Abstract
Pathogenic mycobacteria survive within macrophages by precluding the fusion of phagosomes with late endosomes or lysosomes. Because the molecular determinants of normal phagolysosome formation are poorly understood, the sites targeted by mycobacteria remain unidentified. We found that Hrs, an adaptor molecule involved in protein sorting, associates with phagosomes prior to their fusion with late endosomes or lysosomes. Recruitment of Hrs required the interaction of its FYVE domain with phagosomal phosphatidylinositol 3-phosphate, but two other attachment sites were additionally involved. Depletion of Hrs by use of small interfering RNA impaired phagosomal maturation, preventing the acquisition of lysobisphosphatidic acid and reducing luminal acidification. As a result, the maturation of phagosomes formed in Hrs-depleted cells was arrested at an early stage, characterized by the acquisition and retention of sorting endosomal markers. This phenotype is strikingly similar to that reported to occur in phagosomes of cells infected by mycobacteria. We therefore tested whether Hrs is recruited to phagosomes containing mycobacteria. Hrs associated readily with phagosomes containing inert particles but poorly with mycobacterial phagosomes. Moreover, Hrs was found more frequently in phagosomes containing avirulent Mycobacterium smegmatis than in phagosomes with the more virulent Mycobacterium marinum. These findings suggest that the inability to recruit Hrs contributes to the arrest of phagosomal maturation induced by pathogenic mycobacteria.
Phagosomes, which are formed by invagination of the plasma membrane, acquire microbicidal properties by a series of fusion and fission events that culminate with the formation of phagolysosomes (7, 33). This sequence of events, collectively known as phagosomal maturation, is often impaired by microorganisms such as pathogenic mycobacteria. By co-opting the cellular machinery responsible for maturation, mycobacteria avoid exposure to the harsh hydrolytic environment of phagolysosomes, thereby managing to survive and multiply within macrophages (27).
Mycobacteria arrest phagosomal maturation at an early stage, precluding the acquisition of late endosomal markers like lysobisphosphatidic acid (LBPA). During the normal course of maturation, this transition was recently shown to require de novo synthesis of phosphatidylinositol 3-phosphate [PI(3)P] by Vps34, the class III phosphatidylinositol 3-kinase (9, 34). However, Fratti et al. (9) reported that phagosomes containing mycobacteria recruit Vps34 normally. Thus, the effects of the microorganism are possibly exerted downstream of the formation of PI(3)P. This phosphoinositide serves as a ligand for a variety of proteins bearing either FYVE or PX domains that may contribute to the maturation of the phagosome. Of these, early endosome antigen 1 (EEA1) was shown to be depleted from mycobacterial phagosomes (9). EEA1 is thought to be important in the homotypic fusion of early endosomes (29), but it is not clear whether it contributes directly to the fusion of phagosomes with late endosomes, the event that is central to mycobacterial survival. It is likely that other mediators of phagosomal maturation are targeted by mycobacteria.
Hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) is emerging as a central coordinator of late endosomal sorting (5, 16, 23-25) and may play a comparable role in phagosomal maturation. Like EEA1, Hrs possesses a FYVE domain capable of interacting with PI(3)P, as well as domains that promote association with ubiquitylated proteins (the UIM motif) and with clathrin (4, 23, 24, 26, 31). Like its Saccharomyces cerevisiae homologue, Vps27p, Hrs is thought to function in the generation of multivesicular bodies, acting in conjunction with the ESCRT-I complex to segregate cargo and induce membrane budding. Given the prominent role of Hrs in the biogenesis of late endosomes, we considered whether it is involved in phagosomal maturation and, more importantly, whether its activity is affected by mycobacteria.
MATERIALS AND METHODS
Reagents and antibodies.
Dulbecco's minimal Eagle's medium and fetal bovine serum were from Wisent Inc. HEPES-buffered RPMI, wortmannin, tubulin antibody, and human immunoglobulin G (IgG) were from Sigma. Latex beads were from Bangs Laboratories. Sheep red blood cells (RBC) and rabbit anti-RBC IgG were from ICN-Cappel. Fluorochrome-conjugated secondary antibodies were all from Jackson ImmunoResearch. LysoTracker (Red DND-99) was from Molecular Probes. Mouse and rat anti-LAMP-1 antibodies were from the Developmental Studies Hybridoma Bank, maintained by the University of Iowa and Johns Hopkins University. Goat anti-EEA1 and anti-c-Myc antibodies were from Santa Cruz Biotechnology. Antibodies to Mycobacterium were from Cygnus Technologies. The preparation of antibodies to LBPA and Hrs has been described elsewhere (15, 24).
Cell culture, transfection, and plasmids.
Culture conditions for macrophage RAW 264.7, COS-IIA, and Chinese hamster ovary cells (CHO-IIA) stably transfected with FcγRIIA receptors have been previously described (34). The generation of the plasmids used for expression of wild-type and mutant forms of either epitope-tagged or yellow fluorescent protein (YFP)-conjugated Hrs is described in detail elsewhere (23, 24, 26). The plasmids encoding the PX domain of p40phox and the 2-FYVE domain of EEA1 were generously provided by M. Yaffe (MIT, Cambridge, Mass.) and L. Cantley (Beth Israel Deaconess Medical Center, Boston, Mass.) and have been described elsewhere (14, 34). The cells were transiently transfected by using FuGENE-6 (Roche Molecular Biochemicals) as suggested by the manufacturer.
Treatment with siRNA.
Small interfering RNA (siRNA) directed toward nucleotides 160 to 180 relative to the start codon of the human Hrs (GenBank accession number NM 004712.31) was purchased from Dharmacon Research (Lafayette, Colo.) as double-stranded, desalted, and gel-purified preparations. The sequence used for siRNA was selected according to the guidelines in reference 8. Transfection of siRNA by use of oligofectamine (Invitrogen) was performed according to the manufacturer's directions by using 240 pmol of siRNA to transfect ∼100,000 COS-IIA cells grown on a coverslip placed within a well of a six-well plate. Cells were grown for 72 h and then processed for immunofluorescence, electron microscopy, or immunoblotting.
Phagocytosis assays and treatment with wortmannin.
Fresh or fixed sheep RBC were opsonized with rabbit anti-sheep RBC antibody (1:50). Latex beads were opsonized with 1 mg of human IgG/ml. Opsonization was for either 1 h at room temperature or overnight at 4°C. Where noted, the cells were treated with 100 nM wortmannin for 30 min prior to phagocytosis. The onset of phagocytosis was synchronized by allowing the particles to bind to cells on ice for 5 min, and ingestion was then initiated by incubation at 37°C. Excess particles were washed away with phosphate-buffered saline (PBS) and, where indicated, the cells were incubated in culture medium at 37°C for the specified additional chase period. To identify adherent particles that were not internalized, the cells were incubated at 4°C with Cy5-labeled donkey anti-rabbit IgG (1:1,000) or Cy5-labeled donkey anti-human IgG (1:1,000) for 10 min.
Culturing and phagocytosis of mycobacteria.
Mycobacterium smegmatis mc2 155 and Mycobacterium marinum 1218R (ATCC 927) were transformed with the plasmid pG13 as described previously (2). Cultures were grown in Middlebrook 7H9 media supplemented with 10% oleic acid-albumin-dextrose-catalase (Difco) and 25 μg of kanamycin (Sigma)/ml. Typically, M. smegmatis cultures were grown for 16 h at 37°C and M. marinum cultures were grown for 36 to 48 h at 30°C. Before use, cultures were washed twice with PBS before being homogenized with 50 strokes on ice, followed by sonication at 60% power (Sonic Dismembrator Model 300; Fisher) for 2 min, followed by centrifugation at low speed to remove aggregates. For phagocytosis assays, mycobacteria were then sedimented onto RAW 264.7 cells by centrifugation (∼10 bacteria/cell). Where required, extracellular bacteria were identified by labeling with anti-Mycobacterium antibodies.
Fluorescence and confocal microscopy.
To estimate phagosomal pH, cells were allowed to internalize particles and then 50 nM LysoTracker Red was added. Labeling was terminated after 5 min by placing the cells on ice. Live cells were analyzed immediately by fluorescence microscopy to determine the percentage of LysoTracker-positive phagosomes. The protocols for immunostaining of EEA1 (34), LAMP-1 (34), and LBPA (15) have been detailed in the respective references. For Hrs staining, the cells were permeabilized with 0.05% saponin and then fixed for 30 min at 4°C. Permeabilized cells were incubated with primary antibody for 1 h, washed extensively, and then incubated with secondary antibodies for 1 h at room temperature and washed again. Coverslips were then mounted and fixed onto glass slides by using a mounting reagent (Dako Corp., Mississauga, Canada). The antibody dilutions used were EEA1 (1:50), LAMP-1 (1:4), myc (1:200), Mycobacterium (1:40), LBPA (1:400), and Hrs (1:200). Both live and fixed samples were analyzed by using the LSM 510 laser scanning confocal microscope (Zeiss) or a DMIRE2 epifluorescence microscope (Leica) with a 100× oil immersion objective. Digital images were prepared by using AdobePhotoshop6 and Adobe Illustrator10 (Adobe Systems Inc.).
Electron microscopy.
Control and siRNA-treated cells were fixed with 2% glutaraldehyde in 0.1 M Sorenson's phosphate buffer (pH 7.2) for 5 min before the coverslip was scraped off and the cultures were subjected to centrifugation. Fixation was continued at room temperature for two additional hours. Cells were then postfixed in 1% OsO4 in phosphate buffer at room temperature for 2 h. Cells were stained en bloc for 1 h with 1% uranyl acetate in H2O followed by dehydration and embedding in Epon resin (EMbed-812; Electron Microscopy Sciences). Sections (70 to 80 nm thick) were collected on copper grids, stained with uranyl acetate and lead citrate, and viewed by using a Philips CM100 electron microscope, and images were captured by a Kodak Megaplus Camera, model 1.6i.
Other methods.
Phagosomes were isolated by the method of Desjardins et al. (6) from RAW 264.7 cells grown on 14-cm-diameter petri dishes to 80 to 90% confluence. The protein concentration of the phagosomal preparation was determined with the bicinchoninic acid assay (Pierce), with albumin as a standard. Isolated phagosomes or whole-cell extracts were solubilized in Laemmli's sample buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto polyvinylidene difluoride membranes. The membranes were blocked overnight with 5% milk in PBS and 0.05% Tween 20. Antibodies to Hrs and tubulin were used at a 1:1,000 dilution, and LAMP-1 antibody was used at a 1:10 dilution. Immunoreactive bands were visualized by ECL (Amersham, Piscataway, N.J.).
RESULTS
Distribution of Hrs during phagocytosis.
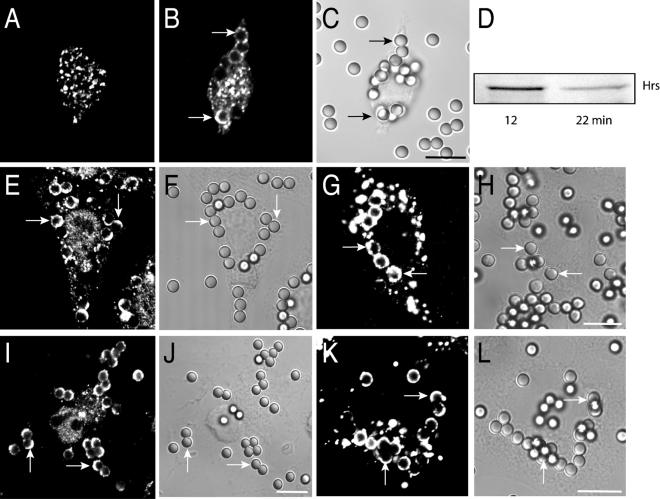
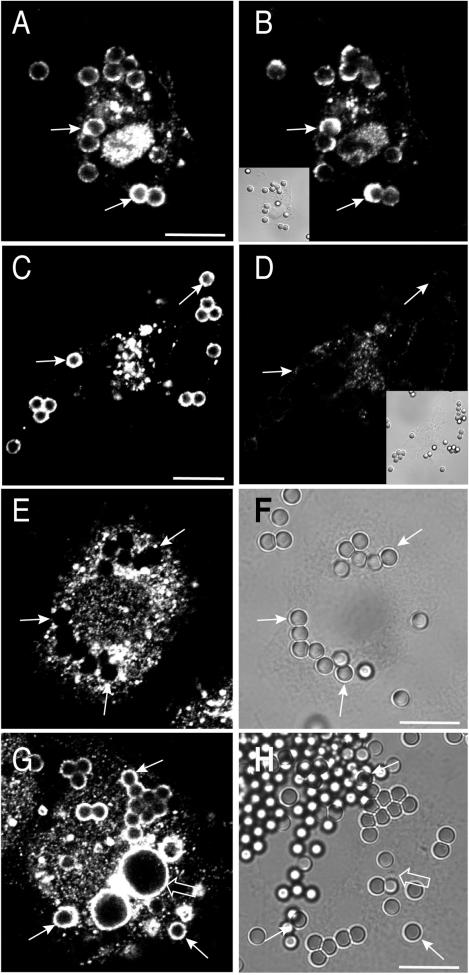
Immunofluorescence analysis revealed that in quiescent RAW264.7 macrophages Hrs is found exclusively in intracellular vesicular structures (Fig. 1A), as described for other cells (21, 31). The distribution of Hrs overlaps substantially with that of EEA1, a marker of a subset of early endosomes (not shown). When particles are engulfed, a fraction of the Hrs redistributes to the phagosome (Fig. 1B), showing a unique pattern. Unlike other phagosomal constituents described heretofore, Hrs was rarely found to be homogeneously dispersed throughout the phagosomal membrane and was instead distributed in a patchy or half-moon pattern. After 12 min of phagocytosis, 66% of the phagosomes were Hrs positive, and of these 45.7% ± 5.7% had a half-moon appearance. A similar nonhomogeneous distribution was observed with engineered phagocytes (Fig. 1E and I), i.e., CHO or COS cells that were conferred phagocytic capability by stable transfection with FcγRIIA receptors (13). After a 25-min phagocytic pulse, Hrs was detected in 69.8% of phagosomes in CHO cells and in 52.8% ± 4.2% of these Hrs was distributed nonhomogeneously. The specificity of the immunostaining of the endogenous Hrs was verified by analyzing the localization of Hrs tagged with either the myc epitope or with YFP that was heterologously transfected into phagocytic cells. As shown in Fig. 1G and K, the tagged Hrs showed a distribution virtually identical to that of the endogenous protein. The association of Hrs with engulfed particles was also verified by immunoblotting purified phagosomes obtained from RAW 264.7 cells. As shown in Fig. 1D, Hrs accumulates transiently in phagosomes, being readily detectable at early times (e.g., 12 min after initiation of phagocytosis) and diminishing in density thereafter. Digital fluorescence imaging of live RAW 264.7 cells transiently transfected with YFP-Hrs was used to confirm this observation. Hrs was never found in the cups lining nascent phagosomes and was detectable only after phagosomal sealing had been completed. When cells were fixed and stained by using phalloidin, F-actin dissociation from the phagosome was found to precede Hrs accumulation (not shown). Hrs was clearly detectable on phagosomes as early as 5 to 8 min and was largely depleted by 18 min.
FIG. 1.
Immunostaining of endogenous and ectopically expressed Hrs. (A and B) Immunostaining of endogenous Hrs in quiescent RAW 264.7 cells (A) and in cells allowed to ingest IgG-opsonized latex beads for 10 min prior to fixation and permeabilization (B). (C) The corresponding differential interference contrast (DIC) image. (D) Detection of Hrs in phagosomes isolated from cells following a 12-min phagocytosis pulse or a 12-min pulse followed by a 10-min chase (22 min), as indicated. Identical amounts of protein (10 μg) were loaded in each lane. (E to H) Distribution of Hrs in CHO-IIA cells following ingestion of latex beads for 20 min. (E and F) Immunostaining of endogenous Hrs and the corresponding DIC image, respectively. (G and H) Distribution of myc-Hrs following transient transfection and the corresponding DIC image, respectively. (I to L) Distribution of Hrs in COS-IIA cells following ingestion of latex beads for 20 min. (I and J) Immunostaining of endogenous Hrs and the corresponding DIC image, respectively. (K and L) Distribution of myc-Hrs following transient transfection and the corresponding DIC image, respectively. The arrows point to Hrs-positive phagosomes. The images and the blot are representative of at least four experiments of each type. Bars, 10 μm.
Wortmannin impairs the association of Hrs with phagosomes.
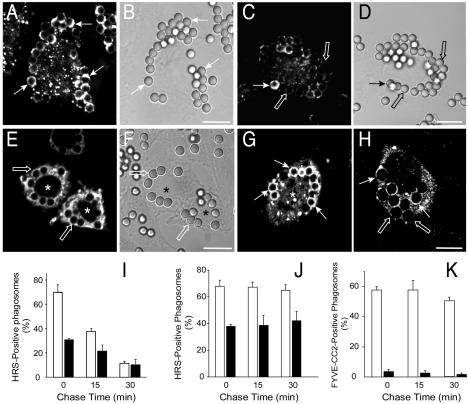
The discontinuous pattern of Hrs on the phagosomal membrane differs significantly from the homogeneous distribution of PI(3)P, gleaned by use of a chimeric construct of two FYVE domains and green fluorescent protein (GFP) (34). This appeared to contravene the notion that binding to the phosphoinositide is an important determinant of Hrs recruitment. To test this premise, cells were treated with wortmannin, a phosphatidylinositol 3-kinase inhibitor shown to deplete cellular PI(3)P by inhibition of Vps34 (36). Under conditions where PI(3)P is no longer detectable on phagosomes, wortmannin treatment reduced but did not abolish the binding of Hrs. Similar results were obtained for FcγRIIA-expressing CHO-IIA cells (Fig. 2A to D), for RAW 264.7 macrophages (not illustrated), and also when endogenous and ectopically expressed Hrs were analyzed (Fig. 2I and J, respectively). Immediately after a 20-min phagocytosis pulse, 69.8% ± 6.5% of the phagosomes had Hrs in otherwise untreated CHO-IIA cells but only 31.3% ± 0.9% did after treatment with wortmannin. The fractions of positive phagosomes dropped to 38.6% ± 1.8% and 22% ± 5.6%, respectively, after a 15-min chase period (Fig. 2I). These findings imply that the interaction of its FYVE domain with PI(3)P is not the sole determinant of Hrs association with phagosomes.
FIG. 2.
Effects of wortmannin on Hrs recruitment to the phagosomes and localization of Hrs mutants. CHO-IIA were allowed to internalize 3-μm-diameter latex beads for 20 min and then fixed and subjected to immunostaining. Where indicated, the cells were pretreated with 100 nM wortmannin or were transfected with epitope-tagged Hrs mutants. (A to D) Distribution of wild-type Hrs in control (A and B) and wortmannin-treated (C and D) cells. (E and F) Distribution of HrsR183A. (G) Distribution of HrsFYVE+CC. (H) Distribution of HrsΔUIM. The images in panels A, C, E, G, and H are representative confocal fluorescence images. The images in panels B, D, and F are the differential interference contrast images corresponding to those in panels A, C, and E, respectively. Transfected cells are indicated by asterisks. The solid arrows point to Hrs-positive phagosomes, while open arrows indicate Hrs-deficient phagosomes. Bars, 10 μm. Images are representative of at least four experiments of each type. (I to K) Quantification of the effect of wortmannin on the phagosomal acquisition of wild-type or mutant Hrs. The cells were allowed to internalize opsonized beads for 20 min (chase time, 0 min) and then chased for the times indicated in the graph. Shown are endogenous Hrs (I), transfected wild-type Hrs (myc-tagged) (J), and HrsFYVE+CC (K). Empty columns, control cells; black columns, wortmannin-treated cells. Data are means ± standard errors of three separate experiments (200 cells were counted in each).
Transfection of mutant forms of Hrs was used to define the domains required for its association to phagosomes. Engineered phagocytes were used for these experiments because the efficiency of ectopic expression of Hrs and its mutants in RAW 264.7 cells was very low (<1%), precluding the accumulation of large sets of statistically meaningful data. The point mutant HrsR183A was used to evaluate the role of the FYVE domain in phagosomal association. This mutation was reported to abolish PI(3)P binding while maintaining the overall conformation of Hrs (11, 20). The accumulation of HrsR183A on phagosomal membranes was considerably reduced in comparison with the wild-type construct (cf. panels A and E of Fig. 2) under conditions where the two forms of the molecule were expressed to comparable levels. Thus, an intact FYVE domain is essential for optimal phagosomal association. Though necessary, the FYVE domain is not sufficient for binding; recruitment of the isolated FYVE domain of Hrs to the phagosome was negligible (not shown). By contrast, the HrsFYVE-CC construct, containing both the coiled-coil (CC) and FYVE domains of Hrs, was targeted efficiently to phagosomes (Fig. 2G). Binding is not due solely to the CC domain, since exposure of the cells to wortmannin virtually eliminated the interaction (Fig. 2K). It is noteworthy that the extent of the displacement of HrsFYVE-CC from the phagosome induced by wortmannin was much greater than that noted for full-length Hrs. We interpret these findings to mean that yet another domain of the protein, distinct from the CC and FYVE domains, is involved in tethering Hrs to phagosomes. The contribution of this third domain is most apparent following inhibition of Vps34.
Hrs possesses a region capable of interacting with ubiquitylated proteins, namely the UIM motif. To test whether this motif contributes to the association of Hrs with the phagosomal membrane, we measured the interaction of UIM-deficient Hrs (HrsΔUIM) with phagosomes. Typical results are illustrated in Fig. 2H. UIM-deficient Hrs bound to phagosomes considerably less efficiently than did its wild-type counterpart. Densitometric analysis of the digital images indicated that omission of the UIM depressed binding by ∼75%. Together, these data indicate that at least three regions of Hrs, namely the FYVE, UIM, and CC domains, are required for optimal binding of Hrs to phagosomes.
Overexpression of Hrs alters phagosomal maturation.
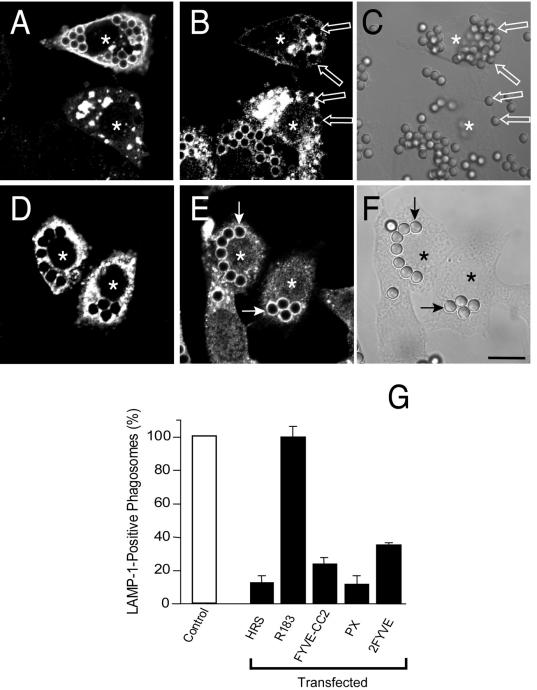
We next proceeded to evaluate the possible role of Hrs in phagosomal maturation. To this end, we initially planned to use a dominant-negative strategy by transfection of mutant forms of Hrs. However, during the course of control experiments, it became obvious that even the overexpression of wild-type Hrs altered the kinetics of phagosomal maturation. This is apparent when comparing the rate of dissociation of endogenous and ectopically expressed Hrs. (Fig. 2I and J). While wortmannin inhibits association to a similar extent in the two cases, Hrs dissociates much more slowly when overexpressed. Although a systematic quantitation was not performed, it appeared as if the delay in dissociation was proportional to the level of overexpression (not shown). The reduced dissociation of Hrs was accompanied by arrested maturation, as indicated by the inability of the phagosomes to acquire LAMP-1, a marker of late endocytic compartments (Fig. 3A to C).
FIG. 3.
Effects of Hrs overexpression on LAMP-1 acquisition by phagosomes. CHO-IIA transfected with myc-tagged wild-type Hrs (A to C) or HrsR183A (D to F) were allowed to interact with opsonized latex beads for 20 min, and after unbound beads were washed, the phagosomes were allowed to mature for 60 min. The cells were then fixed and immunostained with myc (A and D) and LAMP-1 (B and E) antibodies. C and F are the corresponding DIC images. Transfected cells are identified by asterisks. The solid and open arrows point to LAMP-1-positive and -negative phagosomes, respectively. Images are representative of four experiments of each type. Bars, 10 μm. (G) Quantification of the effects of various constructs on LAMP-1 acquisition by phagosomes. CHO-IIA cells either were left untransfected (empty column) or were transfected with wild-type Hrs, HrsR183A, HrsFYVE+CC, the PX domain of p40phox or two tandem FYVE domains of EEA1, as specified. LAMP-1 acquisition by phagosomes was determined as above. Data are means ± standard errors of three experiments (200 cells counted in each).
We speculated that overexpression of Hrs may exert this inhibitory effect by scavenging an inordinately large fraction of PI(3)P. To test this hypothesis, cells were transfected with comparable amounts of HrsR183A, which lacks the ability to bind the inositide. As illustrated in Fig. 3D to F, expression of large amounts of HrsR183A had no discernible effect on maturation, supporting the notion that scavenging of PI(3)P is responsible for the effects of excess Hrs. Accordingly, we found that overexpression of the HrsFYVE-CC construct and of two other constructs capable of binding PI(3)P (the PX domain of p40phox and a tandem 2-FYVE domain of EEA1) had sizable inhibitory effects (Fig. 3G).
Effect of depletion of endogenous Hrs.
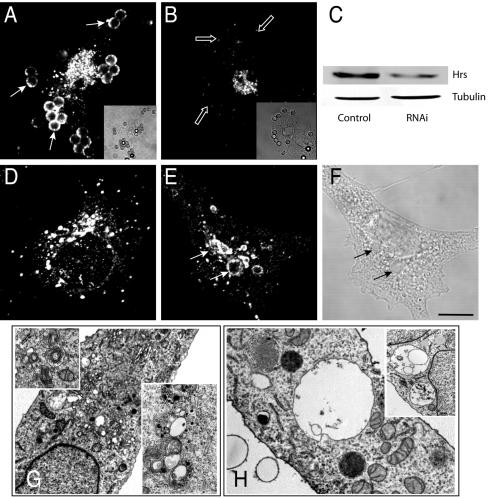
While the above results confirm the importance of PI(3)P availability for completion of phagosomal maturation, they also preclude the use of dominant-negative constructs to analyze the putative role of Hrs in the process. As an alternative, we attempted to deplete the endogenous Hrs by using siRNA. We initially confirmed that synthesis of the protein was arrested by a 72-h treatment with the siRNA. As shown in Fig. 4A and B, Hrs was extensively depleted in phagocytic COS-IIA cells, as observed by immunostaining. Unlike the untreated cells, which were all immunoreactive, approximately 20 to 30% of the cells stained only marginally with the antibody to Hrs. This estimate was confirmed by immunoblots of whole-cell extracts, which showed a 70% decrease in the total Hrs content of the population (mean from three experiments). The loss was specific, since the levels of tubulin were unaffected (Fig. 4C) and the cells retained their ability to perform phagocytosis at normal rates. The siRNA was ineffective towards RAW 264.7 cells, likely because of their refractoriness to transfection. All subsequent experiments were therefore performed with the readily transfectable COS-IIA cells.
FIG. 4.
Effects of siRNA on endogenous Hrs. COS-IIA cells either were mock transfected (A, D, and Control lane in panel C) or were transfected with Hrs siRNA for 72 h. (A and B) The cells were allowed to ingest particles before immunostaining of the endogenous Hrs. Solid and open arrows point to Hrs-positive or negative phagosomes, respectively. (C) Lysates of mock-transfected cells (control) or cells transfected with Hrs siRNA were subjected to immunoblotting with antibodies to Hrs or tubulin. (D and E) EEA1 immunostaining. (F) Differential interference contrast image corresponding to that shown in panel E. Arrows point to EEA1-positive large vacuoles. Bar, 10 μm. (G and H) Transmission electron micrographs. Magnified insets reveal multivesicular bodies in mock-transfected cells (G) and spacious vacuoles in Hrs-depleted cells (H).
The cells treated with Hrs siRNA exhibited large vacuoles that were frequently EEA1 positive (Fig. 4E and F). Ultrastructural analysis indicated that the vacuoles were often empty or contained infrequent smaller vacuoles within their lumen but rarely small vesicles. Overall, the number of dense multivesicular structures was smaller in siRNA-treated cells than in controls (cf. Fig. 4G and H). This phenotype resembles that reported for murine embryonic cells lacking Hrs (16), suggesting that siRNA-treated cells are suitable models to study the consequences of selective ablation of Hrs.
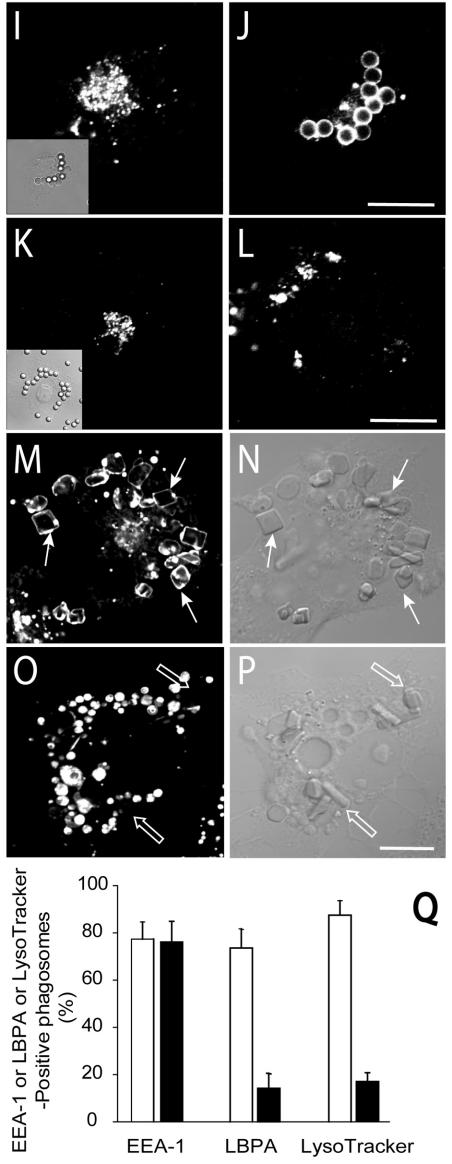
The effects of Hrs depletion on phagosomal maturation were studied next. In cells treated with Hrs siRNA, phagocytosis proceeded normally and at early times acquisition of EEA1 by the phagosomes was indistinguishable from that of controls (76% ± 7% and 77% ± 6%, respectively, after 20 min; Fig. 5A to D and Q). However, EEA1 dissociation from phagosomes was impaired in Hrs-depleted cells. EEA1 was clearly enriched in 95.9% ± 4.2% of the phagosomes of Hrs-deficient cells even after 3 h, a time when control phagosomes are entirely devoid of EEA1 (Fig. 5E to H). This suggested that maturation was arrested at the early phagosomal stage in Hrs-depleted cells. This notion was confirmed by analyzing the presence of LBPA and the extent of acidification of phagosomes of control and depleted cells. While the vast majority (∼80%) of the phagosomes acquire LBPA after 90 min in control cells, only 14% ± 8% of the phagosomes contained the lysolipid in Hrs-depleted cells (Fig. 5I to L and Q). Moreover, LysoTracker, a fluorescent acidotropic probe accumulated distinctly in phagosomes of normal cells, but poorly, if at all, in phagosomes of Hrs-deficient cells (Fig. 5M to P). Nearly 90% of normal phagosomes accumulated LysoTracker, but this figure dropped to only 18% ± 8% following elimination of Hrs (Fig. 5Q). This implies that the pH of Hrs-depleted phagosomes is higher than that of normal phagosomes. Failure to acidify was not due to an effect of Hrs ablation on the vacuolar ATPases responsible for endomembrane acidification, since multiple acidic vesicles and vacuoles were observable in the depleted cells (Fig. 5O). Instead, the impaired acidification likely reflects the inability of ATPase-enriched vesicles to merge with the phagosomes. Jointly, these results suggest that Hrs is essential for the progression of early phagosomes to phagolysosomes.
FIG. 5.
Effects of Hrs depletion by siRNA on phagosomal maturation. COS-IIA cells were either mock transfected (A, B, E, F, I, J, M, and N) or were transfected with Hrs siRNA for 72 h (C, D, G, H, K, L, O, and P). (A to D) Cells were fixed and permeabilized 20 min after initiation of phagocytosis and immunostained for EEA1 (A and C) and Hrs (B and D). Corresponding differential interference contrast (DIC) images are shown in insets. (E and G) Cells were allowed to internalize particles for 20 min, followed by a 3-h chase at 37°C. Next, the cells were fixed, permeabilized, and immunostained for EEA1 (E and G). Panels F and H show the corresponding DIC images. (I to P) Cells were allowed to internalize particles for 20 min followed by a 90-min chase at 37°C. (I and K) Hrs immunostaining. Insets show DIC images. (J and L) LBPA immunostaining. (M and O) Accumulation of LysoTracker. Panels N and P show the corresponding DIC images. (Q) Quantification of the effects of Hrs depletion on EEA1, LBPA, and LysoTracker acquisition by phagosomes from experiments like those illustrated in panels A to P. Staining was measured at the following times after phagocytosis: EEA1, 20 min; LBPA, 90 min; and LysoTracker, 90 min. Open columns, mock-transfected cells; black columns, cells transfected with Hrs siRNA. Data are means ± standard errors of four separate experiments (200 cells were counted in each).
Hrs acquisition by mycobacterium-containing phagosomes is impaired.
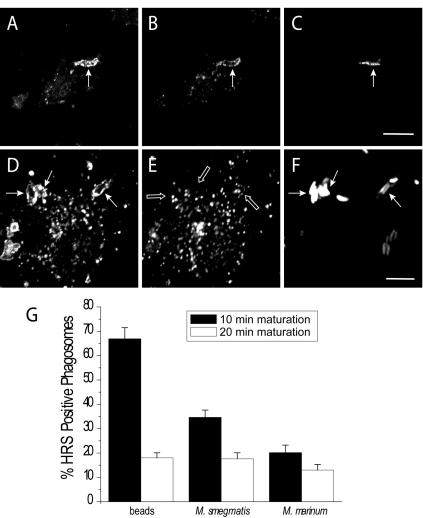
The stage at which maturation is arrested by depletion of Hrs is similar to that noted earlier for phagosomes containing virulent mycobacteria. It is therefore conceivable that the failure of mycobacterium-containing vacuoles to progress to phagolysosomes is related to impaired Hrs recruitment or function. This hypothesis was tested by comparing the Hrs content of phagosomes formed by cells challenged with either mycobacteria of different degrees of virulence or inert particles. RAW 264.7 macrophages were exposed either to M. marinum, a pathogen of fish and frogs that is phylogenetically related to M. tuberculosis (32) and affects maturation in a similar manner (2), or to the usually avirulent M. smegmatis. Latex beads opsonized with IgG were used as an inert control. Internalization was allowed to occur for various periods of time, and after fixation and permeabilization, the presence of Hrs in phagosomes was assessed by immunofluorescence. Care was taken to exclude extracellular, adherent bacteria or beads from the quantitation (see Materials and Methods). As shown in Fig. 6, up to 66.1% of the beads acquired Hrs after 10 min and the fraction of Hrs-positive particles decreased thereafter. As was found for latex beads, the distribution of Hrs on phagosomes containing M. smegmatis was nonhomogeneous (Fig. 6B). By contrast, only 20% of the phagosomes containing M. marinum were found to contain Hrs 10 min after internalization. The slower kinetics of phagocytosis of the bacteria cannot account for this difference, because the fraction of Hrscontaining phagosomes decreased further with longer times. When the period of exposure to the bacteria was shortened to less than 10 min, the efficiency of phagocytosis was reduced to the point where statistical evaluation was compromised. Nevertheless, it was qualitatively apparent that the fraction of phagosomes with Hrs was not higher at earlier times. A significantly higher fraction of phagosomes formed by ingestion of the avirulent M. smegmatis had clearly detectable Hrs (Fig. 6). These findings demonstrate a positive correlation between the Hrs content of phagosomes and their ability to complete maturation.
FIG. 6.
Acquisition of Hrs by phagosomes containing beads or mycobacteria. (A to F) RAW 264.7 cells were allowed to ingest M. smegmatis (A to C) or M. marinum (D to F) transformed by pG13, and the presence of EEA1 (A and D) or Hrs (B and E) on phagosomes was assessed by immunostaining. The location of the bacteria was visualized by detecting endogenously expressed GFP fluorescence (C and F). Images are representative of five experiments. (G) RAW 264.7 cells were allowed to ingest IgG-opsonized latex beads, M. smegmatis, or M. marinum, and the presence of Hrs on phagosomes was assessed by immunostaining. Extracellular adherent bacteria were identified by using antibodies and were excluded from the calculations. The presence of Hrs on phagosomes was assessed 10 min (black bars) or 20 min (open bars) after internalization. Data show the percentages of Hrs-positive phagosomes and are means ± standard errors of three independent experiments (80 to 100 phagosomes for each condition). Size bar, 5 μm.
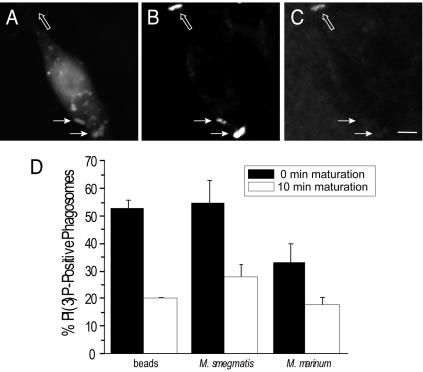
Because the FYVE domain was shown, as reported above, to be important for Hrs recruitment to phagosomes, defective acquisition of its ligand, PI(3)P, may account for the reduced recruitment of Hrs. This hypothesis was tested by directly measuring the accumulation of PI(3)P on phagosomes by use of two tandem FYVE domains fused to GFP, as described earlier (34). The results of these experiments are shown in Fig. 7. As reported earlier, PI(3)P is readily detected in phagosomes formed upon ingestion of inert particles (latex beads). The phosphoinositide is most abundant early after phagocytosis and decreases in abundance after 10 to 20 min. Similar results were obtained when visualizing phagosomes formed by engulfment of avirulent M. smegmatis. However, the fraction of PI(3)P-positive phagosomes declined significantly when they were generated by engulfment of M. marinum (from >50 to 33%). As before, this is unlikely to reflect delayed internalization kinetics, since the fraction of PI(3)P-containing phagosomes was lower at later times (Fig. 7). The paucity of PI(3)P may account, at least partly, for the reduced Hrs content of M. marinum-containing phagosomes.
FIG. 7.
Accumulation of PI(3)P by phagosomes. RAW 264.7 cells were allowed to ingest IgG-opsonized latex beads, M. smegmatis, or M. marinum, and the presence of PI(3)P on phagosomes was assessed by transfection of 2-FYVE-GFP and fluorescence microscopy. Extracellular adherent bacteria (open arrows) were identified by using anti-Mycobacterium antibodies prior to permeabilization, followed by a Cy5-conjugated secondary antibody, and were excluded from the calculations. Total bacteria (arrows) were visualized by using anti-Mycobacterium antibodies after permeabilizing the cells, followed by a Cy3-conjugated secondary antibody. A typical experiment using M. smegmatis is illustrated in panels A to C, showing the distribution of PI3P (A) and the location of total bacteria (B) and identifying extracellular bacteria (C). A summary of similar data from multiple experiments is presented in panel D. The presence of PI(3)P on phagosomes (D) was assessed after a 20-min pulse of phagocytosis (black bars) or 10 min thereafter (open bars). Extracellular particles were removed after the 20-min pulse to preclude continued internalization during the 10-min chase. Data show the percentages of PI(3)P-positive phagosomes and are means ± standard errors of three independent experiments. Size bar, 5 μm.
DISCUSSION
We report here that Hrs is recruited to phagosomes, where it plays an essential role in the maturation sequence. Interaction between the FYVE domain of Hrs and PI(3)P on the early phagosomal membrane contributes to the recruitment process. However, several lines of evidence indicate that other factors are also involved: (i) treatment with wortmannin under conditions that deplete PI(3)P to undetectable levels only partially reduced the binding of Hrs to phagosomes (Fig. 2); (ii) similarly, the recruitment of HrsR183A, which lacks the ability to bind PI(3)P, was reduced but not abolished; (iii) association of the isolated FYVE domain of Hrs to the phagosome was negligible; and (iv) the distribution of Hrs and that of the phosphoinositide on the phagosomal membrane were clearly different, since while PI(3)P is distributed homogeneously, Hrs displays a half-moon or patchy appearance. To our knowledge, this is the first evidence of the existence of microdomains on the phagosomal membrane. These results bear some resemblance to the findings of Raiborg and collaborators (24), who reported that the PI(3)P ligand, EEA1, and Hrs localize to distinct regions of Rab5Q79L-induced giant endosomes.
In addition to the FYVE domain, we found that the CC domain of Hrs is required for optimal association with phagosomes and that another region of the molecule must also contribute to the process. Interaction of the UIM motif of Hrs with ubiquitylated proteins (22, 23, 28) on the phagosomal membrane appears to provide this additional attachment site. Ubiquitylation of the Fcγ receptors that mediate phagocytosis, and possibly also that of other proteins associated with the receptor complex, has been reported to occur shortly after ligand binding. The E3 ubiquitin ligase Cbl is known to associate with the receptor complex and is likely to be responsible, at least in part, for the observed ubiquitylation. Whether additional ubiquitylation occurs at later stages of maturation remains undefined. Because Hrs does not associate with the phagosomes immediately after they seal, we propose that interaction of the UIM with ubiquitylated proteins is insufficient to recruit and maintain Hrs on the phagosome. This likely requires the subsequent synthesis of PI(3)P, and a three-point attachment is completed by association of the CC domain with an as-yet-undefined partner on the phagosomal membrane.
We initially attempted to use expression of dominant-negative mutants to evaluate the contribution of Hrs to phagosomal function. Indeed, truncated Hrs mutants were found to interfere with phagosomal maturation. However, the inhibitory mechanism was found to be trivial, consisting namely of scavenging of phagosomal PI(3)P. For this reason, we were compelled to use siRNA to deplete the endogenous Hrs in order to assess its function. The phenotype of our Hrs-depleted cells resembled those, published recently, of cells of flies and mice lacking a functional HRS gene (16, 17). More importantly, we found that the elimination of Hrs is associated with defective phagosomal maturation; acquisition of LBPA and luminal acidification were severely impaired. The latter likely reflects a reduced density of V-ATPases on the phagosomal membrane.
In control cells, a large fraction of the cellular LBPA redistributes to the phagosomes during maturation (Fig. 5I and J). This implies that fusion with existing LBPA-containing compartments is an important contributor to the acquisition of the lysolipid by the phagosome. The nearly complete elimination of LBPA from phagosomes in Hrs-depleted cells therefore implies that their ability to fuse with late endosomes or lysosomes was altered. The paucity of V-ATPases is consistent with this notion. This is not attributable to depletion of LBPA-containing or acidic late endosomes or lysosomes, since these were readily apparent in seemingly normal numbers in cells lacking Hrs (Fig. 5). One must therefore postulate that the ability of phagosomes to fuse with these compartments depends on Hrs, which may induce sorting of fusogenic proteins into active patches on the phagosomal membrane. This extends the repertoire of functions attributed to Hrs, which was heretofore thought to act mainly in membrane fission events. Hrs may exert its effects on fusion by binding to SNAP-25 (3), a known component of the fusogenic machinery of cells (30, 35).
The phenotype of phagosomes in Hrs-depleted cells bears a striking resemblance to that of phagosomes induced by virulent mycobacteria. Both are deficient in LBPA and show incomplete acidification, appearing to be arrested at an early stage of maturation. This raises the possibility that impairment of Hrs function is part of the mechanism used by mycobacteria to prevent phagolysosome formation. Indeed, we find that recruitment of Hrs is considerably lower in the case of virulent mycobacteria than in that of opsonized latex particles or avirulent bacteria. Several putative targets can be envisaged to account for the defective recruitment, based on the preceding discussion. First, the PI(3)P content of the phagosome may be reduced in mycobacterial phagosomes. This possibility was tested experimentally by using a fluorescent probe to detect this inositide. As shown in Fig. 7, fewer phagosomes were found to contain detectable amounts of PI(3)P in the case of virulent bacteria than in their avirulent counterparts or inert particles. At first glance, this finding seems to be at odds with the observation of Fratti et al. (9) that Vps34 is recruited normally to mycobacterial phagosomes. However, the activity of the kinase may be altered by the bacterium. Alternatively, pathogenic mycobacteria may promote phosphoinositide 3-phosphatase activity or release molecules that compete with binding of FYVE domain-containing molecules such as Hrs or the 2-FYVE-GFP probe to PI(3)P. In addition, the bacteria may interfere with Hrs recruitment by blocking the association of the CC domain with its target or by reducing the interaction between the UIM motif and phagosomal ubiquitylated substrates.
Multiple mechanisms have been put forward to account for the effects of mycobacteria on phagosome maturation: reduced calcium responsiveness (18, 19), impairment of EEA1 recruitment (9), and enhanced SNARE degradation (10), along with the defective Hrs recruitment reported here. These are not mutually exclusive and may act jointly to ensure intraphagosomal survival of the bacteria. Other intracellular parasites, like Salmonella and Shigella, are known to target multiple mechanisms of the host cell, often using redundant effectors (1, 12). In the case of mycobacteria, Hrs appears to be an attractive target, inasmuch as this protein plays an essential role in the early-to-late phagosome transition, the very step affected by the parasite.
Acknowledgments
M. marinum and pG13 were kind gifts to S.G. from Lucia Parker.
This work was supported by the Canadian Institutes of Health Research, the Canadian Arthritis Society, and the National Sanatorium Association. O. V. Vieira was the recipient of a postdoctoral fellowship from PRAXIS XX1 (BPD/22039/99). C.C.S. is the recipient of a graduate studentship from the Canadian Institutes of Health Research. H.S. was supported by the Norwegian Cancer Society and the Novo Nordisk Foundation. S.G. is the current holder of the Pitblado Chair in Cell Biology.
REFERENCES
- 1.Adam, T. 2001. Exploitation of host factors for efficient infection by Shigella. Int. J. Med. Microbiol. 291:287-298. [DOI] [PubMed] [Google Scholar]
- 2.Barker, L. P., K. M. George, S. Falkow, and P. L. Small. 1997. Differential trafficking of live and dead Mycobacterium marinum organisms in macrophages. Infect. Immun. 65:1497-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bean, A. J., R. Seifert, Y. A. Chen, R. Sacks, and R. H. Scheller. 1997. Hrs-2 is an ATPase implicated in calcium-regulated secretion. Nature 385:826-829. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, N., A. Horman, and P. Woodman. 2002. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 157:91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin, L. S., M. C. Raynor, X. Wei, H. Q. Chen, and L. Li. 2001. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J. Biol. Chem. 276:7069-7078. [DOI] [PubMed] [Google Scholar]
- 6.Desjardins, M., J. E. Celis, G. van Meer, H. Dieplinger, A. Jahraus, G. Griffiths, and L. A. Huber. 1994. Molecular characterization of phagosomes. J. Biol. Chem. 269:32194-32200. [PubMed] [Google Scholar]
- 7.Desjardins, M., L. A. Huber, R. G. Parton, and G. Griffiths. 1994. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J. Cell Biol. 124:677-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 9.Fratti, R. A., J. M. Backer, J. Gruenberg, S. Corvera, and V. Deretic. 2001. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J. Cell Biol. 154:631-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fratti, R. A., J. Chua, and V. Deretic. 2002. Cellubrevin alterations and Mycobacterium tuberculosis phagosome maturation arrest. J. Biol. Chem. 277:17320-17326. [DOI] [PubMed] [Google Scholar]
- 11.Gaullier, J. M., E. Ronning, D. J. Gillooly, and H. Stenmark. 2000. Interaction of the EEA1 FYVE finger with phosphatidylinositol 3-phosphate and early endosomes. Role of conserved residues. J. Biol. Chem. 275:24595-24600. [DOI] [PubMed] [Google Scholar]
- 12.Holden, D. W. 2002. Trafficking of the Salmonella vacuole in macrophages. Traffic 3:161-169. [DOI] [PubMed] [Google Scholar]
- 13.Indik, Z. K., J. G. Park, S. Hunter, M. Mantaring, and A. D. Schreiber. 1995. Molecular dissection of Fc gamma receptor-mediated phagocytosis. Immunol. Lett. 44:133-138. [DOI] [PubMed] [Google Scholar]
- 14.Kanai, F., H. Liu, S. J. Field, H. Akbary, T. Matsuo, G. E. Brown, L. C. Cantley, and M. B. Yaffe. 2001. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 3:675-678. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, T., M. H. Beuchat, M. Lindsay, S. Frias, R. D. Palmiter, H. Sakuraba, R. G. Parton, and J. Gruenberg. 1999. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1:113-118. [DOI] [PubMed] [Google Scholar]
- 16.Komada, M., and P. Soriano. 1999. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 13:1475-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd, T. E., R. Atkinson, M. N. Wu, Y. Zhou, G. Pennetta, and H. J. Bellen. 2002. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108:261-269. [DOI] [PubMed] [Google Scholar]
- 18.Malik, Z. A., G. M. Denning, and D. J. Kusner. 2000. Inhibition of Ca(2+) signaling by Mycobacterium tuberculosis is associated with reduced phagosome-lysosome fusion and increased survival within human macrophages. J. Exp. Med. 191:287-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik, Z. A., C. R. Thompson, S. Hashimi, B. Porter, S. S. Iyer, and D. J. Kusner. 2003. Cutting edge: Mycobacterium tuberculosis blocks Ca(2+) signaling and phagosome maturation in human macrophages via specific inhibition of sphingosine kinase. J. Immunol. 170:2811-2815. [DOI] [PubMed] [Google Scholar]
- 20.Misra, S., and J. H. Hurley. 1999. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell 97:657-666. [DOI] [PubMed] [Google Scholar]
- 21.Mu, F. T., J. M. Callaghan, O. Steele-Mortimer, H. Stenmark, R. G. Parton, P. L. Campbell, J. McCluskey, J. P. Yeo, E. P. Tock, and B. H. Toh. 1995. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 270:13503-13511. [DOI] [PubMed] [Google Scholar]
- 22.Polo, S., S. Sigismund, M. Faretta, M. Guidi, M. R. Capua, G. Bossi, H. Chen, P. De Camilli, and P. P. Di Fiore. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416:451-455. [DOI] [PubMed] [Google Scholar]
- 23.Raiborg, C., K. G. Bache, D. J. Gillooly, I. H. Madshus, E. Stang, and H. Stenmark. 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4:394-398. [DOI] [PubMed] [Google Scholar]
- 24.Raiborg, C., K. G. Bache, A. Mehlum, E. Stang, and H. Stenmark. 2001. Hrs recruits clathrin to early endosomes. EMBO J. 20:5008-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raiborg, C., K. G. Bache, A. Mehlum, and H. Stenmark. 2001. Function of Hrs in endocytic trafficking and signalling. Biochem. Soc. Trans. 29:472-475. [DOI] [PubMed] [Google Scholar]
- 26.Raiborg, C., B. Bremnes, A. Mehlum, D. J. Gillooly, A. D'Arrigo, E. Stang, and H. Stenmark. 2001. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 114:2255-2263. [DOI] [PubMed] [Google Scholar]
- 27.Russell, D. G. 2001. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2:569-577. [DOI] [PubMed] [Google Scholar]
- 28.Shih, S. C., D. J. Katzmann, J. D. Schnell, M. Sutanto, S. D. Emr, and L. Hicke. 2002. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4:389-393. [DOI] [PubMed] [Google Scholar]
- 29.Simonsen, A., R. Lippe, S. Christoforidis, J. M. Gaullier, A. Brech, J. Callaghan, B. H. Toh, C. Murphy, M. Zerial, and H. Stenmark. 1998. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394:494-498. [DOI] [PubMed] [Google Scholar]
- 30.Sollner, T., S. W. Whiteheart, M. Brunner, H. Erdjument-Bromage, S. Geromanos, P. Tempst, and J. E. Rothman. 1993. SNAP receptors implicated in vesicle targeting and fusion. Nature 362:318-324. [DOI] [PubMed] [Google Scholar]
- 31.Stenmark, H., R. Aasland, B. H. Toh, and A. D'Arrigo. 1996. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem. 271:24048-24054. [DOI] [PubMed] [Google Scholar]
- 32.Tonjum, T., D. B. Welty, E. Jantzen, and P. L. Small. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 36:918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Underhill, D. M., and A. Ozinsky. 2002. Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 20:825-852. [DOI] [PubMed] [Google Scholar]
- 34.Vieira, O. V., R. J. Botelho, L. Rameh, S. M. Brachmann, T. Matsuo, H. W. Davidson, A. Schreiber, J. M. Backer, L. C. Cantley, and S. Grinstein. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber, T., B. V. Zemelman, J. A. McNew, B. Westermann, M. Gmachl, F. Parlati, T. H. Sollner, and J. E. Rothman. 1998. SNAREpins: minimal machinery for membrane fusion. Cell 92:759-772. [DOI] [PubMed] [Google Scholar]
- 36.Yano, H., S. Nakanishi, K. Kimura, N. Hanai, Y. Saitoh, Y. Fukui, Y. Nonomura, and Y. Matsuda. 1993. Inhibition of histamine secretion by wortmannin through the blockade of phosphatidylinositol 3-kinase in RBL-2H3 cells. J. Biol. Chem. 268:25846-25856. [PubMed] [Google Scholar]