The nuclear face of the nuclear pore complex (NPC) interfaces with chromatin, transcription, and transport intermediates. A novel architecture for the nuclear face of the trypanosome NPC provides insights into NPC function and evolution.
Abstract
The nuclear pore complex (NPC) has dual roles in nucleocytoplasmic transport and chromatin organization. In many eukaryotes the coiled-coil Mlp/Tpr proteins of the NPC nuclear basket have specific functions in interactions with chromatin and defining specialized regions of active transcription, whereas Mlp2 associates with the mitotic spindle/NPC in a cell cycle–dependent manner. We previously identified two putative Mlp-related proteins in African trypanosomes, TbNup110 and TbNup92, the latter of which associates with the spindle. We now provide evidence for independent ancestry for TbNup92/TbNup110 and Mlp/Tpr proteins. However, TbNup92 is required for correct chromosome segregation, with knockout cells exhibiting microaneuploidy and lowered fidelity of telomere segregation. Further, TbNup92 is intimately associated with the mitotic spindle and spindle anchor site but apparently has minimal roles in control of gene transcription, indicating that TbNup92 lacks major barrier activity. TbNup92 therefore acts as a functional analogue of Mlp/Tpr proteins, and, together with the lamina analogue NUP-1, represents a cohort of novel proteins operating at the nuclear periphery of trypanosomes, uncovering complex evolutionary trajectories for the NPC and nuclear lamina.
INTRODUCTION
The nuclear periphery is a specialized subcompartment of the nucleus, whose components mediate a multitude of key cellular processes, including transcription, regulation of the cell cycle, and nucleocytoplasmic trafficking. The sole route for bidirectional molecular exchange across the nuclear envelope (NE; Gerace and Burke, 1988; Akey and Goldfarb, 1989; Silver, 1991) is represented by nuclear pore complexes (NPCs), large (∼50 MDa in yeast, ∼100 MDa in vertebrates) macromolecular assemblies composed of ∼30 different proteins or nucleoporins forming an octagonally symmetrical framework that spans the NE (Akey and Goldfarb, 1989; Yang et al., 1998). Although the primary function of the NPC is nucleocytoplasmic transport, several nucleoporins, particularly those comprising the nuclear basket, have additional functions, both at the nuclear periphery and within the nucleoplasm. The nuclear basket is highly dynamic, with components interacting with numerous molecules and structures, including lamina proteins, nuclear transport machinery, and proteins involved in DNA repair and cell cycle checkpoints (Goldberg and Allen, 1996; Galy et al., 2000; Iouk et al., 2002).
Given that, in the context of eukaryotic diversity, yeast and vertebrates are closely related, our understanding of the evolution of the NPC and the functions of individual nucleoporins remains somewhat limited. Despite conservation of core elements, differences between species in nuclear organization, coupled with distinct mechanisms governing key cellular processes, suggests a high degree of structural and functional plasticity at the nuclear periphery between lineages (Allen and Douglas, 1989; Goldberg and Allen, 1996; DeGrasse et al., 2009; Fiserova et al., 2009; Tamura et al., 2010; Field et al., 2014). Trypanosomatids are highly divergent from animals and fungi and possess unusual mechanisms regulating gene expression and cell cycle progression. These protozoa are members of the Excavata supergroup, which includes many pathogens and environmentally important species, and likely diverged very early from the main eukaryotic lineage shortly after the last eukaryotic common ancestor (LECA); these deep taxonomic divisions reflect fundamental differences in cellular architecture and ancestry (Cavalier-Smith and Chao, 2010; Adl et al., 2012).
The nuclear periphery and lamina in higher eukaryotes are classically associated with transcriptional repression, although, significantly, heterochromatin does not extend to the immediate NPC vicinity, which remains transcriptionally active (Swift, 1959; Watson, 1959; Krull et al., 2010). The Drosophila nuclear basket nucleoporins Nup153 and Megator are mobile within the nucleoplasm, and their location changes coincide with involvement in transcriptional regulation and cell cycle progression (Aitchison et al., 1995; Mishra et al., 2010; Vaquerizas et al., 2010). Similarly, the mammalian nuclear basket protein Tpr forms dynamic filaments extending to the nucleolus (Cordes et al., 1997; Fontoura et al., 2001). Hence the nuclear basket is dynamic, and components form physical connections between the nuclear interior and NPC. In addition to functioning in transcriptional processes, nuclear basket nucleoporins also have varied roles in mitosis; Tpr, Megator, Arabidopsis NUA, and Mlp1/2 interact with components of the spindle checkpoint at the nuclear periphery. At the onset of mitosis, these nucleoporins recruit the spindle checkpoint proteins Mad1 and Mad2 to the kinetochores, where they delay anaphase until the sister chromatids correctly align (Iouk et al., 2002; Scott et al., 2005; Lee et al., 2008; Lince-Faria et al., 2009; Nakano et al., 2010; Ding et al., 2013). Moreover, Mlp2 directly binds to the yeast spindle organizer, the spindle pole body (SPB), and contributes to its assembly (Niepel et al., 2005, 2013), and Drosophila Megator, in complex with Skeletor and Chromator, forms a structure that likely supports and stabilizes the mitotic spindle (Walker et al., 2000; Qi et al., 2004; Johansen and Johansen, 2007). Together these findings suggest that the individual functions of basket nucleoporins at mitosis have diverged since the LECA and provide further evidence to suggest that the functions of peripheral nuclear proteins are plastic.
We previously identified 22 Trypanosoma brucei nucleoporins by proteomics, revealing that nucleoporin secondary structural architecture and domain organization are well conserved compared with yeast and Metazoa (DeGrasse et al., 2009). Furthermore, the karyopherins, responsible for mediating many nucleocytoplasmic translocation pathways, are also broadly conserved in most lineages, suggesting that core mechanisms mediating protein and mRNA nucleocytoplasmic transport are also conserved (O'Reilly et al., 2011; Field et al., 2014). Significantly, ∼240 chromosomes/DNA elements must be faithfully segregated during mitosis in T. brucei, a number far outweighing the observable kinetochores, suggesting unusual chromosome segregation mechanisms (Solari, 1995; Ersfeld and Gull, 1997). Although the trypanosome spindle microtubules are highly oriented and anchor at distinct spindle attachment sites (SASs), definitive SPB components have yet to be described. Further, in higher eukaryotes the LINC protein complex spans the NE to connect the nuclear lamina and cytoskeleton (Lee et al., 2002; Crisp et al., 2006; Stewart-Hutchinson et al., 2008) and participates in regulating nuclear positioning and anchoring of the SPB to the nucleus (Hagan and Yanagida, 1995; Malone et al., 1999; Jaspersen et al., 2002, 2006; Malone et al., 2003). Of significance, no components of the LINC complex have been identified in trypanosomes (Field et al., 2012). Transcription is predominantly polycistronic in trypanosomatids, with recent data suggesting that mRNA copy number may relate to cistron structure (Kelly et al., 2012). Deep sequencing demonstrated that most of the genome is transcribed (Siegel et al., 2010; Archer et al., 2011; Kolev et al., 2012) and that RNA polymerase start and stop sites are marked by specific histone modifications (Siegel et al., 2009). A diverse family of RNA-binding proteins (RBPs) containing the RRM motif have a major role in RNA copy number control, and several are implicated as developmental program master regulators (Estevez, 2008; Archer et al., 2009; Stern et al., 2009). Our recent description of NUP-1, a trypanosomal lamina component highly distinct from metazoan lamins, but which nevertheless performs analogous functions, led us to reconsider our understanding of lamina origins and question the functional diversity of components at the trypanosome nuclear periphery (DeGrasse et al., 2009; DuBois et al., 2012; Alsford et al., 2012; Field et al., 2012).
TbNup110 and TbNup92, two trypanosome NPC-associated proteins, are predicted to have predominantly coiled-coil structure (DeGrasse et al., 2009). TbNup92 locates at the nuclear periphery at interphase and associates with distal poles of the NE during mitosis. We proposed that TbNup110 and TbNup92 represent the Mlp/Tpr proteins of trypanosomes, and here we investigate the evolutionary relationship of TbNup92 and its functions. Our data indicate distinct evolutionary origins for TbNup92 and the Mlps despite TbNup92 performing similar roles to Mlps in spindle association and chromosome segregation. However, we find no evidence for transcriptional barrier activity, consistent with trypanosome genome organization.
RESULTS
TbNup92 is a coiled-coil protein containing a breast cancer–susceptibility protein C-terminal domain
Earlier proteomic analysis of purified trypanosome nuclear envelopes, followed by in situ tagging and localization, identified 22 nucleoporins with highly divergent primary structures (DeGrasse et al., 2009). Of these, TbNup110 and TbNup92 are predicted to possess extensive coiled-coil secondary structures similar to those predicted for nuclear basket nucleoporins Mlp/Tpr but distinct from either the β-propeller/α-solenoid of scaffold nucleoporins or the disordered structure of FG-repeat nucleoporins (Strambio-de-Castillia et al., 1999; Kosova et al., 2000; Niepel et al., 2013). Although nuclear basket nucleoporins are commonly involved in cell cycle checkpoint functions during mitosis, diversity in composition and architecture of nuclear periphery components has led to functional diversity across various lineages. Further, TbNup92 migrates to the distal poles of the nucleus during mitosis, highly suggestive of association with the SPB, as well as being disposed at the nuclear face of the NPC. On the basis of these criteria, we suggested that TbNup110 and TbNup92 are trypanosome orthologues of Mlp1 and Mlp2, respectively (DeGrasse et al., 2009).
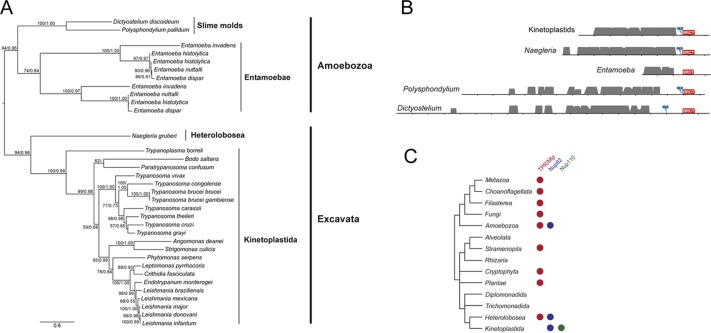
To test this more exhaustively, we analyzed the distribution of genes encoding TbNup110 and TbNup92 orthologues across eukaryotes to determine the evolutionary relationship between these gene products and the Mlp/Tpr proteins. We found a single orthologue of both TbNup110 and TbNup92 in all kinetoplastids with available genome data, including the newly identified early-branching trypanosomatid Paratrypanosoma (Flegontov et al., 2013) and the free-living bodonid Bodo saltans. Despite the apparent structural similarities, sequence searches failed to identify obvious Mlp1, Mlp2, or Tpr orthologues in these genomes (Figure 1; Neumann et al., 2010; Field et al., 2014). Although these data initially suggested that orthologue identification failure could simply arise from extreme divergence, we noted that TbNup110 and TbNup92 are significantly shorter than their putative yeast and metazoan counterparts, and both lack the central Tpr/Mlp1/Mlp2-like domain, a prominent characteristic of nuclear basket nucleoporins. In addition, the predicted coiled-coil patterns of the TbNup92 and TbNup110 differ from those of Mlps and Tpr (Supplemental Figure S1); TbNup92 lacks a major region of coiled-coil discontinuity described for Mlp1 and Mlp2, which is suggested to serve as a hinge and forms the NPC-binding site of Mlp1 (Niepel et al., 2013). By contrast, TbNup110 consists of discrete coiled-coil segments separated by major gaps, suggesting more flexible structure (Supplemental Figure S1). Although no conserved domains have been identified in TbNup110, TbNup92 does possess a breast cancer–susceptibility protein C-terminal (BRCT) domain (residues 642–712; Figure 1), which is typically found in proteins involved in DNA repair and the cell cycle checkpoint. Consequently, HMMER homology search for TbNup92 homologues identified various BRCT-domain containing proteins. For most, the BRCT domain was the only region similar to TbNup92, but several returned sequences did have a predicted architecture similar to that of TbNup92 and were returned TbNup92 as top hit by reverse-blast, indicating true TbNup92 orthology. One TbNup92 orthologue was found in the amoeboflagellate Naegleria gruberi, an excavate distantly related to kinetoplastids, and the others in slime molds and entamoebids (group Amoebozoa). A phylogenetic tree of these sequences placed the Naegleria sequence in the expected position as sister lineage to kinetoplastids and also resolved Amoebozoa as a monophyletic clade (Figure 1). Of interest, the sequences of Naegleria and Polysphondylium possess the K-K/R-X-K/R consensus sequence of the monopartite classic nuclear localization signal (NLS; Chelsky et al., 1989; Hodel et al., 2001; Lange et al., 2007; Yang et al., 2011), which is shared with the TbNup92 kinetoplastid orthologues (Figure 1 and Supplemental Figure S2A). Such an NLS is also present in the Dictyostelium orthologue but not in entamoebids (Figure 1).
FIGURE 1:
Phylogenetic analysis and domain structure of TbNup92. (A) Maximum-likelihood phylogenetic tree of TbNup92 homologues identified in kinetoplastids and other eukaryotes. Numbers at nodes are bootstrap/SH-like aLRT values. (B) Domain architecture of TbNup92. The predicted coiled-coil regions of TbNup92 are in gray. The height of the gray areas reflects the likelihood of the prediction as displayed in the Coils/Pcoils prediction tool, scanning window 28 (see Supplemental Figure S1 for more details). In case of kinetoplastids, the alignment of Nup92 sequences of all kinetoplastids was used for the prediction. Position of the C-terminal BRCT domain (in red) was identified by the NCBI conserved domain search. Presence of the classic monopartite nuclear localization signal that is conserved among most TbNup92 sequences is highlighted by a blue mark. (C) Coulson plot showing the distribution of nuclear basket proteins that were described for opisthokonts (Mlp/Tpr) and T. brucei (TbNup92 and TbNup110) among eukaryotic groups as revealed by homology searches.
Because both an Mlp/Tpr and a TbNup92 orthologue are present in Naegleria and amoebozoans (Figure 1), they cannot sensu stricto be fully equivalent, that is, true orthologues. However, the predicted presence of extensive coiled coil in both Mlp/Tpr and TbNup92/110 leaves open the possibility of distant evolutionary relationship between these proteins. Moreover, contrary to TbNup92, that TbNup110 orthologues are kinetoplastid restricted reinforces the view that TbNup92 and TbNup110 represent a family of proteins distinct from Mlp/Tpr (Figure 1C and Supplemental Figure S2B). As a means to explore this relationship in further detail, we investigated whether TbNup92 assumed similar or distinct functions to yeast and vertebrate Mlp/Tpr proteins.
TbNup92 displays cell cycle–dependent positioning
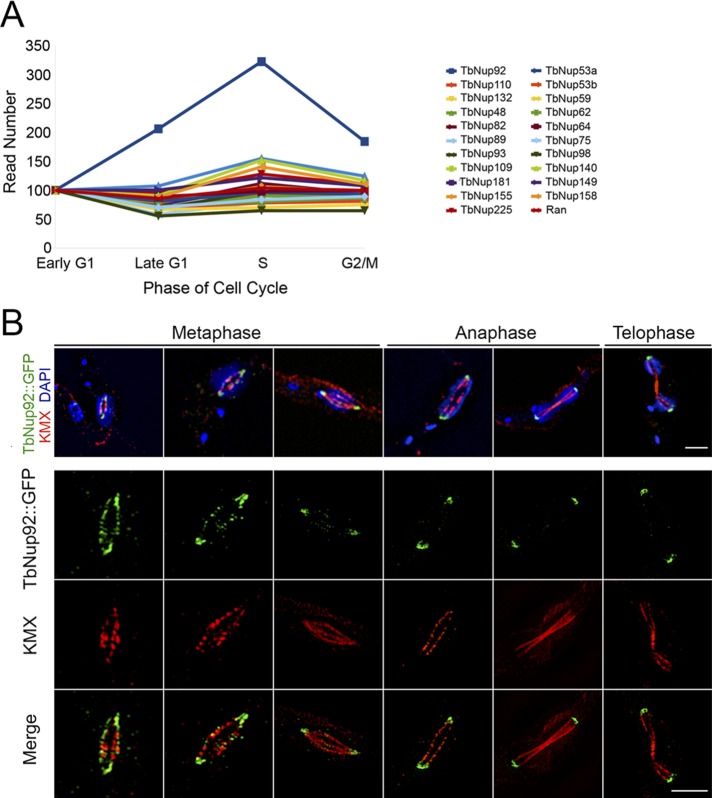
Whole-transcriptome profiling of T. brucei throughout the cell cycle (Archer et al., 2011) revealed no changes to transcript levels for most nucleoporins (Figure 2A). In contrast, TbNup92 mRNA copy number is substantially up-regulated during S phase (Figure 2A), which corresponds to redistribution of TbNup92 protein.
FIGURE 2:
Cell cycle positioning and relationship with mitotic spindle. (A) Whole-transcriptome profiling of procyclic-form T. brucei (Archer et al., 2011) shows that most nucleoporins maintain a steady rate of gene expression throughout the cell cycle (normalized to G1). In contrast, TbNup92 expression is substantially up-regulated at S phase, which coincides with the redistribution of TbNup92 during mitosis. (B) TbNup92::GFP cells were fixed and probed with anti-GFP (green) and anti–β-tubulin (KMX) antibodies for the spindle microtubules (red). Cells were imaged by superresolution microscopy, and the central slice from the z-stack is illustrated. At the onset of mitosis, TbNup92 localizes to the spindle attachment site and along the polymerizing spindle microtubules. At early anaphase, TbNup92 concentrates at the posterior and anterior poles, which likely corresponds to the localization of the SPB.
Trypanosome cells may be conveniently categorized within the cell cycle by staining the nucleus and kinetoplast (the mitochondrial DNA); the kinetoplast elongates and divides just before the nucleus enters S phase (Sherwin and Gull, 1989). We examined the distribution of TbNup92 throughout the cell cycle by genomically tagging the C-terminus of a single allele with green fluorescent protein (GFP) in PCF cells, counterstaining with 4′,6-diamidino-2-phenylindole (DAPI), and conducting an analysis by confocal and superresolution microscopy. Whereas the majority of T. brucei nucleoporins remain anchored at the NPC through mitosis, TbNup92 showed distinct localizations at interphase and mitosis (Figures 2 and 3). By comparison between intranuclear tubulin, revealed with an anti–β-tubulin antibody (KMX) to highlight the spindle microtubules (Figure 2B), and TbNup92, we observed that TbNup92 localizes to SASs, that is, poles close to the nuclear envelope, and displays a punctate distribution along the length of the spindle microtubules at the onset of mitosis. This cell cycle–dependent localization of TbNup92 is remarkably similar to that of Drosophila Megator (Qi et al., 2004). As cells progress into anaphase, TbNup92 is mostly concentrated at the SAS at the posterior and anterior poles of the dividing nuclei. It is most probable that TbNup92 is anchored at, or interacts with, components forming a spindle anchor at G2/mitosis.
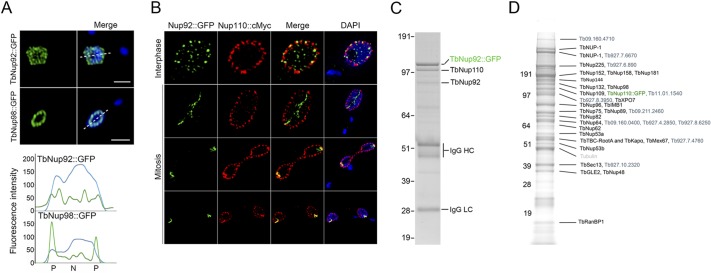
FIGURE 3:
TbNup92 interacts with NPC components and is distributed within the nucleoplasm. (A) GFP (top and bottom, green) and DAPI (top and bottom, blue) signals were recorded across optical sections of the center of nuclei (dashed line) and imaged by confocal microscopy. Whereas TbNup98::GFP signal was restricted to the nuclear periphery, TbNup92::GFP fluorescence was documented at both the nuclear periphery and within the nucleoplasm. (B) Fixed cells expressing TbNup92::GFP and TbNup110::cMyc were probed with anti-GFP and anti-cMyc antibodies. Cells were imaged by superresolution microscopy. Shown are the optical sections of the center of nuclei taken from image series along the z-axis. TbNup92 is juxtaposed to TbNup110 at the nuclear periphery and is also present within the nucleoplasm. In all images, DAPI was used to visualize DNA (blue). Scale bar, 2 μm. (C) Pullouts using TbNup92::GFP as the handle were done under the following buffer conditions: 20 mM HEPES, pH 7.4, 20 mM NaCl, 50 mM citrate, 0.5% Triton, 0.1% Tween, and protease inhibitors. Mass spectrometry of the bands reveal that TbNup92 interacts with the nuclear basket nucleoporin TbNup110. (D) For TbNup110::GFP pullouts, the following conditions were used: 20 mM HEPES, pH 7.4, 250 mM sodium citrate, 0.5% Triton, 0.5% N,N-bis-(3-d-gluconamidopropyl)deoxycholamide, and protease inhibitors. TbNup110 interacts with a number of core components of the T. brucei NPC.
We also examined the location of TbNup92 at interphase, as it was apparent that at G1 there was a substantial presence within the nucleoplasm, albeit with clear exclusion from the nucleolus (Figure 3, A and B). The fluorescence intensities of GFP and DAPI across a central nuclear slice stained for TbNup92::GFP or TbNup98::GFP, an FG-repeat containing nucleoporin, were determined (Figure 3A). The DAPI signal is relatively constant throughout the nucleoplasm, with a decrease corresponding to the nucleolus, whereas two clear maxima are found for TbNup98, reflecting disposition at the nuclear envelope. In contrast, a more homogeneous level of fluorescence was found for TbNup92 across the entire nucleus, confirming a presence at the nuclear periphery but also substantially within the nucleoplasm. Using TbNup110::cMyc as an NPC marker, we also find that TbNup92 is juxtaposed to TbNup110 that is localized on the nuclear face of the NPC (Figure 3B). Of interest, we documented a small but significant concentration of TbNup110::cMyc at the SAS that colocalized with TbNup92::GFP (Supplementary Movie S1). To test whether this cell cycle–dependent positioning was unique to the basket nucleoporins, we followed the distribution of the FG-repeat nucleoporin TbNup98, which is a core component of the T. brucei NPC (Obado, Field, Rout, and Chait, unpublished data). Consistent with previous findings, TbNup98 maintained a peripheral punctate distribution throughout the cell cycle and did not concentrate at the SAS at mitosis, indicating that redistribution to the SAS was specific to nuclear basket components (DeGrasse et al., 2009; Supplemental Figure S3).
Given the mobility of TbNup92, we tested the association of TbNup92 with the NPC by affinity isolation of TbNup92::GFP fusion protein from cryomilled cells, followed by mass spectrometric identification of coisolating proteins (Figure 3C). This confirmed a highly specific association between TbNup92 and TbNup110, although the absence of more extensive interactions with the NPC suggests that TbNup92 is peripherally associated and not a core component (Supplementary Table S1). Further, confirmation of connectivity between TbNup92 and the NPC was provided by affinity isolation with TbNup110::GFP as affinity handle. TbNup110 associates with a large number of nucleoporins, indicating more extensive contacts between TbNup110 and the NPC (Figure 3D). TbNup92 is therefore associated peripherally with the NPC, probably via TbNup110, itself more strongly anchored to the NPC.
TbNup92 is required for faithful mitotic segregation of sister chromatids
We initially used RNA interference (RNAi)–mediated knockdown to examine TbNup92 function. We validated the efficacy and specificity of RNAi by quantitative reverse transcriptase PCR (qRT-PCR) and found 60% specific reduction to TbNup92 mRNA level at 24 h postinduction (Supplemental Figure S4A). No significant proliferative defects were observed after induction over a 7-d period (Supplemental Figure S4B), although a small alteration to progression through the cell cycle was observed (Supplemental Figure S4C). Cells in which the kinetoplast had divided but not the nucleus (2K1N) decreased between 6 and 24 h postinduction, whereas cells in which both nucleus and kinetoplast completed mitosis but cytokinesis remained to be achieved increased. These data suggest a minor delay to entry into G1/S phase.
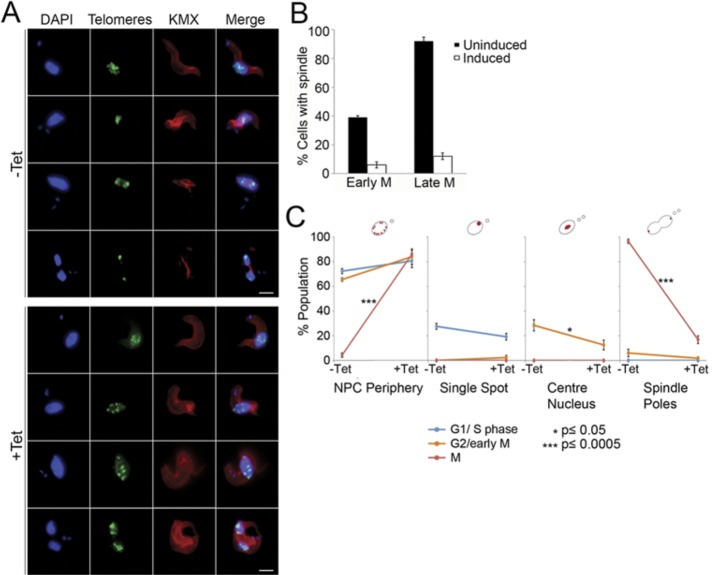
We inspected TbNup92-knockdown cells for their ability to position chromosomes, using fluorescence in situ hybridization (FISH) telomere labeling together with KMX antibody staining. Unexpectedly we found that the anti–β-tubulin antibody in most (84%) mitotic cells failed to detect a spindle, suggesting that the spindle was in some manner abnormal, preventing immunochemical detection (Figure 4, A and B). Of interest, this failure to detect the mitotic spindle coincided with telomere misplacement (Figure 4, A, bottom, and C). Rather than aligning at the center of the mitotic nucleus before separating and migrating to the poles of the separating daughter nuclei, the telomeres in TbNup92-knockdown cells remained distributed around the nuclear periphery throughout the cell cycle. These data, combined with the finding that TbNup92 localizes along the polymerizing spindle microtubules, imply that, like Drosophila Megator, TbNup92 may fulfill an important role in spindle formation and/or maturation, as well as ensure correct attachment of chromosomes and their subsequent segregation; analysis of a knockout cell indicated that this did not result in changes to ploidy of specific chromosomal regions (see later discussion). A similar defect in spindle formation in trypanosomes was described by us recently in which knockdown of TbRHP, a Rho-like GTPase (TbRHP), also led to perturbation of KMX spindle staining (Abbasi et al., 2011). Although a spindle, albeit with altered structure, must be present, these data suggest that in trypanosomes, reactivity to KMX may be compromised. However, TbNup92 is differentiated from TbRHP, as mitosis and cell division in TbRHP were compromised, whereas in the present case knockdown did not result in a significant cell cycle defect.
FIGURE 4:
TbNup92 is required for maintaining the correct distribution of telomeres during mitosis. TbNup92 RNAi was induced in BSF cells. (A) Uninduced and induced cells were fixed and probed with anti–β-tubulin antibody (KMX; red) and FISH for the telomeric repeat regions (green). DAPI was used to visualize DNA (blue). Scale bar, 2 μm. The spindle microtubules were undetectable by KMX in knockdown cells, and this coincided with mislocalization of the telomeres during mitosis. (B) Quantitation of the presence or absence of spindle microtubules in interphase and mitotic cells (N = 100). Plotted data represent the average of three replicate experiments. (C) The nuclear localization of telomeres was scored in 100 cells at each stage of the cell cycle. Average scores of triplicate experiments are plotted (*p ≤ 0.05; ***p ≤ 0.0005). Consistent with failure to detect the mitotic spindle in mitotic knockdown cells, the telomeres often failed to condense at the center of the nucleus. At anaphase/telophase, rather than concentrating as single punctum at the spindle poles, the telomeres were most likely to be distributed within the nucleoplasm of TbNup92-depleted cells.
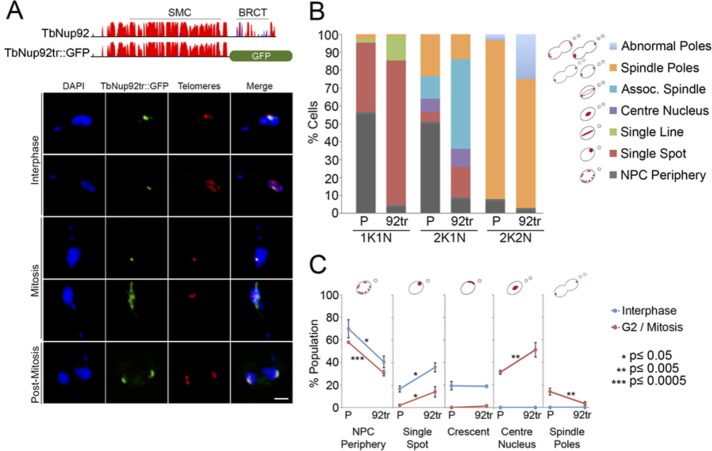
The TbNup92 BRCT domain is required for distribution during interphase
To investigate the role of the BRCT domain, we fused GFP in situ to the TbNup92 open reading frame (ORF) to eliminate the BRCT-containing C-terminus (residues 640–813; Figure 5A) and removed the second allele of TbNup92 by replacing the ORF with a neomycin resistance gene, to create cell line TbNup92tr::KO. Proliferation of TbNup92tr::KO cells was reduced ∼35% compared with parental cells (Supplemental Figure S5A), and similar to knockdown, the cell line accumulated 1K1N cells, suggesting that TbNup92tr::KO cells experienced delays in progressing through G1/S phase (Supplemental Figure S5B). These data suggest that, although there is a fitness cost, T. brucei is viable with expression of a single, mutated TbNup92 allele.
FIGURE 5:
The C-terminal BRCT domain is required for the interphase distribution of TbNup92. (A) Top, predicted secondary structure of TbNup92. The y-axis indicates the confidence score of the predicted secondary structure. Coiled coils are in red and the β-sandwich in blue. TbNup92 contains a central SMC domain and a C-terminal BRCT domain. A single allele of TbNup92 was truncated by replacing residues 639–813 with a GFP tag. The second allele of TbNup92 was replaced with a neomycin resistance gene. Bottom, the nuclear localization of TbNup92 is disrupted in cells expressing TbNup92 lacking the C-terminus containing the BRCT domain (TbNup92tr::GFP, green). Cells were costained with a FISH probe for the telomeres (red). DAPI was used to visualize DNA (blue). Scale bar, 2 μm. (B) The localization of TbNup92::GFP and TbNup92tr::GFP was documented in 100 1K1N, 2K1N, and 2K2N cells. Mean scores of triplicate experimental replicates are shown. Loss of the C-terminus containing the BRCT domain prevents TbNup92 from redistributing to the nuclear periphery and nucleoplasm during interphase. At mitosis, TbNup92tr::GFP is more likely to be distributed along the spindle microtubules, and this coincides with the mislocalization of TbNup92tr::GFP at the spindle poles during late mitosis. (C) A telomere FISH probe was used to visualize the telomeric repeats in TbNup92::GFP- and TbNup92tr::GFP-tagged cells. The localization of the telomeres in 100 interphase and 100 mitotic cells was scored. The mean scores from triplicate experimental replicates are plotted (*p ≤ 0.05; **p ≤ 0.005; ***p ≤ 0.0005). At interphase, the telomeres in TbNup92tr::GFP cells are more likely to be localized as single puncta at the periphery compared with those in TbNup92::GFP cells. In mitotic cells, the telomeres remain fixed at the center of the nucleus for a longer period in TbNup92tr::GFP cells before eventually migrating to the spindle poles.
BRCT domain deletion disrupted TbNup92 localization throughout the cell cycle (Figure 5A, bottom, and B). During G1/S phase, TbNup92tr::GFP predominantly localized as a single punctum at the nuclear periphery (p ≤ 0.005), indicating that the C-terminal BRCT domain is required for the interphase distribution of TbNup92 at the nuclear periphery and within the nucleoplasm. Similar to yeast Mlp2, which localizes in the same hemisphere of the nucleus as the SPB (Niepel et al., 2005), TbNup92tr::GFP often localized as a nuclear periphery crescent (p ≤ 0.005). However, with no cytological markers for the SPB, we were unable to confirm the location of the SPB in these cells. Despite the altered distribution of TbNup92tr::GFP compared with intact TbNup92, affinity isolation using TbNup92tr::GFP as the handle revealed that the protein still interacted with TbNup110 (unpublished data). Clearly the context of interactions between TbNup92 and TbNup110 is complex, so that, for example, a population of TbNup110 is targeted to the SAS, and the BRCT domain does not appear to be essential for TbNup92 and TbNup110 interaction. The modulation, therefore, of interactions between these proteins may rely on posttranslational modifications—for example, O-GlcNAc, phosphorylation, ubiquitylation/sumoylation, or other moiety—and is consistent with the known prolific modifications of mammalian and yeast NPC components and the known phosphorylation of TbNup92 (Nett et al., 2009).
Association of TbNup92 with the basket nucleoporin TbNup110 (Figure 3) led us to ask whether localization of TbNup110 at the NPC is relies on TbNup92. TbNup110 was genomically tagged at the C-terminus with a hemagglutinin (HA) epitope in TbNup92tr::KO cells and imaged by confocal microscopy (Supplemental Figure S5C). The peripheral punctate distribution of TbNup110 was unaffected, indicating that deletion of the BRCT domain of TbNup92 does not affect TbNup110 positioning. Of interest, the telomeres were almost twice as likely to be distributed as a single puncta at the nuclear periphery and often colocalized with TbNup92tr::GFP (p ≤ 0.05) compared with parental cells (Figure 5, A, bottom, and C), suggesting that whereas TbNup92tr::GFP does not appear to influence NPC positioning, full-length TbNup92 is required for correct distribution of telomeres at the interphase nucleus periphery.
At prophase/early metaphase, TbNup92 is present along the spindle microtubules for a short time. We found that TbNup92tr::GFP displayed increased residence along the length of the spindle microtubules (p ≤ 0.005; Figure 5, A, bottom, and B), and this was accompanied by a significant increase in the frequency of cells with telomeres localized toward the center of the nucleus (p ≤ 0.005; Figure 5, A and C), suggestive of a delay in progressing past metaphase. Together these data support a role for TbNup92 in spindle formation/maturation and possibly also the correct timing of events at metaphase/anaphase. TbNup92tr::GFP eventually localized to the spindle attachment site at anaphase, albeit with abnormalities in the distribution of TbNup92tr::GFP in 24% of cells (Figure 5B); the TbNup92tr::GFP signal at the poles was often diffuse or asymmetrical in intensity, whereas some cells displayed multiple TbNup92tr::GFP foci at either pole, all suggestive of a failure in correct targeting of truncated TbNup92 protein.
The SMC domain of TbNup92 is required for localization to the SAS
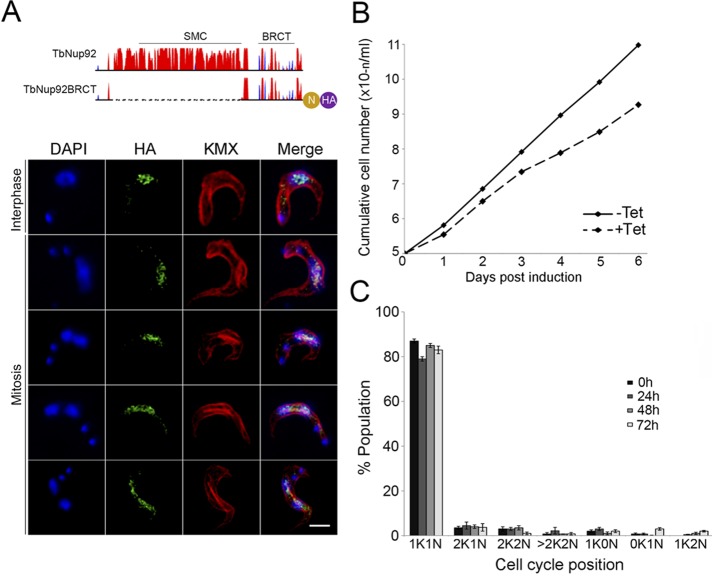
Given a clear role for the BRCT domain in targeting TbNup92, we sought a role for the central SMC domain. A synthetic gene, TbNup92BRCT, was designed that contained the first 30 amino acids of TbNup92, immediately followed by the final 193 amino acids, eliminating the predicted SMC domain. An NLS derived from NUP-1 and a 3×HA epitope were included at the C-terminus (Figure 6A and Supplemental Figure S6). TbNup92BRCT was expressed in bloodstream from cells under the control of an inducible promoter.
FIGURE 6:
The central SMC domain of TbNup92 is required for the localization of TbNup92 to the SAS at mitosis. (A) Top, predicted secondary structure of TbNup92. The y-axis indicates the confidence score of the predicted secondary structure. Coiled coils are in red and the β-sandwich in blue. TbNup92::NLS::HA lacking the central SMC domain (residues 210–545) was cloned into pDex-577 (Kelly et al., 2007) and overexpressed in BSF cells (TbNup92BRCT) under the control of a tetracycline-inducible promoter. Bottom, cells were probed with anti-HA (green) and KMX (red) to visualize TbNup92BRCT and the spindle microtubules, respectively. DAPI was used to visualize DNA (blue). Scale bar, 2 μm. Deletion of the central SMC domain prevented TbNup92 from localizing at the spindle attachment site at mitosis. (B) Overexpression of TbNup92BRCT resulted in a slight population growth defect over a 6-d period. (C) Examination of DAPI-stained parasites revealed no significant changes to the distribution of parasites through the cell cycle after the overexpression of TbNup92BRCT for a 72-h period (N = 100). The mean scores are plotted.
After induction, the distribution of TbNup92BRCT was determined throughout the cell cycle (Figure 6A, bottom). During interphase, TbNup92BRCT was distributed within the nucleoplasm and at the nuclear periphery, similar to full-length TbNup92. However, as cells entered mitosis, TbNup92BRCT failed to migrate to the spindle poles and instead remained distributed throughout the nucleus. Given that full-length TbNup92 localized to the SAS during mitosis, this failure to redistribute led us to ask whether the spindle microtubules were still forming and, if so, whether they were normally distributed. Cells were stained with the anti–β-tubulin antibody KMX (Figure 6A, bottom, red) to highlight the spindle microtubules; despite the unusual distribution of TbNup92BRCT, the localization of the spindle microtubules in these cells was indistinguishable from that in uninduced cells.
Overexpression of TbNup92BRCT resulted in a minor proliferation defect over a 6-d period (Figure 6B) but without an obvious cell cycle arrest or increase in unusual cell types (Figure 6C). These findings were supported by fluorescence-activated cell sorting (FACS) cytometry (unpublished data), where the population karyotypes were largely indistinguishable out to 3 d of induction. Given that these cells were still expressing full-length TbNup92, the severity of the phenotype is likely attenuated, but these data indicate that TbNup92BRCT elicits a distinct phenotype to TbNup92tr::KO. In summary, in contrast to the C-terminal BRCT domain that likely has functions important for interphase, the central SMC domain of TbNup92 does not influence the localization of TbNup92 during interphase but is required for the redistribution of TbNup92 to the spindle attachment site during mitosis.
TbNup92 is nonessential and not required for accurate positioning of the NPC
The absence of a severe proliferative defect after TbNup92 RNAi suggested that the protein is nonessential. However, the effect on spindle structure and chromosome positioning point toward an important role in mitosis. Further, because TbNup92 is apparently associated with the nucleocytoplasmic face of the NPC at interphase, we wanted to determine whether the protein plays a role in NPC positioning or segregation.
We chose to create a TbNup92-null cell line by replacing both alleles of TbNup92 with neomycin and hygromycin resistance genes. To confirm deletion of both TbNup92 alleles, we prepared indexed DNA libraries from genomic DNA of TbNup92∆ cells and resequenced the genome using Illumina multiplex sequencing (BGI, Beijing, China). The resequenced genome was mapped to the reference 427 T. brucei genome, and although the read depth was relatively constant across the entire genome, no reads were generated that mapped to the TbNup92 ORF (Supplemental Figure S7A). In addition, TbNup92 mRNA was essentially undetectable in the knockout cell line (Supplemental Figure S7B), confirming deletion of the TbNup92 ORF from the genome. Transfection efficiency, even with PCF cells, was extremely low (∼3 × 10−8) and no TbNup92∆ clones were attained in BSF cells, which have considerably lower transfection efficiencies, despite repeated attempts.
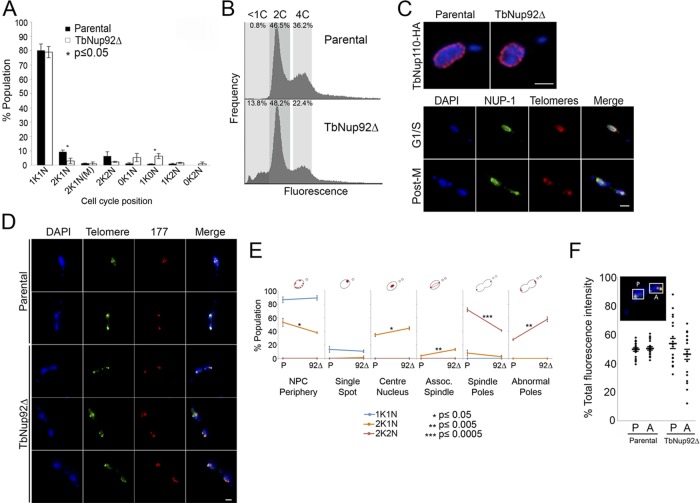
TbNup92∆ cells display a substantial proliferative defect (Supplemental Figure S7C), although the population was still viable, suggesting that TbNup92 is sensu stricto nonessential for PCF cells. The growth phenotype was reflected in delays in progression through the cell cycle (Figure 7A). A significant decrease in the number of 2K1N (p ≤ 0.05) and an increase in cells with abnormal numbers of nuclei or kinetoplasts were observed, indicating that TbNup92∆ cells are delayed at G1/S phase. Further, the TbNup92∆ population contained a number of cells with unusual DNA content (13.8%) compared with the parental wild-type population (0.8%; Figure 7B).
FIGURE 7:
TbNup92 is required for the correct segregation of chromosomes at mitosis. (A) Parental and TbNup92Δ cells were fixed and stained with DAPI to allow the categorization of cells into distinct stages of the cell cycle (N = 100). Mean scores are plotted. TbNup92Δ cells experienced delays in progressing past G1/S phase (1K1N). Accumulation of unusual cell types (0K1N, 1K0N, 1K2N, 0K2N) suggests that TbNup92Δ cells also experience errors in completing mitosis and cytokinesis. (B) Parental and TbNup92Δ cells were fixed and stained with propidium iodide and subject to FACS analysis to determine the karyotype of the population. Cells with <2C DNA content were recorded in the TbNup92Δ population, indicating that there are errors in successfully segregating the chromosomes at mitosis. (C) Top, parental and TbNup92Δ cells expressing TbNup110::HA were fixed and probed with anti-HA to visualize TbNup110. The localization of TbNup110 at the nuclear periphery was indistinguishable in both parental and TbNup92Δ cell lines. Bottom, cells were fixed and stained using the anti–NUP-1 repeat region antibody (green) and FISH for telomeres. The distribution of NUP-1 in TbNup92Δ cells was unaffected throughout the cell cycle. (D–F) Cells were fixed and stained with FISH probes for telomeric repeats (green) and minichromosome 177–base pair repeats (red). (D) The distribution of the telomeres was indistinguishable from the 177–base pair repeat regions in both the parental and TbNup92Δ cells. (E) The localization of telomeres was scored in 100 1K1N, 2K1N, and 2K2N cells. The experiment was repeated in triplicate, and mean scores are plotted (*p ≤ 0.05; **p ≤ 0.005; ***p ≤ 0.0005). In 2K1N TbNup92Δ cells, the telomeres were more likely to be distributed at the center of the nucleus or along the spindle microtubules. During late mitosis, multiple telomeric foci were observed in TbNup92Δ cells. (F) The fluorescence intensity of telomeres at the posterior and anterior poles of 20 dividing nuclei (imaged using a wide-field microscope) were recorded. In the parental cells, the telomere fluorescence at the poles was almost equal, confirming that there is equal division of sister chromatids to the spindle poles. In TbNup92Δ cells, evidence of aneuploidy is apparent, since the fluorescence intensity of telomeres at the poles was markedly variable. For all images, DAPI was used to visualize DNA (blue). Scale bar, 2 μm.
Consistent with observations in TbNup92tr::KO cells, the distribution of TbNup110::HA at the nuclear periphery was indistinguishable in both parental and TbNup92∆ cells (Figure 7C). First, this confirms that both TbNup110 targeting and NPC positioning do not require TbNup92. Second, this is consistent with the absence of a significant interaction between TbNup92 and NUP-1, the only known component of the trypanosome lamina and required for NPC positioning (DuBois et al., 2012). Furthermore, we costained TbNup92Δ cells with the telomere FISH probe and an antibody against the NUP-1 repeat. The distribution of the repeats was indistinguishable from that in the parental cells (Figure 7C), additional confirmation that TbNup92 is nonessential for connections between the NPC and the lamina.
In trypanosomes, the megabase and minichromosomes separate by two distinct mechanisms during mitosis (Gull et al., 1998). Visualization of both megabase and minichromosomes highlighted clear abnormalities in chromosome distribution in mitotic TbNup92∆ cells (Figure 7, D and E). Orthologues of various proteins involved in the metaphase–anaphase checkpoint have been identified in trypanosomes, including Mad2, CDK1, cyclin, APC/C, separase, and cohesin (Mottram et al., 2003; Gluenz et al., 2008), suggesting that the functioning of the checkpoint machinery is likely to be conserved in T. brucei (Gluenz et al., 2008). At late G2/early mitosis, the telomeres and minichromosomes were significantly more central within the nucleus (p ≤ 0.05) or aligned with the spindle microtubules (p ≤ 0.005) than in the parental cells, indicating premature entry into mitosis (Figure 7, D and E). This phenotype is typical of cells in which correct functioning of the metaphase–anaphase checkpoint machinery is compromised. Further evidence suggesting that TbNup92 plays a role in a metaphase–anaphase checkpoint comes from the finding that chromosomes displayed a staggered, nonuniform migration to the SAS in TbNup92Δ cells (Figure 7, D and E). The fluorescence intensity of telomeres at the posterior and anterior poles of dividing nuclei (typical of anaphase) was determined for both parental wild-type 427 and TbNup92Δ cells; fluorescence at the poles of dividing nuclei in parental cells was similar, suggesting that chromosomal segregation is equal between the two daughter nuclei, but segregation is unequal in TbNup92Δ cells (Figure 7F). Ignoring the highly repetitive telomeric and centromeric regions, analysis of the resequenced genome from the TbNup92Δ cells shows that read depth across the genome was relatively uniform; therefore, although there is no evidence for major ploidy alterations to specific genomic regions in the TbNup92∆ cells, both FACS and telomeric FISH analysis suggest defects to correct chromosomal segregation and therefore are evidence for aneuploidy, although at a low level within the population and apparently not within specific regions of the genome.
Phenotypic reversion in long-term culture of TbNup92∆ cells
We noticed that TbNup92∆ cells, after continual culture for ∼400 generations, no longer displayed a growth phenotype (Supplemental Figure S8A). With the exception of a small proportion of zoids (1K0N cells), cell cycle progression was normal and the telomeres segregated with wild-type fidelity (Supplemental Figure S8, B–D). Resequencing and comparison of the TbNup92∆ and the revertant line (TbNup92∆rev) genomes revealed no obvious rearrangements, but a large number of single nucleotide polymorphisms were found (Supplemental Figure S9). Despite 333 nonsynonymous homozygous and 11,426 nonsynonymous heterozygous differences between these genomes, none mapped to ORFs that are potentially associated with TbNup92 function (e.g., TbNup110 or other nucleoporins). This genomic plasticity may contribute to TbNup92 nonessentiality, but, of importance, it suggests considerable selective pressure to regain accurate chromosomal segregation.
TbNup92 does not demarcate areas of high transcriptional activity
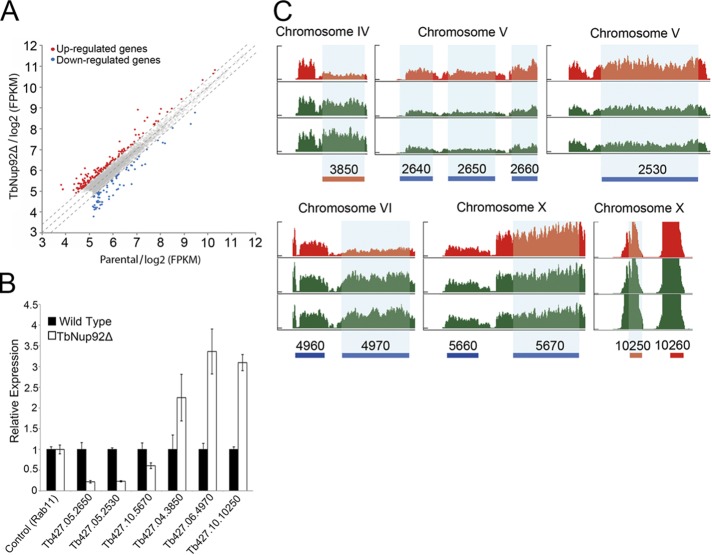
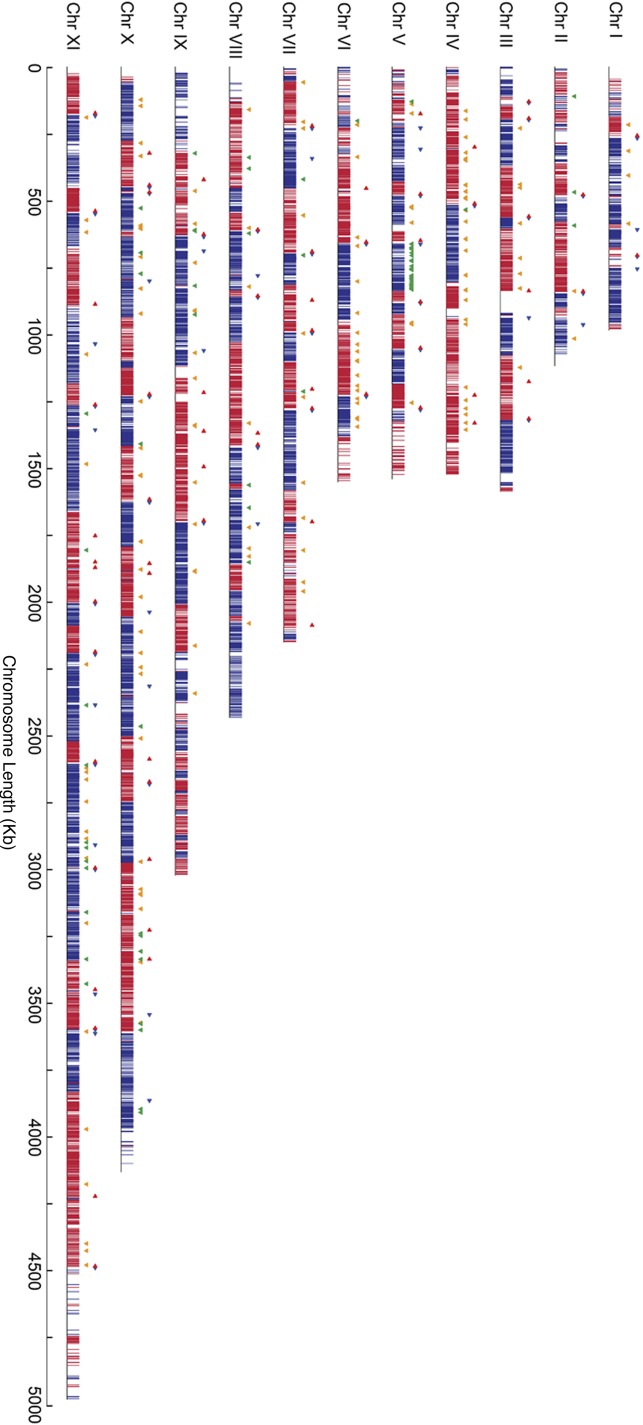
NUP-1 is involved in regulation of VSG genes at the nuclear periphery; knockdown leads to increased transcription from silent telomeric VSG loci (DuBois et al., 2012). We asked whether TbNup92 plays a role in promoting localized transcription, similar to Drosophila Megator and yeast Nup153. We compared genome-wide transcript levels between parental and TbNup92Δ cells by RNA sequencing (RNA-seq), mapping to the 427 genome (see Materials and Methods and Supplemental Table S2). mRNA abundance from 157 genes was significantly up-regulated (p < 0.05), whereas 75 genes were significantly down-regulated (p < 0.05) in TbNup92∆ cells (Figures 8A and 9 and Supplemental Table S2); several of the more prominently altered transcripts were validated by qRT-PCR (Figure 8, B and C). We did observe an overrepresentation of Gene Ontology (GO) terms associated with mRNA turnover (Supplemental Table S2), which included ALBA 1 and 3 and ZFP-1, with known roles in the control of surface antigen expression mediated via modulations of mRNA copy number (Hendriks et al., 2001; Walrad et al., 2009, 2012; Mani et al., 2011). There was no evidence for differential expression across entire cistrons, with the exception of 31 down-regulated genes localized to a specific chromosome V polycistronic transcription unit (Figure 9). There is no evidence for specific functions, the presence of gene clusters, or unusual DNA elements within this cistron. Further, we asked whether there was a positional effect of TbNup92 knockout on transcript levels relative the predicted transcriptional start sites (TSSs; Siegel et al., 2009; Supplemental Figure S10). The distributions of the positions of all genes and differentially expressed genes relative to the TSS are not significantly different (p = 0.31). Hence TbNup92 is unlikely to play a major role in control of transcription, but differential transcript levels of nonclustered individual genes may indicate a role in mRNA processing/packaging.
FIGURE 8:
TbNup92 does not influence polycistronic transcription units. (A) Transcript abundances of individual genes in parental and TbNup92Δ cells based on FKPM. All FPKMs of annotated transcripts were normalized by quantile normalization. Of 824 genes, the expression of 157 genes was significantly up-regulated, and 75 genes were significantly down-regulated after deletion of TbNup92. (B) The relative expression of the most up-regulated (Tb427.04.3850, Tb427.06.4970, Tb427.10.10250) and down-regulated (Tb427.05.2650, Tb427.05.2530, Tb427.10.5670) genes identified by RNA-seq were verified by qRT-PCR. Expression levels were normalized to Rab11. (C) RNA-seq profiles of the most up-/down-regulated genes (highlighted by vertical light blue bar) in parental cells (red) and two separate TbNup92Δ clones (green; p < 0.05).
FIGURE 9:
Genome-wide RNA-seq profiles after deletion of TbNup92. Differentially expressed genes after deletion of TbNup92 are shown along each chromosome. Blue and red bars represent the forward and reverse strands, respectively. A total of 157 genes were significantly (p < 0.05), >1.33-fold up-regulated (orange triangles), and 75 genes were significantly (p < 0.05) >1.33-fold down-regulated (green triangles) after deletion of TbNup92. The predicted transcription start sites (Siegel et al., 2009) are depicted by blue and red triangles according to their positions on the forward and reverse strands, respectively. TbNup92 does not appear to influence cistron-wide gene expression.
DISCUSSION
The NPC is highly conserved through evolution, albeit with considerable sequence divergence (DeGrasse et al., 2009). The nuclear face of the NPC interacts with chromatin, the transcriptional apparatus, the lamina. and the mitotic apparatus (Ishii et al., 2002; Taddei et al., 2006; Zuccolo et al., 2007; Nakano et al., 2010; Strambio-De-Castilla et al., 2010; Vaquerizas et al., 2010). This feature of the NPC may therefore experience greater selective pressure, reflecting the diversity of transcriptional mechanisms—for example, promoter-based versus polycistronic modes, open versus closed mitosis, and many other nuclear functions. Here we investigated the evolutionary history and functions of TbNup92, a trypanosome NPC-associated protein.
TbNup92 possesses a BRCT domain, a predicted unbroken coiled coil, and a molecular weight of <100 kDa, differentiating its architecture from that of the Mlps, Tpr, and NUA. Orthologues of both Mlp or Tpr and TbNup92 are found in several genomes, and this, together with distinct structural features, suggests that TbNup92 is evolutionarily distinct from the Mlps or Tpr. This may represent an example of convergent evolution or reflect that the Mlps, Tpr, and TbNup92 represent distinct descendants from a very ancient common ancestor that diverged before the LECA, that is, during the transition period following eukaryogenesis. Of significance, the root of eukaryotes may sit quite close to the separation between trypanosomes and most other lineages, including Naegleria. The presence of TbNup92 in Naegleria and the Amoebazoa is highly significant, in that it embraces lineages on both sides of the putative root, indicating an origin before their division but with apparent extensive secondary losses, counting for the absence from the vast majority of extant taxa. The patchy distribution of TbNup92, as well as lamins, now known to be in Amoebazoa as well as Metazoa, plus the likely widespread but patchy presence of LINC complex proteins, all suggest significant plasticity within the proteins that subtend the nuclear envelope and significant complexity in the evolution of these systems both before and after the LECA.
Of significance, TbNup92 associates solely with TbNup110 and no other NPC components, whereas TbNup110 interacts with many core nucleoporins. This is highly distinct from Mlp1 or Mlp2, both of which display high connectivity with the NPC (Niepel et al., 2013) and suggests a peripheral association for TbNup92 with the NPC. The presence of an NLS and nucleoplasmic and spindle localizations all argue for a nuclear location for TbNup92 and hence association with the NPC nuclear basket.
BRCT domains are typically involved in cell cycle checkpoints, DNA transcription, DNA replication, and DNA repair (Huyton et al., 2000; Glover et al., 2004; Mohammad and Yaffe, 2009), and in TbNup92 the BRCT C-terminal domain is necessary and sufficient for nuclear peripheral and nucleoplasmic targeting. Conversely, the central SMC domain is necessary for localization to the spindle attachment sites. TbNup92 has a strong presence at the nuclear periphery, suggesting that TbNup92 anchors at the NPC, similar to both Mlp and Tpr, and may form filaments projecting into the nucleoplasm, so far only reported for Tpr (Cordes et al., 1997; Fontoura et al., 2001). Although it is unclear whether TbNup92 oligomerizes, as is frequently observed for coiled-coil proteins, we note that the flexible hinge within Mlp1 could be recapitulated by the formation of TbNup92 dimers to create a nuclear basket with overall higher structure rather homologous with yeast and metazoa.
Deletion of the BRCT domain also delayed migration of TbNup92 to the spindle poles. Overall, the effect of this deletion affects telomere positioning, delaying telomere redistribution at metaphase, as well as during the mitosis-to-interphase transition, and evidence of chromosome segregation defects for both megabase and minichromosomes were clear from the TbNup92 knockout. Further, evidence for roles in spindle formation and/or spindle anchoring for TbNup92 arises from both targeting to the SAS at metaphase and inaccurate telomere positioning in late mitotic cells. The latter could arise via errors in SAS formation or in construction of the spindle itself, especially as TbNup92 locates along the entire assembling spindle at early metaphase, before redistribution to the SAS at anaphase. Therefore these data suggest that TbNup92 has a role in chromosome segregation. Of significance, Mlp2 knockouts accumulate multiple intranuclear SPBs (Niepel et al., 2005), whereas Tpr interacts with dynein and dynactin, which recruit chromosomes also to the spindle poles (Sharp et al., 2000; Ligon et al., 2001); hence TbNup92 and Mlp/Tpr likely share roles in the correct construction of spindle-anchoring structures, as well as in migration of chromosomes to the spindle poles.
Evidence for a role for TbNup92 in metaphase-to-anaphase transition stems from the observation that in TbNup92tr::KO cells there is an increase in frequency of telomeres at the center of the mitotic nucleus, that is, a delay in proceeding past metaphase. Mlp and Tpr are required to maintain the peripheral distribution of Mad1/2 and Mps1 during interphase and redistribution to kinetochores at mitosis. Knockouts of Mlp1/2 and Tpr affect the dynamics of exit from metaphase (Iouk et al., 2002; Scott et al., 2005; Lee et al., 2008; Lince-Faria et al., 2009; Nakano et al., 2010; Ding et al., 2013). Therefore the similar effects on metaphase kinetics obtained by manipulating TbNup92 and Mlp/Tpr expression may indicate a role for TbNup92 in anchoring and targeting checkpoint proteins. Although the components of the trypanosome kinetochore, for example, are unknown, many of the components of the spindle checkpoint machinery, such as Mad2, plus the cohesins are present in trypanosomes (Gull et al., 1998; Ogbadoyi et al., 2000; Mottram et al., 2003; Gluenz et al., 2008).
Nuclear basket proteins in animals, plants, and fungi bind chromatin and influence transcription. Drosophila Nup153 and Megator demarcate regions of high transcriptional activity (Vaquerizas et al., 2010). Similar functions have been suggested for Mlp1/Mlp2 (Casolari et al., 2004), together with roles in mRNA export (Ishii et al., 2002; Casolari et al., 2004). There was no evidence for a role in cocistronic expression changes for TbNup92, with the exception of a gene cluster on chromosome V. Considered together with polycistronic transcription in trypanosomes, it appears unlikely that TbNup92 interacts with specific regions of the genome for the purpose of increasing transcriptional activity, and hence TbNup92 lacks a clear boundary function similar to Megator/Mlp. Regulation of trypanosome gene expression is heavily reliant on mRNA turnover, reflected in the expansion of genes encoding for RNA-binding proteins in the T. brucei genome (De Gaudenzi et al., 2005); of significance, TbNup92-knockout cells differentially expressed genes with GO terms associated with RNA turnover. Genes encoding RNA-binding proteins ALBA-1, ALBA-3, and ZFP-1, which influence expression of GPEET and EP1 procyclins (Hendriks et al., 2001; Walrad et al., 2009, 2012; Mani et al., 2011), were up-regulated in the TbNup92 knockout, and furthermore GPEET and EP procyclins were up-regulated, suggesting that differential expression of ALBA-1, ALBA-3, and ZFP-1 may account for increased procyclin expression, raising the possibility that TbNup92 could associate with a particular subset of RNA-binding proteins.
In conclusion, we demonstrate that TbNup92 is associated with the trypanosome NPC and plays similar roles to Mlp/Tpr in terms of spindle association and fidelity of chromosome segregation, but with absence of evidence for a role in the creation of transcriptional boundaries. The absence of orthology between TbNup92 and Mlp/Tpr and the restricted taxonomic distribution of the former echo the apparent distinct origin of the trypanosome nuclear lamina component NUP-1 from metazoan lamins. This suggests that a highly divergent scaffold is present at the trypanosome nuclear envelope, with significant implications for the origins of chromosomal segregation and gene expression mechanisms.
MATERIALS AND METHODS
Bioinformatics
Conserved protein domains were predicted by a National Center for Biotechnology Information (NCBI) conserved domain search. The probability that a certain region adopts a coiled-coil conformation was predicted by the COILS/PCOILS tool (http://toolkit.tuebingen.mpg.de/pcoils/) using either a single sequence or the protein alignment as query. The presence of Nup92 and Nup110 orthologues in the genomes of other kinetoplastids was determined by blast searches in various protein and nucleotide databases (http://tritrypdb.org/tritrypdb/, www.genedb.org, NCBI GenBank, and our in-house database of kinetoplastid genomes). Julius Lukes (Cˇeské Budeˇjovice, Czech Republic) kindly provided access to sequence data for Paratrypanosoma confusum. The presence of TbNup92 and TbNup110 homologues in other eukaryotes was searched by HMMER (Finn et al., 2011) in the UniProtKB database using the kinetoplastid sequence alignment as query. The candidate homologues were reverse blasted against the protein database of T. brucei and inspected for the presence of coiled coils and conserved domains. Protein sequences were aligned using MergeAlign (Collingridge and Kelly, 2012; Katoh et al., 2005). Phylogenetic trees were constructed in PhyML 3.1 (Guindon et al., 2010) using the default settings and the robustness of individual branches evaluated by SH-like approximated likelihood ratio test and bootstrap after 100 iterations.
Cell culture
BSF T. brucei brucei MITat 1.2 (M221 strain) and PCF T. b. brucei MITat 1.2 (Lister 427) were grown as previously described (Brun and Schonenberger, 1979; Hirumi and Hirumi, 1994). Single marker BSF (SMB) cell lines were used for expression of tetracycline-inducible constructs. Expression of plasmid constructs was maintained using antibiotic selection at the following concentrations: G418 and hygromycin B at 1 μg/ml and phleomycin at 0.1 μg/ml for BSF, and G418 at 20 μg/ml and hygromycin B at 25 μg/ml for PCF.
In situ genomic tagging
TbNup92, TbNup98, and TbNup110 ORFs were tagged using pMOTag4G, pMOTag23H (Oberholzer et al., 2006), and a modified pMOTag3G in which the GFP epitope was replaced with a 13xcMyc epitope (pMOTag313M) as templates. We used the following primers (all primer sequences are given in the 5’-to-3’ direction): TbNup110FOR, GAAAAGGCGATGCGACTACTGCACGTCAACAAGCAACTTGTGGAGAGAGTCAAAACCAGTCGAACTGAAGGAGAATCCCAGTCCAGTGGTACCGGGCCCCCCCTCGAG; TbNup110REV, GAGCATACGTACACGTACACGTACACGTACACGAATTGTCATACAACCTGACTAGCAGACGTAAGGCGCTACGAACCTTTACTGTGGTTCAAACAAAAATGGCGGCCGCTCTAGAACTAGTGGAT; TbNup92FOR, ACTCGGGCAGTAACTTGGGATAAGTTGGTTGAGCTCATTTACCCCGTTAAAATCGGTGTCGTGGCAGAGGAAGTAAAGCAGCCACAGGGTACCGGGCCCCCCCTCGAG; TbNup92REV, AATATATTATGCTCCCTCTGCAATTGGGGGGGGGGAGAGGAAAGAGGGGGAGAGAGGCTCTAAAGGATGAGATGAAATACCGAACGTTGGCGGCCGCTCTAGAACTAGTGGAT; TbNup98FOR, TGGGAATGCTTCAGCAAGTGGTGAAAAGAACAATGCTCCACGGAATCCCTTCTCATTTGGTGCCTCTTCTGGGAATGCTGGTACCGGGCCCCCCCTCGAG; and TbNup98REV, ACTAAAGAAGGGTAGAAAACAAAGAAAACACCAAATAAGGTACCTGACGCAGCGGCAACACCACGTCGACTTGCTGGCGGCCGCTCTAGAACTAGTGGAT.
To truncate a single allele of TbNup92 by replacing the C-terminal part of the protein (residues 640–813) that includes the BRCT domain with a GFP tag, we used the following primers: TbNup92truncFOR, ATACAATCGGCTAATGTGCCTCCATCCGCCAGTAAGCAAGCGAAGAGGAGCCGATCCGTCGAGCAACGTGTTTTTACAATCAGCGGTGGTACCGGGCCCCCCCTC; TbNup92truncREV, and CGTGGGGACCGGTGAGTTGGATTTACACTCGGCAACTGTGGCGTACGGAAGCTTGTGAATTTTCTCAAGTAACTCCGTTCCGTCAAATGGCGGCCGCTCTAGAACTAGTGGAT. Linear PCR products were purified by ethanol precipitation. For PCFs, electroporation was performed with 15 μg of DNA using a Bio-Rad Gene Pulser II (1.5 kV and 25 μF). Electroporation of BSFs was carried out using an Amaxa Nucleofector (T-cell nucleofection buffer, program X-001). Positive clones were assayed for correct insertion and expression using Western blotting and immunofluorescence microscopy as described.
Immunofluorescence microscopy
Cells were prepared as previously described (DuBois et al., 2012). Antibodies were used at the following concentrations: rabbit anti-GFP, 1:3000; goat anti-rabbit immunoglobulin G (IgG) Alexa Fluor 488 (Molecular Probes, Paisley, UK), 1:1000; mouse anti-HA (Santa Cruz Biotechnology, Heidelberg, Germany), 1:1000; mouse anti-cMyc (Sigma-Aldrich, Dorset, UK), 1:1500; mouse anti-tubulin clone KMX-1 (Millipore, Watford, UK), 1:2000; goat anti-mouse IgG Alexa Fluor 568 (Molecular Probes), 1:1000; goat anti-mouse IgG Alexa Fluor 594 (Molecular Probes), 1:1500; and polyclonal rabbit anti–NUP-1, 1:750 (DuBois et al., 2012). Wide-field epifluorescence images were acquired using a Nikon Eclipse E600 epifluorescence microscope equipped with a Hamamatsu ORCA charge-coupled device camera, and data were captured using MetaMorph (Universal Imaging, Marlow, UK). Confocal microscopy images were acquired with a Leica TCS-NT confocal microscope with a 100×/1.4 numerical aperture objective. Images were processed with Huygens deconvolution software (Scientific Volume Imaging, Hilversum, Netherlands) and Photoshop (Adobe Systems, UK). Three-dimensional structured illumination microscopy was performed on a DeltaVision OMX V3 (Applied Precision, Preston, UK). Images were acquired in conventional mode and superresolution mode with a 60×/1.42 numerical aperture. Images were deconvolved using softWoRx 5.0 software (Applied Precision). Quantitation was performed on raw images gathered under nonsaturating conditions using ImageJ (Rasband and Bright, 1995) and Fiji (Schindelin et al., 2012) software.
Fluorescence in situ hybridization
The telomere PNA FISH kit (DAKO, Ely, UK) was used to highlight telomeric repeats according to the manufacturer's instructions. Combined visualization of telomeres and the minichromosome 177–base pair repeat regions was carried out as previously described (DuBois et al., 2012) using the telomere PNA probe and a digoxigenin-labeled 177–base pair fragment specific to the minichromosomal repeats. Cells were imaged as described.
Western immunoblotting
Proteins from whole-cell lysates (107 cells/lane) were resolved by 12.5% SDS–PAGE. Proteins were transferred to polyvinylidene fluoride membranes (Millipore) and blocked, and antigens were visualized using standard methods. For visualization, monoclonal mouse anti-GFP (Roche) at 1:1000 or monoclonal mouse anti-HA (Sigma-Aldrich) at 1:10,000 in TBST (Tris-buffered saline + 0.05% Tween 20) incubated for 60 min at ambient temperature was used. Primary antibody binding was detected using secondary anti-IgG horseradish peroxidase (HRP) conjugates (Sigma-Aldrich) at 1:10,000 in TBST for 45 min. Detection of HRP-conjugated secondary antibodies was by chemiluminescence using luminol (Sigma-Aldrich) and x-ray film (Kodak).
Gene knockout
To generate gene knockout constructs, a ∼1-kb DNA fragment from the 5’-untranslated region (UTR) of the Nup92 locus was amplified by PCR using the following primers: Nup925’_FOR, CGGGTACCATATGATGAGCGCACGAGCTTCTT; and Nup925’_REV, CGCTCGAGTGAATCAATGCACTTACATTTAAACAC. The 5’-UTR PCR products were digested with KpnI and XhoI restriction enzymes and cloned into KpnI- and XhoI-restricted pXS5:NEO and pXS5:HYG plasmids to create pXS5_N925’UTR:NEO and pXS5_N925’UTR:HYG, respectively. The 3’-UTR sequence immediately downstream of the Nup92 ORF (∼1 kb) was amplified by PCR using the following primers: Nup923’_FOR, CGACTAGTAATATATTATGCTCCCTCTGCTTGG; and Nup923’_REV, CGGAGCTCGTTACAAGAAATATATACATCCCGTG. The 3’-UTR PCR products were digested with SpeI and SacI restriction enzymes and cloned into pXS5_N925’UTR:NEO and pXS5_N925’UTR:HYG to generate pXS5_N925’3’UTR:NEO and pXS5_N925’3’UTR:HYG, respectively. Each plasmid was sequenced for correct insertion of the constructs. The pXS5_N925’3’UTR:NEO construct was linearized with KpnI and SacI before being transfected into procyclic-form trypanosomes to replace a single copy of TbNup92 in the genome under the selection of G418 sulfate antibiotics. To target the second TbNup92 gene, the pXS5_N925’3’UTR:HYG construct was linearized with KpnI and SacI and transfected into the heterozygous TbNup92-knockout cell line under the selection of G418 and hygromycin antibiotics. Positive clones were confirmed by whole-genome resequencing and qRT-PCR to measure TbNup92 mRNA levels.
Generation and overexpression of the TbNup92 C-terminal BRCT domain
The C-terminal BRCT domain of TbNup92 (TbNup92:BRCT) was overexpressed in trypanosomes under the control of an inducible T7 promoter. The construct consists of the first 90 bases of the TbNup92 ORF immediately followed by the final 579 bases of the ORF. A NLS (derived from NUP-1; DuBois et al., 2012) and 3×HA epitopes were incorporated at the C-terminus. The entire ORF was synthesized from synthetic oligonucleotides by Invitrogen Life Technologies and subsequently cloned into SpeI- and BamHI-restricted, pDex-577–inducible expression vector (Kelly et al., 2007) to create pDex-577_BRCT. The construct was sequenced for correct insertion into the plasmid. pDEX-577_BRCT was linearized with NotI and transfected into SMB cells. For inducible expression of TbNup92:BRCT, cells were induced with 0.1 μg/ml tetracycline for 6 h.
Real-time quantitative reverse transcriptase PCR
RNA was purified from cell lysates using an RNeasy Mini Kit (Qiagen, Manchester, UK) according to the manufacturer's instructions. cDNA was synthesized from 1 μg of purified RNA using Superscript III Reverse Transcriptase (Invitrogen) accompanied with RNase OUT (Invitrogen). qRT-PCR using cDNA templates was performed with iQ-SYBRGreen Supermix and MiniOpticon Real-Time PCR Detection System (Bio-Rad, Hemel Hempstead, UK). Fluorescence was quantified after each cycle, and the results were analyzed using MiniOpticon software (Bio-Rad) with Rab11 as a housekeeping gene to normalize RNA input. The following primer pairs were used: Nup92qRTFOR, GAGGGTTTTGTTTCGTCCAG, and Nup92qRTREV, CCAAGTTACTGCCCGAGTGT; Rab11qRTFOR, ATCGGCGTGGAGTTTATGAC, and Rab11qRTREV, GTGGTAAATCGAACGGGAGA; EP2ProcyclinFOR, CAGGACGAAGTTGAGCCTG, and EP2ProcyclinRev, TGCAAGTGTCCTGTCGCC; Tb427.04.3850FOR, CACGAACGCATTGTTGACTC, and Tb427.04.3850REV, CCCCTTTGAAACACTGAAATG; Tb427.06.4970FOR, GCAACAACTGGAGGATGAGG, and Tb427.06.4970REV, TGAGTTTGAATCCCCGTCTT; Tb427.05.2650FOR, ACGGAAAGAATTGGGAGGAT, and Tb427.05.2650REV, ACTGGGCTGTTTCCACTGAG; Tb427.05.2530FOR, TGGGTTATCCGTGGAAAAGA, and Tb427.05.2530REV, GCTGCAATCCTGTTGATCCT; and Tb427.10.5670FOR, CTACACAATGTGCCGAATGC, and Tb427.10.5670REV, CCATTTCACGAAACAGGTTG.
Next-generation sequencing
DNA was extracted from TbNup92Δ cells using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's instructions. For each extraction, 5 × 107 cells were used, and DNA was eluted to a final concentration of 100 ng/μl. DNA was sequenced by 76–base pair paired-end Illumina sequencing at the Beijing Genomics Institute (www.genomics.cn/en/). Reads were clipped based on phred score >20 using the fastx program. Read errors and ambiguous bases were then corrected using the ALLPATHS find read errors algorithm (MacCallum et al., 2009), with two cycles of read error correction and the default settings for k-mer size. Duplicate reads and reads with a postclipped length of <20 nucleotides were discarded using custom Perl scripts. The clipped, corrected, and filtered reads were then assembled using Velvet (Zerbino and Birney, 2008) and multiple k-mer sizes (k-mer = 31, 41, 51, and 61), using the T. brucei 427 genome sequence as a scaffold. The resulting contigs from all assemblies were then postassembled using CAP3 (Huang and Madan, 1999) to yield a final genome assembly.
For RNA-seq, RNA was extracted and cDNA synthesized as described. Sheared cDNA was sequenced by 76–base pair paired-end Illumina sequencing. Paired-end reads were mapped to the T. brucei 427 strain reference genome sequence using Burrows–Wheeler Aligner (Li and Durbin, 2010). The mapped reads were further filtered by SAMtools (Li et al., 2009), allowing the read to be mapped in proper pair by a minimum 20 MAPQ (MAPing Quality). After filtering, the paired reads were aligned to annotated transcripts. Transcript abundances were calculated based on (fragments per kilobase of transcript per million mapped reads (FPKM; Trapnell et al., 2010). All FPKMs of annotated transcripts were normalized by quantile normalization. Low FPKM values (less than the median of FPKM values for all annotated transcripts) were removed. Transcripts that are annotated as rRNA or tRNA were excluded. To calculate the statistical significance for transcript level changes, the observed log-fold expression changes (FPKMtreat/FPKMwt), were tested against an empirical null distribution using a t test. The empirical null distribution of the log-fold changes were computed from all possible permutations of the samples as described in Budhraja et al. (2003). A p value for the comparison of the distributions of the positions of all genes and differentially expressed genes relative to the cistron transcription start sites was calculated based on the two-sample Kolmogorov–Smirnov test using the kstest2 MATLAB routine.
Flow cytometry
For FACS analysis, cells were prepared as previously described (Hammarton et al., 2003). Briefly, cells were fixed in 70% methanol in phosphate-buffered saline (PBS) and incubated at 4°C for 1 h. Cells were then washed in PBS and resuspended in 30 μg/ml propidium iodide in PBS and incubated at 37°C for 45 min. FACS was performed with a Cyan ADP MLE FACScan (Becton Dickinson, GE) using detector PE Texas red. Analysis was carried out using Dako Cytomation FACS software.
Interactome analysis
Protein–protein interactions between TbNup92 and other trypanosome proteins were analyzed by immunoisolation after cryomilling of parasites. Full details of the methodology are to be published elsewhere (Obado, Field, Rout, Chait, in preparation). In brief, ∼1010 procyclic-form trypanosomes harboring TbNup110::GFP or TbNup92::GFP were lysed by mechanical milling in a Retsch Planetary Ball Mill PM200 using liquid nitrogen cooling (Retsch, United Kingdom). Aliquots of powder were thawed in the presence of solubilization buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, 20 mM NaCl, 50 mM citrate, 0.5% Tween) and TbNup92::GFP/TbNup110::GFP were isolated using llama anti-GFP antibodies coupled to Dynabeads. All washes were in the same buffer. Affinity-isolated proteins of TbNup92 were reduced in the presence of 20 mM dithiothreitol (DTT) and alkylated after the addition of 50 mM iodoacetamide. Proteins were separated by SDS–PAGE and identified by mass spectrometry (Kalkum et al., 2003; DeGrasse et al., 2009). Protein bands were excised and destained in 50 mM ammonium bicarbonate/50% acetonitrile (ACN). Proteins were then trypsin digested in-gel (100–200 ng/band, depending on band intensity) and peptides extracted using a solution containing C18 beads (POROS) in 5% formic acid/0.1% trifluoroacetic acid. The beads were washed extensively using 0.5% acetic acid, and peptides were eluted from the beads sequentially using 40% ACN, 0.5% acetic acid, and then 80% ACN/0.5% acetic acid. The ACN is more volatile than acetic acid and was evaporated using a SpeedVac (Thermo, Pittsburgh, PA). A pressure Baum was used to load the acetic acid–peptide mixture onto a nano–liquid chromatography column (PicoTip, New Objective) packed with 3-μm C18 beads. Liquid chromatography–tandem mass spectrometry was performed on a Velos Orbitrap (Thermo Electron, Waltham, MA) and proteins identified on the Global Proteome Machine database (www.thegpm.org) essentially as described previously (DeGrasse et al., 2009).
Affinity isolates of TbNup110 were reduced in 50 mM DTT and alkylated using 75 mM iodoacetamide before fractionation by SDS–PAGE. Protein gels were stained using GelCode Blue (Thermo Scientific). Protein bands were excised and identified using a SCIEX prOTOF 2000 matrix-assisted laser desorption/ionization orthogonal time-of-flight (DeGrasse et al., 2009) Proteomics Mass Spectrometer (PerkinElmer, Akron, OH). Peak lists were submitted to ProFound and searched against an in-house–curated T. brucei database using data from GeneDB (www.genedb.org).
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust (Program Grant 082813 to M.C.F.), the Medical Research Council (studentship to J.M.H.), the Marie Curie Fund (Fellowship 300013 to L.K.), and the National Institutes of Health (R21 AI096069 and U54 GM103511 to B.T.C., J.A., and M.P.R., U01 GM098256 to M.P.R., P50 GM076547 to J.A., and P41 GM103314 to B.T.C.).
Abbreviations used:
- BRCT
C-terminal domain of breast cancer susceptibility protein
- BSF
bloodstream form
- FPKM
fragment per kilobase of transcript per million reads
- LECA
last eukaryotic common ancestor
- NE
nuclear envelope
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- PCF
procyclic form
- RBPs
RNA-binding proteins
- SAS
spindle attachment site
- SPB
spindle pole body
- TSS
transcription start site
- VSG
variant surface glycoprotein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-12-0750) on March 5, 2014.
The authors have no conflict of interest to declare.
REFERENCES
- Abbasi K, DuBois KN, Dacks JB, Field MC. A novel Rho-like protein TbRHP Is involved in spindle formation and mitosis in trypanosomes. PLoS One. 2011;6:e26890. doi: 10.1371/journal.pone.0026890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol. 1995;131(6 Pt 2):1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW, Goldfarb DS. Protein import through the nuclear pore complex is a multistep process. J Cell Biol. 1989;109:971–982. doi: 10.1083/jcb.109.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JL, Douglas MG. Organization of the nuclear pore complex in Saccharomyces cerevisiae. J Ultrastruct Mol Struct Res. 1989;102:95–108. doi: 10.1016/0889-1605(89)90047-5. [DOI] [PubMed] [Google Scholar]
- Alsford S, DuBois K, Horn D, Field MC. Epigenetic mechanisms, nuclear architecture and the control of gene expression in trypanosomes. Expert Rev Mol Med. 2012;29:e13. doi: 10.1017/erm.2012.7. [DOI] [PubMed] [Google Scholar]
- Archer SK, Inchaustegui D, Queiroz R, Clayton C. The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One. 2011;6:e18425. doi: 10.1371/journal.pone.0018425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Luu VD, de Queiroz RA, Brems S, Clayton C. Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog. 2009;5:e1000565. doi: 10.1371/journal.ppat.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R, Schonenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Budhraja V, Spitznagel E, Timothy Schaiff W, Sadovsky Y. Incorporation of gene-specific variability improves expression analysis using high density DNA microarrays. BMC Biol. 2003;28:1. doi: 10.1186/1741-7007-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE. Phylogeny and evolution of Apusomonadida (Protozoa: Apusozoa): new genera and species. Protist. 2010;161:549–576. doi: 10.1016/j.protis.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Chelsky D, Ralph R, Jonak G. Sequence requirements for synthetic peptide mediated translocation to the nucleus. Mol Cell Biol. 1989;9:2487–2492. doi: 10.1128/mcb.9.6.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge PW, Kelly S. MergeAlign: improving multiple sequence alignment performance by dynamic reconstruction of consensus multiple sequence alignments. BMC Bioinformatics. 2012;13:117. doi: 10.1186/1471-2105-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gaudenzi J, Frasch AC, Clayton C. RNA-binding domain proteins in kinetoplastids: A comparative analysis. Eukaryot Cell. 2005;4:2106–2114. doi: 10.1128/EC.4.12.2106-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, et al. Cancer-specific requirement for BUB1B/BUBR1 in human brain tumor isolates and genetically transformed cells. Cancer Discov. 2013;3:198–211. doi: 10.1158/2159-8290.CD-12-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois KN, et al. NUP-1 is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 2012;10:e1001287. doi: 10.1371/journal.pbio.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersfeld K, Gull K. Partitioning of large and minichromosomes in Trypanosoma brucei. Science. 1997;276:611–614. doi: 10.1126/science.276.5312.611. [DOI] [PubMed] [Google Scholar]
- Estevez AM. The RNA-binding protein TbDRBD3 regulates the stability of a specific subset of mRNAs in trypanosomes. Nucleic Acids Res. 2008;36:4573–4586. doi: 10.1093/nar/gkn406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Horn D, Alsford S, Koreny L, Rout MP. Telomeres, tethers and trypanosomes. Nucleus. 2012;3:478–486. doi: 10.4161/nucl.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Koreny L, Rout MP. Enriching the pore: splendid complexity from humble origins. Traffic. 2014;15:141–156. doi: 10.1111/tra.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER Web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 2009;59:243–255. doi: 10.1111/j.1365-313X.2009.03865.x. [DOI] [PubMed] [Google Scholar]
- Flegontov P, et al. Paratrypanosoma is a novel early-branching trypanosomatid. Curr Biol. 2013;23:1787–1793. doi: 10.1016/j.cub.2013.07.045. [DOI] [PubMed] [Google Scholar]
- Fontoura BMA, Dales S, Blobel G, Zhong HL. The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc Natl Acad Sci USA. 2001;98:3208–3213. doi: 10.1073/pnas.061014698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- Gerace L, Burke B. Functional organization of the nuclear envelope. Annu Rev Cell Biol. 1988;4:335–374. doi: 10.1146/annurev.cb.04.110188.002003. [DOI] [PubMed] [Google Scholar]
- Glover JNM, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Gluenz E, Sharma R, Carrington M, Gull K. Functional characterization of cohesin subunit SCC1 in Trypanosoma brucei and dissection of mutant phenotypes in two life cycle stages. Mol Microbiol. 2008;69:666–680. doi: 10.1111/j.1365-2958.2008.06320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Allen TD. The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J Mol Biol. 1996;257:848–865. doi: 10.1006/jmbi.1996.0206. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Gull K, Alsford S, Ersfeld K. Segregation of minichromosomes in trypanosomes: implications for mitotic mechanisms. Trends Microbiol. 1998;6:319–323. doi: 10.1016/s0966-842x(98)01314-6. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene SAD1 (+) associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarton TC, Clark J, Douglas F, Boshart M, Mottram JC. Stage-specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J Biol Chem. 2003;278:22877–22886. doi: 10.1074/jbc.M300813200. [DOI] [PubMed] [Google Scholar]
- Hendriks EF, Robinson DR, Hinkins M, Matthews KR. A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J. 2001;20:6700–6711. doi: 10.1093/emboj/20.23.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Axenic culture of African trypanosome bloodstream forms. Parasitol Today. 1994;10:80–84. doi: 10.1016/0169-4758(94)90402-2. [DOI] [PubMed] [Google Scholar]
- Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J Biol Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyton T, Bates PA, Zhang XD, Sternberg MJE, Freemont PS. The BRCA1 C-terminal domain: structure and function. Mutat Res. 2000;460:319–332. doi: 10.1016/s0921-8777(00)00034-3. [DOI] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: The nuclear pore connection. Cell. 2002;109(5):551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- Jasperson SL, Giddings TH, Winey M. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol. 2002;159:945–956. doi: 10.1083/jcb.200208169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Martin AE, Glazko G, Giddings TH, Morgan G, Mushegian A, Winey M. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J Cell Biol. 2006;174:665–675. doi: 10.1083/jcb.200601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen KM, Johansen J. Cell and molecular biology of the spindle matrix. Int Rev Cytol. 2007;263:155–206. doi: 10.1016/S0074-7696(07)63004-6. [DOI] [PubMed] [Google Scholar]
- Kalkum M, Lyon GJ, Chait BT. Detection of secreted peptides by using hypothesis-driven multistage mass spectrometry. Proc Natl Acad Sci USA. 2003;100:2795–2800. doi: 10.1073/pnas.0436605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, Kramer S, Schwede A, Maini PK, Gull K, Carrington M. Genome organization is a major component of gene expression control in response to stress and during the cell division cycle in trypanosomes. Open Biol. 2012;2:120033. doi: 10.1098/rsob.120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, et al. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol. 2007;154:103–109. doi: 10.1016/j.molbiopara.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ramey-Butler K, Cross GAM, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova B, Pante N, Rollenhagen C, Podtelejnikov A, Mann M, Aebi U, Hurt E. Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with Nic96p. J Biol Chem. 2000;275:343–350. doi: 10.1074/jbc.275.1.343. [DOI] [PubMed] [Google Scholar]
- Krull S, Doerries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29:1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Starr D, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon LA, Karki S, Tokito M, Holzbaur ELF. Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat Cell Biol. 2001;3:913–917. doi: 10.1038/ncb1001-913. [DOI] [PubMed] [Google Scholar]
- Lince-Faria M, Maffini S, Orr B, Ding Y, Florindo C, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J Cell Biol. 2009;184:647–657. doi: 10.1083/jcb.200811012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum I, et al. ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads. Genome Biol. 2009;10:R103. doi: 10.1186/gb-2009-10-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Mani J, Guttinger A, Schimanski B, Heller M, Acosta-Serrano A, Pescher P, Spath G, Roditi I. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS One. 2011;6:e22463. doi: 10.1371/journal.pone.0022463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BMA, Dasso M. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol. 2010;12:164–169. doi: 10.1038/ncb2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair. 2009;8:1009–1017. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram JC, Helms MJ, Coombs GH, Sajid M. Clan CD cysteine peptidases of parasitic protozoa. Trends Parasitol. 2003;19:182–187. doi: 10.1016/s1471-4922(03)00038-2. [DOI] [PubMed] [Google Scholar]
- Nakano H, Funasaka T, Hashizume C, Wong RW. Nucleoporin translocated promoter region (Tpr) associates with dynein complex, preventing chromosome lagging formation during mitosis. J Biol Chem. 2010;285:10841–10849. doi: 10.1074/jbc.M110.105890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett IR, Martin DM, Miranda-Saavedra D, Lamont D, Barber JD, Mehlert A, Ferguson MA. The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol Cell Proteomics. 2009;8:1527–1538. doi: 10.1074/mcp.M800556-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteosome. Mol Biol Cell. 2013;24:3920–3938. doi: 10.1091/mbc.E13-07-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Strambio-de-Castillia C, Fasolo J, Chait BT, Rout MP. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol. 2005;170:225–235. doi: 10.1083/jcb.200504140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer M, Morand S, Kunz S, Seebeck T. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol. 2006;145:117–120. doi: 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Ogbadoyi E, Ersfeld K, Robinson D, Sherwin T, Gull K. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma. 2000;108:501–513. doi: 10.1007/s004120050402. [DOI] [PubMed] [Google Scholar]
- O'Reilly AJ, Dacks JB, Field MC. Evolution of the karyopherin-beta family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PLoS One. 2011;6:e19308. doi: 10.1371/journal.pone.0019308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HY, et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol Biol Cell. 2004;15:4854–4865. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS, Bright DS. NIH Image—a public domain image processing program for the Macintosh. Microbeam Analysis. 1995;4:137–149. [Google Scholar]
- Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4362–4374. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat Cell Biol. 2000;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- Sherwin T, Gull K. The cell-division cycle of Trypanosoma brucei-brucei—timing of event markers and cytoskeletal modulations. Philos Trans R Soc Lond B Biol Sci. 1989;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang XN, Dewell S, Cross GAM. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang XN, Dewell S, Cross GAM. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver PA. How proteins enter the nucleus. Cell. 1991;64:489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- Solari AJ. Mitosis and genome partition in trypanosomes. Biocell. 1995;19:65–84. [PubMed] [Google Scholar]
- Stern MZ, Gupta SK, Salmon-Divon M, Haham T, Barda O, Levi S, Wachtel C, Nilsen TW, Michaeli S. Multiple roles for polypyrimidine tract binding (PTB) proteins in trypanosome RNA metabolism. RNA. 2009;15:648–665. doi: 10.1261/rna.1230209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Swift H. Studies on nuclear fine structure. Brookhaven Symp Biol. 1959;12:134–152. [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–U174. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. nuclear pore proteins Nup153 and Megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DL, Wang D, Jin Y, Rath U, Wang YM, Johansen J, Johansen KM. Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. J Cell Biol. 2000;151:1401–1411. doi: 10.1083/jcb.151.7.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walrad P, Paterou A, Acosta-Serrano A, Matthews KR. Differential trypanosome surface coat regulation by a CCCH protein that co-associates with procyclin mRNA cis-elements. PLoS Pathog. 2009;5:e1000317. doi: 10.1371/journal.ppat.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walrad PB, Capewell P, Fenn K, Matthews KR. The post-transcriptional trans-acting regulator, TbZFP3, co-ordinates transmission-stage enriched mRNAs in Trypanosoma brucei. Nucleic Acids Res. 2012;40:2869–2883. doi: 10.1093/nar/gkr1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML. Further observations on the nuclear envelope of the animal cell. J Biophys Biochem Cytol. 1959;6:147–156. doi: 10.1083/jcb.6.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang FM, Lin YC, Hu MC. Identification of two functional nuclear localization signals mediating nuclear import of liver receptor homologue-1. Cell Mol Life Sci. 2011;68:1241–1253. doi: 10.1007/s00018-010-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.