Abstract
The Rad6-Rad18 ubiquitin-conjugating enzyme complex of Saccharomyces cerevisiae promotes replication through DNA lesions via three separate pathways that include translesion synthesis (TLS) by DNA polymerases ζ (Polζ) and Polη and postreplicational repair mediated by the Mms2-Ubc13 ubiquitin-conjugating enzyme and Rad5. Here we report our studies with a proliferating cell nuclear antigen (PCNA) mutation, pol30-119, which results from a change of the lysine 164 residue to arginine. It has been shown recently that following treatment of yeast cells with DNA-damaging agents, the lysine 164 residue of PCNA becomes monoubiquitinated in a Rad6-Rad18-dependent manner and that subsequently this PCNA residue is polyubiquitinated via a lysine 63-linked ubiquitin chain in an Mms2-Ubc13-, Rad5-dependent manner. PCNA is also modified by SUMO conjugation at the lysine 164 residue. Our genetic studies with the pol30-119 mutation show that in addition to conferring a defect in Polζ-dependent UV mutagenesis and in Polη-dependent TLS, this PCNA mutation inhibits postreplicational repair of discontinuities that form in the newly synthesized strand across from UV lesions. In addition, we provide evidence for the activation of the RAD52 recombinational pathway in the pol30-119 mutant and we infer that SUMO conjugation at the lysine 164 residue of PCNA has a role in suppressing the Rad52-dependent postreplicational repair pathway.
Proliferating cell nuclear antigen (PCNA) is the eukaryotic sliding clamp required for processive DNA synthesis. The Saccharomyces cerevisiae POL30 gene encoding PCNA is essential for cell viability (7), and conditional lethal pol30 mutations confer defects in DNA replication (1, 3). PCNA is loaded onto the template-primer junctions of DNA by replication factor C in an ATP-dependent reaction. DNA polymerase δ (Polδ) then binds PCNA and carries out processive DNA synthesis (6, 23). In reconstituted systems containing viral origin sequences, PCNA and Polδ carry out replication of both the leading and lagging strands (26, 38). In addition to its essential role in DNA replication, PCNA has been shown to be required for various DNA repair processes, including nucleotide excision repair (NER), base excision repair, and mismatch repair (23); more recently, PCNA has been shown to interact physically and functionally with the various translesion synthesis (TLS) polymerases of yeasts and humans (9-11, 13).
Genetic studies of S. cerevisiae have indicated that PCNA is involved in RAD6-dependent error-free postreplicative bypass of UV-damaged DNA (35). Postreplicative bypass processes come into play when the DNA replicational machinery encounters a DNA lesion in the template strand and is unable to replicate past the lesion. Replication of damaged DNA templates can occur by error-free damage avoidance processes in which the undamaged complementary sequence is used to accomplish replication through the damaged site (14) or it may involve TLS by a specialized DNA polymerase across from the lesion. The S. cerevisiae RAD6 and RAD18 genes are required for the error-free as well as mutagenic modes of damage bypass (25, 30). Rad6, a ubiquitin-conjugating enzyme, exists in vivo in a tight complex with Rad18, a DNA binding protein (4, 5). Mutations in RAD6 and RAD18 confer a high degree of sensitivity to UV light, and they engender a defect in the replication of UV-damaged DNA (28). Also, UV-induced mutagenesis does not occur in either rad6 or rad18 mutants (2, 8, 25).
The Rad6-Rad18-mediated ubiquitin conjugation promotes replication through DNA lesions via three different pathways: the Polζ- and Polη-dependent TLS pathways and the Rad5-, Mms2-Ubc13-dependent postreplicational repair pathway (34). The Rev3 and Rev7 proteins together comprise DNA Polζ (27), which promotes TLS by extending from the nucleotides inserted opposite the lesion site by another DNA polymerase (29). For certain DNA lesions (such as, for example, UV lesions and abasic sites), Polζ promotes mutagenic TLS through the lesion (12, 17, 20), whereas for a lesion such as thymine glycol, it promotes error-free replication through the lesion (22).
The RAD30 gene of yeast encodes Polη, which promotes error-free replication through cyclobutane pyrimidine dimers. Polη is unique among eukaryotic DNA polymerases in its proficient ability to replicate through a cis-syn thymine-thymine dimer (19, 21, 39, 40), and genetic studies of yeasts as well as of humans have indicated a major role for Polη in promoting error-free replication through the cyclobutane pyrimidine dimers formed at TC and CC sites (32, 41).
The RAD5, MMS2, and UBC13 genes function in a third Rad6-Rad18-dependent lesion bypass pathway. Deletion of any of these genes inactivates the postreplicational repair of discontinuities that form in the DNA synthesized from UV-damaged DNA templates (34). This process might involve a copy-choice type of DNA synthesis, wherein the newly synthesized daughter strand of the undamaged complementary sequence is used as the template for bypassing the lesion. Rad5 is a member of the SWI/SNF family of ATPases, and it has a C3HC4 motif characteristic of ubiquitin ligase (E3) proteins (18). Mms2 associates with Ubc13, a ubiquitin-conjugating (E2) enzyme, and the Mms2-Ubc13 complex assembles polyubiquitin chains linked through lysine 63 of ubiquitin (16). Rad5 physically interacts with Rad18 and Ubc13 (36).
More recently, it has been shown that in yeast cells treated with DNA-damaging agents, PCNA becomes monoubiquitinated at its lysine 164 residue via the action of Rad6-Rad18; subsequently, this PCNA residue is polyubiquitinated via a lysine 63-linked ubiquitin chain in an Mms2-, Ubc13-, and Rad5-dependent reaction (15). Epistasis analyses with a pol30 mutation in which the lysine 164 residue has been changed to arginine and which disables the ubiquitin conjugation reaction have indicated a role for PCNA ubiquitination in Rad6-Rad18-dependent repair processes (15, 33).
In studies that were carried out in our laboratory with pol30-119, a mutant of yeast PCNA that was initially isolated in Connie Holms laboratory (1) and which is due to a change of lysine 164 to arginine, we had observed results similar to those that have been reported by Jentsch and colleagues (15, 33). In view of the finding that the pol30 K164R mutation inactivates PCNA ubiquitination, we have reexamined our data and we report these results here. Our studies confirm the previously reported observations with this pol30 mutation, and they extend them in several ways. In particular, we find that in addition to conferring a defect in Polζ-dependent UV mutagenesis and in Polη-dependent TLS, the pol30-119 mutation impairs the postreplicational repair of discontinuities that form in the newly synthesized DNA across from UV lesions. Even though the pol30-119 mutation apparently impairs all three Rad6-Rad18-dependent lesion bypass processes, intriguingly, its level of UV sensitivity is much less than that of the rad6Δ and rad18Δ mutants. Our genetic data support the inference that the Rad52-dependent postreplicational repair pathway is activated in the pol30-119 mutant and that that accounts for the higher level of UV resistance of this PCNA mutant compared to that of the rad6Δ or rad18Δ mutants. PCNA is additionally modified by SUMO conjugation at the lysine 164 and lysine 127 residues (15); our studies point to an inhibitory role of SUMO conjugation at the lysine 164 residue of PCNA for Rad52-dependent postreplicational repair.
MATERIALS AND METHODS
Strains and plasmids.
Mutations in the POL30 gene which encodes PCNA were obtained by PCR mutagenesis and plasmid shuffle and screened for cold sensitivity (14°C) and for sensitivity to methyl methanesulfonate (MMS) as described previously (1). One of the mutations, pol30-119, had no effect on growth at any of the temperatures, but it conferred MMS and UV sensitivity. All yeast strains used for epistasis analysis and for alkaline sucrose gradient sedimentation are isogenic to EMY74.7 (MATa his3-Δ1 leu2-3-112 trp1Δ ura3-52). The one-step gene replacement method was used to generate genomic deletions of the RAD1, RAD5, RAD6, RAD52, REV3, and RAD30 genes as described previously (34, 35). An SacI, PvuI digest of plasmid pCH1654 (containing the pol30-119 mutant gene in which arginine replaces the lysine 164 residue) was used to generate an integrant of the pol30-119 mutation at the POL30 site in the genome. The various deletion mutations were subsequently generated in the pol30-119 strain. The wild-type EMY74.7 strain and its pol30-119 derivative were used to measure UV-induced CAN1S-to-can1r forward mutation frequencies. Ethidium bromide mutagenesis was used to obtain [rho0] derivatives which lack mitochondrial DNA, yielding isogenic strains YR1-65 (rad1Δ) and YR1-131 (rad1Δ pol30-119) used for alkaline sucrose gradient sedimentation.
UV survival and UV mutagenesis.
UV irradiation and determination of UV-survival and UV-induced mutation frequencies were done as previously described (35).
Sedimentation in alkaline sucrose gradients of nuclear DNA from UV-irradiated cells.
Strains grown overnight at 30°C in synthetic complete medium lacking uracil but supplemented with uridine were UV irradiated at a confluence rate of 0.1 J/m2/s and pulse labeled for 15 min with [3H]uracil. Following UV irradiation, cells were washed and resuspended in high uracil medium and incubated for an additional 30 min or 6 h. Conversion of cells to spheroplasts, alkaline sucrose sedimentation, and processing of samples were done as described previously (34, 35).
RESULTS
The pol30-119 mutation.
Mutations of the POL30 gene were obtained using PCR mutagenesis and plasmid shuffle (1). A number of mutations were identified by their sensitivity to MMS and by a cold-sensitive growth phenotype at 14°C. One of the mutants, pol30-119, exhibits normal growth, cell viability, and cell cycle progression phenotypes at 14, 30, and 37°C. The pol30-119 mutant also grows normally in 0.1 M hydroxyurea-containing medium, indicating a normal synthetic-phase (S-phase) checkpoint. The pol30-119 mutation, however, confers sensitivity to UV light and MMS. The pol30-119 mutation is due to the change of lysine 164 to arginine. This residue is located at the monomer-monomer interface of PCNA, and it is conserved in PCNA from diverse sources, including yeasts, Drosophila spp., frogs, mice, and humans.
Epistasis of pol30-119 with mutations in genes belonging to the RAD6 group.
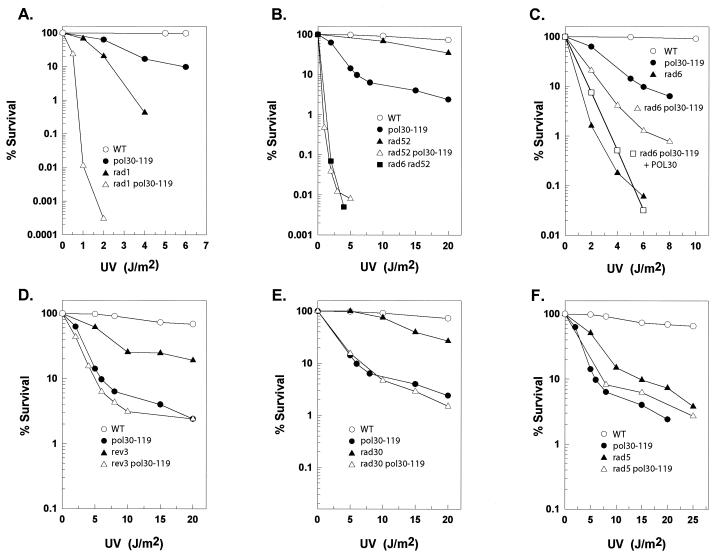
In yeast, genes belonging to three epistasis groups function in the repair of UV-damaged DNA (30). Genes in the RAD3 group are required for the removal of damage by NER, those in the RAD6 group promote error-free and mutagenic bypass of DNA lesions during replication, and those in the RAD52 group mediate double-strand break repair and genetic recombination. To determine the epistasis relationships of pol30-119 with mutations in genes belonging to these groups, we combined the pol30-119 mutation with the rad1Δ mutation defective in NER, the rad6Δ mutation defective in replicational bypass of DNA lesions, or the rad52Δ mutation defective in double-strand break repair and genetic recombination and compared the UV sensitivity of these double mutants with that of the respective single mutants. The pol30-119 mutation exhibits a synergistic increase in UV sensitivity in combination with the rad1Δ mutation (Fig. 1A) and the rad52Δ mutation (Fig. 1B), suggesting that pol30-119 impairs a repair pathway distinct from the NER and genetic recombination pathways. The UV sensitivity of the pol30-119 rad6Δ mutant, however, was no greater than that of the rad6Δ strain (Fig. 1C). The epistasis of the rad6Δ mutation over pol30-119 suggested that this PCNA mutation inactivates RAD6-dependent lesion bypass processes.
FIG. 1.
Epistasis analysis of the pol30-119 mutation. Survival curves after UV irradiation of wild-type (WT) strain EMY74.7, its isogenic pol30-119 derivative, and isogenic derivatives carrying deletion mutations of different RAD genes and of the REV3 gene are shown. Survival curve results represent averages of at least two different experiments for each strain. □ (C), rad6Δ pol30-119 strain carrying the wild-type POL30 gene on the CEN ARS TRP1 plasmid pBL230.
To identify which of the Rad6-Rad18-dependent pathways is specifically inactivated by the pol30-119 mutation, we examined the epistasis relationships of the pol30-119 mutation with the rev3Δ, rad30Δ, and rad5Δ mutations. Surprisingly the pol30-119 mutation exhibited an epistatic relationship with all three deletion mutations, for in each case the UV sensitivity of the double mutant was no greater than that of the pol30-119 mutation (Fig. 1D to F). These epistatic relationships suggested that the pol30-119 mutation interferes with all three Rad6-Rad18-dependent lesion bypass processes.
Defective UV mutagenesis in the pol30-119 mutant.
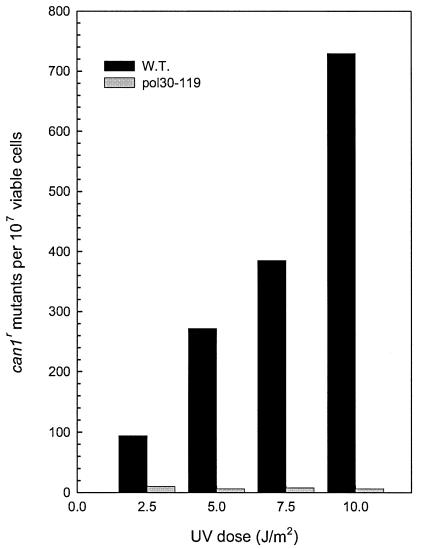
The epistasis of pol30-119 with the rev3Δ mutation suggested a role of PCNA in REV3-dependent mutagenic bypass of UV lesions. To verify this, we compared the frequency of UV-induced mutations in pol30-119 with that in the wild-type strain. As shown in Fig. 2, the frequency of CAN1S-to-can1r forward mutations fell precipitously in the pol30-119 strain, indicating a defect in UV mutagenesis.
FIG. 2.
Effect of pol30-119 on UV-induced CAN1S-to-can1r mutations. The results represent an average of five or more experiments for the wild-type (W.T.) and pol30-119 mutant strains.
Impaired postreplicational repair in the pol30-119 mutant.
Even though postreplicational repair of UV-damaged DNA is inhibited in both the rad5Δ and mms2Δ mutants, the rad5Δ mutation confers a much higher level of UV sensitivity than the mms2Δ mutation (34). This observation has suggested an additional role for Rad5 in a process other than postreplicational repair, raising thereby the possibility that the epistasis of pol30-119 over rad5Δ was due to the inactivation of this other Rad5-dependent repair process. To ascertain whether PCNA ubiquitination was in fact necessary for postreplicational repair, we carried out experiments to directly check for this.
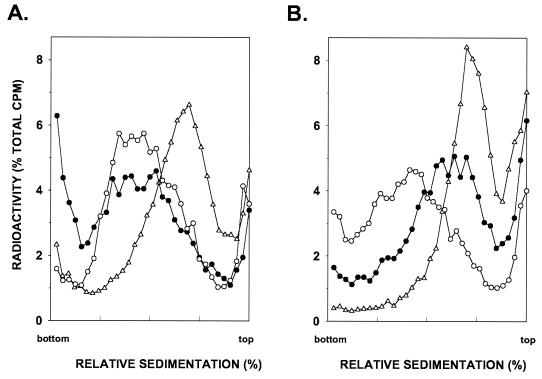
To examine the effect of the pol30-119 mutation on postreplicational repair, we determined the size of newly synthesized DNA in the rad1Δ and rad1Δ pol30-119 strains following UV irradiation. Because of the lack of NER in the rad1Δ strain, UV damage persists in DNA and replication of such DNA requires the various lesion bypass processes. The rad1Δ and rad1Δ pol30-119 strains were UV irradiated at 2.5 J/m2, and the size of newly synthesized DNAs was examined by alkaline sucrose gradient sedimentation following pulse labeling of DNA with [3H]uracil for 15 min and an additional 30-min chase in medium containing a high concentration of unlabeled uracil. In both the rad1Δ and rad1Δ pol30-119 strains, DNA sediments towards the top in alkaline sucrose gradients (indicative of discontinuities in the newly synthesized strand) (Fig. 3). In the unirradiated rad1Δ and rad1Δ pol30-119 cells, however, the size of DNA synthesized following the 15-min pulse and 30-min chase periods was the same as in uniformly labeled cells (data not shown), indicating that this period is sufficient to synthesize normally sized DNA in unirradiated cells. Previously, we had shown that the size of newly synthesized DNA in the rad1 mutant decreases with increasing UV dose and correlates with the average distance between photoproducts present in the parental DNA (28). These discontinuities presumably reflect gaps that form in the newly synthesized strand across from the damage site in the template strand. In rad1Δ cells incubated for 6 h following UV irradiation, daughter strands attain the same size as in unirradiated cells, indicating that postreplicative gap-filling processes have restored normal size to daughter strands (Fig. 3A). By contrast, the rad1Δ pol30-119 strain is unable to restore normally sized DNA in UV-irradiated cells following a 6-h incubation period (Fig. 3B). Thus, the pol30-119 mutation impairs the efficiency of postreplicational repair of UV-damaged DNA.
FIG. 3.
Sedimentation in alkaline sucrose gradients of nuclear DNA from cells incubated for different periods following UV irradiation with 2.5 J/m2. The rad1Δ (A) and rad1Δ pol30-119 (B) strains (YR1-65 and YR1-131, respectively) were UV irradiated and then pulse labeled with [3H]uracil for 15 min followed by a 30-min (▵) or 6-h (•) chase in high-uracil medium before conversion to spheroplasts and sedimentation of DNA in alkaline sucrose gradients. Unirradiated cells pulse labeled for 15 min and chased for 6 h in high-uracil medium are indicated (○).
Requirement of RAD52 for the suppression of UV sensitivity of rad6Δ by pol30-119.
Although the various genetic observations strongly suggest that pol30-119 impairs all three Rad6-Rad18-dependent pathways of lesion bypass, intriguingly, this mutation confers a much lower degree of UV sensitivity than the rad6Δ or rad18Δ mutations. This observation suggested either that the pol30-119 mutation imparts only a partial deficiency in Rad6-Rad18-dependent bypass processes or that an alternate lesion bypass pathway becomes activated in this PCNA mutant. To examine this point further, we first determined whether the pol30-119 mutation suppresses the UV sensitivity of the rad6Δ or rad18Δ mutants (since that would imply the activation of an alternate Rad6-Rad18-independent pathway of lesion bypass). As shown in Fig. 1C, the rad6Δ pol30-119 mutant is not as UV sensitive as the rad6Δ mutation, which implies that the pol30-119 mutation suppresses the repair defectiveness of the rad6Δ mutation. Similar results were obtained with the rad18Δ mutation (data not shown). Introduction of the wild-type POL30 gene on an ARS CEN TRP1 plasmid increased the UV sensitivity of the rad6Δ pol30-119 mutant to the rad6Δ level. We conclude from these observations that in the pol30 K164R mutant an alternate repair pathway becomes activated and that in the presence of wild-type PCNA this alternate pathway remains relatively dormant.
In addition to Rad6-Rad18-dependent postreplicational repair, the Rad52-dependent recombinational pathway contributes to the postreplicational repair of UV-damaged DNA (28). If suppression in the pol30-119 mutant were due to the activation of Rad52-dependent postreplicational repair, we would expect the UV sensitivity of the pol30-119 rad52Δ double mutant to be the same as that of the rad6Δ rad52Δ mutant (since both the Rad6- and Rad52-dependent lesion bypass pathways would then be inactivated in the pol30-119 rad52Δ mutant). From the very similar UV sensitivities of the pol30-119 rad52Δ and rad6Δ rad52Δ double mutants (Fig. 1B), we conclude that activation of the Rad52 pathway underlies the reduced UV sensitivity of the pol30-119 mutant and the suppression of the UV sensitivity that occurs in the rad6Δ or rad18Δ mutants when they are combined with the pol30-119 mutation. The implications of these results for the effects of SUMO modification of the lysine 164 residue of PCNA on Rad52-dependent postreplicational repair are discussed below.
DISCUSSION
It has been shown previously that in vivo, the Rad6 and Rad18 proteins mediate the monoubiquitination of PCNA at the lysine 164 residue and that Mms2-Ubc13 in conjunction with Rad5 (15) modulates the polyubiquitination of this residue through lysine 63-linked chains of ubiquitin. Here we show that the pol30-119 mutation, which results from a change of the lysine 164 residue of PCNA to arginine, confers a defect in the Rad6-Rad18-dependent lesion bypass processes. In addition to exhibiting epistasis with the rad6Δ and rad18Δ mutations, the pol30-119 mutation displays epistatic interactions with the rev3Δ, rad30Δ, and rad5Δ mutations, each of which inactivates an alternate Rad6-Rad18-dependent lesion bypass processes. In addition, we show an impairment of UV mutagenesis and inhibition of postreplication repair in the pol30-119 mutant. The results of epistasis analyses and UV mutagenesis confirm the observations that have been reported recently with regard to this pol30 mutant (15, 33). Our finding that there is a very considerable inhibition of postreplicational repair in the pol30-119 mutant could not have been predicted from the epistasis results with rad5Δ, however, since in addition to its involvement in postreplicational repair, Rad5 is likely involved in another repair process as well (34). Without a direct demonstration of a postreplicational repair defect, therefore, one could not be certain which of the Rad5 functions was inactivated by pol30-119. Thus, in addition to confirming the observations that have been reported previously with the K164R mutation of PCNA, our studies extend them and provide further support to the conclusion that PCNA ubiquitination is a prerequisite for all three Rad6-Rad18-dependent lesion bypass processes.
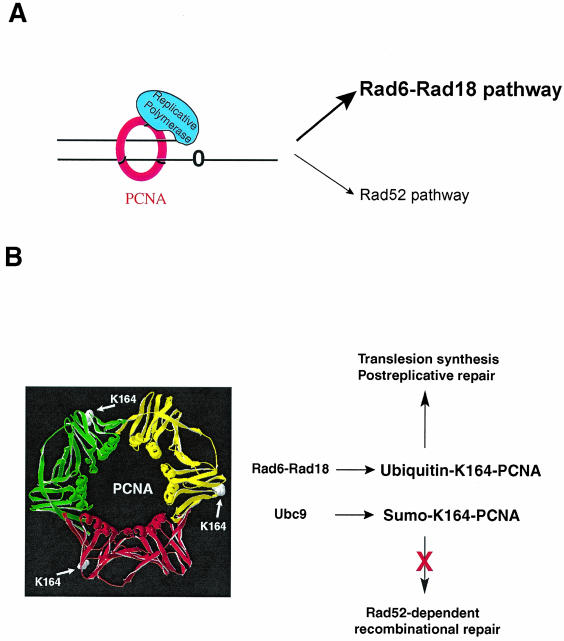
If all three Rad6-Rad18-dependent lesion bypass pathways are inactivated by the pol30-119 mutation, how do we then account for the much higher level of UV resistance of the pol30-119 mutant compared to that of the rad6Δ and rad18Δ mutants? Although Rad6-Rad18-dependent lesion bypass is the predominant way by which replication through DNA lesions is accomplished in yeast cells, the Rad52 system does contribute to lesion bypass (28), albeit in a subsidiary way (Fig. 4A). Under certain circumstances, however (as in the absence of the Srs2 DNA helicase, for example, which actively disrupts the Rad51-nucleoprotein filament and whose presence therefore would inhibit the Rad52-dependent recombinational bypass of DNA lesions), the Rad52 pathway becomes activated (24, 37). Consequently, the srs2Δ mutation suppresses the DNA damage sensitivity of the rad6Δ and rad18Δ mutants (31). Here we provide genetic evidence that the pol30-119 mutation suppresses the UV sensitivity of the rad6Δ and rad18Δ mutants and that this suppression is mediated via the Rad52 pathway.
FIG. 4.
Roles of Rad6-Rad18-mediated ubiquitin conjugation and Ubc9-mediated SUMO attachment to the lysine 164 residue of PCNA. (A) In wild-type yeast cells, Rad6-Rad18-dependent processes play a predominant role in lesion bypass whereas Rad52-dependent recombinational bypass plays a relatively minor role. (B) Rad6-Rad18-dependent ubiquitin conjugation at the lysine 164 residue of PCNA promotes TLS by DNA Polζ and Polη and postreplicational repair of discontinuities that form in the newly synthesized DNA across from UV lesions. Ubc9-mediated SUMO modification of lysine 164 of PCNA is proposed to inhibit Rad52-dependent lesion bypass.
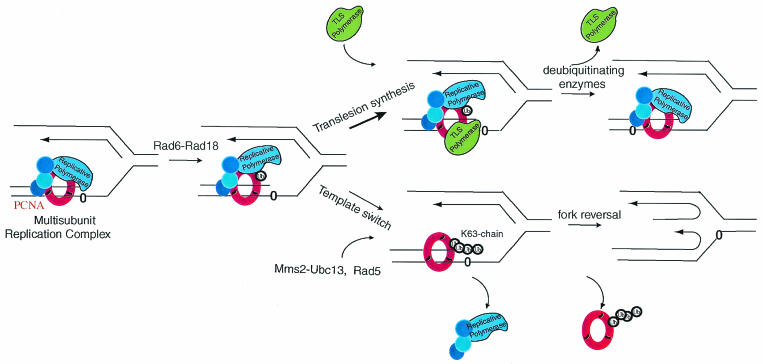
SUMO is conjugated to PCNA at the lysine 164 residue and also at lysine 127; lysine 164, however, is the primary site of this modification (15). On the basis of the observation that the pol30 K127R K164R mutation (in which both the lysine residues subject to SUMO modification have been changed to arginine) confers a somewhat higher level of UV resistance than the pol30 K164R mutation (in which SUMO conjugation can still occur at the K127 site), it has been suggested that SUMO conjugation at lysine 127 is inhibitory for DNA repair (33). Since the difference in the UV survival rates of the K127R K164R and K164R mutants is very slight, however, the inhibitory effect of SUMO conjugation at K127 on DNA repair is likely to be minimal. From our observation indicating that the Rad52 pathway is activated in the pol30 K164R mutant, we infer a role for SUMO modification of lysine 164 in the inhibition of Rad52-dependent lesion bypass. Thus, we suggest that in wild-type yeast cells, whereas ubiquitin conjugation at the lysine 164 residue of PCNA activates all three Rad6-Rad18-dependent lesion bypass processes, SUMO modification of this PCNA residue inactivates Rad52-dependent lesion bypass (Fig. 4B). SUMO modification of PCNA, which is prevalent during the S phase (15), might be a device used for keeping the Rad52 recombinational pathway in check during the S phase because of the attendant risk of chromosome rearrangements. Our results, taken together with previously published observations (15, 33), support the conclusion that attachment of a single ubiquitin molecule at lysine 164 of PCNA activates TLS by Polζ and Polη whereas polyubiquitination of this PCNA residue through a lysine 63-linked ubiquitin chain activates a template switch mechanism (Fig. 5). It is conceivable that in one PCNA trimer, ubiquitin and SUMO modifications can occur on the K164 residues of two different monomers and that that accounts for the simultaneous activation and inhibition of Rad6-Rad18- and Rad52-dependent lesion bypass processes, respectively.
FIG. 5.
A model for the role of Rad6-Rad18-dependent monoubiquitination (Ub) and Mms2-Ubc13-, Rad5-dependent polyubiquitination at the lysine 164 residue of PCNA in TLS and in template switching, respectively. For simplicity, the possible SUMO modification of lysine 164 in one of the PCNA monomers is not shown. See Discussion for further details. 0, DNA lesion.
In Fig. 5, we present a model to explain the possible role of ubiquitin modification of PCNA in TLS and in template switching. During normal replication, the PCNA-bound replicative polymerase is at the primer template junction. Monoubiquitination of the lysine 164 residue of PCNA disrupts the replication ensemble so that the polymerase can no longer access the primer-template junction, and this PCNA modification enables the entry of a TLS polymerase into the replication ensemble. Since the TLS polymerases are able to bind PCNA in the absence of any ubiquitin modification, this modification may have no effect on PCNA binding by the TLS polymerases; instead, it could be important for disrupting the association of some PCNA-bound protein that is a part of the replication ensemble and which is otherwise inhibitory to the binding of PCNA by the TLS polymerases. After the lesion has been bypassed, deubiquitination of the monoubiquitinated lysine 164 residue of PCNA could lead to the repositioning of the replicative polymerase at the primer-template junction and to the exit of the TLS polymerase from the replicational complex. For template switching to occur, we envision that PCNA polyubiquitination destabilizes the replication ensemble to such an extent as to cause its dissociation from the replication fork, thereby providing access of the primer end to the various proteins required for template switching and synthesis.
Acknowledgments
The pol30-119 mutation was made and identified by Neelam Amin in Connie Holm's laboratory. We are grateful to them both for providing it to us. We also thank Peter Burgers for providing us with plasmid pBL230 containing the wild-type POL30 gene.
This work was supported by grant GM19261 from the National Institutes of Health.
REFERENCES
- 1.Amin, N. S., and C. Holm. 1996. In vivo analysis reveals that the interdomain region of the yeast proliferating cell nuclear antigen is important for DNA replication and DNA repair. Genetics 144:479-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. D., D. N. Chadee, and B. A. Kunz. 1994. Roles of the yeast RAD18 and RAD52 DNA repair genes in UV mutagenesis. Mutat. Res. 315:281-293. [DOI] [PubMed] [Google Scholar]
- 3.Ayyagari, R., K. J. Impellizzeri, B. L. Yoder, S. L. Gary, and P. M. J. Burgers. 1995. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and repair. Mol. Cell. Biol. 15:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. [DOI] [PubMed] [Google Scholar]
- 5.Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. [DOI] [PubMed] [Google Scholar]
- 6.Bambara, R. A., R. S. Murante, and L. A. Henricksen. 1997. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 272:4647-4650. [DOI] [PubMed] [Google Scholar]
- 7.Bauer, G. A., and P. M. J. Burgers. 1990. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 18:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassier-Chauvat, C., and F. Fabre. 1991. A similar defect in UV-induced mutagenesis conferred by the rad6 and rad18 mutations of Saccharomyces cerevisiae. Mutat. Res. 254:247-253. [DOI] [PubMed] [Google Scholar]
- 9.Haracska, L., R. E. Johnson, I. Unk, B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 21:7199-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haracska, L., R. E. Johnson, I. Unk, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc. Nat. Acad. Sci. USA 98:14256-14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haracska, L., C. M. Kondratick, I. Unk, S. Prakash, and L. Prakash. 2001. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell 8:407-415. [DOI] [PubMed] [Google Scholar]
- 12.Haracska, L., I. Unk, R. E. Johnson, E. Johansson, P. M. J. Burgers, S. Prakash, and L. Prakash. 2001. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 15:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haracska, L., I. Unk, R. E. Johnson, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2002. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol. Cell. Biol. 22:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins, N. P., K. Kato, and B. Strauss. 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101:417-425. [DOI] [PubMed] [Google Scholar]
- 15.Hoege, C., B. Pfander, G.-L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645-653. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, R. E., L. Haracska, S. Prakash, and L. Prakash. 2001. Role of DNA polymerase η in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 21:3558-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, R. E., S. T. Henderson, T. D. Petes, S. Prakash, M. Bankmann, and L. Prakash. 1992. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 12:3807-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science 283:1001-1004. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. E., M. T. Washington, L. Haracska, S. Prakash, and L. Prakash. 2000. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406:1015-1019. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, R. E., M. T. Washington, S. Prakash, and L. Prakash. 2000. Fidelity of human DNA polymerase η. J. Biol. Chem. 275:7447-7450. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, R. E., S.-L. Yu, S. Prakash, and L. Prakash. 2003. Yeast DNA polymerase zeta (ζ) is essential for error-free replication past thymine glycol. Genes Dev. 17:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelman, Z., and J. Hurwitz. 1998. Protein-PCNA interactions: a DNA-scanning mechanism? Trends Biol. Sci. 23:236-238. [DOI] [PubMed] [Google Scholar]
- 24.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence, C. W. 1982. Mutagenesis in Saccharomyces cerevisiae. Adv. Genetics 21:173-254. [DOI] [PubMed] [Google Scholar]
- 26.Murakami, Y., and J. Hurwitz. 1993. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J. Biol. Chem. 268:11008-11017. [PubMed] [Google Scholar]
- 27.Nelson, J. R., C. W. Lawrence, and D. C. Hinkle. 1996. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272:1646-1649. [DOI] [PubMed] [Google Scholar]
- 28.Prakash, L. 1981. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184:471-478. [DOI] [PubMed] [Google Scholar]
- 29.Prakash, S., and L. Prakash. 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16:1872-1883. [DOI] [PubMed] [Google Scholar]
- 30.Prakash, S., P. Sung, and L. Prakash. 1993. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 27:33-70. [DOI] [PubMed] [Google Scholar]
- 31.Schiestl, R. H., S. Prakash, and L. Prakash. 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124:817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stary, A., P. Kannouche, A. R. Lehmann, and A. Sarasin. 2003. Role of DNA polymerase η in the UV mutation spectrum in human cells. J. Biol. Chem. 278:18767-18775. [DOI] [PubMed] [Google Scholar]
- 33.Stelter, P., and H. D. Ulrich. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425:188-191. [DOI] [PubMed] [Google Scholar]
- 34.Torres-Ramos, C., S. Prakash, and L. Prakash. 2002. Requirement of RAD5 and MMS2 for post replication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:2419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torres-Ramos, C. A., B. L. Yoder, P. M. J. Burgers, S. Prakash, and L. Prakash. 1996. Requirement of proliferating cell nuclear antigen in RAD6-dependent postreplicational DNA repair. Proc. Nat. Acad. Sci. USA 93:9676-9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulrich, H. D., and S. Jentsch. 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19:3388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 38.Waga, S., and B. Stillman. 1994. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 369:207-212. [DOI] [PubMed] [Google Scholar]
- 39.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 2000. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Nat. Acad. Sci. USA 97:3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washington, M. T., L. Prakash, and S. Prakash. 2003. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase η. Proc. Nat. Acad. Sci. USA 100:12093-12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, S.-L., R. E. Johnson, S. Prakash, and L. Prakash. 2001. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21:185-188. [DOI] [PMC free article] [PubMed] [Google Scholar]