Abstract
Selenocysteine (Sec) is naturally incorporated into proteins by recoding the stop codon UGA. Sec is not hardwired to UGA, as we found the Sec insertion machinery to be able to site-specifically incorporate Sec directed by 58 of the 64 codons. For 15 sense codons, complete conversion of the codon meaning from canonical amino acid to Sec was observed along with a 10-fold increase in selenoprotein yield compared to Sec insertion at the three stop codons. This high-fidelity sense-codon recoding mechanism was demonstrated for Escherichia coli formate dehydrogenase and recombinant human thioredoxin reductase and confirmed by independent biochemical and biophysical methods. Although Sec insertion at UGA is known to compete against protein termination, it is surprising that the Sec machinery has the ability to outcompete abundant aminoacyl-tRNAs in decoding sense codons. The findings have implications for the process of translation and the information storage capacity of the biological cell.
Keywords: genetic code, sense codon recoding, RNA engineering, selenocysteine, synthetic biology
Sense codon recoding is supposed to be impossible. Indeed, the fact that a codon can have more than one meaning was a dogma-breaking finding[1] and is one reason why the mechanism of selenocysteine (Sec) insertion into proteins provoked intense biochemical investigation over the last three decades. In addition, selenium is an essential micronutrient in humans.[2] Selenium in proteins is found in the form of Sec in enzymes that maintain the cell’s redox balance, defending the cell against reactive oxygen species. Diseases involving Sec biosynthesis or selenoprotein malfunction have only recently surfaced because defects in these pathways are devastatingly detrimental to proper neuronal function and development.[3]
The promise of sense codon recoding is being actively pursued as a means to further expand the genetic code and synthesize proteins with multiple non-canonical amino acids (ncAAs).[4] Nature expanded the genetic code with two amino acids (Sec and pyrrolysine) by recoding or reassignment of stop codons.[5] Thus, an efficient, naturally evolved machinery already exists that directs recoding of particular UGA stop codons to Sec (Scheme 1A). Although Sec is not found in all organisms (notably lacking in Plants, Fungi, and most Archaea), the 21st amino acid is genetically encoded in all three domains of life. Sec is biosynthesized on its tRNA,[6] and translational recoding of UGA requires the resulting Sec-tRNASec product, a specialized elongation factor (SelB in E. coli), and a downstream mRNA hairpin motif known as the Sec insertion sequence (SECIS).
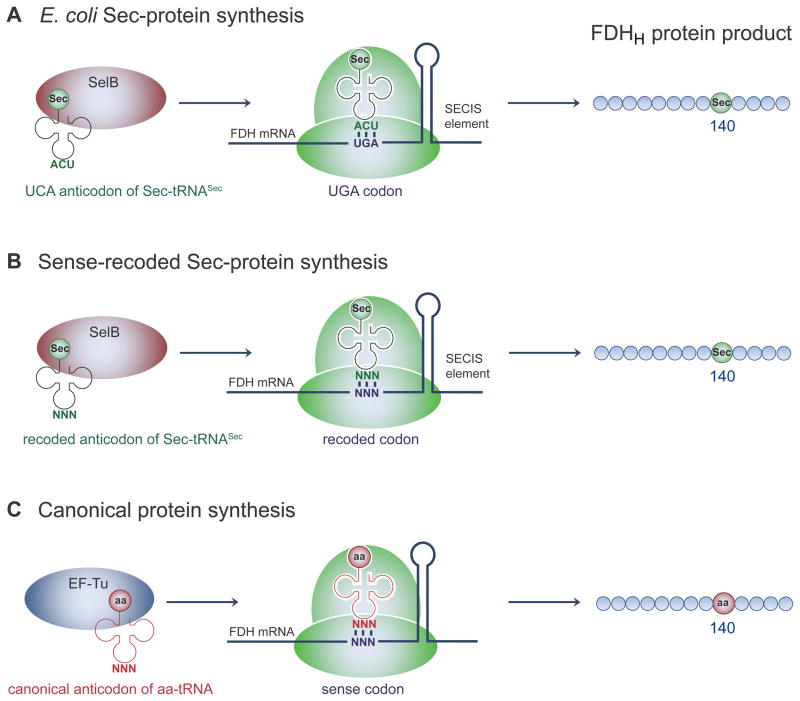
Scheme 1.
Codon recoding with Sec. A) Synthesis of the E. coli Sec-containing formate dehydrogenase (FDH) from the wild type fdhF gene requires recoding the UGA stop codon at position 140 to Sec by Sec-tRNASec, elongation factor SelB, and the SECIS mRNA hairpin. Replacing the UGA with any of the 64 NNN triplets at codon 140 in fdhF and co-expressing a cognate Sec-tRNASecNNN is expected to yield SelB and SECIS-dependent sense codon-recoded Sec-containing FDHH (B) that may compete with canonical AA insertion (C) directed by native aminoacyl-tRNAs and EF-Tu dependent protein synthesis. In this case (C), SECIS is still present in the fdhF mRNA, but SECIS is not expected to interact with EF-Tu directed decoding.
We investigated the possibility to site-specifically reassign multiple sense codons using the Sec machinery. Previous attempts to encode Sec with the Leu UUA codon[7] and the Trp UGG codon[8] produced lower selenoprotein yields compared to UGA-encoded Sec and suggested significant canonical AA contamination. To systematically investigate the recoding capacity of the Sec machinery, we created a library of E. coli formate dehydrogenase (FDHH) mutants. Each gene variant had one of the 64 codons encoding a critical Sec residue (Sec140). In nature, FDHH is part of the membrane associated formate-hydrogen lyase (FHL) complex that decomposes formate to H2 and CO2 under fermentative conditions. The FHL complex shuttles electrons from formate to hydrogenase 3 that reduces protons to hydrogen molecules during anaerobic respiration.[9] Because the enzymatic activity of FDHH is dependent on the active site residue Sec140, which coordinates an active site molybdopterin cofactor[10], sense-codon recoding is easily monitored in vivo and in vitro using the artificial electron acceptor benzyl viologen (BV). Active FDHH reduces BV to a purple color[11] that is clearly visualized in living cells or monitored spectroscopically with purified FDHH.
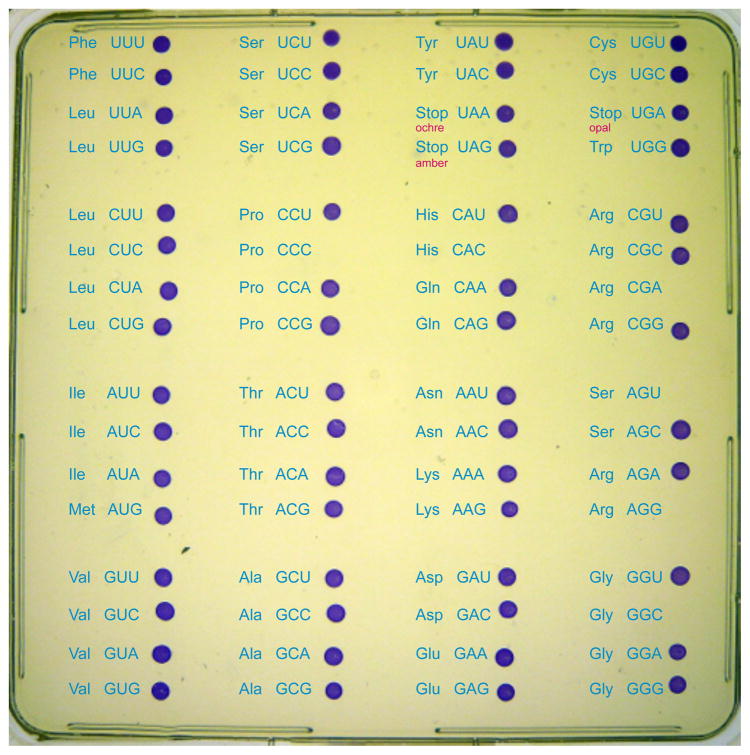
Each FDHH gene variant (fdhF 140NNN) was co-expressed with selA, selB, and a selC mutant (expressing tRNASecNNN) with the cognate anticodon (Scheme 1). We anticipated that theses constructs could give rise to two different protein products. Sec-tRNASecNNN and SelB should compete with canonical aminoacyl-tRNANNN (AA-tRNANNN) and EF-Tu, which could give rise to Sec-containing FDHH (Scheme 1B), canonical AA containing FDHH (Scheme 1C), or a mixture of both protein species. The plasmid-borne FDHH and sel genes complemented an E. coli ΔselAΔselBΔfdhF deletion strain (MH5[12]) that is otherwise unable to produce selenoproteins. Quite unexpectedly, the in vivo assay shows that the Sec recoding machinery successfully alters the meaning of all 3 stop codons and nearly all sense codons to Sec (Figure 1). For 58 of the possible 64 codons, the coloration obtained by Sec-dependent BV reduction is as intense as for the UGA (wild type) Sec codon. Although this assay is qualitative, it demonstrates for the first time, that most codons are recodable. At least one codon for each of the 20 canonical amino acids is recodable to Sec, so no particular AA-tRNA species is able to completely outcompete Sec insertion. For example, the meaning of all six Leu codons, five out of six Ser codons, and four out of six Arg codons was re-assigned to Sec.
Figure 1.
Recoding the genetic code with Sec. The canonical genetic code table is overlaid on a single agar plate spotted with 64 E. coli FDHH variants. In each case, an E. coliΔselAΔselBΔfdhF deletion parent strain was complemented with E. coli selA, selB, and each of the 64 fdhF 140NNN codon mutants and tRNASecNNN variants with the respective cognate anticodon. The capacity of each strain to recode the indicated codon to Sec is evidenced by an in vivo BV reduction assay in which purple colored cells express active Sec-containing FDHH.
Only six codons proved refractory to Sec insertion as indicated by the lack of FDHH activity. Strikingly, four of these codons (CGA, AGU, AGG and GGC) were found in the NGN boxes of the codon table. SDS-gels and western blot analysis (Figures S1, S2) show that full length FDHH of 80 kDa is produced for all codons except the arginine codons AGG and CGA. These codons show a truncated FDHH product at 15.5 kDa, suggesting that protein synthesis terminates at position 140. These data indicate that canonical AA-tRNA completely outcompetes Sec insertion (Scheme 1C) at AGU, GGC, CAC, and CCC, but the meaning of AGG and CGA is converted to stop by an unknown mechanism.
The Sec machinery has the inherent capability to recode nearly all codons (Figure 1), but this in vivo assay does not provide quantitative information regarding the degree of recoding. Under anaerobic conditions, 64 N-terminal His-tagged FDHH proteins were over-expressed from the fdhF 140NNN gene variants and purified (Figures S1, S2). The Sec insertion ratio was quantitated by comparison of the initial velocity of BV reduction in sense-codon recoded versus wild-type FDHH variants in vitro (Table S1). Although the same concentration of purified FDHH protein is used in each assay, the specific activity values differ, which reveals the amount of active (i.e., Sec140) FDHH versus the amount of inactive (i.e., canonical AA140) FDHH protein in each preparation. The assays confirmed Sec incorporation in FDHH for 58 codons with relative FDHH specific activities ranging from 12–100% of the wild type. Selenoprotein yield was greater for most of the recoded sense codons compared to Sec insertion at UGA or the other stop codons, UAA and UAG (Table S2).
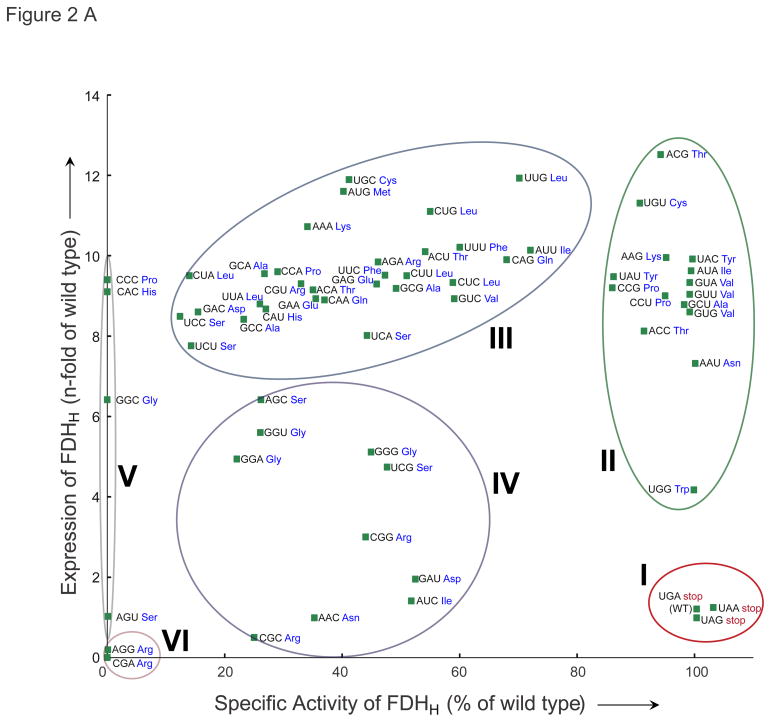
Plotting FDHH protein yield versus specific activity (Figure 2A) revealed six distinct types of recoding behavior. Including the three stop codons (Figure 2, group I), 18 codons led to fully active FDHH (~100% specific activity), indicating complete conversion of the codon meaning to Sec (groups I and II). For the two other stop codon variants (fdhF 140UAG and fdhF 140UAA), specific activity and protein yield was nearly identical to UGA-encoded Sec. In contrast, 15 codons (group II) are also recoded at high-fidelity, but they produce significantly more pure selenoprotein compared to UGA-encoded Sec. Relative FDHH specific activities for codon group II were in the range of 86–100% with 7–13 fold more Sec-containing FDHH production. FDHH variant 140ACG (a Thr codon) showed the highest expression level, while the highest specific activity (i.e., highest fidelity Sec recoding) was observed for the Tyr codon 140UAC. MS/MS analysis confirmed Sec incorporation at position 140 for the Tyr UAC codon (Figure 2B), and no Tyr containing peptide was identified. (Table S2A, B).
Figure 2.
Quantitation of codon recoding with Sec. A) The relative yield of the 64 FDHH 140NNN variants (wild type FDHH 140UGA = 1) is plotted versus specific activity (relative to wild type FDHH 140UGA = 100%). Activity, yield values and standard deviations are shown in Table S1. Different levels of specific activity observed for the 64 FDHH variants are the result of the partial incorporation of the respective canonical AAs (see Table S2). B) MS/MS identification of UAC-encoded Sec. The spectrum of the Sec-containing tryptic peptide, peptide sequence, and position of the observed b- and y-ions are indicated (see also Table S2). Sec (U) was identified by the Mascot protein identification software as a Se-IAN-Cys modification of Cys; selenium alkylation (Se-CH2-CONH2).
For a total of 11 codons, MS/MS analysis identified only the Sec-containing peptide (Table S2, Figure S3). To independently confirm this high-fidelity Sec insertion, purified FDHH variants were analyzed by inductively coupled plasma (ICP)-MS, which provides an accurate estimation of the Se content of the protein sample. These data show 99.9% Sec insertion at UGA in FDHH, confirm complete recoding for UAC (98.3% Sec), and slightly reduced Sec-insertion at UAU (79.3%). The ICP-MS data correlate well with the FDHH specific activities for all other samples tested (Tables S1, S3). Sense codon recoding with Sec requires expression of all the normal Sec insertion components including an intact SECIS, SelB, and Sec-tRNASec formation on a tRNA with a ‘sense-cognate’ anticodon (Supporting Text). Sec-tRNASecUCA (wild type) is unable to decode 140UAC or 140UAU with Sec. In the absence of any component, approximately the same amount of full-length FDHH is produced, but it is totally inactive and contains Tyr140 (Figure S4).
Forty codons displayed ambiguous decoding at position 140 of FDHH. Of these, 30 codons (group III) enabled partial recoding (12–72% Sec) as indicated by specific BV reduction activity (Figure 2, Table S1). These codons produce two protein variants, one containing Sec and one containing canonical AA at position 140, which results in full-length but inactive protein (Figure S1, S2). MS/MS confirmed ambiguous decoding for the Glu codon GAA, and for the three codons (Phe UUC, Leu CUC, Ala GCC) Sec insertion at 140 was clearly identified (Figure S3). These variants displayed markedly increased FDHH protein yields (8–12 fold) compared to UGA-encoded Sec. The 10 group IV codons show 22–52% Sec incorporation and smaller increases in FDHH yield (1–6 fold). Confirming the initial result (Figure 1), four codons were refractory to Sec-recoding (group V) and two Arg codons (CGA and AGG, group VI) produced little protein (Figure 2, Table S1), all of which is truncated at position 140 (Figure S1).
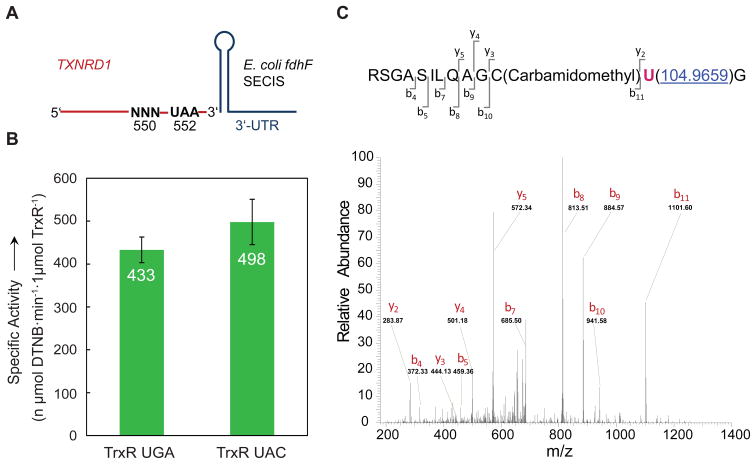
To test the generality of this novel sense-codon recoding mechanism, we expressed a recombinant human selenoprotein, thioredoxin reductase (TrxR) with native UGA-encoded or UAC-encoded Sec (Figure 3A). Thioredoxin reductase is an essential selenoprotein involved in maintaining proper redox balance in human cells. The role of Se is also important in this enzyme as the Sec-containing TrxR is more resistant to oxidative inactivation compared to a Cys-ortholog.[13] Because of its ability to enhance cell survival by defending the cell against reactive oxygen species, TrxR1 over-expression is associated with numerous cancer cell lines, oncogenesis, and metastasis in lymph nodes.[14] TrxR is an emerging target for novel cancer therapeutics.[15] For these reasons, the ability to more efficiently produce TrxR is a significant advance for the field of redox biology, which is expected to impact future therapeutic development. Similar to our observations of UAC-encoded Sec in FDHH, TrxR 550UAC leads to 4-fold higher yield of pure selenoprotein compared to UGA-encoded selenoenzyme. The specific activities are indistinguishable for TrxR produced from these two constructs (Figure 3B). No Tyr contamination was detected by MS/MS analysis (Figure 3C), and ICP-MS analysis independently confirmed 98.5% Sec incorporation in response to UGA and 95.1% Sec insertion for UAC in TrxR (Table S3).
Figure 3.
Recoding the Tyr codon UAC to Sec in human TrxR1. A) Schematic representation of the expression construct for human TXNRD1. An E. coli fdhF SECIS element is attached directly after the UAA codon of TXNRD1 in the 3′ untranslated region (UTR). B) In vitro activity assay of recombinant human TrxR 550UGA and TrxR 550UAC co-expressed with tRNASecUCA and tRNASecGUA, respectively. Error bars show standard deviations of quadruplicate experiments. C) MS/MS identification of Sec incorporation into TrxR 550UAC. The spectrum of the Sec-containing peptide, the peptide sequence and the positions of the observed b- and y-ions are indicated. Sec was identified by MASCOT as a Se-IAN-Cys modification of Cys; selenium alkylation (Se-CH2-CONH2).
It remains unclear why certain codons are more ‘recodable’ than others. It is reasonable to assume that codon usage might correlate with recodability, because translation is believed to be less efficient with lower abundant codons. The intracellular concentration of AA-tRNA[16] is another likely explanation. Perhaps codons read by less abundant AA-tRNA species are more easily recoded. Despite the logic of these ideas, there is no general correlation between recodability and codon usage or AA-tRNA concentration (Figure S5). There are potential complicating factors; perhaps the Sec-tRNASecNNN species are not decoded on the ribosome with equal efficiency due to compatibility of the tRNA body and anticodon loop. There is also the question whether the nature of the mRNA codon (position 140) affects the structural integrity of the SECIS element. Furthermore, differences between AA-tRNA levels may not sufficient enough to fundamentally alter competition with Sec-tRNASecNNN. Based on crystal structures, a model of the ribosome bound Sec-decoding complex (i.e., ribosome, SelB, Sec-tRNASec, mRNA with SECIS) indicates extensive interactions between SelB and the 30S subunit in regions that are known to alter the accuracy of mRNA decoding.[17] Kinetic and structural studies of sense-codon decoding with Sec will help define the mechanism discovered here.
In the normal Sec-decoding situation, the release factor (RF2 in E. coli) competes with Sec insertion,[18] leading to truncated FDHH (Figure S4). Encoding Sec with sense codons escapes competition with RFs, which leads to the significantly enhanced selenoprotein yield for all but 10 codons. It is unclear why nature selected UGA as the ‘Sec’ codon when most other codons lead to more selenoprotein production. Perhaps the scarcity of Se in the environment induced organisms to naturally limit the level of selenoprotein synthesis with RF competition. RF interaction may, nevertheless, affect the recodability of certain sense codons. Due to its similarity with UGA, UGG is a hotspot for premature termination by RF2,[19] which may explain why this codon produces less Sec-containing FDHH than all other group II codons (Figure 2).
Encoding Sec with sense codons will become a useful method for efficiently producing selenoproteins; yet this discovery opens the door to recode many codons, not only with Sec, but possibly also with a diverse array of ncAAs of biomedical and biotechnological interest. The human proteome contains many essential modified proteins that arise by posttranslational modification (PTM), and mis-modified proteins may lead to defects in cellular signaling and protein aggregates that form the molecular basis for diseases. In order to biochemically define the role of PTMs and to produce therapeutic agents that specifically target mis-modified proteins, there is an urgent need to synthesize proteins containing several different, distinct ncAAs. Progress in the last decade allowed site-specific incorporation of 1 or 2 ncAAs into recombinant proteins by reassignment of nonsense codons.[4b] In vitro protein synthesis methods were recently enhanced by creating dual-meaning initiation and sense codons to synthesize potential chemotherapeutic peptides with 2 ncAAs,[20] and amino acid starvation methods permitted production of proteins with 3 ncAAs for biological imaging applications.[21] Furthermore, unlike the Sec-machinery that only recodes codons associated with SECIS, current genetic code expansion methods are not site-specific because they lead to global reassignment of stop codons; this contaminates the natural proteome with ncAAs, resulting in growth defects.[22]
In order to synthesize proteins with more than 23 amino acids, additional ‘recodable’ codons are needed.[23] We anticipate that mutagenesis of the Sec insertion machinery will enable protein synthesis with multiple ncAAs, which will require engineering SelB, SECIS, and also orthogonal tRNA synthetases to acylate tRNASec and tRNASec-like molecules with ncAAs. The fact that SepCysS forms Cys-tRNASec, which actively decodes UGA with Cys instead of Sec, is an encouraging example.[24]
The fact that the Sec insertion machinery has the intrinsic capability to alter the meaning of 58 of the 64 codons indicates that the biological cell will be able to genetically encode far more amino acids than previously recognized. Although the Sec insertion machinery recodes some codons poorly, it is possible that the recoding fidelity may be enhanced through selection. Given that certain codons are recodable at high-fidelity, it is conceivable that in nature Sec is encoded by codons other than UGA, suggesting the existence of unknown selenoproteins. Indeed, SECIS-like structures have been identified bioinformatically and dismissed as evolutionary remnants.[25] If other RNA signals, like SECIS, exist in nature or if they can be engineered in the laboratory, there is potentially no limit to the number of amino acids that the cell can encode and no reason to stop at 20 or even 23.
Supplementary Material
Acknowledgments
We thank Hans Aerni, Måns Ehrenberg, Ilka Heinemann, and Eric Westhof for insightful comments and discussion. We are also grateful for the dedicated efforts of Kathryn Stone and Jean Kanyo at the W.M. Keck MS facility at Yale, and Zhengrong Wang and Ying Kiu from the Kline Geology Lab at Yale for assistance with ICP-MS analysis. This work was supported by grants to D.S. from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy (DE-FG02-98ER20311; for funding the genetic experiments), the National Institute of General Medical Sciences (GM22854), and by DARPA contract N66001-12-C-4211. M.J.B. was a Feodor Lynen Postdoctoral Fellow of the Alexander von Humboldt Foundation (Bonn, Germany). J.M.L.H. was supported by a graduate fellowship from Harvard Medical School.
Footnotes
Author Contributions
M.J.B, D.S., and P.O. designed research; M.J.B., J.M.L.H., and P.O. performed research; M.J.B., J.M.L.H., D.S. and P.O. analyzed data; M.J.B., G.M.C., D.S., and P.O. wrote the paper.
References
- 1.Zinoni F, Birkmann A, Leinfelder W, Böck A. Proc Natl Acad Sci USA. 1987;84:3156–3160. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rayman MP. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 3.a) Agamy O, Ben Zeev B, Lev D, Marcus B, Fine D, Su D, Narkis G, Ofir R, Hoffmann C, Leshinsky-Silver E, Flusser H, Sivan S, Söll D, Lerman-Sagie T, Birk OS. Am J Hum Genet. 2010;87:538–544. doi: 10.1016/j.ajhg.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Seeher S, Mahdi Y, Schweizer U. Cur Protein Pept Sci. 2012;13:337–346. doi: 10.2174/138920312801619448. [DOI] [PubMed] [Google Scholar]
- 4.a) Krishnakumar R, Prat L, Aerni HR, Ling J, Merryman C, Glass JI, Rinehart J, Söll D. Chembiochem. 2013 doi: 10.1002/cbic.201300444. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Donoghue PO, Ling J, Wang YS, Söll D. Nat Chem Biol. 2013;9:594–598. doi: 10.1038/nchembio.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambrogelly A, Palioura S, Söll D. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 6.Yuan J, Donoghue PO, Ambrogelly A, Gundllapalli S, Sherrer RL, Palioura S, Simonovic M, Söll D. FEBS Lett. 2010;584:342–349. doi: 10.1016/j.febslet.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry MJ, Harney JW, Ohama T, Hatfield DL. Nucleic Acids Res. 1994;22:3753–3759. doi: 10.1093/nar/22.18.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Croitoru V, Rutishauser D, Cheng Q, Arnér ES. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawers G. Antonie van Leeuwenhoek. 1994;66:57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- 10.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. Science. 1997;275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 11.Axley MJ, Grahame DA, Stadtman TC. J Biol Chem. 1990;265:18213–18218. [PubMed] [Google Scholar]
- 12.Aldag C, Bröcker MJ, Hohn MJ, Prat L, Hammond G, Plummer A, Söll D. Angew Chem Int Ed. 2013;52:1441–1445. doi: 10.1002/anie.201207567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snider GW, Ruggles E, Khan N, Hondal RJ. Biochemistry. 2013;52:5472–5481. doi: 10.1021/bi400462j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YC, Prabhu KS, Mastro AM. Nutrients. 2013;5:1149–1168. doi: 10.3390/nu5041149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Holmgren A. Antioxid Redox Signal. 2012;17:1738–1747. doi: 10.1089/ars.2012.4650. [DOI] [PubMed] [Google Scholar]
- 16.a) Dong H, Nilsson L, Kurland CG. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]; b) Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshizawa S, Rasubala L, Ose T, Kohda D, Fourmy D, Maenaka K. Nat Struct Mol Biol. 2005;12:198–203. doi: 10.1038/nsmb890. [DOI] [PubMed] [Google Scholar]
- 18.Mansell JB, Guevremont D, Poole ES, Tate WP. EMBO J. 2001;20:7284–7293. doi: 10.1093/emboj/20.24.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. Proc Nat Acad Sci USA. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sund J, Ander M, Aqvist J. Nature. 2010;465:947–950. doi: 10.1038/nature09082. [DOI] [PubMed] [Google Scholar]
- 20.Goto Y, Iseki M, Hitomi A, Murakami H, Suga H. ACS Chem Biol. 2013 doi: 10.1021/cb400549p. [DOI] [PubMed] [Google Scholar]
- 21.Lepthien S, Merkel L, Budisa N. Angew Chem Int Ed. 2010;49:5446–5450. doi: 10.1002/anie.201000439. [DOI] [PubMed] [Google Scholar]
- 22.Heinemann IU, Rovner AJ, Aerni HR, Rogulina S, Cheng L, Olds W, Fischer JT, Söll D, Isaacs FJ, Rinehart J. FEBS Let. 2012;586:3716–3722. doi: 10.1016/j.febslet.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, Isaacs FJ. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan J, Hohn MJ, Sherrer RL, Palioura S, Su D, Söll D. FEBS Let. 2010;584:2857–2861. doi: 10.1016/j.febslet.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Romero H, Salinas G, Gladyshev VN. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.