-SPE recycling step on a second Sep-Pak C18 cartridge (SPE2) [13]. Utilisation of a recycling method overcomes the problem of limited solubility of derivatised sterols/oxysterols in highly aqueous solvents necessary to trap the analytes on the trap column.
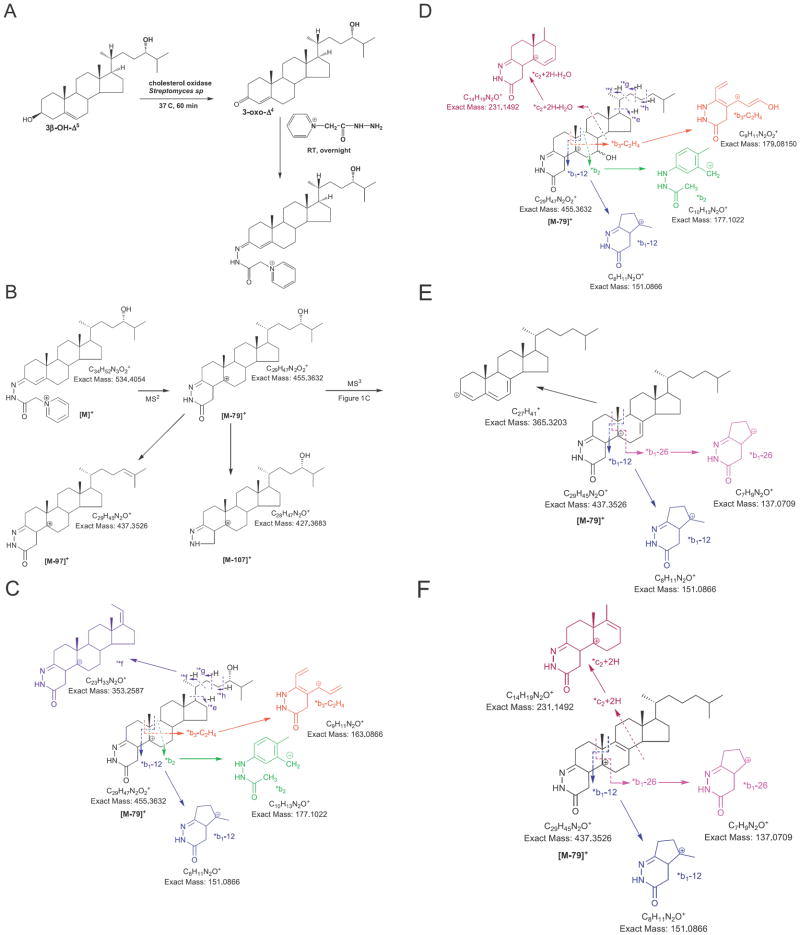
Fig. 1.
Charge-tagging of sterols/oxysterols. (A) Oxidation of 3β-hydroxy-5-ene steroids to 3-oxo-4-ene equivalents followed by derivatisation with GP reagent. (B) MS2 fragmentation of GP-tagged steroids. (C) MS3 fragmentation of the [M−79]+ ion from B. In A, B and C, 24S-hydroxycholesterol is the exemplar. (D) The effect of 7-hydroxylation on MS3 fragmentation. (E) MS3 fragmentation of GP-tagged 7-DHC. (F) MS3 fragmentation of GP-tagged 8-DHC.
2.5. LC–ESI-MS and MSn analysis
The LC–MS(MSn) system consisted of an LTQ Orbitrap XL or an LTQ Orbitrap Velos (Thermo Fisher Scientific, UK) equipped with an ESI probe coupled to a Dionex Ultimate 3000 HPLC system (Dionex, UK) and was operated essentially as described previously [13]. Chromatographic separation was performed using a Hypersil Gold RP column (50 mm × 2.1 mm, 1.9 μm, Thermo-Scientific) at room temperature. Mobile phase A consisted of 0.1% formic acid in 33.3% methanol, 16.7% acetonitrile. Mobile phase B consisted of 0.1% formic acid in 63.3% methanol, 31.7% acetonitrile. For the separation of derivatised oxysterols we used two different LC gradient methods. The first one provides a separation of the majority of oxysterols and sterols, but cannot resolve to base line 24S-, 25-, 24R- and (25R)26-hydroxycholesterol or 24S,25-epoxycholesterol and (24Z)26-hydroxydesmosterol. The second, longer gradient was developed in order to provide a more accurate quantification of these oxysterols. Note in this article we use the nomenclature recommended by Lipid Maps where hydroxylation of the terminal carbon of cholesterol introducing 25R stereochemistry is at C-26 [18]. Thus, (25R)26-hydroxycholesterol is named 26-hydroxycholesterol. It should be noted that many articles refer to this molecule as 27-hydroxycholesterol. The shorter LC gradient (method 1) was as follows. After 1 min at 20% B, the proportion of B was raised to 80% B over the next 7 min and maintained at 80% B for further 5 min, before returning to 20% B in 0.1 min. The column was re-equilibrated for a further 3.9 min, giving a total run of 17 min. The flow rate was 200 μL/min, and the eluent was directed to the ESI source of the LTQ-Orbitrap mass spectrometer. The longer LC gradient (method 2) was as follows. After 1 min at 20% B, the proportion of B was raised to 55% B over the next 19 min using gradient curve 8 (Chromeleon software, Dionex). The proportion of B was then increased to 80% over 10 min, before returning to 20% B in 0.1 min. The column was re-equilibrated for a 3.9 min giving a total run time of 34 min. The flow rate was 200 μL/min, and the eluent was directed to the ESI source of the LTQ-Orbitrap mass spectrometer.
GP derivativatised sterols/oxysterols derived from brain were injected on to the LC column in 60% methanol, 0.1% formic acid. GP-tagged steroids are stable in 100% methanol, the stability of GP-tagged steroids in 60% methanol 0.1% formic acid is limited and the analytes diluted to 60% methanol solution were injected within 24 h.
The LTQ-Orbitap was operated utilising three scan events. First, a Fourier transform (FT)MS scan in the Orbitrap over the m/z range 400–605 at 30,000 resolution (full width at half-maximum height, FWHM, definition) was performed, followed by data dependent MS2 ([M]+→) and MS3 ([M]+ → [M−79]+→) events in the LTQ linear ion trap (LIT). These MSn scans in the LIT were performed in parallel to acquisition of the high-resolution FTMS scan by the Orbitrap. A precursor-ion inclusion list was defined according to the m/z of the [M]+ ions of expected sterols/oxysterols so that MS2 was preferentially performed on these ions in the LIT if their intensity exceeded a preset minimum. If a fragment-ion corresponding to the neutral loss of 79 Da (loss of pyridine, Fig. 1B) from the precursor-ion was observed in the MS2 event and the signal was above a preset minimum, MS3 was performed on this fragment.
2.6. Quantification of sterols/oxysterols
Sterols/oxysterol were quantified by the stable isotope dilution method. The internal standard used for quantification of oxysterols was 24R/S-[2H6]hydroxycholesterol, while sterols were quantified against [2H7]cholesterol. Previous studies have shown that once GP-tagged sterols/oxysterols with a 3-oxo-4-ene structure give a similar response upon analysis by LC–ESI-MS [19]. This allows the general use of 24R/S-[2H6]hydroxycholesterol and [2H7]cholesterol as internal standards for oxysterols and sterols, respectively. While isotope dilution gives quantitative values in brain for the native molecules of these two surrogates, values for other oxysterols and sterols are formally quantitative estimates.
3. Results
In brain of normal adult animals the level of cholesterol exceeds that of the most abundant oxysterol by a factor of more than 500, and of minor oxysterols by more than 1,000,000. Thus, even a minor degree of autoxidation of cholesterol can lead to the artefactual formation of oxysterols at levels equivalent or greater than those found endogenously. Similarly, 7-DHC is also susceptible to autoxidation, even more so than cholesterol [20]. So to minimise the possibility of autoxidation during sample work-up cholesterol and other sterols of similar polarity were separated from oxysterols at an initial stage of sample preparation. To achieve this, the brain extract was fractionated on the first Sep-Pak C18 column (SPE1). Oxysterols eluted in fraction SPE1-FR1, while cholesterol and similarly hydrophobic sterols eluted in fraction SPE1-FR3. Following derivatisation and subsequent purification on a second Sep-Pak C18 column GP-tagged molecules were submitted to LC–ESI-MS(MSn) analysis. Sterols in brain are known to be present almost exclusively in their free non-esterified form [11]; this alleviates the need for base hydrolysis at elevated temperature and removes a potential source of autoxidation. In our experience the greatest danger of autoxidation presents itself when cholesterol rich samples are heated in air [21]. Here we avoid this possibility by conducting all sample work-up at room temperature, separating cholesterol from oxysterols and storing all samples at −20 °C or below. In our experience when these precautions are taken autoxidation is minimised and there is no evidence for artefact oxysterols showing side-chain oxidation [19,21]. This was confirmed here by searching for autoxidation products of [2H7]cholesterol. Only trace levels of B-ring autoxidation products were observed.
GP charge-tagged compounds were separated by RP-LC linked to the ESI source of an LTQ-Orbitrap mass spectrometer. The eluting compounds were identified using LC retention time, exact mass measurement, MS3 fragmentation profile and comparison to authentic standards where the standards were available. In the absence of authentic standards “presumptive identifications” were made based on these parameters. The FTMS high-resolution scan generates spectra with mass accuracy, better than 5 ppm, allowing the creation of reconstructed of ion chromatograms (RICs) for selected m/z values. Subsequent MS2 ([M]+→) and MS3 ([M]+ → [M−79]+→) spectra were preferentially recorded on target ions on an include list. MS3 spectra were recorded only on productions formed as a result of the neutral loss of 79 Da from the [M]+ ion in the MS2 event (Fig. 1B). The loss of 79 Da is characteristic of GP-tagged molecules and provides an additional step in the identification and purification processes.
In our mouse model of SLOS, cholesterol synthesis should be impaired by the heterozygous T93M “knock-in” mutation and deletion of exons 3, 4 and 5 in Dhcr7. The enzyme Dhcr7 reduces the Δ7 double bond in 7-DHC and 7-DHD, leading to formation of cholesterol and desmosterol via the Kandustch–Russell and Bloch pathways, respectively. To investigate the effect of these genetic manipulations in our SLOS mouse model we quantified the substrates and products of Dhcr7, i.e. 7-DHC, 7-DHD, cholesterol and desmosterol. We also measured the levels of 8-DHC and 8-DHD which are isomersisation products of 7-DHC and 7-DHD [22], respectively.
3.1. 7-DHC, 8-DHC, 7-DHD, 8-DHD, desmosterol and cholesterol
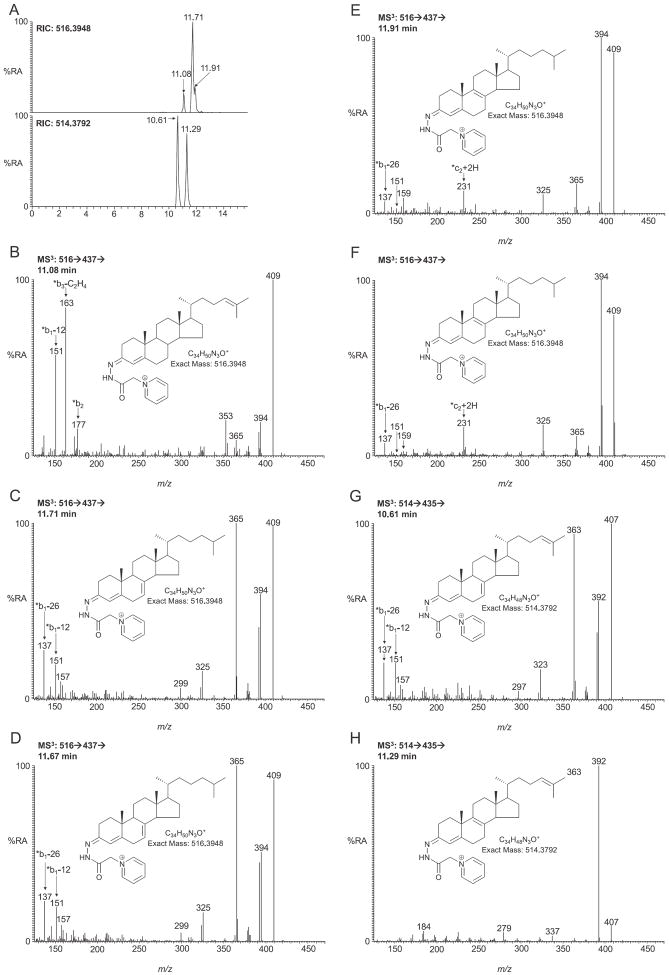
GP-tagged DHCs give an [M]+ ion of m/z of 516.3948. The RIC for this m/z from newborn SLOS mouse brain is shown in Fig. 2A (upper panel). The chromatogram was acquired using the short LC gradient (method 1). The MS3 ([M]+ → [M−79]+→) spectrum of the major peak eluting at 11.71 min (Fig. 2C) shows low mass fragment ions at m/z 137 (*b1-26) and 151 (*b1-12) characteristic of the GP-derivatised 3-oxo-4,7-diene structure (generated by cholesterol oxidase treatment of 7-DHC followed by GP derivatisation, Fig. 1E) [3]. A spectrum of the authentic GP-tagged standard is shown for comparison in Fig. 2D. Other ions characteristic of DHCs include m/z 409 formed by the loss of the CO group from [M−79]+ ion, m/z 394 formed by the additional loss of NH, and m/z 365 corresponding to the carbocation of the triply unsaturated sterol (Fig. 1E). The level of 7-DHC in brain of SLOS mice was measured to be 316 ± 75 ng/mg (n = 4, mean ± SE). As is evident in Fig. 2A, a later eluting component appears as a shoulder to the peak corresponding to 7-DHC. This is identified as 8-DHC (cholesta-5,8(9)-dien-3β-ol) (Fig. 2E), the spectrum of the authentic standard is shown in Fig. 2F. The MS3 spectrum is quite different to that of the 7-DHC isomer, with far less abundant ions at m/z 365, 151 and 137, but a more abundant ion at m/z 231 (*c2 + 2H, Fig. 1F). Previously we surmised this compound to be cholesta-4,6-dien-3β-ol, possibly formed as an artefact of our derivatisation methodology from 7-DHC [13], but analysis of authentic standards of 7-DHC and newly the available 8-DHC standard confirm its identity as endogenous 8-DHC. The level of this isomer was estimated to be 134 ± 34 ng/mg in SLOS animals. As 8-DHC is believed to be formed from 7-DHC [22], their combined abundance, 450 ng/mg, is representative of the immediate cholesterol precursors in the Kandutsch–Russell pathway. For comparison the equivalent value in the wild type mouse is 252 ng/mg. In the chromatogram of the SLOS mouse an earlier eluting peak (11.08 min in Fig. 2A) corresponding to another dehydrocholesterol isomer was observed. The retention time, exact mass and MS3 spectrum define this peak to correspond to desmosterol (Fig. 2B, a library of MS3 spectra of common GP-tagged sterols is available at http://sterolanalysis.org.uk/). Unsurprisingly, in light of the requirement of the Dhcr7 enzyme to produce desmosterol via the Bloch pathway the desmosterol level is diminished in the SLOS mouse (130 ± 30 ng/mg) compared the wild type (440 ± 88 ng/mg). In the Bloch pathway the precursor of desmosterol is 7-DHD. This metabolite following GP-tagging has an m/z of 514.3792. Two components from SLOS mouse brain elute with this m/z (Fig. 2A, lower panel). The MS3 spectrum of the component eluting at 10.61 min gives a spectrum similar to that of 7-DHC but with high mass fragment ions, resulting from the loss of the derivatising group, displaced down by two mass units (Fig. 2G and C). The A/B ring fragment ions are identical to those observed in the MS3 spectrum of 7-DHC (i.e. m/z 137, 151) indicating that the extra unsaturation is elsewhere, most probably at C-24. The latter chromatographic peak eluting at 11.29 min gives an MS3 spectrum more in line with that predicted for 8-DHD (cholesta-5,8(9),24-trien-3β-ol) (Fig. 2H). However, authentic standards are not available for 7-DHD and 8-DHD to confirm these presumptive identifications. The levels of these two presumptively identified molecules in the SLOS mouse are 136 ± 31 ng/mg and 192 ± 55 ng/mg, respectively. The combined level of 7-DHD and 8-DHD is 328 ng/mg in the SLOS mouse compared to 144 ng/mg in the wild type. As was the case with immediate precursors of cholesterol in the Kandutsch–Russell pathway, there is an elevation in the level of the immediate precursors of desmosterol in the Bloch pathway. The SLOS mouse is viable, hence retains some Dhcr7 enzymatic activity allowing the formation of cholesterol. Its level in the SLOS mouse is 1531 ± 241 ng/mg compared to 2351 ± 255 ng/mg in the wild type mouse.
Fig. 2.
LC–MS(MS)n analysis of DHCs and DHDs from newborn Dhcr7Δ3-5/T93M mouse brain following GP charge-tagging. (A) Upper panel, LC–MS RIC for m/z 516.3948 ± 10 ppm corresponding to DHCs. Lower panel, LC–MS RIC for m/z 514.3792 corresponding to DHDs. The short gradient was employed. MS3 (516 → 437→) spectra of the peaks eluting at (B) 11.08 min corresponding to desmosterol (cholesta-5,24-dien-3β-ol); (C) 11.71 min corresponding to 7-DHC (cholesta-5,7-dien-3β-ol); (D) authentic 7-DHC; (E) 11.91 min corresponding to 8-DHC (cholesta-5,8(9)-dien-3β-ol); (F) authentic 8-DHC. MS3 (514 → 435→) spectra of peaks eluting at (G) 10.61 min corresponding to 7-DHD (cholesta-5,7,24-trien-3β-ol) and (H) 11.29 min corresponding to 8-DHD (cholesta-5,8(9),24-trien-3β-ol). Authentic standards were not available for 7- or 8-DHD, hence identifications are presumptive. Shown as insets in each spectrum are the structures of the GP-tagged molecules.
3.2. 24S,25-Epoxycholesterol and cholesterol and desmosterol derived oxysterols
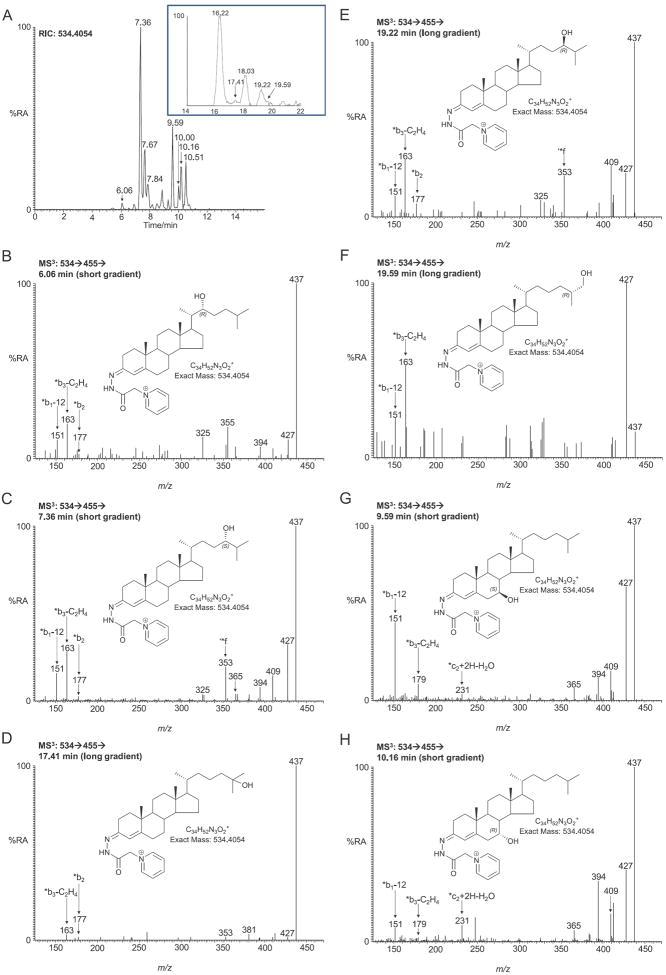
The availability of cholesterol in the SLOS mouse brain suggests that a normal profile of cholesterol derived oxysterols may be observed, but at lower levels than in the wild type animal. Similar to the wild type mouse, the pattern of monohydroxycholesterols in the brain of the newborn SLOS mouse is dominated by 24S-hydroxycholesterol (Fig. 3A). The chromatogram (short gradient) of GP-tagged monohydroxycholesterols (RIC m/z 534.4054) shows two closely eluting peaks at 7.36 and 7.67 min identified as syn and anti conformers of GP-derivatised 24S-hydroxycholesterol. Both peaks give an identical MS3 ([M]+ → [M−79]+→) spectrum showing a characteristic triad of low mass fragment ions at m/z 151 (*b1-12), 163 (*b3-C2H4) and 177 (*b2), and a distinctive ion at 353 (*f, Figs. 3C and 1C). The enzyme responsible for the formation of 24S-hydroxycholesterol from cholesterol is cytochrome P450 46a1 (Cyp 46a1 in mouse and CYP46A1 in human) and is expressed in normal brain exclusively in neurons [23]. In the SLOS newborn mouse the concentration of 24S-hydroxycholesterol in brain was determined to be 0.346 ± 0.041 ng/mg. This value is about 70% that found in the wild type newborn animal (0.510 ± 0.034 ng/mg [13]). This lower level of 24S-hydroxycholesterol in SLOS brain is statistically significant (P < 0.05) and is a reflection of a lower cholesterol concentration in SLOS mouse brain than in the wild type. Application of the short chromatographic gradient as utilised in Fig. 3A only partially resolves 24S-hydroxycholesterol and 25-hydroxycholesterol, 24R-hydroxycholesterol and 26-hydroxycholesterol making quantification of the latter three compounds difficult. To resolve this issue we developed a longer chromatographic gradient giving better separation of these isomers (Fig. 3A, inset). A great advantage of utilising the GP-tag with MS3 fragmentation is that the resulting spectra allow isomer differentiation. For example, the MS3 spectrum of 25-hydroxycholesterol (Fig. 3D) is completely dominated by the fragment ion at m/z 437 resulting from facile dehydration of the [M−79]+ ion and is quite different from MS3 spectra of the other isomers. 25-Hydroxycholesterol is formed from cholesterol either in a reaction catalysed by cholesterol 25-hydroxylase (Ch25h) [24] or as a side-product of Cyp46a1 or Cyp27a1 catalysed hydroxylation of cholesterol [25,26]. The level of 25-hydroxycholesterol in the SLOS mouse brain was at about the detection limit of our methodology (0.006 ± 0.002 ng/mg). 26-Hydroxycholesterol is formed in a Cyp27a1 catalysed reaction [27], and in SLOS mouse brain the level of this oxysterol was determined to be 0.025 ± 0.017 ng/mg (Fig. 3F). As well as finding 24S-hydroxycholesterol in brain, we also find low levels of the 24R-isomer (0.048 ± 0.008 ng/mg) (Fig. 3E). We also identified low levels of 22R-hydroxycholesterol (0.010 ± 0.002 ng/mg) giving a peak eluting at 6.06 min in the short gradient (Fig. 3B). This is generated from cholesterol via oxidation by Cyp11a1 (P450SCC) [28]. The combined levels of the side-chain hydroxycholesterols in the SLOS mouse is 0.435 ng/mg, this compares with 0.594 ng/mg in the wide type mouse [13], i.e. about 70%.
Fig. 3.
LC–MS(MS)n analysis of monohydroxycholesterols from newborn Dhcr7Δ3-5/T93M mouse brain following GP charge-tagging. (A) LC-MS RIC for m/z 534.4054 ± 10 ppm using the short gradient. Shown in the inset is the chromatographic separation of 24S-, 25-, 24R and 26-hydroxycholesterols using the longer gradient. MS3 (534 → 455→) spectra of peaks eluting at (B) 6.06 min (short gradient) corresponding to 22R-hydroxycholesterol (cholest-5-ene-3β,22R-diol); (C) 7.36 min (short gradient, 16.22 min longer gradient) corresponding to 24S-hydroxycholesterol (cholest-5-ene-3β,24S-diol); (D) 17.41 min (longer gradient) corresponding to 25-hydroxycholesterol (cholest-5-ene-3β,25-diol); (E) 19.22 min (longer gradient, 7.84 min short gradient) corresponding to 24R-hydroxycholesterol (cholest-5-ene-3β,24R-diol); (F) 19.59 min (longer gradient) corresponding to (25R)26-hydroxycholesterol (cholest-(25R)-5-ene-3β,26-diol); (G) 9.59 min (short gradient) corresponding to 7β-hydroxycholesterol (cholest-5-ene-3β,7β-diol) and (H) 10.16 min (short gradient) corresponding to 7α-hydroxycholesterol (cholest-5-ene-3β,7α-diol). Other peaks eluting with the short gradient displayed in chromatogram 3A correspond to the second conformers of 24S-hydroxycholesterol (7.67 min) and of 7β-hydroxycholesterol (10.00 min) and to 6β-hydroxycholesterol (10.51 min). The peak at 18.03 min eluting with the longer gradient corresponds to the second conformer of 24S-hydroxycholesterol. Shown as insets in each spectrum are the structures of the GP-tagged molecules.
We also identified monohydroxycholesterols where hydroxylation is in the B-ring at positions 7β-, 7α- and 6β (0.244 ± 0.078, 0.132 ± 0.051 and 0.129 ± 0.045 ng/mg, respectively, Fig. 3G and H, see Fig. 1D). These may be autoxidation products of cholesterol, formed during sample storage, or they may be formed endogenously by reaction of cholesterol with reactive oxygen species (ROS), or alternatively they may be formed extra-cerebrally and cross the BBB from the circulation into brain. 6β-Hydroxycholesterol is formed from 5,6-epoxycholesterol following acid hydrolysis and subsequent dehydration [19]. The level of 7-oxocholesterol was below the limit of detection and no evidence of 4α- or 4β-hydroxycholesterols was found.
24S,25-Epoxycholesterol is formed in parallel to cholesterol via a shunt of the mevalonate pathway. The RIC of m/z 532.3898, the appropriate m/z of the GP-tagged 24S,25-epoxycholesterol, from SLOS mouse brain samples revealed two peaks eluting at 6.68 and 6.94 min corresponding to the syn and anti conformers of GP-tagged 24S,25-epoxycholesterol (0.024 ± 0.004 ng/mg) (Fig. 4A). The MS3 spectra (Fig. 4B) are identical to those of the authentic standard. The peak eluting at 7.72 min was identified as GP-tagged 24-oxocholesterol (0.035 ± 0.006 ng/mg). The presence of a 24-oxo group is characterised by an abundant [M−79−CO]+ ion (m/z 425) in the MS3 spectrum (Fig. 4C). 24-Oxocholesterol is formed from 24S,25-epoxycholesterol during derivatisation. Analysis of the newborn SLOS mouse brain also revealed a peak with retention time 7.98 min which gave an MS3 spectrum corresponding to GP-tagged 22-oxocholesterol (0.118 ± 0.028 ng/mg) (Fig. 5C). The peak in the RIC of m/z 532.3898 eluting at 7.36 min (0.062 ± 0.011 ng/mg) was in our earlier publication presumptively identified to correspond to GP-tagged 23-hydroxydesmosterol [13]. This was based on an MS3 ([M]+ → [M−79]+→) spectrum being completely dominated by the [M−79−H2O]+ ion at m/z 435 (Fig. 5B) and an [M]+ → [M−97]+ → MS3 spectrum indicating that the facile hydroxy group is located α to the side-chain double bond in desmosterol. Other compounds identified in SLOS new born mouse brain include (24Z)26-hydroxydesmosterol eluting at 7.10 min (0.016 ± 0.003 ng/mg, Fig. 5A). Pikuleva and Javitt have shown previously that CYP27A1 can hydroxylate desmosterol at position C-26 [29]. Close inspection of the RIC m/z 532.3898 reveals further peaks at 8.60 and 9.33 min that give MS3 spectra compatible with GP-tagged 7β- and 7α-hydroxydesmosterols, respectively (0.024 ± 0.005 ng/mg and 0.027 ± 0.013 ng/mg), although authentic standards are not available to confirm the identifications (Fig. 5D and E). These oxysterols could be formed in Cyp7a1 catalysed reactions with desmosterol as the substrate [30], or via ROS or autoxidation. While the RIC for m/z 532.3898 from the wild type and SLOS mouse are similar up to 9.5 min (cf. Figs. 4A and 3A in Meljon et al. [13]), the late regions of the chromatograms are quite different between the two genotypes with the appearance of major new peaks at 9.64 min and 10.10 min in the RIC from the SLOS animals (Fig. 4A, see 7-DHC derived oxysterols in Section 3.3).
Fig. 4.
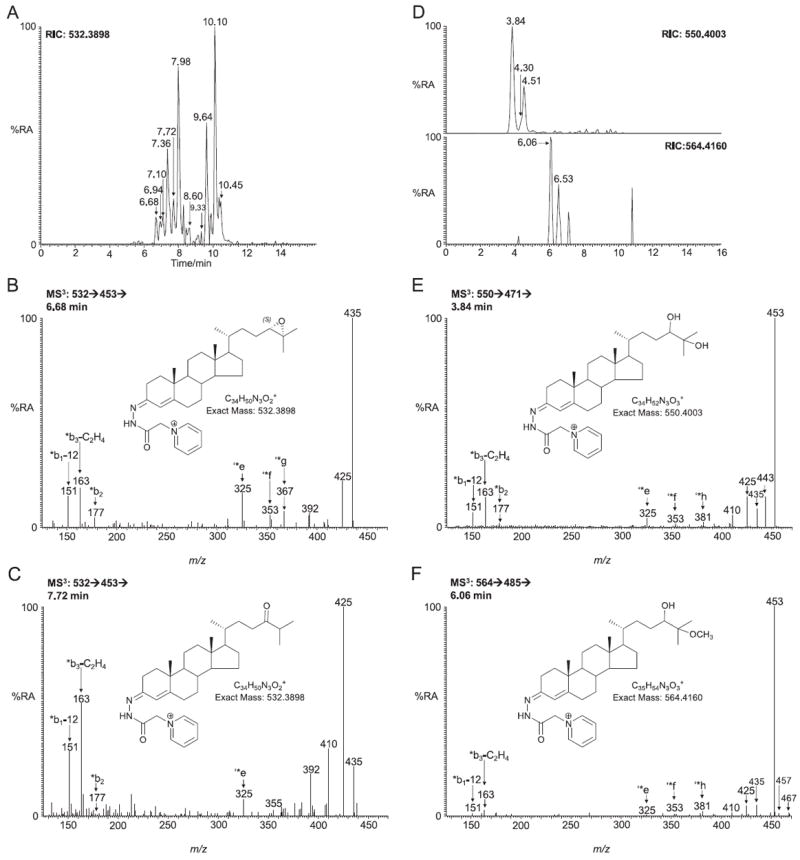
LC–MS(MS)n analysis for 24S,25-epoxycholesterol and other oxysterols of similar m/z from newborn Dhcr7Δ3-5/T93M mouse brain following GP charge-tagging. The short gradient was employed throughout. (A) LC-MS RIC of m/z 532.3898 ± 10 ppm. MS3 (532 → 453→) spectra of peaks eluting at (B) 6.68 min corresponding to 24S,25-epoxycholesterol (3β-hydroxycholest-5-en-24S,25-epoxide) and (C) 7.72 min corresponding to 24-oxocholesterol (3β-hydroxycholest-5-en-24-one). (D) Upper panel, LC–MS RIC of m/z 550.4003 ± 10 ppm corresponding to dihydroxycholesterols. Lower panel, LC–MS RIC of m/z 564.4160 ± 10 ppm corresponding to hydroxymethoxycholesterol. (E) MS3 (550 → 471→) spectrum of 24,25-dihydroxycholesterol (cholest-5-ene-3β,24,25-triol) eluting at 3.84 min. (F) MS3 spectrum of 24-hydroxy-25-methoxycholesterol (3β,24-dihydroxycholest-5-ene-25-methoxide) eluting at 6.06 min (the spectrum could also correspond to the 25-hydroxy-24-methoxy isomer). 24-Oxocholesterol, 24,25-dihydroxycholesterol and 24-hydroxy-25-methoxycholesterol (and/or its 25-hydroxy-24-methoxy isomer) are all formed from 24S,25-epoxycholesterol during GP-derivatisation. Shown as insets in each spectrum are the structures of the GP-tagged molecules.
Fig. 5.
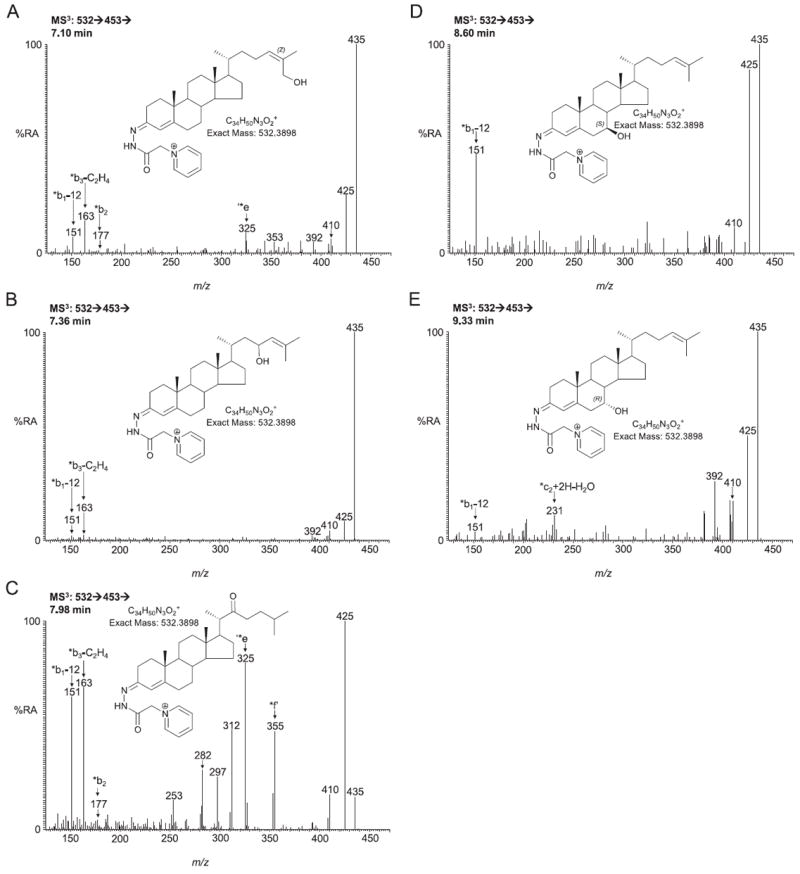
MS3 (532→453→) spectra of other GP-tagged oxysterols eluting in chromatogram 4A. (A) (24Z)26-Hydroxydesmosterol (cholesta-5,24(Z)-diene-3β,26-diol) eluting at 7.10; (B) 23-hydroxydesmosterol (cholesta-5,24-diene-3β,23-diol) eluting at 7.36 min; (C) 22-oxocholesterol (3β-hydroxycholest-5-en-22-one) eluting at 7.98 min; (D) 7β-hydroxydesmosterol (cholesta-5,24-diene-3β,7β-diol) eluting at 8.60 min and (E) 7α-hydroxydesmosterol (cholesta-5,24-diene-3β,7α-diol) eluting at 9.33 min. Authentic standards are only available for (24Z)26-hydroxydesmosterol and 22-oxocholesterol. All other oxysterols are presumptively identified. Shown as insets in each spectrum are the structures of the GP-tagged molecules.
24S,25-Epoxycholesterol is unstable in acid solution and is susceptible to both hydrolysis and methanolysis during the derivatisation process [31]. The RIC of m/z 550.4003 (Fig. 4D upper panel), appropriate for the hydrolysis product, revealed two peaks eluting at 3.84 min and 4.51 min identified as the syn and anti conformers of GP-tagged 24,25-dihydroxycholesterol (0.598 ± 0.128 ng/mg). The MS3 spectra are identical to those of the authentic standard (Fig. 4E). Close inspection of the RIC for m/z 550.4003 shown in Fig. 4D reveals a shoulder on the leading edge of the peak at 4.51 min. The MS3 spectrum suggests the presence of 20,22-dihydroxycholesterol (0.024 ± 0.007 ng/mg). Analysis of the RIC of m/z 564.4160 (Fig. 4D lower panel), appropriate for the methanolysis product of 24S,25-epoxycholesterol showed two peaks with retention times and MS3 fragmentation profiles identical to those obtained during the analysis of the GP-tagged reference standard (Fig. 4F). The [M−79−32]+ ion is characteristic of the presence of a methoxy group. The level of the methanolysis product in SLOS brain was determined to be 0.297 ± 0.103 ng/mg. The combined concentration of native 24S,25-epoxycholesterol, its isomer 24-oxocholesterol, its hydrolysis product 24,25-dihydroxycholesterol and its methanolysis product 24-hydroxy-25-methoxycholesterol equals to 0.920 ng/mg. Considering that the level of cholesterol in the newborn SLOS mouse is only about 65% that of the wild type, it was expected that the 24S,25-epoxycholesterol ratio between the mice would give a similar percentage. In fact, the level in the SLOS mouse is about 80% of that in the wild type. One plausible explanation for the higher than expected level of 24S,25-epoxycholesterol in the SLOS mouse is the down regulation of Cyp7b1, the enzyme responsible for 24S,25-epoxycholesterol metabolism. Björkhem et al. have noted down regulation of CYP7B1 in human SLOS patients [2].
3.3. 7-DHC derived oxysterols
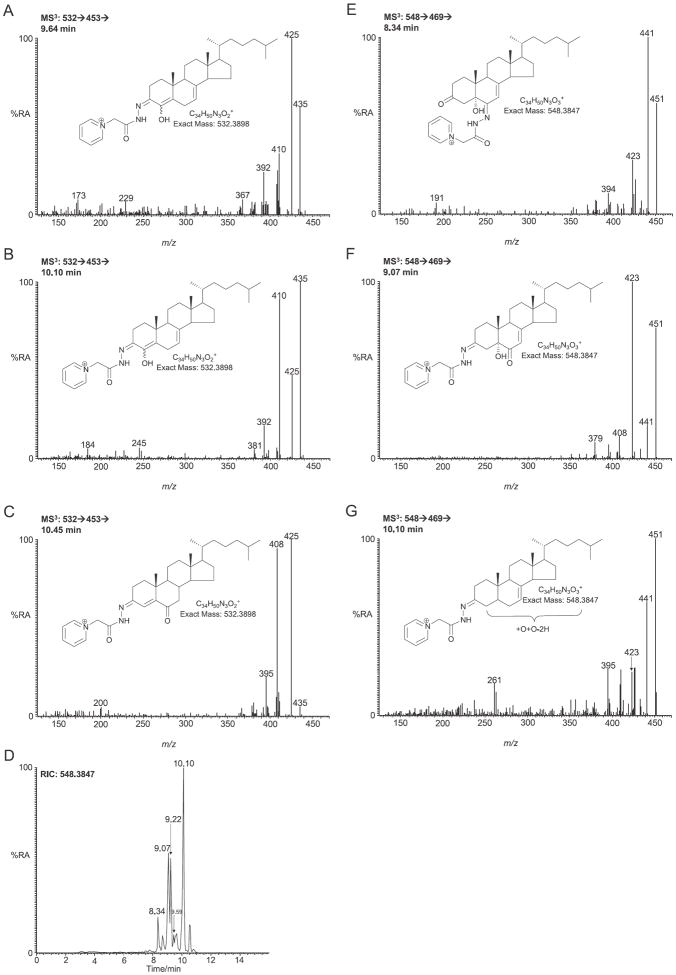
In recent studies performed on developing mouse embryos from another SLOS mouse model, the Dhcr-7-KO mouse which lacks the exon 8 coding sequence in Dhcr7 (Dhcr7Ex8 also called Dhcr7tm1Gst/j), in which essentially no cholesterol is synthesised in embryonic brain, three novel monohydroxydehydrocholesterol isomers were reported to be present in embryonic brain [32,33]. These were identified by high performance (HP)LC purification and NMR analysis to be 4α-, 4β- and 24-hydroxy-7-dehydrocholesterols [33]. In our SLOS mouse model these three novel metabolites may account for the late eluting peaks in the of RIC m/z 532.3898 (Fig. 4A). The components eluting at 9.64 and 10.10 min both give MS3 ([M]+ → [M−79]+→) spectra compatible with an A/B ring system containing two double bonds, and an extra alcohol group (besides that at C-3 oxidised here by cholesterol oxidase to a 3-oxo group), possibly the 4α- and 4β-hydroxy-7-dehydrocholesterol isomers reported by Xu et al. (0.168 ± 0.039 ng/mg, Fig. 6A and B). The abundance of these metabolites is significantly (P < 0.05) elevated in the SLOS mouse, however, no authentic standard is available to confirm their identity. The latest eluting peak (10.45 min) gives a spectrum indicative of GP-tagged cholest-4-ene-3,6-dione (0.040 ± 0.012 ng/mg, Fig. 6C). No evidence was found for a metabolite giving MSn spectra appropriate to 24-hydroxy-7-dehydrocholesterol. Xu et al. also identified 3β,5α-dihydroxycholest-7-en-6-one in the brain extract from embryonic Dhcr-7-KO mouse and suggested its formation was through 5α,6α-epoxycholest-7-en-3β-ol and subsequently cholest-7-en-3β,5α,6β-triol [33]. Following treatment with cholesterol oxidase and GP tagging 3β,5α-dihydroxycholest-7-en-6-one will give an [M]+ ion at m/z 548.3847. The RIC for this mass from brain extracts of our Dhcr7Δ3-5/T93M SLOS mouse is shown in Fig. 6D. The chromatogram shows a myriad of late eluting metabolites. The MS3 spectra of three distinct compounds are shown in Fig. 6E–G. The component eluting at 8.34 (Fig. 6E) gives a pattern of MS3 fragment ions dominated by successive losses of 18 Da (H2O), 28 Da (CO) and 46 Da (H2O + CO) from the [M−79]+ ion. The pattern indicates the presence of a free hydroxy and a carbonyl group attached to the sterol skeleton. The spectrum is compatible with that predicted for GP tagged 3β,5α-dihydroxycholest-7-en-6-one following treatment with cholesterol oxidase where the GP-tag is on C-6 (0.299 ± 0.042 ng/mg). The components eluting at 9.07 (Fig. 6F) and 9.59 min give similar MS3 spectra, this time dominated by the loss of 18 Da (H2O) and 46 Da (H2O + CO) from the [M−79]+ ion, with only a minor loss of 28 Da (CO). Following treatment with cholesterol oxidase 3β,5α-dihydroxycholest-7-en-6-one is converted to 5α-hydroxycholest-7-ene-3,6-dione and this latter pair of peaks probably correspond to syn and anti confirmers of the molecules with the derivatising group at C-3 (1.343 ± 0.368 ng/mg). The final major peak at 10.10 min gives a different MS3 spectrum to that of the earlier isomers, but the parent structure is not immediately obvious (0.602 ± 0.266 ng/mg, Fig. 6G).
Fig. 6.
Oxysterols derived from 7-DHC or cholesterol possibly via free radical oxidation/autoxidation. MS3 spectra (532 → 453→) of GP-tagged oxysterols eluting in chromatogram 4A at (A) 9.64 min and (B) 10.10 min possibly corresponding to 4β-hydroxy-7-dehydrocholesterol (cholesta-5,7-diene-3β,4β-diol) and its 4α isomer, respectively; and (C) 10.45 min corresponding to cholest-4-ene-3,6-dione. (D) RIC of m/z 548.3847 ± 10 ppm. MS3 (548 → 469→) spectra of GP-tagged oxysterols eluting at (E) 8.34 min and (F) 9.07 min possibly both corresponding to 3β,5α-dihydroxycholest-7-en-6-one and (G) 10.10 min probably corresponding to an additional dihydroxycholestenone isomer. Authentic standards are not available for 4-hydroxy-7-dehydrocholesterol or dihydroxycholestenone isomers hence their identifications are presumptive. Shown as insets in each spectrum are the structures of the GP-tagged molecules.
4. Discussion
The Dhcr7Δ3-5/T93M SLOS mouse model is viable [9,10], this is in contrast to two other SLOS models with a null mutation in the Dhcr7 gene in which the mutant progeny die within one day of birth [7,8]. The Dhcr7Δ3-5/T93M mouse is heterozygous with one null allele and one T93 M mutant allele on Dhcr7. The T93M mutation mimics a relatively well known human mutation in DHCR7, and provides mice with a mild SLOS phenotype [9]. In the present study we investigated the sterol and oxysterol content of newborn brain of the Dhcr7Δ3-5/T93M SLOS mouse to ascertain whether the routes of sterol metabolism were the same as in the wild type mouse. Unsurprisingly, the cholesterol and desmosterol levels were reduced in the SLOS mouse compared to the wild type, while their respective precursors 7-DHC (and 8-DHC) and 7-DHD (and 8-DHD) were increased. This is consistent with earlier studies by Marcos et al. and Correa-Cerro et al. who also observed a reduction in cholesterol and an elevation in DHC in 1-day-old SLOS mouse brain [9,10].
The genotype of SLOS is well established [1,5], however, the aetiology of the phenotype is less clear. A lack of cholesterol (or desmosterol) during development is almost certainly the explanation for the phenotypic trait of dysmorphia. However, contributions towards dysmorphia and mental retardation may be caused, at least in part, by a reduction in cholesterol metabolites required for normal development, or a build up of toxic metabolites of 7-DHC (and/or 7-DHD). In brain of the healthy animal the level of cholesterol is tightly regulated [11] at the points of synthesis [34] and of metabolism [14,23]. The enzymes responsible for the first steps of cholesterol metabolism are mostly CYPs (Cyps in mouse) which have broad substrate specificity, this leads to the possibility that not only cholesterol but also 7-DHC and also 8-DHC may act as a substrate with the consequent production of 7-DHC or 8-DHC derived metabolites. This is similarly true for 7-DHD and 8-DHD. Thus, in the present study we have profiled the oxysterol content of brain from the SLOS newborn mouse in attempt to identify metabolites that differ from the wild type. We find a reduction in the level of essentially all cholesterol-derived enzymatically formed oxysterols in the SLOS mouse, in particular 24S-hydroxycholesterol (reduced significantly to about 70% of the wild type, P < 0.05) which is the major oxysterol in brain (Fig. 7). This is not surprising as the major factor affecting the formation of this oxysterol is the amount of available cholesterol substrate [35], which is reduced in the brain of the SLOS mouse to about 65% of the wild type. There are some increases in cholesterol-derived oxysterols formed via ROS in the SLOS mouse compared to the wild type, i.e. 7β- and 6β-hydroxycholesterol (P < 0.05), but this could be a consequence of ex vivo autoxidation facilitated by the enhanced abundance of 7-DHC and an increased propensity for free-radical propagation reactions. However, data generated by observing ex vivo [2H7]cholesterol oxidation in the samples would argue against this. No evidence was found for the presence of 24-hydroxy-7-dehydrocholesterol in either the SLOS or wild type mouse, although the desmosterol derived oxysterols (24Z)26- and 7α-hydroxydesmosterols were observed from both genotypes. This is in contrast to data from Korade et al. who report the formation of 24-hydroxy-7-dehydrocholesterol in brain of the Dhcr-7-KO (Dhcr7Ex8) mouse [32], but in agreement with that of Björkhem et al. who failed to find 7-DHC metabolites in serum of SLOS patients [2]. 24S,25-Epoxycholesterol is formed in a shunt of the mevalonate pathway, also requiring a functional Dhcr7 enzyme. In the SLOS mouse the total amount of 24S,25-epoxycholesterol was reduced compared to the wild type, although not to significance. The reduction was less than that of cholesterol and desmosterol suggesting that perhaps the rate of metabolism of 24S,25-epoxycholesterol is also reduced. This would be consistent with the situation in human where the expression of CYP7B1 is reduced in SLOS patients [2]. CYP7B1 is the enzyme which 7α-hydroxylates 24S,25-epoxycholesterol. An absence of Dhcr7 in the shunt pathway should lead to 24S,25-epoxy-7-dehydrocholesterol as a final product. No evidence for this metabolite was found in our SLOS mouse model.
Fig. 7.
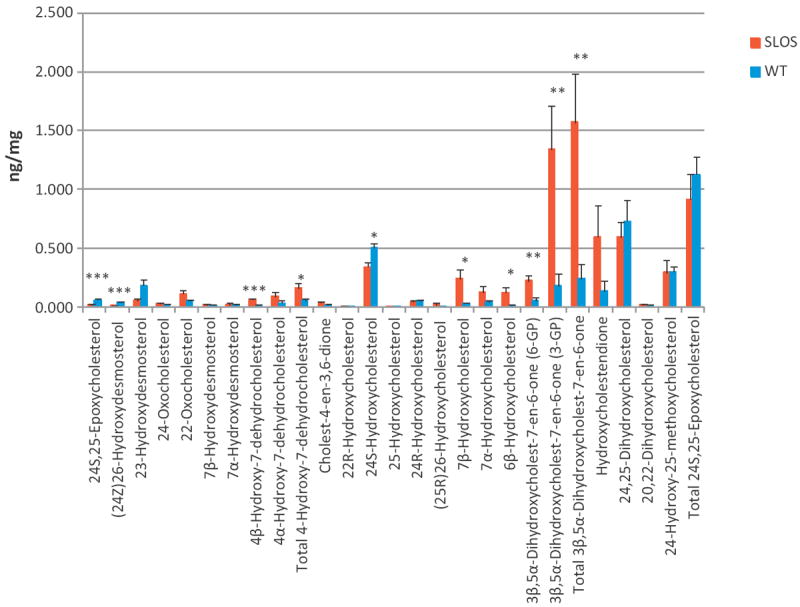
Levels of oxysterols in brain of newborn Dhcr7Δ3-5/T93M and phenotypically normal Dhcr7+/T93M control mice. Data for Dhcr7Δ3-5/T93M mice (n = 4) is in red and that for Dhcr7+/T93 control mice (n = 6) in blue. Statistical analysis was performed by Student’s t-test, P < 0.05 was considered a statistically significant difference (*), P < 0.01 (**), P < 0.001 (***). Data represent mean ± SE.
Recent work from Porter and colleagues on the Dhcr-7-KO mouse (Dhcr7Ex8) which does not survive beyond first day indicates that 3β,5α-dihydroxycholest-7-en-6-one is a major oxysterol in brain [32,33]. Although the authentic standard was unavailable to us, we do observe oxysterols of the appropriate m/z in brain extracts from our Dhcr7Δ3-5/T93M SLOS mouse model. Whether this metabolite is formed endogenously or during sample handling/storage is unclear. This could be clarified by addition of high-purity isotope-labelled 7-DHC and 8-DHC during brain homogenisation. Unfortunately, at the time of our study such standards were not available to us. However, we plan to explore this conundrum in future studies. Two other oxysterols observed by Porter and colleagues are 4α- and 4β-hydroxy-7-dehydrocholesterol [32]. Again in our Dhcr7Δ3-5/T93M SLOS mouse model peaks of the appropriate m/z are observed in brain extracts. Porter et al. suggested that these compounds are formed from 7-DHC in a reaction catalysed by Cyp3a4. If this were the case, it would be expected that 4β-hydroxycholesterol, an enzymatic product of Cyp3a4 oxidation of cholesterol [36], would be observed in brain of our SLOS mouse and the wild type animal also. In fact, 4β-hydroxycholesterol was not detected by us. An alternative explanation is that 4α- and 4β-hydroxy-7-dehydrocholesterols are ROS or autoxidation products of 7-DHC.
In conclusion, in newborn brain from the Dhcr7Δ3-5/T93M mouse model of SLOS there is a reduced level of cholesterol and desmosterol and elevated level of 7-DHC and 7-DHD. In concert to the reduction in cholesterol there is also a reduction of cholesterol derived oxysterols, but an elevation in 7-DHC derived oxysterols. It is not clear at present whether these latter oxysterols are formed by autoxidation, via ROS, or enzymatically. Future studies are planned to clarify this question. It is significant that the major reaction used in terminal cholesterol metabolism in normal brain, 24-hydroxylation, does not appear to be a quantitatively important direct route for metabolism of DHC.
Acknowledgments
Work in Swansea was supported by funding from the UK Research Council BBSRC (BBC5157712, BBC5113561, BBI0017351 to WJG, BBH0010181 to YW). AM is supported by a studentship from BBSRC. The EPSRC National Mass Spectrometry Service Centre is warmly acknowledged for providing access to the LTQ-Orbitrap mass spectrometer. Work at Children’s Hospital Oakland Research Institute (CS and GW) was supported by a National Institutes of Health grant HD053036. Members of the European Network for Oxysterol Research are thanked for informative discussions.
Footnotes
Conflict of interest statement
The authors have no conflicting financial interests.
References
- 1.Kelley RI, Herman GE. Inborn errors of sterol biosynthesis. Annu Rev Genomics Hum Genet. 2001;2:299–341. doi: 10.1146/annurev.genom.2.1.299. [DOI] [PubMed] [Google Scholar]
- 2.Björkhem I, Starck L, Andersson U, Lütjohann D, von BS, Pikuleva I, et al. Oxysterols in the circulation of patients with the Smith–Lemli–Opitz syndrome: abnormal levels of 24S- and 27-hydroxycholesterol. J Lipid Res. 2001;42:366–71. [PubMed] [Google Scholar]
- 3.Griffiths WJ, Wang Y, Karu K, Samuel E, McDonnell S, Hornshaw M, et al. Potential of sterol analysis by liquid chromatography–tandem mass spectrometry for the prenatal diagnosis of Smith–Lemli–Opitz syndrome. Clin Chem. 2008;54:1317–24. doi: 10.1373/clinchem.2007.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelley RI. Diagnosis of Smith–Lemli–Opitz syndrome by gas chromatography/mass spectrometry of 7-dehydrocholesterol in plasma, amniotic fluid and cultured skin fibroblasts. Clin Chim Acta. 1995;236:45–58. doi: 10.1016/0009-8981(95)06038-4. [DOI] [PubMed] [Google Scholar]
- 5.Porter FD, Herman GE. Malformation syndromes caused by disorders of cholesterol synthesis. J Lipid Res. 2011;52:6–34. doi: 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kratz LE, Kelley RI. Prenatal diagnosis of the RSH/Smith–Lemli–Opitz syndrome. Am J Med Genet. 1999;82:376–81. [PubMed] [Google Scholar]
- 7.Fitzky BU, Moebius FF, Asaoka H, Waage-Baudet H, Xu L, Xu G, et al. 7-Dehydrocholesterol-dependent proteolysis of HMG-CoA reductase suppresses sterol biosynthesis in a mouse model of Smith–Lemli–Opitz/RSH syndrome. J Clin Invest. 2001;108:905–15. doi: 10.1172/JCI12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassif CA, Zhu P, Kratz L, Krakowiak PA, Battaile KP, Weight FF, et al. Biochemical, phenotypic and neurophysiological characterization of a genetic mouse model of RSH/Smith–Lemli–Opitz syndrome. Hum Mol Genet. 2001;10:555–64. doi: 10.1093/hmg/10.6.555. [DOI] [PubMed] [Google Scholar]
- 9.Correa-Cerro LS, Wassif CA, Kratz L, Miller GF, Munasinghe JP, Grinberg A, et al. Development and characterization of a hypomorphic Smith–Lemli–Opitz syndrome mouse model and efficacy of simvastatin therapy. Hum Mol Genet. 2006;15:839–51. doi: 10.1093/hmg/ddl003. [DOI] [PubMed] [Google Scholar]
- 10.Marcos J, Shackleton CH, Buddhikot MM, Porter FD, Watson GL. Cholesterol biosynthesis from birth to adulthood in a mouse model for 7-dehydrosterol reductase deficiency (Smith–Lemli–Opitz syndrome) Steroids. 2007;72:802–8. doi: 10.1016/j.steroids.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietschy JM, Turley SD. Thematic review series: brain Lipids, Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;45:1375–97. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Tint GS, Yu H, Shang Q, Xu G, Patel SB. The use of the Dhcr7 knockout mouse to accurately determine the origin of fetal sterols. J Lipid Res. 2006;47:1535–41. doi: 10.1194/jlr.M600141-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meljon A, Theofilopoulos S, Shackleton CH, Watson GL, Javitt NB, Knolker HJ, et al. Analysis of bioactive oxysterols in newborn mouse brain by LC/MS. J Lipid Res. 2012;53:2469–83. doi: 10.1194/jlr.D028233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lütjohann D, Breuer O, Ahlborg G, Nennesmo I, Sidén A, Diczfalusy U, et al. Cholesterol homeostasis in human brain: evidence for an age-dependent flux of 24S-hydroxycholesterol from the brain into the circulation. Proc Natl Acad Sci U S A. 1996;93:9799–804. doi: 10.1073/pnas.93.18.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLachlan J, Wotherspoon AT, Ansell RO, Brooks CJ. Cholesterol oxidase: sources, physical properties and analytical applications. J Steroid Biochem Mol Biol. 2000;72:169–95. doi: 10.1016/s0960-0760(00)00044-3. [DOI] [PubMed] [Google Scholar]
- 16.DeBarber AE, Sandlers Y, Pappu AS, Merkens LS, Duell PB, Lear SR, et al. Profiling sterols in cerebrotendinous xanthomatosis: utility of Girard derivatization and high resolution exact mass LC–ESI-MS(n) analysis. J Chromatogr B Anal Technol Biomed Life Sci. 2011;879:1384–92. doi: 10.1016/j.jchromb.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberg-Larsen H, Strand MF, Grimsmo A, Olsen PA, Dembinski JL, Rise F, et al. High sensitivity measurements of active oxysterols with automated filtration/filter backflush-solid phase extraction-liquid chromatography–mass spectrometry. J Chromatogr A. 2012;1255:291–7. doi: 10.1016/j.chroma.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–61. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Karu K, Turton J, Wang Y, Griffiths WJ. Nano-liquid chromatography–tandem mass spectrometry analysis of oxysterols in brain: monitoring of cholesterol autoxidation. Chem Phys Lipids. 2011;164:411–24. doi: 10.1016/j.chemphyslip.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Korade Z, Porter NA. Oxysterols from free radical chain oxidation of 7-dehydrocholesterol: product and mechanistic studies. J Am Chem Soc. 2010;132:2222–32. doi: 10.1021/ja9080265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths WJ, Crick PJ, Wang Y. Methods for oxysterol analysis: past, present and future. Biochem Pharmacol. 2013 doi: 10.1016/j.bcp.2013.01.027. http://dx.doi.org/10.1016/j.bcp.2013.01.027. in press. [DOI] [PubMed]
- 22.Paik YK, Billheimer JT, Magolda RL, Gaylor JL. Microsomal enzymes of cholesterol biosynthesis from lanosterol. Solubilization and purification of steroid 8-isomerase. J Biol Chem. 1986;261:6470–7. [PubMed] [Google Scholar]
- 23.Lund EG, Guileyardo JM, Russell DW. cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc Natl Acad Sci U S A. 1999;96:7238–43. doi: 10.1073/pnas.96.13.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund EG, Kerr TA, Sakai J, Li WP, Russell DW. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J Biol Chem. 1998;273:34316–27. doi: 10.1074/jbc.273.51.34316. [DOI] [PubMed] [Google Scholar]
- 25.Ali Z, Heverin M, Olin M, Acimovic J, Lövgren-Sandblom A, Shafaati M, et al. On the regulatory role of side-chain hydroxylated oxysterols in the brain. Lessons from CYP27A1 transgenic and cyp27a1−/− mice. J Lipid Res. 2013 doi: 10.1194/jlr.M034124. http://dx.doi.org/10.1194/jlr.M034124. in press. [DOI] [PMC free article] [PubMed]
- 26.Mast N, Norcross R, Andersson U, Shou M, Nakayama K, Björkhem I, et al. Broad substrate specificity of human cytochrome P450 46A1 which initiates cholesterol degradation in the brain. Biochemistry. 2003;42:14284–92. doi: 10.1021/bi035512f. [DOI] [PubMed] [Google Scholar]
- 27.Andersson S, Davis DL, Dahlbäck H, Jörnvall H, Russell DW. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–9. [PubMed] [Google Scholar]
- 28.Heyl BL, Tyrrell DJ, Lambeth JD. Cytochrome P-450scc-substrate interactions. Role of the 3 beta- and side chain hydroxyls in binding to oxidized and reduced forms of the enzyme. J Biol Chem. 1986;261:2743–9. [PubMed] [Google Scholar]
- 29.Pikuleva I, Javitt NB. Novel sterols synthesized via the CYP27A1 metabolic pathway. Arch Biochem Biophys. 2003;420:35–9. doi: 10.1016/j.abb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 30.Heverin M, Meaney S, Brafman A, Shafir M, Olin M, Shafaati M, et al. Studies on the cholesterol-free mouse: strong activation of LXR-regulated hepatic genes when replacing cholesterol with desmosterol. Arterioscler Thromb Vasc Biol. 2007;27:2191–7. doi: 10.1161/ATVBAHA.107.149823. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Sousa KM, Bodin K, Theofilopoulos S, Sacchetti P, Hornshaw M, et al. Targeted lipidomic analysis of oxysterols in the embryonic central nervous system. Mol Biosyst. 2009;5:529–41. doi: 10.1039/b819502a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korade Z, Xu L, Mirnics K, Porter NA. Lipid biomarkers of oxidative stress in a genetic mouse model of Smith–Lemli–Opitz syndrome. J Inherit Metab Dis. 2013;36:113–22. doi: 10.1007/s10545-012-9504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, Korade Z, Rosado DA, Jr, Liu W, Lamberson CR, Porter NA. An oxysterol biomarker for 7-dehydrocholesterol oxidation in cell/mouse models for Smith–Lemli–Opitz syndrome. J Lipid Res. 2011;52:1222–33. doi: 10.1194/jlr.M014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res. 2009;50(Suppl):S15–27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell DW, Halford RW, Ramirez DM, Shah R, Kotti T. Cholesterol 24-hydroxylase: an enzyme of cholesterol turnover in the brain. Annu Rev Biochem. 2009;78:1017–40. doi: 10.1146/annurev.biochem.78.072407.103859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodin K, Andersson U, Rystedt E, Ellis E, Norlin M, Pikuleva I, et al. Metabolism of 4 beta -hydroxycholesterol in humans. J Biol Chem. 2002;277:31534–40. doi: 10.1074/jbc.M201712200. [DOI] [PubMed] [Google Scholar]