Abstract
NK cells are innate immune lymphocytes that express a vast repertoire of germ-line encoded receptors for target recognition. These receptors include inhibitory and activating proteins, among the latter of which is CD16, a low affinity binding Fc receptor. Here, we show that human NK cells expand in response to stimulation with various tumor cell lines. We further demonstrate that the tumor-derived expansion of NK cells is accompanied by rapid, cell-dependent, changes in CD16 expression levels. We show that in NK cells expanded in response to the EBV-transformed cell line 721.221, CD16 is shed and therefore approximately half of the expanded 721.221-derived NK-cell population does not express CD16. We also show, in contrast, that in response to 1106mel cells, CD16 expression is maintained on the cell surface of the expanded NK cells due to an antibody-dependent mechanism. Our results may provide a basis for the selective expansion of NK cells that may be used for tumor immunotherapy.
Keywords: Antibodies, CD16, human NK cells, NK-cell expansion
Introduction
NK cells are experts in the destruction of tumor and virally infected cells [1, 2]. They are considered to be a component of the innate immune system and the specificity their effector functions relies on an integrated processing of signals, transduced by activating, and inhibitory receptors [3]. Inhibitory receptors recognize molecules such as MHC class I proteins that are expressed on healthy cells to protect these cells from NK-cell attack. The NK activating receptors, which include NKG2D, NKp46, NKp44, NKp80, DNAM1, 2B4, and CD16, recognize pathogen-derived ligands, stress-induced molecules, tumor ligands, and self-ligands [3]. To trigger naive NK-cell activity, cross-linking of two or more activating receptors is required; the only exception to this is CD16 (FCIIIγRA), in which cross-linking of only this receptor is sufficient to cause NK-cell degranulation [4]. CD16 is also unique in its expression pattern and NK cells from the peripheral blood consist of two distinct populations, CD56Dim CD16+ (>90% of the cells) and CD56Bright CD16− [5]. Decidual NK cells and NK cells present in the lymph nodes exhibit the mirror image of this pheno-type, consisting almost exclusively of CD56Bright CD16− NK cells. CD16A, the isoform expressed on NK cells, has two extracellular Ig domains, a short cytoplasmic tail and a transmembrane domain that enables its association with adaptor proteins. The Fc portion of IgGs binds to CD16 via its second, membrane-proximal Ig domain, which is recognized by the mAb 3G8 [6] and promotes signal transduction [7]. The distal Ig domain of CD16 is recognized by mAb B73.1 [8]. Recently, this domain was shown to be important for the association between CD16 and CD2, and patients with a mutation in this domain manifest impaired NK-cell cytotoxicity with no effect on antibody-dependent cell-mediated cytotoxicity (ADCC) [9].
Recent works demonstrated that activation of NK cells is followed by rapid and efficient cleavage of CD16 from the surface of activated NK cells. These reports show that CD16 is cleaved from the cell surface by membrane-type 6 matrix metalloproteinase (MMP) [10] and by ADAM-17 [11]. The cleavage of CD16 by MMPs was additionally showed in the process of ADCC.
Here, we demonstrate that human NK cells expand in response to various tumor stimulations and that the expanded NK cells show dramatic changes in CD16 expression depending on the tumor cell line they react to.
Results
Human NK cells expand in response to selected tumor cell lines
To test whether human NK cells expand in response to various tumor cell lines, we incubated freshly isolated NK cells in the presence of human (Fig. 1A and B) and mouse (Fig. 1C) tumor cell lines. The tumor cells were irradiated (to prevent proliferation) and the incubation was done for 5 days in the presence of IL-2 and human sera. The expanded NK cells were identified by physical parameters (FSC and SSC, marked by a grey polygon) and by being positive for CD56 expression and negative for CD3 expression (Supporting Information Fig. 1A). As can be seen, extensive NK-cell expansion was observed in response to only one adherent, MHC class I negative tumor cell line; 1106mel (Fig. 1A) and two EBV-transformed B-cell lines (721.221 and 8866). Other hematological cell lines such as: K562, Jurkat, U937, BJAB, and NB4 also supported the expansion of the NK cells (data not shown). In contrast, little or no expansion (at least fivefold increase compared to input) was detected when NK cells were incubated with tumor cell lines of various origins (the embryonic kidney line 293T, the mammary gland adenocarcinoma MCF7, the choriocarcinoma Jeg3, the cervical adenocarcinoma HeLa, and the lung carcinoma A549, Fig. 1B). Moreover, no expansion was observed when human NK cells were incubated with the mouse cell lines RMAs and BW (Fig. 1C).
Figure 1.
Human NK cells are expanded in response to selected human tumor cell lines. A total of 3 × 105 NK cells were cultured with (A, B) human and (C) mouse cell lines (indicated at the top of the dot plot). Following 5 days in culture the cells were tested for the presence of NK cells by size and granularity (FSC, SSC) then examined for being positive for CD56 expression and negative for CD3. Data shown are representative of three independent experiments. (D) NK-cell numbers following 5 days of incubation with 721.221 or 1106mel cells are shown as mean + SD of four samples from a single experiment representative of three independent experiments.
We decided to concentrate, in the following experiments, on studying the 721.221-expanded and the 1106mel-expanded NK cells because of their different origin (EBV-transformed B cells −721.221 and melanoma-1106mel) and since both tumor lines are class I negative. To test whether the NK-cell expansion observed with 721.221 and with 1106mel cells resulted from increased proliferation or from inhibition of apoptosis we double stained the expanded NK cells with CD56 together with Annexin V (Supporting Information Fig. 1B). Negligible fraction of NK cells positive for Annexin V expression were noted with both cell lines, suggesting that the NK-cell expansion was due to increased proliferation. Although NK cells expanded to a higher extent when incubated with 721.221 cells (average of 5 × 106 cells, 5 days following the incubation) as compared with 1106mel cells (average of 3.7 × 106 cells) (Fig. 1D) this difference was not statistically significant (p = 0.08).
CD16 expression is maintained on the surface of NK cells expanded in response to 1106mel cells
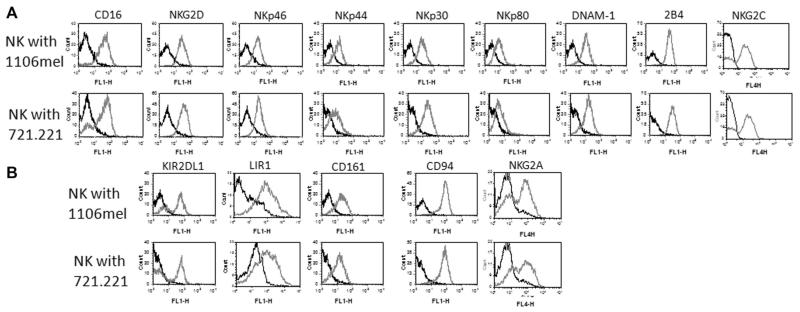
We next analyzed the receptor repertoire of the expanded NK-cell populations. Freshly isolated NK cells were incubated with 721.221 or 1106mel cells and as can be seen in Figure 2, a similar expression pattern of the activating receptors: NKG2D, NKp46, NKp44, NKp80, DNAM1, 2B4 (Fig. 2A), and NKG2C (its expression varied between various donors, Fig. 2A and Supporting Information Fig. 2) and the inhibitory receptors KIR2DL1, LIR1, CD161, CD94, and NKG2A (Fig. 2B), was noted with the exception of CD16. The expanded NK cells cultured with 721.221 cells had a CD16 negative population, while practically all NK cells expanded in response to 1106mel cells were CD16+.
Figure 2.
Analysis of NK-cell receptor repertoire during NK-cell expansion. NK cells were cultured with 1106mel (top) or with 721.221 (bottom). After 10 days in culture the cells were stained for the various NK-cell (A) activating receptors and (B) inhibitory receptors (gray-line histograms). The black-line histograms represent staining with the secondary mAb only. Data shown are representative of three independent experiments performed.
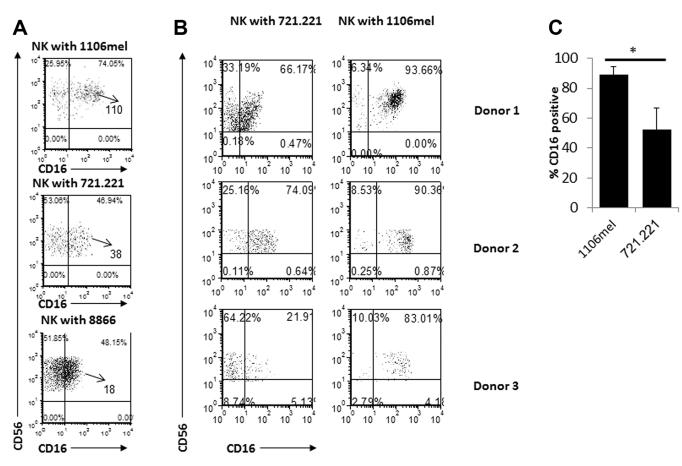
To further investigate the differences observed in CD16 expression in the expanded NK-cell populations we also incubated freshly isolated NK cells, for 5 days, with the RPMI-8866 (8866) cell line that is commonly used to grow NK cells in vitro [12, 13]. As expected, the 8866 cells supported the NK-cell expansion and similarly to the 721.221-expanded NK cells, around half of the 8866-expanded NK cells were CD16 negative (Fig. 3A). In addition the intensity of the CD16 expression as determined by its MFI was higher on the expanded 1106mel-NK cells as compared with that of the 721.221-NK cells and to the 8866-NK cells (Fig. 3A). A similar reduction of CD16 expression was noted in NK cell expanded in response to other cells such as K562 or U937 (data not shown). To verify that the above observations are not specific to a particular donor, we repeated the coculturing experiments with NK cells isolated from various donors (three of them are presented in Fig. 3B). Even though a certain degree of heterogeneity was observed among the different donors, in all cases larger percentages of NK cells expressing CD16 were detected in NK cells expanded in response to 1106mel cells (Fig. 3B). Figure 3C summarizing the CD16+ percentage of NK cells, from all the donors, cocultured with the either 721.221 or 1106mel. As can be seen significantly more CD16+ percentage of NK cells were obtained following the cocultured with 1106mel cells for 5 days (Fig. 3C).
Figure 3.
Analysis of CD16 levels during NK-cell expansion. (A) NK cells were cocultured with 1106mel, 721.221, and 8866 cell lines for 5 days and then analyzed for CD16 expression. The numbers in the quadrants indicates percentages and the MFI are marked by an arrow. (B) NK cells were obtained from three different donors, incubated with 1106mel or with 721.221. The numbers in the quadrants indicates percentages. (A, B) Data shown are representative of two independent experiments. (C) The percentages of CD16+ NK cells following 5 days incubation with 721.221 or 1106mel were calculated after flow cytometric analysis and shown as mean SD of seven donors. *p < 0.02, by two tailed Student’s t-test.
CD16 expression is downregulated on the 721.221-expanded NK cells
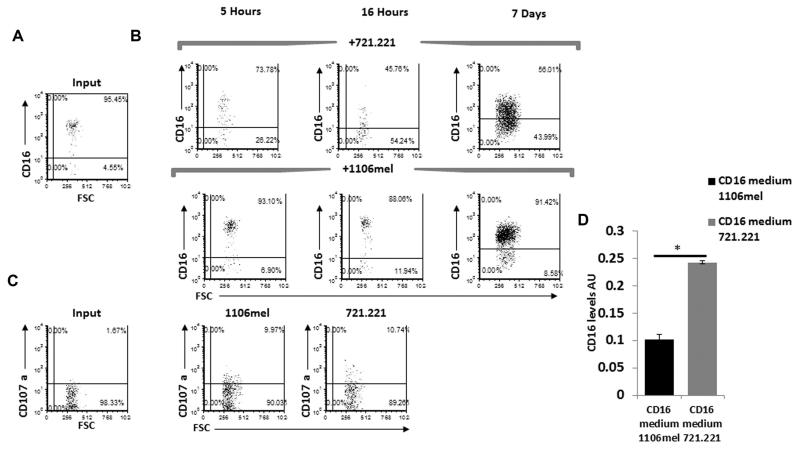
Two NK-cell populations are found in the peripheral blood: the majority (around 90%) express CD56 in intermediate levels and are CD16+, while the remaining NK cells are CD56 bright but do not express CD16 [5]. Thus, to investigate whether the CD16− fraction of the 721.221-expanded NK cells resulted from expansion of the CD56Bright NK-cell population we stained the 721.221-expanded NK cells for CD56 expression and observed that all of them express CD56 at intermediate levels. This suggests that in the presence of 721.221, but not in the presence of 1106mel, CD16 expression is downregulated. To further investigate this issue and to determine the kinetics of the CD16 downregulation we cocultured the NK cells with 721.221 or with 1106mel cells for various time points (5, 16, 24, 48, 96 h, and 7 days, Fig. 4A, B and Supporting Information Fig. 3). We initially determine the CD16 expression immediately after the isolation of NK cells, before incubating them with the targets cells, and in agreement with the literature [5] we observed that CD16 is expressed on approximately 90–95% of the NK cells (Fig. 4A, input). Interestingly, as early as 5 h after the addition of the different targets to the NK cells we already detected a significant difference between the NK cells incubated with 721.221 cells (Fig. 4B top) and those cultured with 1106mel cells (Fig. 4B bottom). After 16 h of coculturing, CD16 expression was downregulated even further in the presence of 721.221 cells, while practically no change in CD16 expression was observed in the presence of 1106mel cells (Fig. 4B). No significant difference in the expression levels of CD16 was detected 24–96 h of coculturing (compared to the first time point, 5 h) of the NK cells with 1106mel or with 721.221 (Supporting Information Fig. 3). Vigorous NK-cell expansion was detected after a week following in the presence of both 721.221 and 1106mel cells (Fig. 4B) and the differences in the CD16 expression were maintained. Around half of the expanded 721.221-NK cells did not express CD16, while around 90% of the expanded 1106mel-NK cells expressed CD16. The CD16 MFI levels however of the 1106mel-expanded NK cells were reduced at that point (Fig. 4B). Two weeks following the incubation all the expanded NK cells (both the 721.221- and the 1106mel-expanded NK cells) were CD16+ and expressed CD16 in intermediate levels (data not shown). Similar results were obtained with additional donors (data not shown).
Figure 4.
CD16 expression is downregulated on expanded NK cells incubated with 721.221, but not with 1106mel cells. (A) Freshly isolated NK cells were stained for CD16 expression (input). (B) NK cells from (A) were incubated with 721.221 (top) and with 1106mel (bottom) for five and 16 h or for 7 days and the expression levels of CD16 were determined. (C) Levels of CD107a expression determined prior to incubation with target cells (input, left dot plots) or following 5-h incubation of the NK cells with 721.221 or with 1106mel. The numbers in the quadrants indicates percentages. Data shown are representative of three experiments performed. (D) CD16 levels in the supernatants of NK cells (incubated either with 721.221 cells or with 1106mel) were quantified by ELISA after 16 h. Data are shown as mean + SD from a single experiment representative of five performed. *p < 0.001, two tailed Student’s t-test.
To test whether the downregulation of CD16 observed in the presence of 721.221 cells correlated with differences in the sensitivity of the target cells to killing by NK cells we performed NK cytotoxicity assays. For that we measure the extent of NK-cell degranulation (as indicated by cell surface staining of CD107a). As can be seen in Fig. 4C NK-cell degranulation was similar in the presence of 721.221 or 1106mel cells (around 10%), while in the absence of these target cells around 2% degranulation was observed (Fig. 4C, input). Thus, the extent of NK-cell degranulation is not the cause of CD16 down regulation.
Previous reports show that CD16 is cleaved from the cell surface by MMP-6 [10] and by metalloprotease-17 (ADAM17) [11]. To investigate whether CD16 was cleaved from the surface of expanded 721.221-NK cells we examined by ELISA assays whether the supernatants of the expanded 721.221 cells contain increased levels of soluble CD16. As can be seen in Fig. 4D in the presence of 721.221 significantly more CD16 was present in the medium, suggesting the shedding of CD16 might explain why in the presence of 721.221 less CD16 is found on the expanded NK cells.
Antibodies are essential for maintaining the CD16 expression on the expanded 1106mel-NK cells
To test why CD16 expression is maintained on the 1106mel-expanded NK cells we studied whether antibodies or CD2 are involved. This is because CD16 is a low affinity Fc receptor [8] and because it was shown that the 1106mel line is recognized by CD16 in a CD2-dependent manner [9, 14]. We initially started investigating whether antibodies are involved and since the coculturing of NK cells with the target cells is done in medium supplemented with 10% human sera, we hypothesized that the immunoglobulins found in the human sera might play a role in the CD16 expression levels. To test this, 721.221 cells (Fig. 5A) or 1106mel cells (Fig. 5B) were incubated with sera obtained from four different healthy donors and stained with fluorescently labeled anti-human antibodies. Importantly, both cells do not express Fc receptors. As can be seen, while little or no recognition of the 721.221 cells was detected (Fig. 5A) the 1106mel cells were recognized by all sera tested (Fig. 5B). Similar results were obtained when sera from other donors were used or when commercially available sera were used (data not shown).
Figure 5.
CD16 is maintained of the surface of 1106mel-expanded NK cells due to antibody recognition. (A, B) Sera from four different donors were incubated with (A) 721.221 cells or (B) with 1106mel cells. Detection of the sera binding to the cells was performed by anti-human IgG (black-line histograms). Staining with 10% fetal calf serum was used as negative control (gray-filled histograms). (C) 1106mel cells were stained either with complete human serum (red line) or with IgG-depleted serum (blue line). (D) NK cells were incubated with 1106mel in the presence of the complete sera (left) or following depletion of IgG (right). Data shown are representative of three independent experiments. (E) Freshly isolated NK cells were incubated either with a control mAb or with the anti-CD16 mAbs (3G8) and then cocultured with irradiated 1106mel cells. Five days later, the cells were stained for CD16 expression. The numbers in the figure indicates the percentages of CD16+ cells. Data shown are representative of two independent experiments. (F) CD16 levels in the supernatants of NK cells (incubated with 1106mel in presence or absence of 3G8 mAb) were quantified by ELISA five days following the coculturing. Data are shown as mean + SD of four replicates from one of two experiments performed. ** p < 0.02 (by two tailed Student’s t-test). (G) Freshly isolated NK cells were cultured with 221/CEA in the absence (left) or in the presence (middle) of polyclonal anti-CEA Abs or with 1106mel (right) and stained 5 days later for CD16 expression. The numbers in the figure indicates the percentages of CD16+ cells. (H) CD16 levels in the supernatants of NK cells (incubated with 221/CEA in presence or absence of anti-CEA polyclonal Abs) were quantified by ELISA after 5 days and shown as mean + SD of four samples/replicates from two experiments. *p < 0.04, (by two tailed Student’s t-test).
To demonstrate that the antibodies in the human sera are responsible for the maintenance of CD16 on the expanded-1106mel NK-cell population we used protein-G beads to deplete the IgGs from the sera. After the verification that the IgG-depleted sera was indeed unable to recognize the 1106mel cells (Fig. 5C), we repeated the coculturing experiments with 1106mel cells and with freshly isolated NK cells in the presence of either complete medium or the IgG-depleted medium. Interestingly, no NK-cell expansion was observed in absence of IgGs (Fig. 5D). Since CD16 is known to promote proliferation in presence of hIL-2 [15], we wanted to see whether the depletion of IgG from serum will affects the survival or the levels of CD16 on the surface of NK cells. For that purpose we monitored the CD16 levels on NK cells cocultured with 1106mel (in the presence or absence of IgG in the sera) during different time points (24, 48, and 72 h) (Supporting Information Fig. 4A). Surprisingly after 24 h of coculturing the MFI levels of CD16 on NK cells in absence of IgG were higher compared to the MFI of CD16 on NK cells in the presence of 1106mel cells and IgGs. Forty-eight hours following the coculturing experiments the CD16 levels were similar on NK in both conditions. In the absence of IgGs, after 72 h following the coculturing the NK-cell numbers were reduced and a CD16 negative population started to emerge (Supporting Information Fig. 4A). Additionally, the MFI of CD16 on the CD16+ NK cells was significantly lower compared to CD16+ NK cells cocultured in presence of IgG. The differences in the surface expression of CD16 inversely correlated with its levels in the supernatants and significantly higher amount of CD16 were found in the supernatants of IgG- expanded NK cells (Supporting Information Fig. 4B)
The CD16 protein has two distinct-binding sites that are involved in its binding to the Fc portion of antibodies (this site is recognized by the 3G8 mAb [6]) and in the CD2-dependent binding to 1106mel cells (this site is recognized by the B73.1 mAb [9, 14]. To demonstrate that the Fc-binding site of CD16 is indeed involved in the maintenance of CD16 expression on the expanded 1106mel-NK-cell population, we incubated freshly isolated NK cells that were treated or not with the 3G8 mAb and with 1106mel cells in the presence of human sera. The addition of 3G8 prior to the coculture with 1106mel resulted in a reduced CD16+ fraction of the NK cells (Fig. 5E). The detection of CD16 expression following the 3G8 blocking was by directly labeled B73.1 mAb that recognize site different than the 3G8 mAb [6]. The difference in the surface levels of CD16 inversely correlated with levels of cleaved CD16, as the addition of 3G8 led to significantly higher CD16 levels in the supernatants (Fig. 5F).
Because the above observations indicates that antibodies bound to CD16 are responsible for the maintenance of CD16 expression on the 1106mel-expanded NK cells, we next questioned whether the expansion of the CD16+ NK cells would also occur with antibodies bound to 721.221 cells. Freshly isolated NK cells were cocultured with either 721.221 transfectants that express the carcinoembryonic antigen (CEA) (221/CEA, used here because we had polyclonal antibodies against CEA), with 221/CEA cells that were pre incubated with anti-CEA polyclonal antibodies, or with 1106mel cells. As can be seen in Figure 5G, when NK cells were cocultured with the 221/CEA cells precoated with antibodies (in the presence of anti CEA plolyclonal Abs) a significant increase in the percentages of the CD16+ population was observed. As above, the levels of cleaved CD16 in the supernatants were reduced in the presence of polyclonal anti CEA antibodies (Fig. 5H).
CD16 positive NK cells that are expanded in response to 721.221 cells are efficient effectors
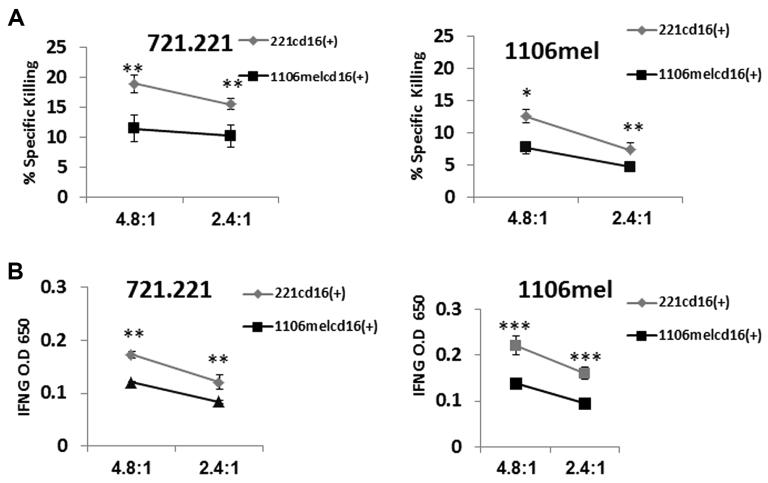
Finally, we investigated how the main effector functions of the expanded NK cells (cytotoxicity and cytokine secretion) are affected by the targets they have responded to. For that purpose NK cells cocultured with 1106mel or with 721.221 cells were used in killing assays against the targets they were incubated with and against the other target. To accurately compare the effector function of the expanded NK-cell population, to test whether CD16 expression is associated with NK-cell functionality and because around half of the expanded NK cells lost CD16 expression in the presence of 721.221 cells (Fig. 6) we sorted the positive CD16 fraction from NK cells expanded either with 1106mel (1106melcd16(+)) or with 721.221(221cd16(+)). As can be seen in Figure 6 the CD16+ NK cells expanded in the presence of 721.221 cells demonstrated superior effector functions (cytotoxicity, Fig. 6A) and cytokine secretion (Fig. 6B) against both 721.211 and 1106mel targets as compared to the 1106mel-expanded NK cells.
Figure 6.
721.221-expanded NK cells are superior effectors over 1106mel expanded cells. (A, B) NK cells were incubated either with 721.221 cells (221cd16(+)) or with 1106mel cells (1106melcd16+) and sorted based on the CD16 expression. The CD16+ fraction was used in (A) a killing assay and (B) IFN-γ secretion assay against 721.221 and 1106mel targets. The E:T ratios are indicated at the X axis. Data are shown as mean + SD of three samples from one of two independent experiments*p value < 0.05, **p value < 0.01 ***p value < 0.001 (by two tailed Student’s t-test).
Discussion
The activation of NK cells is a complex-regulated process and during this process NK cells are not only activated to kill the target cells but they can also be expanded. It was recently shown that some tumor cells support NK-cell expansion [16, 17]. However, in most of these cases the expanded NK cells did not show significant-cell dependent changes in their NK-cell receptor repertoire. Here, we initially observed that NK cells efficiently expanded in response to various hematological tumor cell lines. We further observed that the NK-cell expansion is “missing self”-independent as NK-cell expanded in response to tumor cells that are either positive or negative for MHC class I expression. The reasons for such efficient expansion might be the coactivating signals supplied by the hematological cells [17]. These might include ligands such as CD58 (CD2-ligand), CD48 (2B4 ligand), and the ligands for NKG2D (MICA, MICB, and ULBPs).
The expanded NK cells expressed similar levels of all NK-cell receptors tested, except for CD16. In the presence of 721.221 cells CD16 was downregulated on the expanded NK cells and in the presence of 1106mel cells, CD16 levels were maintained on the expanded NK-cell populations. The changes in CD16 expression are rapid, starting as early as 5 h post coincubation and lasting up until day 14. Prolonged culture of the NK cells for 2 weeks resulted in a uniform CD16 expression on the entire NK-cell population regardless of the type of the cells they responded to (721.221 or 1106mel cells).
CD16 is probably the best example of an NK-cell receptor linking the innate and the adaptive immune responses. In fact, it is the existence of this receptor on NK cells that suggests that the functions of NK cells are not limited to attacking hazardous cells immediately. This is because a major property of CD16 is the binding of antibodies that are generated late during the immune response, when the adaptive immunity is activated. The fact that CD16 is maintained on the 1106mel-expanded NK cells when they are being activated by antibodies is also interesting, in light of previous observations demonstrating that CD16 is the only receptor found on NK cells able to cause direct cytotoxicity by naïve NK cells with no additional stimulation. We suggest that this mechanism of CD16 maintenance on the surface of NK cells in the presence of polyclonal Abs was developed to ensure that in such situations, when antibodies are present against the developing tumors/viruses, CD16 will be available for mediating killing. It will be interesting to determine in the future the identity of the ligand/s present on 1106mel cells that are recognized by the antibodies present in human sera. On the other hand we demonstrate that when antibodies against the tumors are not present, CD16 expression is down regulated on the expanded NK cells. The reduction of CD16 expression on the 721.221-expanded NK cells was associated with increased amount of CD16 present in the supernatants of the NK cells. These observations are in line with two other publications. Peruzzi et al. showed that activation of NK cells by various means, including IL-2 is accompanied by recruitment of MMP25 and subsequent cleavage of CD16 [10]. Zhou et al. demonstrated, using three clinically used mAbs, that during ADCC CD16 is cleaved from the surface of NK by MMPs [18]. Interestingly, we observed that in presence of anti CEA polyclonal antibodies or when human sera was used that binds 1106mel cells (Fig. 5), the CD16 levels were not reduced but were actually maintained on the cell surface. Several differences in the experimental system used by Zhou et al. and ours: Probably the most significant one is that in our system polyclonal antibodies were used, whereas in the other system mAbs were used.
Finally our findings have practical consequence for the use of NK cells in tumor therapy. If antibodies against the tumor cells are present in the patients it might be beneficial to expand the NK cells in the presence of 1106mel cells or similar cells. On the other hand if no antibodies are present it might be better to expand the NK cells in the presence of tumors cells such as 721.221 or similar cells
Material and methods
Cells, NK-cell isolation, and coculturing experiments
PBMCs were derived from healthy donors. NK cells were isolated using the autoMACS instrument (Miltenyi Inc.) according to the manufacturer instructions. A total of 2.5×105 freshly isolated NK cells were grown with 2 × 105 – 2 × 106 of the various tumor cell lines tested, in the presence of human sera and 400 units/ml recombinant IL-2. NK cells purity (>95%, CD56+, CD3−) was confirmed prior to the incubation with the various tumors. The cell lines used in this work were: the MHC class I-negative 721.221 (221), 721.221 cells stably transfected with the CEA protein (221/CEA), the MHC class I negative 1106mel cells, the EBV transformed RPMI 8866 (8866), the embryonic kidney line 293T, the mammary gland adenocarcinoma MCF7, the choriocarcinoma Jeg3, the cervical adenocarcinoma HeLa and the lung carcinoma A549, the mouse MHC class I negative cell line RMAS and the MHC class I positive BW cells. The tumor cells were irradiated by 6000 Rad prior to incubation with NK cells
Cytotoxicity assay
The in vitro cytotoxic activity of NK cells against the various targets was assessed in 5 h 35S release assays as previously described [19]. Briefly, the various targets were labeled by 35S methionine overnight washed twice and plated at 5 × 103 target cells per well in 96 U-shaped plate. NK cells were next added to the targets in the indicated E:T ratios. The spontaneous release was measured from wells with no effectors, while the total labeling of the targets was achieved by resuspending the target cells in 0.1M NaOH. The specific killing percentage was calculated as follows (value spontaneous)/(total spontaneous) × 100
CD107a mobilization assay
A total of 15 × 103 NK cells were incubated with the indicated cell lines in medium containing 10% human serum and sera and 400 units/mL recombinant IL-2 for 5 h or 16 h at and effector to target (E:T) of 1:5. The allophycocyanin-conjugated anti-CD107a mAb and the PE conjugated anti-CD56 mAb were added prior to the addition of the targets. The analysis was performed on CD56+ cells.
Antibodies
The following antibodies were used: CD56-allophycocyanin, CD3-FITC, NKG2D-PE, CD16-FITC(3G8), CD16–PE B73.1,CD16-purified (3G8), CD16-biotin (B73.1), NKp30-allophycocyanin, NKp44-allophycocyanin, NKp-46 purified, DNAM1-FITC, NKp80-purified, CD94-FITC, KIR1-purified, LIR1-PE,CD161-purifed, IFN-γ-purified, IFN-γ-biotin, anti-mouse biotin,strepavidin-HRP, Annexin V allophycocyanin (BioLegend), CD107a BD (bioscience).
Human serum and IgG depletion
For NK-cell cultivation, male AB+ human serum (Sigma Aldrich) was used. For the depletion of IgG from the serum protein G sepharose (Santa Cruz Biotechnology, Santa Cruz, CA) were used. Equal volume of serum and the Protein-G beads were mixed and incubated for 2 h at RT. The tubes were then centrifuged for 10 min at 4000 RPM and the serum was collected. The IgG-depleted sera, was then passed through a 0.2 micron filter (BD) prior the addition into coculturing experiments.
Supplementary Material
Acknowledgments
This study was supported by grants from the Israeli Science Foundation, The ICRF professorship grant, by the Rosetrees trust, by the I-CORE by the GIF foundation and by the ERC advanced grant. O.M is a Crown professor of Molecular Immunology
Abbreviations
- ADCC
antibody-dependent cell-mediated cytotoxicity
- CEA
carcinoembryonic antigen
- MMP
matrix metalloproteinase
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher’s web-site
References
- 1.Raulet DH, Guerra N. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nat. Rev. Immunol. 2009;9:568–580. doi: 10.1038/nri2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.French AR, Yokoyama WM. Natural killer cells and viral infections. Curr. Opin. Immunol. 2003;15:45–51. doi: 10.1016/s095279150200002x. [DOI] [PubMed] [Google Scholar]
- 3.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 2006;6:520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 4.Cassatella MA, Anegon I, Cuturi MC, Griskey P, Trinchieri G, Perussia B. Fc gamma R(CD16) interaction with ligand induces Ca2 +mobilization and phosphoinositide turnover in human natural killer cells. Role of Ca2+ in Fc gamma R(CD16)-induced transcription and expression of lymphokine genes. J. Exp. Med. 1989;169:549–567. doi: 10.1084/jem.169.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 6.Perussia B, Trinchieri G. Antibody 3G8, specific for the human neutrophil Fc receptor, reacts with natural killer cells. J. Immunol. 1984;132:1410–1415. [PubMed] [Google Scholar]
- 7.O’Shea JJ, Weissman AM, Kennedy IC, Ortaldo JR. Engagement of the natural killer cell IgG Fc receptor results in tyrosine phosphorylation of the zeta chain. Proc. Natl. Acad. Sci. USA. 1991;88:350–354. doi: 10.1073/pnas.88.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perussia B, Trinchieri G, Jackson A, Warner NL, Faust J, Rumpold H, Kraft D, et al. The Fc receptor for IgG on human natural killer cells: phenotypic, functional, and comparative studies with monoclonal antibodies. J. Immunol. 1984;133:180–189. [PubMed] [Google Scholar]
- 9.Grier JT, Forbes LR, Monaco-Shawver L, Oshinsky J, Atkinson TP, Moody C, Pandey R, et al. Human immunodeficiency-causing mutation defines CD16 in spontaneous NK cell cytotoxicity. J. Clin. Invest. 2012;122:3769–3780. doi: 10.1172/JCI64837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peruzzi G, Femnou L, Gil-Krzewska A, Borrego F, Weck J, Krzewski K, Coligan JE. Membrane-type 6 matrix metalloproteinase regulates the activation-induced downmodulation of CD16 in human primary NK cells. J. Immunol. 2013;191:1883–1894. doi: 10.4049/jimmunol.1300313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romee R, Foley B, Lenvik T, Wang Y, Zhang B, Ankarlo D, Luo X, et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17) Blood. 2013;121:3599–3608. doi: 10.1182/blood-2012-04-425397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golay J, Manganini M, Facchinetti V, Gramigna R, Broady R, Borleri G, Rambaldi A, et al. Rituximab-mediated antibody-dependent cellular cytotoxicity against neoplastic B cells is stimulated strongly by interleukin-2. Haematologica. 2003;88:1002–1012. [PubMed] [Google Scholar]
- 13.Torelli GF, Guarini A, Maggio R, Alfieri C, Vitale A, Foa R. Expansion of natural killer cells with lytic activity against autologous blasts from adult and pediatric acute lymphoid leukemia patients in complete hematologic remission. Haematologica. 2005;90:785–792. [PubMed] [Google Scholar]
- 14.Mandelboim O, Malik P, Davis DM, Jo CH, Boyson JE, Strominger JL. Human CD16 as a lysis receptor mediating direct natural killer cell cytotoxicity. Proc. Natl. Acad. Sci. USA. 1999;96:5640–5644. doi: 10.1073/pnas.96.10.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren HS, Kinnear BF. Quantitative analysis of the effect of CD16 ligation on human NK cell proliferation. J. Immunol. 1999;162:735–742. [PubMed] [Google Scholar]
- 16.Ahn YO, Kim S, Kim TM, Song EY, Park MH, Heo DS. Irradiated and activated autologous PBMCs induce expansion of highly cytotoxic human NK cells in vitro. J. Immunother. 2013;36:373–381. doi: 10.1097/CJI.0b013e3182a3430f. [DOI] [PubMed] [Google Scholar]
- 17.Lim SA, Kim TJ, Lee JE, Sonn CH, Kim K, Kim J, Choi JG, et al. Ex vivo expansion of highly cytotoxic human NK cells by cocultivation with irradiated tumor cells for adoptive immunotherapy. Cancer Res. 2013;73:2598–2607. doi: 10.1158/0008-5472.CAN-12-2893. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, Gil-Krzewska A, Peruzzi G, Borrego F. Matrix metalloproteinases inhibition promotes the polyfunctionality of human natural killer cells in therapeutic antibody-based anti-tumour immunotherapy. Clin. Exp. Immunol. 2013;173:131–139. doi: 10.1111/cei.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J. Exp. Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.