Abstract
Utilization of nonfermentable carbon sources by Kluyveromyces lactis and Saccharomyces cerevisiae requires the Snf1p kinase and the Cat8p transcriptional activator, which binds to carbon source-responsive elements of target genes. We demonstrate that KlSnf1p and KlCat8p from K. lactis interact in a two-hybrid system and that the interaction is stronger with a kinase-dead mutant form of KlSnf1p. Of two putative phosphorylation sites in the KlCat8p sequence, serine 661 was identified as a key residue governing KlCat8p regulation. Serine 661 is located in the middle homology region, a regulatory domain conserved among zinc cluster transcription factors, and is part of an Snf1p consensus phosphorylation site. Single mutations at this site are sufficient to completely change the carbon source regulation of the KlCat8p transactivation activity observed. A serine-to-glutamate mutant form mimicking constitutive phosphorylation results in a nearly constitutively active form of KlCat8p, while a serine-to-alanine mutation has the reverse effect. Furthermore, it is shown that KlCat8p phosphorylation depends on KlSNF1. The Snf1-Cat8 connection is evolutionarily conserved: mutation of corresponding serine 562 of ScCat8p gave similar results in S. cerevisiae. The enhanced capacity of ScCat8S562E to suppress the phenotype caused by snf1 strengthens the hypothesis of direct phosphorylation of Cat8p by Snf1p. Unlike that of S. cerevisiae ScCAT8, KlCAT8 transcription is not carbon source regulated, illustrating the prominent role of posttranscriptional regulation of Cat8p in K. lactis.
From a historical point of view, one of the first observations made on yeasts was their marked differences when regulation of primary carbon metabolism was considered. Hence, baker's yeast (Saccharomyces cerevisiae) was classified as Crabtree positive because of its capacity to ferment glucose into ethanol even in the presence of oxygen, whereas milk yeast (Kluyveromyces lactis), like the majority of yeast species, belongs to the Crabtree-negative group owing to the absence of aerobic production of ethanol (8). Despite this, most of the genes involved in carbon and energy metabolism in K. lactis and S. cerevisiae show a high degree of sequence conservation, and the genes encoding their regulators, although displaying less sequence similarity, are commonly capable of heterospecific complementation as well (38). The molecular bases for the physiological differences between K. lactis and S. cerevisiae are now known: in S. cerevisiae, fermentation is favored as a result of glucose repression of mitochondrial function, whereas in K. lactis, glucose repression is far less pronounced and strain dependent (2). Moreover, a tight regulation of glucose transport, the higher capacity of respiratory enzymes, a lower gene redundancy in K. lactis, and differences in the organization of regulatory circuits are also implicated in the physiological divergence observed between the two yeast species (2).
The regulatory network governing nonfermentable carbon source utilization in the budding yeasts S. cerevisiae and K. lactis provides an example of such a differential configuration, explaining the dissimilar growth properties of the two yeasts. In S. cerevisiae, a specific DNA sequence called the carbon source-responsive element (CSRE) is required to coordinately activate the transcription of the gluconeogenic and glyoxylate genes during nonfermentative growth. This regulatory sequence is present in the promoter regions of many genes that are up-regulated during the diauxic shift (11, 16), including gluconeogenic genes like FBP1 (encoding fructose-1,6-bisphosphatase) (10, 23, 35) or PCK1 (encoding phosphoenolpyruvate carboxykinase) (23, 26) and glyoxylate cycle genes like MLS1 (encoding malate synthase) (5), ICL1 (encoding isocitrate lyase) (18, 31), and ACS1 (encoding acetyl coenzyme A synthase) (19). Two CSRE-binding factors affecting the transcription of gluconeogenic genes have been identified: Cat8p (27) and Sip4p (34). Both proteins are members of the Zn(II)2Cys6 zinc cluster family, and their activity is dependent on a functional Snf1p protein (17, 34), the central kinase required for growth on alternative sugars and nonfermentable carbon sources such as sucrose, glycerol, and ethanol (4, 6, 40). Whereas two-hybrid interaction revealed Sip4p as a direct phosphorylated target of the Snf1p kinase (21), a direct interaction between Cat8p and Snf1p could not be demonstrated (36). However, genetic evidence supports this hypothesis (17).
Functional homologues of Snf1p and Cat8p have been identified in K. lactis. FOG2/KlSNF1 complemented the phenotype of the fog2 mutant (deficient in fermentative and oxidative growth) and shows high sequence similarity to SNF1 (14). KlCAT8 was cloned by high-copy suppression of the klfog2/klsnf1 mutation, suggesting that it acts downstream of KlSnf1p (13). A Klcat8 mutation revealed that KlCat8p is required for efficient utilization of C2 carbon sources like ethanol or acetate but is dispensable for growth on glycerol and for derepression of KlFBP1 and KlPCK1 (13).
To elucidate the signaling events leading to activation of Cat8p, we assayed for two-hybrid interaction between KlSnf1p and KlCat8p. Our data support the view that KlCat8p is a direct target of KlSnf1p kinase activity. Serine 661, located in an Snf1 consensus phosphorylation site, was identified as a regulatory site that controls the KlCat8p activation function, probably via regulated phosphorylation, and corresponding serine 562 in ScCat8p has a similar role.
MATERIALS AND METHODS
Strains, media, and yeast techniques.
Escherichia coli strain DH10B (Gibco BRL) was used for all cloning procedures and was grown on standard Luria-Bertani medium. S. cerevisiae strain BOY (MATα his trp ura3::pSH18-34,6 lexAop-LacZ lexAop-LEU2) (15) was used for one- and two-hybrid tests. Other one-hybrid experiments involved S. cerevisiae strain MAV103 (MATa ura3-52 leu2-3,112 trp1-901 his3Δ200 ade2-101 gal4Δgal80ΔGAL1::lacZ lys2::GAL1-HIS3 SPAL10::URA3) (33). Complementation experiments were performed with S. cerevisiae strains CEN.NB1-1A (MATa ura3-52 his3-Δ1 leu2-3,112 trp1-289 cat8::LEU2 SUC2), CEN.PK2-1C (MATa ura3-52 his3-Δ1 leu2-3,112 trp1-289 SUC2) (29), and Y14311 (BY4742; MATα his3-Δ1 leu2Δ0 lys2Δ0 ura3Δ0 snf1::kanMX4) (Euroscarf). The K. lactis strains used in this study were JA6 (MATα ade1-600 adeT-600 trp1-11 ura3-12) (3) and congenic strains yIG2 (MATα ade1-600 adeT-600 trp1-11 ura3-12 mig1-Δ2::URA3) (12), yIG8 (MATα ade1-600 adeT-600 trp1-11 ura3-12 cat8Δ1) (13), JSD1R4 (13), yGC661A (MATα ade1-600 adeT-600 trp1-11 ura3 cat8-s661a), and yGC661E (MATα ade1-600 adeT-600 trp1-11 ura3 CAT8-S661E) (this study). The yGC661A and yGC661E strains were obtained by two-step gene replacement of the KlCAT8 wild-type allele. The first step was carried out by site-specific integration, in JA6, of the pRS306-CAT8S661A and pRS306-CAT8S661E vectors linearized within the KlCAT8 open reading frame (ORF) by digestion with BlpI. In the second step, eviction of the integrated plasmid was obtained by selection for loss of the vector's ScURA3 marker on minimal medium supplemented with 5-fluoroorotic acid. Replacement of the wild-type allele with the mutated S661A or S661E allele of KlCat8p was verified among 5-fluoroorotic acid-resistant clones by sequencing of PCR-amplified chromosomal DNA and Southern blot analysis.
All yeasts were grown in minimal medium (0.67% Difco yeast nitrogen base without amino acids supplemented with the required amino acids and bases) containing the appropriate carbon source.
Plasmids.
KlCAT8 was amplified from K. lactis genomic DNA with primers uCAT8 (5′-TGCTCTAGAATGGTCGAGAAGAAAGAT-3′) and dCAT8 (5′-ATAGTTTAGCGGCCGCTCAATTTCCATTTTGCCAGCG-3′), adding XbaI and NotI restriction sites to the 5′ and 3′ ends of the ORF, respectively. The PCR fragment was cloned in XbaI/NotI sites of pRS306, pJG45 (15), and pEG202 (15) to give pRS306CAT8, pJG45CAT8, and pEG202CAT8, respectively. pEG202CAT8Δ3204-4335 was obtained by BamHI/NotI restriction of pEG202CAT8, Klenow polymerase fill in, and self-ligation. pGBT9KlCAT8 was constructed by inserting an XbaI/SalI fragment of pEG202CAT8 and an EcoRI/XbaI linker (5′-GAATTCCCGGGGATCCGTCGAC-3′) into the multiple cloning site of pGBT9 (Clontech). pEG202CAT8Δ48-3405 and pGBT9KlCAT8Δ48-3405 were obtained by NcoI restriction of pEG202CAT8 and pGBT9KlCAT8, respectively, and religation.
A fragment of ScCAT8 was amplified from S. cerevisiae genomic DNA with primers prScECO (5′-CCGGAATTCATGGCAAATAA-3′) and prScBAM (5′-CCGGGATCCAAAAGATATAG-3′), respectively, adding EcoRI and BamHI restriction sites to the 5′ and 3′ ends of the fragment. The PCR fragment was cloned into the multiple cloning site of pGBT9 to give pGBT9SCPME. A PvuII/PmeI fragment of yEPCAT8 (containing ScCAT8 received from K. D. Entian) was introduced into pGBT9SCPME BamHI filled in with Klenow polymerase/PmeI to give pGBT9ScCAT8, which contains a fully reconstituted ScCAT8 gene.
KlSNF1 was amplified from K. lactis genomic DNA with primers prSNF1 (5′-GGAATTCATGTCGCACGACCCAAAT-3′) and prSNF4 (5′-CCGCTCGAGTCAACTTCCTTGGCTATT-3′), adding EcoRI and XhoI restriction sites to the 5′ and 3′ ends of the ORF, respectively. The PCR fragment was cloned into the EcoRI/SalI restriction sites pEG202 to give pEG202SNF1. The K64M allele of KlSNF1 was created by mutagenic PCR amplification. Two PCRs were done in parallel on K. lactis genomic DNA with, on the one hand, primers prSNF1 and prSNF3 (5′-GATGATCATGAGTGCCAC-3′), introducing the Met64 codon and a BspHI restriction site, and, on the other hand, primers prSNF4 and prSNF2, the latter being the antisense of prSNF3. The fragments amplified in these two PCRs were pooled and submitted to PCR amplification with prSNF1 and prSNF4. The resulting amplified DNA fragment, harboring the Met64 codon and a BspHI restriction site and flanked by EcoRI and XhoI restriction sites, was cloned into the EcoRI/SalI restriction sites of pEG202 to give pEG202SNF1K64M.
The mutant alleles of KlCAT8 and ScCAT8 were created by the same strategy, with primers listed below, and replaced the corresponding wild-type allele in pRS306CAT8, pGBT9KlCAT8, pGBT9ScCAT8, pGID1 (13), and yEPCAT8. KlCAT8S661A was created with primers pCKD (5′-CAGGTGTCCTGCAGCCGTA-3′) and prCKU (5′-CAGGTGTCCTGCAGCCGTA-3′). KlCAT8S661E was created with primers pr40661ED (5′-ACCACCTAGTACAGCTTCAGGACATCGATGTAACCTTAGT-3′) and pr40661EU (5′-ACTAAGGTTACATCGATGTCCTGAAGCTGTACTAGGTGGT-3′). KlCAT8S871A was created with primers pr40871AU (5′-ATCGGTTGACAGATCTCTGGCATCCTATGTCCTATTACAG-3′) and pr40871AD (5′-CTGTAATAGGACATAGGATGCCAGAGATCTGTCAACCGAT-3′). KlCAT8S871E was created with primers prSSUE (5′-CTCTCGAGTCATATGTCCTA-3′) and prSSDE (5′-TAGGACATATGACTCGAGAG-3′). ScCAT8S562A was created with primers prSC562AU (5′-AGATGTCCTGCAGCCGTA-3′) and prSC562AD (5′-TACGGCTGCAGGACATCT-3′). ScCAT8S562E was created with primers pr562EU (5′-AGACTTCATCGATGTCCTGAGGCCGTATTAAGTGTACACT-3′) and pr562ED (5′-AGTGTACACTTAATACGGCCTCAGGACATCGATGAAGTCT-3′). ScCAT8S803A was created with primers pr803UA (5′-AGGACGTCAGCCTCATATAT-3′) and pr803DA (5′-ATATATGAGGCTGACGTCCT-3′). ScCAT8S802-4A was created with primers pr803UA3 (5′-AGGACGGCAGCCGCATATAT-3′) and pr803DA3 (5′-ATATATGCGGCTGCCGTCCT-3′).
pAHCCAT8 was obtained by inserting an XbaI/NotI fragment of pRS306CAT8 into pAHC (received from T. P. Mäkelä) opened by XbaI/NotI. The promoter of KlCAT8 was amplified from K. lactis genomic DNA with primers pPCAT8up (5′-AACGAGCTCAAATACATAATACTTAGA-3′) and pPCAT8down (5′-AATCTCGAGGTATCTATCTGCTTCAAA-3′), adding SacI and NotI restriction sites to the 5′ and 3′ ends of the promoter. The SacI/NotI PCR fragment was ligated to a SacI/NotI fragment of pAHCCAT8 to give pAHCpCAT8. Hemagglutinin (HA)-pGID1 was obtained by ligating a BamHI/SacI fragment of pAHCpCAT8 filled in with Klenow polymerase to a BamHI/SalI fragment of pGID1 filled in with Klenow.
General DNA and RNA techniques.
Standard DNA manipulations were performed as described by Ausubel et al. (1). PCR amplification reactions were done with 2 U of DNA polymerase from Pyrococcus furiosus (Promega), 50 pmol of each primer, and 1 μg of genomic DNA with the following cycling procedure: 3 min at 95°C; 35 cycles of 1.5 min at 95°C, 1.5 min at 50°C, and 3 min at 72°C; and 10 min at 72°C. Every insert created by PCR was sequenced by automated sequencing (ABI). The procedure used for Northern blot analysis has been described previously. DNA probes for KlACT1, (a 1.1-kb EcoRI fragment of pGICL1) (12), KlICL1 (a 1.2-kb EcoRI fragment received from J. J. Heinisch), and KlCAT8 (a 4.3-kb XbaI-NotI fragment of pRS306CAT8) were 32P labeled with the Random Prime labeling kit (Invitrogen).
β-Galactosidase activity measurements.
S. cerevisiae cells were pregrown in minimal medium containing 2% glucose, diluted to an optical density at 600 nm (OD600) of 0.1 in 15 ml of minimal medium containing the appropriate sugar, and grown until an OD600 of 0.4 was reached. β-Galactosidase activity (Miller units) was determined on permeabilized cells as described by Miller (24).
Immunoblot analysis of HAKlCat8p.
K. lactis cells were pregrown in minimal medium containing 2% glucose to an OD600 of 0.6 and then shifted for 2 h to 2% glucose or 3% ethanol. Cells were collected by rapid filtration on 0.45-μm-pore-size filters (Millipore HLVP04700) and immediately frozen in liquid nitrogen. All of the following steps were performed at 4°C. Cell cakes were transferred to a 14-ml tube containing 1.5 ml of glass beads (Sigma G8772; 425 to 600 μm; acid washed) and 1 ml of 20 mM HEPES (pH 7.5), 0.5 mM EDTA, 100 mM NaCl, 20% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Complete Mini; 1 tablet for 10 ml; Roche). The mixture was vortexed five times for 1 min each time with at least 1 min on ice between each mixings. Beads were removed after centrifugation for 5 min at 5,000 × g, and supernatants were transferred to a 1.5-ml tube. Cell debris were removed by 1 h of ultracentrifugation at 40,000 rpm (70 iTi Beckman rotor). Supernatants were stored at −80°C. Ten micrograms of protein was loaded and separated by sodium dodecyl sulfate-6% polyacrylamide gel electrophoresis. Immunoblot detection was carried out with monoclonal anti-HA (Babco). λ-Phosphatase treatment (200 U/lane) was carried out for 2 h at 37°C. As a control experiment, an inhibitor of λ-phosphatase was added (1 mM Na3VO4/lane).
RESULTS
KlCat8p has transactivation capacity regulated by carbon source.
KlCat8p, a member of the family of Zn(II)2Cys6 transcriptional activators, is able to substitute for ScCat8p in S. cerevisiae and is required in K. lactis for derepression of the isocitrate lyase (KlICL1) and malate synthase (KlMLS1) genes (13, 20) and the tight carbon source regulation of KlACS1, KlACS2, and JEN1 (22). In order to investigate whether KlCat8p itself has transactivation potential, we fused the complete KlCAT8 ORF to the DNA binding domain of LexA and assayed the influence of the fusion gene on expression of a LexA-dependent reporter gene, lacZ, encoding β-galactosidase in the established S. cerevisiae strain BOY (15). As shown in Table 1, LexA-KlCat8p functions as a transcription activator, the transactivation function being moderately regulated by the carbon source: in ethanol-grown cells, β-galactosidase activity was about fourfold higher than in those grow in glucose or galactose, and in those grown in glycerol, β-galactosidase activity was slightly elevated over the level in those grown in glucose. When KlCAT8 was fused to the ScGal4p DNA binding domain with a UASGAL-lacZ reporter gene in strain MAV103, regulation was more pronounced (Table 1): induction was weak in low-glucose medium (2.3-fold) and galactose-raffinose medium (16-fold), moderate in glycerol medium (43-fold), and strong in ethanol medium (600-fold). The differences probably reside in the strength of the promoter governing expression of the KlCat8 fusion proteins: LexA-KlCAT8 is expressed from a wild-type ScADH1 promoter, whereas low expression of Gal4p-KlCat8p is ensured through a truncated and constitutive ScADH1 promoter. The results indicate that KlCat8p is controlled by the carbon source at a posttranscriptional level.
TABLE 1.
Carbon source regulation of transactivation properties of different alleles of KlCat8p fusion proteinsa
| Allele | 2% glucose | 0.05% glucose | 2% galactose-raffinose 1% | 3% glycerol | 3% ethanol |
|---|---|---|---|---|---|
| LexA | 0.20 (0.02) | 0.16 (0.0) | 0.23 (0.07) | 0.12 (0.02) | 0.10 (0.0) |
| LexA KlCat8 | 32 (0.77) | 32 (0.65) | 27 (1.0) | 49 (7.8) | 120 (2.9) |
| LexA KlCat8Δ3204-4335 | 0.28 (0.07) | 0.23 (0.02) | 0.40 (0.1) | 0.05 (0.05) | 0.17 (0.03) |
| LexA KlCat8Δ48-3405 | 170 (26) | 660 (85) | 430 (18) | 220 (20) | 510 (91) |
| Gal4 | 0.03 (0.0) | 0.06 (0.01) | 0.08 (0.01) | 0.0 (0.0) | 0.0 (0.0) |
| Gal4 KlCat8 | 0.06 (0.03) | 0.14 (0.06) | 0.95 (0.4) | 2.6 (0.8) | 36 (7.9) |
| Gal4 KlCat8Δ48-3405 | 3.1 (0.26) | 65 (1.7) | 68 (4.5) | 41 (2.0) | 56 (8.4) |
Strain BOY was transformed with plasmids expressing different alleles of LexA-KlCat8p fusions from the ScADH1 promoter, and strain MAV103 was transformed with plasmids expressing different alleles of Gal4-KlCat8p fusions from a truncated ScADH1 promoter. Transformants were grown on selective culture medium supplemented with the indicated carbon source and assayed for β-galactosidase activity as described in Materials and Methods. β-Galactosidase activities (Miller units) are means of three independent measurements, with standard deviations in parentheses.
The C terminus of KlCat8p contains a transcription activation domain.
When the LexA-KlCat8p fusion protein was truncated at the C terminus by 377 amino acids (LexA-KlCat8Δ3204-4335; Fig. 1), transactivation of the lacZ reporter gene was eliminated (Table 1). Consistently, the LexA-KlCAT8Δ3204-4335 construct was unable to complement a Sccat8 mutant for growth on nonfermentable carbon sources (Fig. 2A), suggesting that the C terminus of KlCat8p contains a transcriptional activation domain like its S. cerevisiae homolog (29). To directly assay the role of the KlCat8p C terminus, we created an internal 1,119-amino-acid deletion of LexA-KlCat8p, fusing in frame the C-terminal 312 amino acids of the protein to the N-terminal 17 amino acids of KlCat8p (Fig. 1). As expected, this construct, which had lost the majority of the KlCAT8 coding sequence, including the region encoding the zinc cluster DNA binding region, could not complement the nonfermentable growth defect of an Sccat8 mutant (Fig. 2A). However, when fused to LexA, the truncated fragment (KlCat8Δ48-3405p) could activate transcription of the LexA-controlled lacZ reporter gene (Table 1). Depending on the carbon source, activation in this case was even 4- to 20-fold higher than with wild-type LexA-KlCat8p although the strong ethanol induction was not observed. Again, similar results were observed when KlCat8Δ48-3405p was fused to the binding domain of ScGal4p: activation was up to 500-fold higher than with wild-type Gal4-KlCat8p, depending on the carbon source. Importantly, the transactivation function was down-regulated by glucose (Table 1). We concluded that the C terminus of KlCat8p functions as an independent transactivation domain when targeted to DNA and that some regions involved in the transactivator's response to glucose are retained when most of the central region is removed.
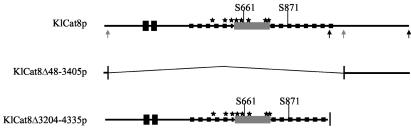
FIG. 1.
Restriction and domain map of KlCat8p and its deletion alleles. The two black boxes represent the zinc cluster and the downstream coiled-coil domains, highly conserved in more than 100 zinc cluster proteins. The grey line represents the middle homology region found in more than 50 zinc cluster proteins. The small black squares cover regions that are highly conserved only among the three known yeast Cat8p homologues. The stars indicate the positions of the eight motifs conserved in zinc cluster proteins as described by Poch (25). Serines 661 and 871 are positioned on the KlCat8p sequence. Grey arrows indicate the sites used in the internal deletion Δ48-3405, and black arrows point to the C-terminal deletion Δ3204-4335.
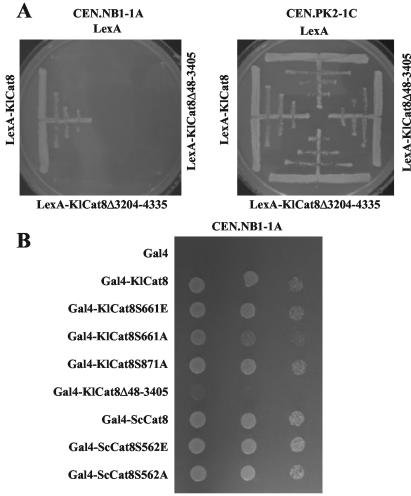
FIG. 2.
Heterospecific complementation of an Sccat8 mutant with deletion and serine mutant alleles of KlCat8p. (A) Strains CEN.NB1-1A (Sccat8) and CEN.PK2-1C (ScCAT8) were transformed with pEG202-based plasmids expressing LexAp, LexA-KlCat8, LexA-KlCat8Δ3204-4335p, and LexA-KlCat8Δ48-3405p and streaked on selective culture medium supplemented with glycerol. (B) Strain CEN.NB1-1A was transformed with pGBT9-based plasmids expressing Gal4p, Gal4-KlCat8p, Gal4-KlCat8Δ48-3405p, Gal4-KlCat8S661Ap, Gal4-KlCat8S871Ap, Gal4-KlCat8S661Ep, Gal4-ScCat8p, Gal4-ScCat8S562Ap, and Gal4-ScCat8S562Ep. Transformants were dropped in three successive 10-fold dilutions on selective culture medium supplemented with glycerol.
We noticed that, when fused to the binding domain of ScGal4p, KlCat8Δ48-3405p was able to faintly complement an sccat8 mutation (Fig. 2B, line 6), despite the deletion of the zinc cluster domain of KlCAT8. It is possible that the zinc cluster domain of ScGal4p present in Gal4-KlCat8Δ48-3405p confers weak CSRE binding activity. Interestingly, it has been reported previously that overexpression of CAT8-INO2TAD from the MET25 promoter could suppress the inability of an snf1Δ strain to grow on galactose (27).
Expression analysis of an in vivo target of KlCat8p: KlICL1.
Activity of the glyoxylate cycle enzyme isocitrate lyase was previously shown to be lost in a klcat8 mutant (13). We analyzed the effect of KlCAT8 disruption on transcription of the isocitrate lyase KlICL1 gene under various growth conditions: total RNA extracted from cells pregrown on high-glucose medium and then shifted for 4 h to high-glucose, galactose-raffinose, glycerol, or ethanol medium was analyzed by Northern blotting. The results, displayed in Fig. 3 (wt and Δ columns, KlICL1 line), show that in wild-type strain JA6, KlICL1 gene expression is undetectable in high-glucose and galactose-raffinose media whereas low-to-moderate expression is observed in glycerol- and ethanol-grown cells, respectively. This transcription was abolished in the Klcat8 mutant strain, confirming that KlICL1 transcription depends on KlCat8p. It should be pointed out that actin mRNAs were undetectable in ethanol-grown Klcat8 cells, probably owing to the absence of growth of the mutant strain on this carbon source.
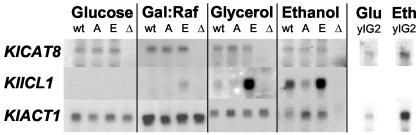
FIG. 3.
Northern blot analysis of KlICL1 and KlCAT8 expression in K. lactis strains harboring various alleles of KlCAT8. Total RNA was extracted from strains JA6 (wt), yGC661A (A), yGC661E (E), yIG8 (Δ), and yIG2 (klmig1) grown in minimal medium containing 2% glucose, 3% galactose-raffinose, or 3% glycerol or ethanol as the carbon source. Each RNA sample (10 μg) was electrophoresed on an agarose gel in the presence of formaldehyde, blotted onto a nylon membrane, and hybridized to KlICL1, KlCAT8, and KlACT1 probes.
Evidence of physical interaction between KlSnf1p and KlCat8p.
The KlCAT8 gene had been isolated as a multicopy suppressor of a Klsnf1 mutant (13). On the basis of findings on the genetic interaction between the S. cerevisiae homologues (17, 28, 29), we hypothesized that KlSnf1p is required for activation of KlCat8p. To address the question of whether KlCat8p is a direct target of the KlSnf1p protein kinase, we tested for the interaction between KlSnf1p and KlCat8p with the two-hybrid system. A LexA-KlSnf1 fusion protein under the control of the constitutive ScADH1 promoter was coexpressed with a VP16-KlCat8 fusion protein conditionally expressed from the galactose-inducible ScGAL1 promoter in S. cerevisiae. The VP16-KlCAT8 and LexA-KlSNF1 fusion genes were shown to be functional by complementation of the respective S. cerevisiae mutants for growth on nonfermentable carbon sources (ethanol and glycerol; data not shown). The two-hybrid interaction between KlCat8p and LexA-KlSnf1p was analyzed in strain BOY containing the LexA-dependent lacZ reporter gene by measuring β-galactosidase activity. In galactose-raffinose-grown cells, expression of LexA-KlSnf1p alone gave a threefold increase in activity over the background level of cells containing none of the fusion proteins or expressing VP16-KlCat8p alone (Table 2). Coexpression of both fusion proteins gave a further fourfold increase over the LexA-KlSnf1p-mediated level. On 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-containing plates (data not shown), these cells stained dark blue whereas the control strains were white (vector control or VP16-KlCat8p control) or light blue (LexA-KlSnf1p control), corresponding to the β-galactosidase activity measurements.
TABLE 2.
Two-hybrid interaction between KlCat8p and either LexA-KlSnf1p or LexA-KlSnf1K64Mpa
| Bait fusion | Prey fusion | β-Galactosidase activity |
|---|---|---|
| LexA | VP16 | 0.3 (0.1) |
| LexA | VP16 KlCat8 | 0.3 (0.1) |
| LexA KlSnf1 | VP16 | 1.0 (0.2) |
| LexA KlSnf1 | VP16 KlCat8 | 3.2 (1.3) |
| LexA KlSnf1K64M | VP16 | 1.8 (0) |
| LexA KlSnf1K64M | VP16 KlCat8 | 6.0 (0.8) |
Strain BOY was transformed with pEG202-based plasmids expressing LexAp, LexA-KlSnf1p, or LexA-KlSnf1K64Mp from the ScADH1 promoter and pJG45-based plasmids expressing VP16p or VP16p-KlCat8p from the ScGAL1 promoter. Double transformants were grown in selective growth medium and assayed for β-galactosidase activity as described in Materials and Methods. β-Galactosidase activities (Miller units) are means of three independent measurements, with standard deviations in parentheses.
Trying to obtain a stronger interaction between the two proteins, we used an approach that had proven successful in analyzing protein binding to the Snf1p kinase from S. cerevisiae. In that case, mutation of lysine 84 to arginine in the highly conserved catalytic core resulted in loss of catalytic activity and in tighter interaction with putative target proteins like ScMig1p (32). When we exchanged the corresponding position in KlSnf1p (K64) for methionine, the mutant protein was unable to complement an Scsnf1 mutant strain (data not shown), indicating that the K. lactis mutant protein had also lost its enzymatic activity. In our two-hybrid test, transformants expressing LexA-KlSnf1K64Mp together with VP16-KlCat8p indeed stained darker blue than those containing wild-type LexA-KlSnf1p (data not shown) and showed a further twofold increase in β-galactosidase activity (Table 2). A slight increase in β-galactosidase activity was also observed with LexA-KlSnf1K64Mp alone. We take these data as evidence of a physical interaction between KlSnf1p and KlCat8p.
Mutation of two putative Snf1p-phosphorylated serines in the KlCat8p sequence.
On the basis of the interaction data shown above and the finding that Cat8p is a phosphoprotein in S. cerevisiae (29), we searched for potential Snf1p phosphorylation sites in KlCat8p. A consensus substrate recognition sequence was determined in vitro for the mammalian Snf1p homologue, AMP-activated protein kinase, with synthetic peptides (7). This consensus site, ΦXRXXSXXΦ, containing an arginine at position −3 and hydrophobic residues at positions −5 and +4 relative to the phosphorylated serine, was found at two sites in the KlCat8p sequence (S661 and S871; Fig. 1 and 4). Serine 661 is located within the so-called “middle homology region” (Fig. 1) described by Poch (25), just outside the C terminus of motif V within a stretch of 13 amino acids (Fig. 4) highly conserved between the known yeast Cat8p homologues. Only in the Candida albicans Cat8p sequence is the potential phosphorylated serine replaced by an alanine. Serine 871 is located downstream of the middle homology region (Fig. 1), in a stretch of 16 amino acids that is moderately conserved in the Saccharomyces and Kluyveromyces Cat8 proteins (Fig. 4). We converted these two phosphorylatable serine residues to alanine and glutamate by site-directed mutagenesis. Alanine replacement mimics a permanent unphosphorylated status, whereas the charge introduced by glutamate substitution is presumed to imitate permanent phosphorylation of the replaced serine.
FIG. 4.
Potential Snf1p recognition sites in homologues of Cat8p. The consensus Snf1p recognition sequence is shown: Φ, hydrophobic residue (L > F = I = M > V in position −5 and L > I > F > M > V in position +4). Potential Snf1p recognition sites in Cat8p homologues are aligned and numbered when possible according to the position of the phosphorylatable serine. The alanine and glutamic acid substitutions resulting from site-directed mutagenesis are underlined.
The complete ORF of all four serine mutant alleles of KlCAT8 was fused to the DNA binding domain of ScGal4p to analyze the effects of serine replacement on KlCat8p transcriptional activity with the UASGAL-lacZ reporter gene. All four serine mutants of Gal4-KlCat8p, expressed from the truncated ScADH1 promoter on a multicopy plasmid, could complement the nonfermentative growth defects of an Sccat8 mutant (Fig. 2B and data not shown). β-Galactosidase activities in transformants of UASGAL-lacZ mutant strain MAV103 expressing the Gal4-KlCat8 protein variants revealed a strong influence of the serine 661 mutations on the transactivation function. Whereas the serine-to-alanine mutation (Gal4-KlCat8S661Ap) reduced activities under all growth conditions, transactivation by Gal4-KlCat8S661Ep was elevated over the Gal4-KlCat8p wild-type level (Table 3). These data indicate that serine 661 has a role in regulating the KlCat8p activation function and strongly suggest that this site is targeted by regulated phosphorylation. Obviously, Snf1p is a strong candidate for the responsible kinase. Remarkably, activity with the Gal4-KlCat8S661Ep variant was 9- to 15-fold higher than with Gal4-KlCat8p on all of the carbon sources tested except ethanol, indicating that these growth conditions lead to maximal activation of KlCat8p. The mutant fusion protein reached the same level on glycerol but not on the other carbon sources tested, indicating that S661E mimics the activated form but additional forms of regulation exist. In particular, activity levels are much lower in high-glucose medium but are still higher for the S661E mutant compared to the wild-type level.
TABLE 3.
Carbon source regulation of transactivation properties of serine mutant alleles of Kl-Cat8p and ScCat8p fusion proteinsa
| Allele | 2% glucose | 0.05% glucose | 2% galactose-1% raffinose | 3% glycerol | 3% ethanol |
|---|---|---|---|---|---|
| Gal4 | 0.03 (0.0) | 0.06 (0.01) | 0.08 (0.01) | 0.0 (0.0) | 0.0 (0.0) |
| Gal4 KlCat8 | 0.06 (0.03) | 0.14 (0.06) | 0.95 (0.4) | 2.6 (0.8) | 36 (7.9) |
| Gal4 KlCat8S661A | 0.01 (0.0) | 0.0 (0.0) | 0.03 (0.05) | 0.66 (0.4) | 13 (1.1) |
| Gal4 KlCat8S661E | 0.54 (0.16) | 10 (0.67) | 14 (4.1) | 23 (0.6) | 25 (0.44) |
| Gal4 KlCat8S871A | 0.08 (0.03) | 0.29 (0.14) | 1.2 (0.03) | 2.5 (0.25) | 49.1 (2.8) |
| Gal4 KlCat8S871E | 0.07 (0.02) | 0.38 (0.11) | 2.2 (0.15) | 5.0 (2.5) | 36 (7.9) |
| Gal4 ScCat8 | 0.25 (0.04) | 0.25 (0.02) | 1.4 (0.07) | 10.5 (0.4) | 43.14 (2.9) |
| Gal4 ScCat8S562A | 0.22 (0.2) | 0.2 (0.02) | 0.48 (0.12) | 3.92 (1.0) | 26.8 (0.82) |
| Gal4 ScCat8S562E | 1.02 (0.04) | 9.6 (0.7) | 6.8 (0.54) | 26 (2.2) | 44 (9.7) |
| Gal4 ScCat8S802-4A | 0.21 (0.24) | 0.28 (0.04) | NDb | ND | 51.2 (5.6) |
Strain MAV103 was transformed with pGBT9-based plasmids expressing different alleles of Gal4-KlCat8p and Gal4-ScCat8p fusions from a truncated ScADH1 promoter. Transformants were grown on selective culture medium supplemented with the indicated carbon sources and assayed for β-galactosidase activity as described in Materials and Methods. β-Galactosidase activities (Miller units) are means of three independent measurements, with standard deviations in parentheses.
ND, not determined.
The mutations at serine 871 had only a marginal effect, if any. Transactivation by Gal4-KlCat8S871E was two- to threefold higher than that by Gal4-KlCat8p on low-glucose, galactose-raffinose, and glycerol media, whereas S871A had no influence (Table 3).
Wishing to investigate the evolutionary significance of this result, we mutated the corresponding serine of the ScCat8 protein to alanine and glutamate and fused the mutant alleles to the DNA binding domain of ScGal4p, giving Gal4-ScCat8S562Ap and Gal4-ScCat8S562Ep, respectively (Fig. 4). These two ScCat8 mutant proteins were shown to be functional in a complementation test (Fig. 2B) and behaved similarly to their K. lactis counterparts, i.e., reduction of transactivation by the alanine mutant on all of the carbon sources tested (1.1- to 2.9-fold, depending on the carbon source; Table 3) and augmentation of transactivation by the glutamate mutant on all of the carbon sources tested except ethanol (2.5- to 38-fold, depending on the carbon source; Table 3). Finally, suppression tests with ScCat8S562 mutants in an Scsnf1 deletion strain (Fig. 5) showed that glutamate mutation of the serine increased the suppression capacity of ScCAT8 whereas a serine-to-alanine mutation nearly abolished the suppression capacity of ScCAT8 when growth on ethanol is considered. Parallel observation of allele specificity has previously been made by Rahner and coworkers: in their study, wild-type ScCAT8 was not able to suppress an snf1Δ mutant strain whereas constitutively active ScCAT8-INO2TAD was (27).
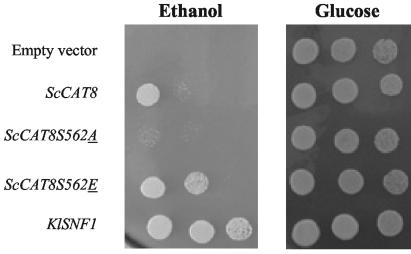
FIG. 5.
Suppression of the ethanol growth defect of an scsnf1Δ strain by overexpression of different alleles of ScCAT8. Y14311 (Δsnf1) was transformed with plasmids YEp351 (empty plasmid) and plasmids YEpCAT8, YEpCAT8S562A, and YEpCAT8S562E, respectively, expressing ScCat8p, ScCat8S562Ap, and ScCat8S562Ep. pGBT9SNF1, expressing KlSnf1p, was transformed into Y14311 as a positive control. Transformants were dropped at three different dilutions onto minimal medium containing 3% ethanol as a carbon source and glucose-rich medium as a positive growth control.
As with KlCat8S871A, mutation of the corresponding serine of the ScCat8 protein (S803) to alanine did not affect its functionality (data not shown) or its transactivation ability (Table 3). One possible explanation for this result may be the presence of multiple serines and threonines in the environment of KlCat8p S871 and ScCat8p S803 (Fig. 4). To rule out this possibility, we mutated three serines located in the vicinity of ScCat8p S803 to alanines (Fig. 4). The resulting mutant protein, ScCat8S802-4Ap, when fused to the DNA binding domain of ScGal4p, activated the transcription of a UASGAL-lacZ reporter gene like the wild-type control (Table 3).
The KlCAT8-S661 mutations affect the growth and KlCat8p-dependent gene expression of K. lactis.
In order to investigate the effects of serine 661 substitutions on in vivo KlCat8p function in K. lactis, we replaced the genomic KlCAT8 gene with the serine mutant alleles KlCAT8-S661A and KlCAT8-S661E.
As a control experiment, analysis of KlCAT8 expression was performed with wild-type and Δklcat8, klcat8-S661A, and KlCAT8-S661E mutant cells grown on various carbon sources. Figure 3 shows that expression of KlCAT8 remains constant and unaffected by the carbon source and by a MIG1 deletion, in contrast to the S. cerevisiae situation, in which ScCAT8 expression is glucose repressed by Mig1p (17). As expected, KlCAT8 mRNAs were undetected in the klcat8 disruptant strain. Finally, expression of the mutant alleles of KlCAT8 was unaffected by the serine-to-alanine and serine-to-glutamate mutations, at both the mRNA and protein levels (Fig. 3 and data not shown).
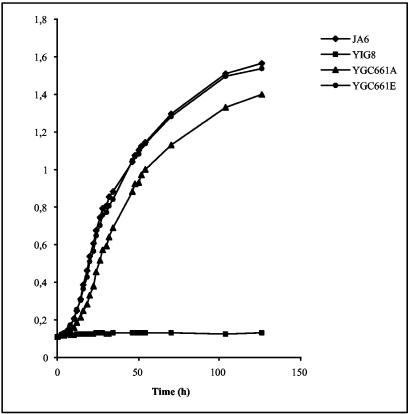
We characterized the growth phenotype of the klcat8-S661A mutant strain in liquid cultures containing various nonfermentable carbon sources (Fig. 6). Congenic wild-type and Δklcat8 mutant K. lactis strains were analyzed as positive and negative controls, respectively. We observed that the growth of a klcat8-S661A mutant strain was reduced compared to that of the wild type on ethanol-containing medium, although not to the same extent as that of the congenic klcat8 mutant strain. This indicates that KlCat8p phosphorylation at serine 661 is required for efficient utilization of this carbon source. As expected, mutation of serine 661 to glutamic acid did not affect growth under our culture conditions (Fig. 6).
FIG. 6.
Effect of a serine 661 mutation in KlCAT8 on growth in ethanol. Strain JA6 (wild type), yIG8 (Δklcat8), yGC661A (klcat8-S661A), and yGC661E (KlCAT8-S661E) cells were pregrown in minimal medium containing 2% glucose and diluted in minimal medium containing 3% ethanol to an OD600 of 0.1, and the OD600 was monitored for 130 h.
In order to ascertain the role of serine 661 phosphorylation of KlCat8p on its in vivo transactivation capacity, we performed a Northern blot analysis and compared the expression of KlICL1 in wild-type and Δklcat8, klcat8-S661A, and KlCAT8-S661E mutant K. lactis strains cultivated in the presence of different carbon sources (Fig. 3). A significant reduction of KlICL1 expression in ethanol-grown klcat8-S661A mutant cells compared to the that in the wild type was observed, although to a lesser extent than in klcat8 mutant cells. On the other hand, an increase in KlICL1 mRNAs was observed with the KlCAT8-S661E mutant grown on ethanol, glycerol, or galactose-raffinose medium, compared to the corresponding wild-type levels. Altogether, these Northern results indicate that phosphorylation of serine 661 is required for full in vivo transcriptional activity of KlCat8p. Consistent results have recently been obtained with cDNA of an S. cerevisiae CAT8-S562E mutant strain grown on galactose and hybridized on DNA microarrays (G. Charbon et al., unpublished data).
KlCAT8 is phosphorylated in a KlSnf1p-dependent fashion.
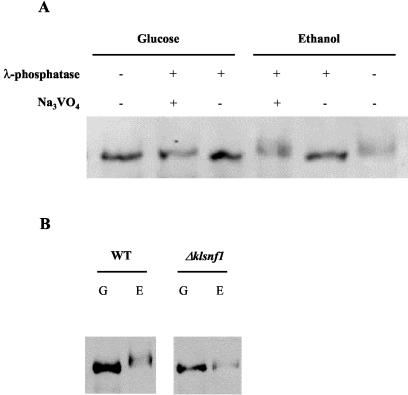
Western blot analysis revealed a multiple-band migration of HA epitope-tagged KlCat8p from crude extracts of ethanol-grown cells (Fig. 7A). This pattern of migration is not observed when cells are grown in 2% glucose. We could establish that this apparent molecular weight shift is due to phosphorylation, since λ-phosphatase treatment of protein extracts turned the multiple-band pattern to a unique band (Fig. 7A). Our results propose KlSnf1p as a potent KlCat8p-kinase candidate. We therefore questioned the requirement for KlSnf1p in the phosphorylation process of KlCat8p by analyzing the migration profile of HA-KlCat8p in a Δklsnf1 background shifted from 2% glucose to 3% ethanol for 2 h. The results presented in Fig. 7B show that almost all, if not all, of the phosphorylation of HA-KlCat8p is absent in the Δklsnf1 mutant, as it migrates at the same level under both glucose and ethanol conditions. This result further strengthens the hypothesis that KlSnf1p is a kinase for KlCat8p. We also looked at the migration profile of HA-KlCat8S661A and observed no clear modification of the phosphorylation pattern in ethanol-grown cells (data not shown), suggesting that residues other than serine 661 are responsible for the mobility shift of KlCat8p.
FIG. 7.
Western blot analysis of KlCat8p posttranslational modification. Cells were pregrown in 2% glucose and shifted for 2 h to 2% glucose (G) or 3% ethanol (E). (A) Wild-type (WT) strain JA6 was transformed with a multicopy plasmid (HApGID1) expressing HA-KlCat8p. (B) Wild-type strain JA6 and Δklsnf1 mutant strain JSD1R4 were transformed with a multicopy plasmid (HApGID1) expressing HA-KlCat8p.
DISCUSSION
Regulation of Cat8 activity by Snf1p-dependent phosphorylation at serine 661/562.
We have identified serine 661 of KlCat8p and corresponding serine 562 of ScCat8p as sites crucial for regulation of Cat8p activity in response to the carbon source. Indeed, mutations to alanine reduce the Cat8 transactivation function whereas mutations to glutamate enforce activity, suggesting that these mutations mimic the constitutive dephosphorylated and phosphorylated states of this site, respectively.
Together with previous observations, our data strongly suggest that, in the wild-type Cat8 protein, S562/S661 is the target of regulated phosphorylation by the Snf1 kinase. First, it is well established that Cat8p activity is controlled by Snf1p: multicopy CAT8 can suppress the growth deficiency on nonfermentable carbon sources of an snf1 mutant of S. cerevisiae (17) or K. lactis (13), and the phosphorylation state of ScCat8p (28, 29) and KlCat8p (Fig. 7) is affected by mutations in SNF1. Second, physical interaction between Snf1p and Cat8p is indicated by the two-hybrid interaction between lexA-KlSnf1p and VP16-KlCat8p (Table 2). Third, these conserved serines are part of a consensus sequence for phosphorylation by Snf1p (and related kinases), which is itself located in the middle homology region, which is known as a regulatory domain of C6 zinc cluster transcription factors (30). Fourth, we have demonstrated a genetic interaction between Sccat8S562E and SNF1 (Fig. 5). Taken together, these data strongly support the view that Cat8p is a direct target of the Snf1 kinase, although the possibility that an intermediate kinase is involved cannot completely be ruled out. To address this question, we tried to reconstitute Snf1 phosphorylation of KlCat8p in vitro. We observed that a Gst-KlCat8 fusion protein produced in E. coli can be phosphorylated with a yeast cell lysate and radiolabeled [32γ]ATP (data not shown). However, no difference could be shown between the mutant and the wild-type Cat8 protein and between an snf1 mutant and a wild-type lysate. This result was not entirely unexpected since the specificity of the phosphotransfer reaction may not be retained in vitro and additional phosphorylation sites are likely to be present in Cat8p.
The serine-to-glutamate mutants, both in the context of the Gal4 fusion protein (Table 3) and in the wild-type context (Fig. 3), indicate that a primary role of residue 661/562 is to allow for inactivation of Cat8p by dephosphorylation. In contrast, the serine-to-alanine mutants (Table 3 and Fig. 3) are still active when Cat8p reaches the highest activity (in ethanol-grown cells), indicating that phosphorylation is not essential to activate Cat8p under these conditions. Therefore, phosphorylation apparently overcomes some form of negative regulation of Cat8p.
Other forms of regulation of Cat8p activity.
Negative regulation of Gal4p-Cat8p is relieved in the Gal4-KlCat8Δ48-3405 deletion (Table 1). Thus, as in other proteins of the zinc cluster family, the middle homology region may have a negative influence on the activation domain (9, 30). We propose that this inhibitory effect is relieved by the Snf1p kinase in the cases of KlCat8p and ScCat8p.
Our results also suggest that serine 562/661 is not the only site responsible for the Snf1-dependent regulation of Cat8p because KlCat8S661Ap remains phosphorylated in an Snf1p-dependent manner (data not shown). S871 in KlCat8p is the only other site conforming to the ΦXRXXSXXXΦ consensus; however, mutation of this serine to alanine or glutamate did not influence the transactivation function. Phosphorylation at additional sites may thus be indirectly influenced by Snf1p or may not be part of a consensus site. The Gal4-KlCat8Δ48-3405 fusion protein had a high activation activity on all of the carbon sources tested except for a high glucose concentration. Thus, repression by glucose must function via a different mechanism than repression by a low glucose or galactose concentration.
Transcription of KlCat8p is constitutive.
In S. cerevisiae, in addition to its carbon source-dependent phosphorylation, Cat8p activity is also regulated at the transcriptional level: Mig1p represses CAT8 transcription under high-glucose conditions (17). In K. lactis, we show that CAT8 transcription is unregulated by the carbon source and by Mig1p (Fig. 3). This was first presumed from the absence of a potential Mig1p binding site in the KlCAT8 upstream region (13) and is consistent with the minor role played by Mig1p in K. lactis (12, 13).
Differences between K. lactis and S. cerevisiae in the regulation of carbon metabolism.
Cat8 is a key regulator of carbon flux in yeast. Transcriptome analysis in S. cerevisiae has shown that all of the key enzymes that replenish carbon for anabolism in response to carbon limitation are regulated by Cat8p at the transcriptional level (16, 39). In striking contrast to that in S. cerevisiae, gluconeogenesis in K. lactis does not require KlCat8p and growth on glycerol is not affected in a Klcat8 mutant (13). The data presented show that not only the lack of CSREs in the control region of key gluconeogenic genes is a major difference between K. lactis and S. cerevisiae, explaining the difference in Cat8p dependence, but there is also a difference in the activity status of Cat8p in glycerol-grown cultures. The activation function of KlCat8p on glycerol is much lower than that of ScCat8p (Table 3), and KlCat8p target genes like KlICL1 are expressed at a low level (Fig. 3). Since the dephosphorylated state of S661 is crucial for low activity, we conclude that KlSnf1p signaling to KlCat8p is downregulated during growth of K. lactis on glycerol, probably reflecting reduced carbon limitation compared to that of S. cerevisiae. The more efficient metabolism of glycerol by K. lactis is reflected in the high growth rates (comparable to those obtained with glucose or lactose) that can be achieved with glycerol as the sole carbon source and is likely to be responsible for reduced Snf1p signaling. This implies that reporter gene expression controlled by Gal4-Cat8p can be used to monitor the activity status of the Snf1-Cat8p signaling chain and thus reflect an important aspect of the nutritional state of the cell. It should be noted, however, that Cat8p action tends to relieve carbon limitation by mobilizing alternative resources such that starvation conditions and activation of Snf1 signaling are normally transient. Moreover, the Snf1 protein kinase has been shown to reside in different cellular compartments in S. cerevisiae (37). Signaling through Cat8p may therefore not reflect the Snf1p activity status in all of these compartments. Subcellular localization of Snf1p is influenced by the β subunit in the trimeric kinase complex, of which three variants exist in S. cerevisiae (37). In K. lactis, only a single β subunit, Fog1p, has been found (14). Fog1p is most closely related to Gal83p, raising the possibility that the nuclear function of Snf1p is one found in K. lactis.
KlMig1p has no detectable influence on KlCat8p regulation, confirming earlier findings about a minor role of Mig1p in K. lactis. The constitutive expression of the KlCAT8 gene indicates that regulation of this transcription factor occurs primarily at the level of activity. This correlates with the ability of K. lactis to quickly adapt to alternative carbon sources and the less pronounced glucose repression.
Acknowledgments
We thank K. D. Entian, M. Vidal, T. P. Mäkelä, and J. J. Heinisch for sharing materials used in this study. Monique Dewez is acknowledged for precious technical assistance.
This work was funded by DFG grant FOR466/1-1 to K.D.B. G. Charbon is supported by the Belgian Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Breunig, K. D., M. Bolotin-Fukuhara, M. M. Bianchi, D. Bourgarel, C. Falcone, I. Ferrero, L. Frontali, P. Goffrini, J. J. Krijger, C. Mazzoni, C. Milkowski, H. Y. Steensma, M. Wesolowski-Louvel, and A. M. Zeeman. 2000. Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzyme Microb. Technol. 26:771-780. [DOI] [PubMed] [Google Scholar]
- 3.Breunig, K. D., and P. Kuger. 1987. Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol. Cell. Biol. 7:4400-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson, M., B. C. Osmond, and D. Botstein. 1981. Mutants of yeast defective in sucrose utilization. Genetics 98:25-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspary, F., A. Hartig, and H.-J. Schüller. 1997. Constitutive and carbon source-responsive promoter elements are involved in the regulated expression of the Saccharomyces cerevisiae malate synthase gene MLS1. Mol. Gen. Genet. 255:619-627. [DOI] [PubMed] [Google Scholar]
- 6.Ciriacy, M. 1977. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol. Gen. Genet. 154:213-220. [DOI] [PubMed] [Google Scholar]
- 7.Dale, S., W. A. Wilson, A. M. Edelman, and D. G. Hardie. 1995. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 361:191-195. [DOI] [PubMed] [Google Scholar]
- 8.De Deken, R. H. 1966. The Crabtree effect: a regulatory system in yeast. J. Gen. Microbiol. 44:149-156. [DOI] [PubMed] [Google Scholar]
- 9.Delaveau, T., A. Delahodde, E. Carvajal, J. Subik, and C. Jacq. 1994. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol. Gen. Genet. 244:501-511. [DOI] [PubMed] [Google Scholar]
- 10.de Mesquita, J. F., O. Zaragoza, and J. M. Gancedo. 1998. Functional analysis of upstream activating elements in the promoter of the FBP1 gene from Saccharomyces cerevisiae. Curr. Genet. 33:406-411. [DOI] [PubMed] [Google Scholar]
- 11.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 12.Georis, I., J.-P. Cassart, K. D. Breunig, and J. Vandenhaute. 1999. Glucose repression of the Kluyveromyces lactis invertase gene KlINV1 does not require Mig1p. Mol. Gen. Genet. 261:862-870. [DOI] [PubMed] [Google Scholar]
- 13.Georis, I., J.-J. Krijger, K. D. Breunig, and J. Vandenhaute. 2000. Differences in regulation of yeast gluconeogenesis revealed by Cat8p-independent activation of PCK1 and FBP1 genes in Kluyveromyces lactis. Mol. Gen. Genet. 1-2:193-203. [DOI] [PubMed] [Google Scholar]
- 14.Goffrini, P., A. Ficarelli, C. Donnini, T. Lodi, P. P. Puglisi, and I. Ferrero. 1996. FOG1 and FOG2 genes, required for the transcriptional activation of glucose-repressible genes of Kluyveromyces lactis, are homologous to GAL83 and SNF1 of Saccharomyces cerevisiae. Curr. Genet. 29:316-326. [PubMed] [Google Scholar]
- 15.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 16.Haurie, V., M. Perrot, T. Mini, P. Jeno, F. Sagliocco, and H. Boucherie. 2001. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 276:76-85. [DOI] [PubMed] [Google Scholar]
- 17.Hedges, D., M. Proft, and K.-D. Entian. 1995. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15:1915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kratzer, S., A. Schöler, and H.-J. Schüller. 1995. Regulatory factors of carbon source-dependent transcription of gluconeogenic genes ICL1 and ACS1 in S. cerevisiae. Yeast S214. 11:
- 19.Kratzer, S., and H.-J. Schüller. 1995. Carbon source-dependent regulation of the acetyl-coenzyme A synthetase-encoding gene ACS1 from Saccharomyces cerevisiae. Gene 161:75-79. [DOI] [PubMed] [Google Scholar]
- 20.Krijger, J.-J. 2002. Carbon source-responsive elements and gene regulation by CAT8 and SIP4 in the yeast Kluyveromyces lactis. PhD thesis. Martin-Luther-Universität, Halle-Wittenberg, Germany.
- 21.Lesage, P., X. Yang, and M. Carlson. 1996. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol. Cell. Biol. 16:1921-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodi, T., M. Saliola, C. Donnini, and P. Goffrini. 2001. Three target genes for the transcriptional activator Cat8p of Kluyveromyces lactis: acetyl coenzyme A synthetase genes KlACS1 and KlACS2 and lactate permease gene KlJEN1. J. Bacteriol. 183:5257-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercado, J. J., and J. M. Gancedo. 1992. Regulatory regions in the yeast FBP1 and PCK1 genes. FEBS Lett. 311:110-114. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Poch, O. 1997. Conservation of a putative inhibitory domain in the GAL4 family members. Gene 184:229-235. [DOI] [PubMed] [Google Scholar]
- 26.Proft, M., D. Grzesitza, and K.-D. Entian. 1995. Identification and characterization of regulatory elements in the phosphoenolpyruvate carboxykinase gene PCK1 of Saccharomyces cerevisiae. Mol. Gen. Genet. 246:367-373. [DOI] [PubMed] [Google Scholar]
- 27.Rahner, A., M. Hiesinger, and H.-J. Schüller. 1999. Deregulation of gluconeogenic structural genes by variants of the transcriptional activator Cat8p in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 34:146-156. [DOI] [PubMed] [Google Scholar]
- 28.Rahner, A., A. Schöler, E. Martens, B. Goolwitzer, and H.-J. Schüller. 1996. Dual influence of the yeast Cat1p (Snf1p) protein kinase on carbon source-dependent transcriptional activation of gluconeogenic genes by the regulatory gene CAT8. Nucleic Acids Res. 24:2331-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Randez-Gil, F., N. Bojunga, M. Proft, and K.-D. Entian. 1997. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol. Cell. Biol. 17:2502-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schjerling, P., and S. Holmberg. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24:4599-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schöler, A., and H.-J. Schüller. 1994. A carbon source-responsive promoter element necessary for activation of the isocitrate lyase gene ICL1 is common to genes of the gluconeogenic pathway in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3613-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vidal, M., R. K. Brachmann, A. Fattaey, E. Harlow, and J. D. Boeke. 1996. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. USA 93:10315-10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincent, O., and M. Carlson. 1998. Sip4, a snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 17:7002-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent, O., and J. M. Gancedo. 1995. Analysis of positive elements sensitive to glucose in the promoter of the FBP1 gene from yeast. J. Biol. Chem. 270:12832-12838. [DOI] [PubMed] [Google Scholar]
- 36.Vincent, O., S. Kuchin, S. Hong, R. Townley, V. Vyas, and M. Carlson. 2001. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:5790-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wésolowski-Louvel, M., K. D. Breunig, and H. Fukuhara. 1996. Kluyveromyces lactis, p. 139-201. In K. Wolf (ed.), Non-conventional yeasts in biotechnology. Springer-Verlag, Heidelberg, Germany.
- 39.Young, E. T., K. M. Dombek, C. Tachibana, and T. Ideker. 2003.. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem., 278:26146-26158. [DOI] [PubMed]
- 40.Zimmermann, F. K., I. Kaufmann, H. Rasenberger, and P. Haubetamann. 1977. Genetics of carbon catabolite repression in Saccharomyces cerevisiae: genes involved in the derepression process. Mol. Gen. Genet. 151:95-103. [DOI] [PubMed] [Google Scholar]