Abstract
The elicitation of cellular antiviral activities is dependent on the rapid transcriptional activation of interferon (IFN) target genes. It is not clear how the interferon target promoters, which are organized into chromatin structures in cells, rapidly respond to interferon or viral stimulation. In this report, we show that alpha IFN (IFN-α) treatment of HeLa cells induced hundreds of genes. The induction of the majority of these genes was inhibited when one critical subunit of the chromatin-remodeling SWI/SNF-like BAF complexes, BAF47, was knocked down via RNA interference. Inhibition of BAF47 blocked the cellular response to viral infection and impaired cellular antiviral activity by inhibiting many IFN- and virus-inducible genes. We show that the BAF complex was required to mediate both the basal-level expression and the rapid induction of the antiviral genes. Further analyses indicated that the BAF complex primed some IFN target promoters by utilizing ATP-derived energy to maintain the chromatin in a constitutively open conformation, allowing faster and more potent induction after IFN-α treatment. We propose that constitutive binding of the BAF complex is an important mechanism for the IFN-inducible promoters to respond rapidly to IFN and virus stimulation.
As an important host defense mechanism against virus infection, the type 1 interferons (IFNs) are rapidly induced to activate antiviral activities in infected and neighboring uninfected cells. Upon the binding of alpha IFN (IFN-α) to its cell surface receptor, the signal transducer and the activator of transcription proteins (STAT1 and STAT2) become tyrosine phosphorylated and associate with p48 (IRF-9/ISGF3γ) to form the trimeric ISGF3 complex. ISGF3 is relocated to the nucleus and binds to the IFN-stimulated response elements (ISREs) to activate the transcription of target genes (7, 23, 36). Viral infection as well as stimulation by double-stranded RNA poly(I)/poly(C) induces type 1 IFN genes (such as IFN-β) and a set of IFN-inducible genes. The antiviral efficiency of this system depends on the rapid transcriptional induction of a number of IFN target genes. It has been suggested that the chromatin structure plays important roles in the regulation of the IFN target genes (3, 5, 29). The chromatin-remodeling BAF or hSWI/SNF complexes (17, 19, 42) have been reported to elevate the basal-level expression and the induction of a subset of IFN-inducible genes (16, 24, 25). However, it is not clear whether the BAF complexes are required for the cellular antiviral activities, and it is not known how the chromatin structure controls the readiness of a promoter to be activated by IFN or viral infection and how the chromatin structure itself is controlled.
The ATP-utilizing chromatin-remodeling complexes are implicated in mediating gene activation in vitro and in vivo by antagonizing chromatin-mediated repression (1, 11, 14, 26, 37, 39, 44). The current view is that upon stimulation, the chromatin-remodeling complexes are recruited to their target promoters, where they alter the local chromatin structure to facilitate subsequent assembly of the transcription machinery and therefore transcriptional activation (2, 6, 8, 22, 30, 32). In order to clarify how the chromatin barrier is overcome for rapid induction upon stimulation, we analyzed the function and mechanism of the BRG1-containing complexes in the IFN-mediated antiviral activities. BRG1 itself, which is the essential ATPase of the complexes, was found to contain nucleosome remodeling activity in an in vitro assay. BAF47 (also known as INI1) is required for the elevated remodeling activity of BRG1 in vitro, and its yeast homologue, Snf5p, coordinates the assembly and remodeling activity of the yeast SWI/SNF complex in vivo (12, 18, 31, 42). Moreover, mutations of BAF47 have been linked to tumor formation in both human and mouse models (13, 21, 33, 40). Therefore, we inhibited the BAF complexes by expressing a small interference RNA (siRNA) targeting BAF47 in HeLa cells. We found that the BAF complex is required for transcriptional induction of the majority of IFN-α- and virus-inducible genes. Our data indicate that the BAF complex plays essential roles in cellular antiviral activities by controlling the chromatin structures of the genes involved in the pathways.
MATERIALS AND METHODS
Cell culture, RNA interference construct, and transfection.
SW-13 cells and HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and 1% penicillin-streptomycin mix and transfected with Superfect transfecting reagent as described previously (25). The RNA interference construct targeting BAF47 was constructed by inserting the cDNA sequence of BAF47 (positions 912 to 934; 5′GGACATGTCAGAGAAGGAGAAC3′) into pBS-U6 as described previously (38). The BAF47 sequence together with the U6 promoter was then subcloned into pREP4-puro, which was generated by deleting the RSK promoter and replacing the hygromycin B selection marker with puromycin in pREP4 (25). For RNA interference assays, HeLa cells were transfected with the siRNA constructs and selected in 1 μg of puromycin/ml for 2 days, followed by DNA microarray, reverse transcription (RT)-PCR, restriction enzyme accessibility, and viral infection analyses.
RT-PCR analysis.
RT-PCR analyses for the experiments shown in Fig. 1, 2, and 3 were performed as described previously (25) by using total RNAs isolated from HeLa cells treated with 500 U of IFN-α per ml (8 h), 10 μg of poly(I)/poly(C) per ml (6 h), or virus (0 to 24 h).
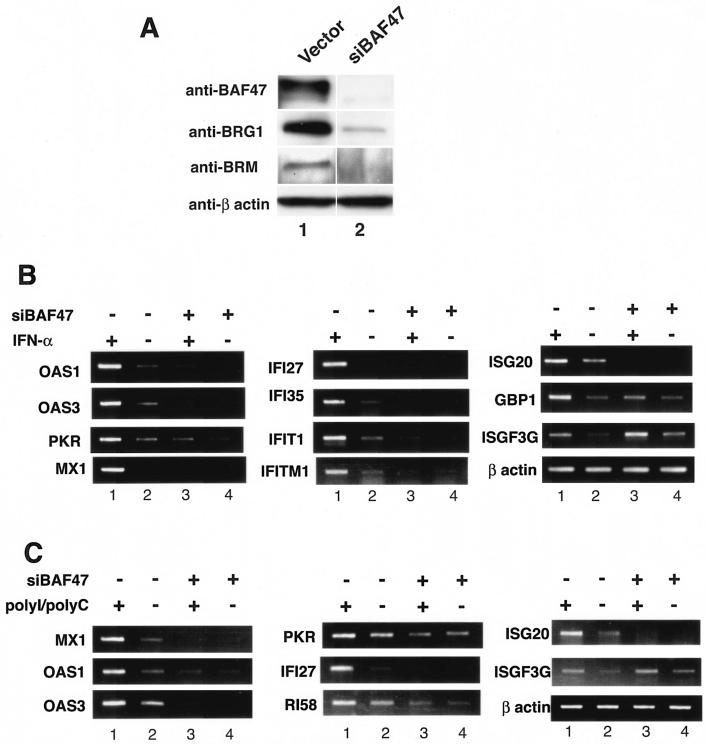
FIG. 1.
The BAF complex is required for cellular responses to IFN-α and poly(I)/poly(C) stimulation. (A) BAF47 was efficiently knocked down by RNA interference. HeLa cells were transfected with a control vector (lane 1) or a siRNA construct targeting BAF47 (lane 2) and selected for 2 days in puromycin. The remaining cells were lysed and analyzed by Western blotting with antibodies against BAF47, BRG1, BRM, or β actin. (B) The BAF complex is required for the induction of IFN target genes by IFN-α. HeLa cells were transfected with the siRNA construct targeting BAF47 (siBAF47) and selected for 2 days with puromycin. Following stimulation with 500 U of IFN-α/ml for 8 h, the total RNAs were isolated for RT-PCR analysis with the primers indicated on the left sides of the panels. (C) The BAF complex is required for the induction of IFN target genes by poly(I)/poly(C). HeLa cells were transfected and selected as described above. Following stimulation with poly(I)/poly(C), total RNAs were isolated and analyzed as described above.
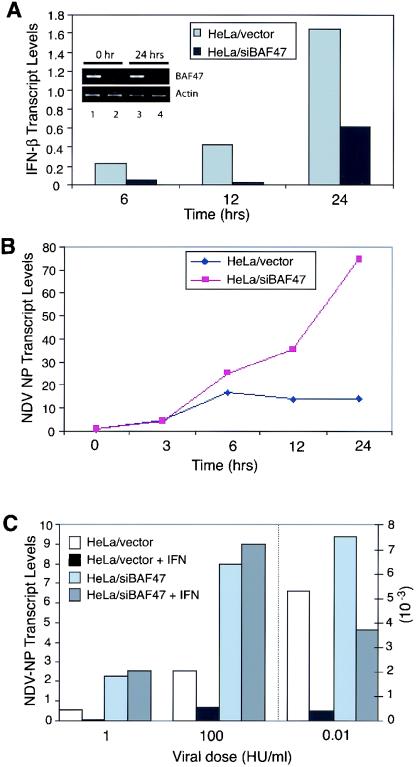
FIG. 2.
The BAF complex is required for the induction of cellular antiviral activities. (A) Expression of IFN-β upon NDV infection. Control or BAF-inhibited (siBAF47) HeLa cells were infected with NDV (100 HU/ml) for 1 h, and IFN-β levels at the indicated time points were measured by real-time PCR. The inset shows the results of RT-PCR analyses of BAF47 and β actin mRNAs after 0 and 24 h of viral infection in HeLa cells transfected with the control vector (lanes 1 and 3) or the siBAF47 construct (lanes 2 and 4). (B) Assessment of NDV replication. Cells were infected with NDV, and NDV NP transcript levels were measured at the indicated time points by real-time PCR. The increase (fold) is shown. (C) Effect of IFN-α on NDV replication. Cells were infected with different doses of NDV (0.01, 1, and 100 HU/ml) in the presence or absence of IFN-α (1,000 U/ml), and viral replication by NDV NP was assessed by real-time PCR at 12 h after infection. Values on the right y axis correspond to transcript levels obtained with 0.01 HU/ml, while those on the left y axis correspond to transcript levels with 1 and 100 HU of NDV/ml.
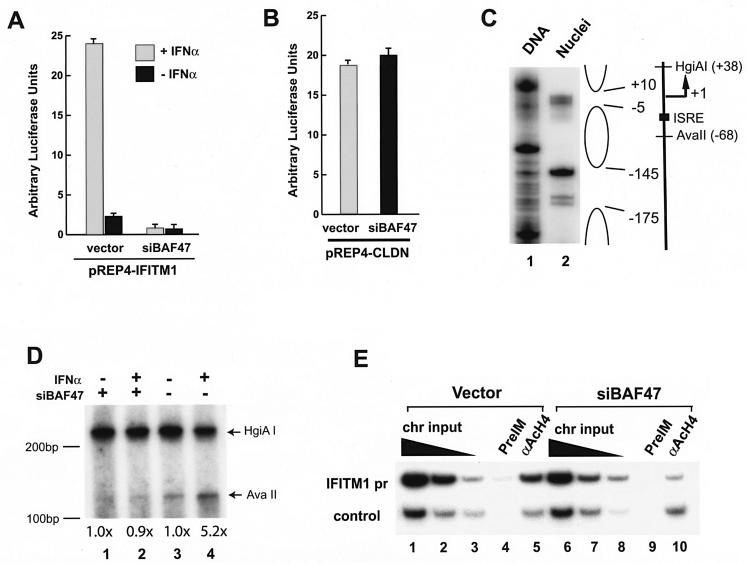
FIG. 3.
The BAF complex regulates IFITM1 promoter activity by modulating its chromatin structure. (A) Inhibiting the BAF complexes abolished the induction of the IFITM1 promoter by IFN-α. HeLa cells were transfected with the IFITM1 promoter reporter construct, pREP4-IFITM1pr-luc, together with a control vector or the siRNA construct targeting BAF47 (siBAF47) for 48 to 72 h. Following treatment with 500 U of IFN-α/ml for 12 h, the luciferase activity was analyzed with a dual luciferase system from Promega. (B) Knockdown of BAF47 did not inhibit claudin promoter activity. HeLa cells were transfected with the claudin promoter reporter construct and analyzed as described for panel A. (C) The IFITM1 promoter has a positioned nucleosome. The genomic DNA or nuclei isolated from HeLa or SW-13 cells was digested with microccocal nuclease. The double-stranded cleavages in the nucleosomal linker regions were detected by ligation-mediated PCR with primers specific for the IFITM1 promoter sequence (PCR primer −273F [5′ACAGTGAGGTCCTGTACTTGCTGG3′] and labeling primer −261F [5′TGTACTTGCTGGCCTGGGGTG3′]). The open oval on the right indicates the regions protected by the nucleosomal structure. The transcription start site (+1) and direction are indicated. The filled square indicates a potential binding site for ISGF3 (ISRE). The recognition sites for the restriction enzymes (AvaII and HgiAI) are indicated. (D) The BAF complex is required for the chromatin remodeling of the IFITM1 promoter upon IFN-α stimulation. HeLa cells were transfected with control vector or siRNA targeting BAF47 (siBAF47) and selected as described in the legend to Fig. 1. After stimulation with 500 U of IFN-α/ml for 2 h, the cells were permeabilized and briefly digested with AvaII enzyme, followed by complete digestion of the purified genomic DNA with HgiAI enzyme. The cleavage sites were detected by linker ligation-mediated PCR with IFITM1 promoter-specific primers. The data were quantified by PhophorImager analysis. The intensity of the bands produced by AvaII digestion after normalization to the HgiAI digestion is indicated below the panel. The experiments were repeated three times, and similar results were obtained. Size markers are indicated on the left side of the panel. (E) Knocking down BAF47 reduced histone H4 acetylation at the IFITM1 promoter. The HeLa cells were transfected with control vector or siRNA targeting BAF47 (siBAF47) and selected as described in the legend to Fig. 1. The chromatin lysates were prepared and immunoprecipitated with preimmune serum and antibodies against tetra-acetylated histone H4 tail as described previously (24). The IFITM1 promoter sequence and the CSF1 upstream sequence (control) in the immunoprecipitated DNA were analyzed by PCR. The chromatin (chr) input was diluted three times at each step.
RT-PCR analyses for the experiments shown in Fig. 5 were performed as described below. SW-13 cells and HeLa cells grown in Dulbecco modified Eagle medium supplemented with 1% fetal calf serum and 1% penicillin-streptomycin mix were treated with 500 U of IFN-α/ml for various times. Total RNAs were extracted with TRIzol reagent (Invitrogen) and reverse transcribed by using the oligo(dT) 12-18 primer with an Invitrogen kit (catalog no. 12371-019). The cDNA (20 ng) was amplified with IFITM1, ISG15, or β actin-specific primers for 12 cycles under PCR conditions of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. One-tenth of the PCR product was slot blotted onto a nylon membrane and detected by hybridization with a 32P-labeled cDNA probe amplified with the same primers. The membranes were visualized by autoradiography or quantified by PhosphorImager analysis. Histograms were plotted after normalization to β actin signals.
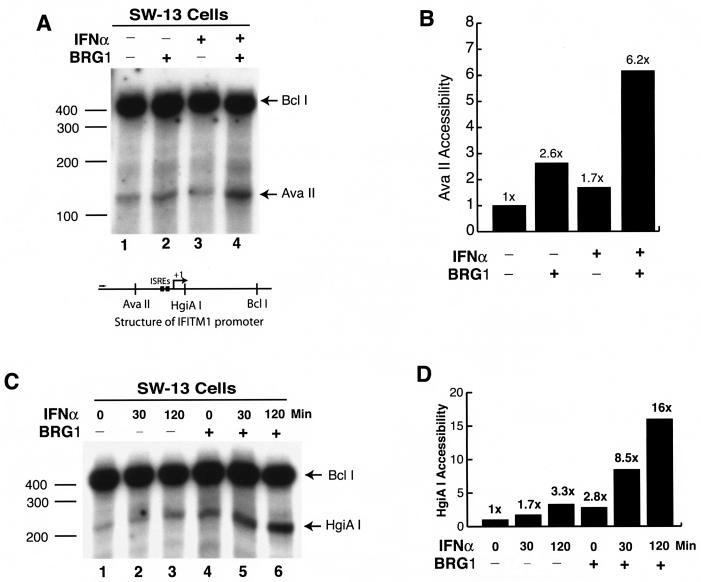
FIG. 5.
Expression of BRG1 controls both basal and induced chromatin remodeling of the IFITM1 promoter. (A) Nuclei isolated from SW-13 cells transfected with pBJ5 or pBJ5-BRG1 for 24 h before treatment with IFN-α for 2 h were briefly digested with AvaII. The purified genomic DNA was digested to completion with BclI. The cleavage sites were detected by linker ligation-mediated PCR with the IFITM1 promoter-specific primers as described in the legend to Fig. 3B. (B) Quantification of the data in panel A by PhosphorImager analysis. (C) Nuclei isolated from SW-13 cells transfected with pBJ5 or pBJ5-BRG1 for 24 h before treatment with IFN-α for various times were briefly digested with HgiAI and analyzed as described for panel A. (D) Quantification of the data in panel C by PhosphorImager analysis. The experiments were repeated three times, with similar results.
ChIP.
The chromatin immunoprecipitation (ChIP) assays were performed as described previously (24). The antibodies used were αBRG1 (19), αAcH4 (41), αPlo II (sc-9001; Santa Cruz), αSTAT (S21220; Transduction Laboratories), and p48 (sc-496; Santa Cruz). For the detection of the ChIP products, IFITM1 promoter primers 5′CCAACACTTAGGAAGTCACTAGTC3′ (−197F) (forward) and 5′CTCCTTTCCCCTGTCGTTTCAGTT3′ (reverse), the IFITM1 3′ untranslated-region primers 5′CGGTCCTGTGACCCCTTAATGGT3′ (forward) and 5′GTTGGGAAGACAGCTTCGACTCC3′ (reverse), and the CSF1 far-upstream-region primers 5′CACTATGTTAGCCAGGATGGTCTC3′ (forward) and 5′CTCTTCCTCCTGATAGCTCCATGA3′ (reverse) were used.
Nucleosome mapping and restriction enzyme accessibility assays.
The nucleosomal structure of the IFITM1 promoter was mapped by using microccocal nuclease and ligation-mediated PCR as described previously (24). The restriction enzyme accessibility assays were performed according to a published procedure (43) with the following modifications. SW-13 cells transfected with pBJ5-BRG1 for 24 h or HeLa cells were treated with 500 U of IFN-α/ml for various times. The nuclei isolated from the cells were digested with 2 μl of HgiAI or AvaII (20 U) for 10 min at 37°C, followed by the addition of 150 μl of stop buffer (10 mM Tris [pH 7.5], 10 mM EDTA, 0.4% sodium dodecyl sulfate, 0.6 mg of proteinase K/ml) and incubation at 50°C for 3 h. The DNA (2 μg) purified by phenol-chloroform extraction and precipitation was digested to completion with 1 μl of BclI (10 units) in 50 μl of the restriction buffer for 2 h at 55°C. Following Klenow enzyme treatment to blunt the DNA ends, the DNA was ligated to a universal linker (28). The cleavage sites were detected by PCR with reaction mixtures containing 10 μl of DNA, 5 μl of 10× PCR buffer [(NH4)2SO4], 5 μl of 25 mM MgCl2, 5 μl of 1 mM deoxynucleoside triphosphates, 2 μl of 2.5 μM long universal primer, 2 μl of 2.5 μM gene-specific primer (IFITM1/−197F), 20 μl of H2O, and 1 μl of Taq polymerase (5 U) under the following conditions: 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by 72°C for 5 min. The PCR products were labeled by the addition of 32P-labeled IFITM1/−170F (5′ACTTGAGTTTCTGATGAGGAAGCC3′) to a concentration of 0.05 μM and subjected to two PCR cycles of 94°C for 2 min, 55°C for 2 min, and 72°C for 2 min. The labeled products were purified and resolved on 9% acrylamide-6 M urea-1× Tris-borate-EDTA gel and visualized by autoradiography and quantitated by PhosphorImager analysis.
Viral infection.
Equal numbers (1 × 105) of control and BAF-inhibited HeLa cells were infected with Newcastle disease virus (NDV) (100 hemolytic units [HU]/ml) for 1 h. Following infection, virus was washed off and cells were incubated in the media described above for the indicated time periods. To study the effect of IFN on virus replication, cells were infected with various doses of virus (0.01, 1, and 100 HU/ml) with or without the addition of IFN-α (1,000 U/ml). IFN was added 18 h prior to infection and was present throughout infection (20). cDNA was prepared by using Superscript II RNase H− reverse transcriptase (Invitrogen) and random primers. The NDV nucleocapsid protein (NP) transcripts were measured as indices of viral replication (34). For real-time PCR, the amplification of cDNA was monitored by using SYBER Green PCR master mix (Applied Biosystems) in combination with the 7000 sequence detection system (ABI-PRISM). Transcript levels were normalized by GAPDH (glyceraldehyde-3-phosphate dehydrogenase), expressed at comparable levels in control and BAF-inhibited cells. Primer sequences used for real-time PCR are available on request.
RESULTS
The BAF complex is required for the basal and induced levels of expression of the majority of the IFN-α-inducible genes.
In order to determine the requirement for the BAF complexes in the IFN signaling pathways, we specifically knocked down BAF47/INI1/hSNF5, a critical subunit of the BRG1- and hBRM-containing complexes, by RNA interference. Transfection of HeLa cells with an episomal BAF47 RNA interference construct for 2 days resulted in complete inhibition of the BAF47 protein (Fig. 1A). Furthermore, both BRG1 and BRM were also significantly down-regulated (Fig. 1A), possibly by the decreased stability in the absence of BAF47. These experiments suggest that the function of the BRG1- and BRM-containing complexes could be severely compromised through the expression of the siRNA targeting BAF47 in the cell. Therefore, mRNA samples were isolated from HeLa cells transfected with the RNA interference construct and were analyzed by DNA microarrays. The analysis revealed that approximately 2.5% of the genes were repressed and that 0.5% were activated more than threefold among the 44,000 cDNA and expressed sequence tag sequences (data not shown). In sharp contrast to the low percentage of affected genes in the whole genome, the expression of more than 90% of the IFN-α-inducible genes was significantly down-regulated in cells expressing the BAF47 siRNA (Fig. 1B, compare lanes 2 and 4, and data not shown). Moreover, upon IFN-α stimulation, the induction of these inducible genes was either dramatically reduced or completely inhibited (Fig. 1B, compare lanes 1 and 3, and data not shown). The strongly inhibited genes included those encoding the well-characterized antiviral proteins such as the double-stranded RNA-dependent protein kinase (PKR), the 2-5A system (OAS1, OAS2, and OAS3), IFITM1, and the Mx proteins (36). The inhibition by the BAF47 siRNA was not caused by nonspecific activity of antisense RNA, since another siRNA construct, which targeted a different region of the BAF47 sequence but was not able to knock down the BAF47 mRNA, failed to inhibit the cellular response to IFN-α stimulation (data not shown). These data strongly suggest the BAF complex is required for the normal function of the IFN signaling pathways.
Knockdown of the BAF complex significantly inhibited the cellular response to poly(I)/poly(C).
To mimic the cellular response to viral infection, control and BAF-inhibited (siBAF47) HeLa cells were treated with double-stranded RNA poly(I)/poly(C) and examined for the expression of IFN-inducible genes. These experiments were of interest, since the above-described microarray analysis found toll-like receptor 3 (TLR3) to be constitutively reduced in BAF-inhibited cells. TLR3 specifically recognizes double-stranded RNA and is required for the poly(I)/poly(C) induction of IFN target genes (4). As shown in Fig. 1C, poly(I)/poly(C) induced the expression of many antiviral genes, such as the Mx1, PKR, OAS3, and IFITM1 genes, in control cells. However, the induction of these genes was dramatically reduced or abolished in cells in which the BAF complex was inhibited. These results indicate that the establishment of antiviral activities is severely impaired when the BAF complex is inhibited.
The BAF complex is required for the elicitation of cellular antiviral activities upon viral infection.
To study whether the inhibition of the BAF complex affects cells' ability to control viral infection, control and BAF-inhibited HeLa cells were infected with NDV. Infected cells were tested first for the induction of the IFN-β gene, an immediate early IFN gene important for establishing an antiviral state in the cells (36). Real-time RT-PCR analysis (Fig. 2A) showed that IFN-β transcript levels were markedly elevated after viral infection in control cells but that transcript levels were much lower in siBAF47 cells at all three time points tested. The inhibition of IFN-β transcription was less severe at the 24-h point than the 6- and 12-h points. Therefore, we examined whether the extent of the BAF47 knockdown diminishes at the 24-h point. As shown in Fig. 2A, no difference in BAF47 knockdown levels was observed during the course of stimulation. These experiments suggest that the BAF complex is required for the rapid and full induction of IFN-β. Consistent with the data for poly(I)/poly(C), the induction of all antiviral proteins shown in Fig. 1C was markedly attenuated or completely blocked in siBAF47 HeLa cells after NDV infection, while these genes were robustly induced in control cells (data not shown). We next assessed whether inhibition of the BAF complex has an effect on viral replication. To this end, the levels of the NDV NP transcripts were measured by real-time RT-PCR at various time points following infection (Fig. 2B). In siBAF47 cells, levels of viral transcripts increased more than 70-fold by 24 h, while the transcripts in control cells increased less than 15-fold over the same period. These results indicate that inhibition of the BAF complex reduces cells' ability to control viral growth, most likely by inhibiting the expression of IFN-β and other antiviral proteins. To examine whether the inhibition of the BAF complex compromises IFN′s antiviral activities, we measured NDV NP transcript levels in cells that had been treated with IFN-α for 18 h prior to NDV infection for 12 h. As seen in Fig. 2C, IFN treatment potently inhibited NDV nucleocapsid transcript expression in control HeLa cells at three doses of NDV, while the treatment did not have discernible inhibitory effects on siBAF47 cells at any viral dose tested. These results indicate that IFN fails to confer antiviral activities on cells in which the BAF complex is inhibited. Similar experiments were performed with vesicular stomatitis virus. Consistent with the data presented above for NDV infection, IFN fully protected control cells from the cytopathic effect of vesicular stomatitis virus. However, it had no protective effects on siBAF47 cells, leading to uncontrolled destruction of the cells (data not shown), confirming that the BAF complex has a critical role in establishing IFN′s antiviral activities.
The BAF complex controls the chromatin accessibility at the IFITM1 promoter.
To elucidate the mechanisms by which the BAF complex mediates the action of IFN-α, we asked whether the BAF complex directly regulates the promoter activity of one IFN-α target gene, the IFITM1 gene. We cloned 200 bp of the IFITM1 promoter region into the chromatin-forming episomal pREP4-luc vector. HeLa cells were cotransfected with the promoter reporter construct and the RNA interference construct targeting BAF47. As shown in Fig. 3A, when cotransfected with a control vector into HeLa cells, the IFITM1 promoter was robustly stimulated by IFN-α. However, siRNA targeting BAF47 strongly inhibited the basal-level activity of the promoter and completely inhibited induction by IFN-α (Fig. 3A), indicating that the BAF complex is required for the activity of the IFITM1 promoter. The expression of siBAF47 had no detectable effect on the activity of the claudin promoter (Fig. 3B), indicating that the inhibition of the IFITM1 promoter was specific.
To clarify how the BAF complex controls the activity of the IFITM1 promoter, we examined the chromatin structure of the endogenous IFITM1 promoter. As shown in Fig. 3C, microccocal nuclease digestion identified a nucleosome between positions −5 and −145 relative to the transcription start site. The ISRE (positions −21 to −35) is located within the nucleosome. In order to determine whether the BAF complex is required to remodel the nucleosomal structure, we probed the chromatin structure by restriction enzyme accessibility assay. Nuclei isolated from HeLa cells were briefly digested with AvaII, which recognizes a sequence beginning at position −68 upstream of the transcription start site (Fig. 3C). Following complete digestion of the purified DNA with HgiAI, the cleavage sites were detected by linker ligation-mediated PCR (28, 43). As shown in Fig. 3D, stimulation of the control HeLa cells with IFN-α for 2 h strongly elevated the accessibility of the promoter to AvaII (compare lanes 3 and 4) about 5.2-fold, indicating that the chromatin was remodeled to a more open structure in response to IFN-α. The BAF47 siRNA abolished the increase in chromatin accessibility (compare lanes 1 and 2), indicating that the BAF complex is required for the chromatin remodeling induced by IFN-α signaling. To determine whether the knockdown of BAF47 changed the levels of histone acetylation at the IFITM1 promoter, we performed ChIP experiments with antibodies against tetra-acetylated histone H4 tail. As shown in Fig. 3E, the IFITM1 promoter band intensity from the cells transfected with the siBAF47 construct was significantly weaker than that from the cells transfected with the control vector (compare lanes 5 and 10, upper panel), while the intensities for the CSF1 gene upstream control sequence were similar (compare lanes 5 and 10, lower panel). Quantification with the PhosphorImager indicates that the IFITM1 promoter signal was threefold higher in the control cells than in the siBAF47 cells. The acetylation levels in the regions 4 kb upstream and 4 kb downstream of the promoter region were not significantly changed by siBAF47 (data not shown), suggesting that the effect of the BAF complex on the acetylation of histone H4 is not global but is localized in the promoter region. These data indicate that the inhibition of BAF47 resulted in lower levels of histone H4 acetylation and a less open chromatin structure at the promoter.
An active BAF complex is required for rapid and full induction of the IFITM1 promoter by IFN-α.
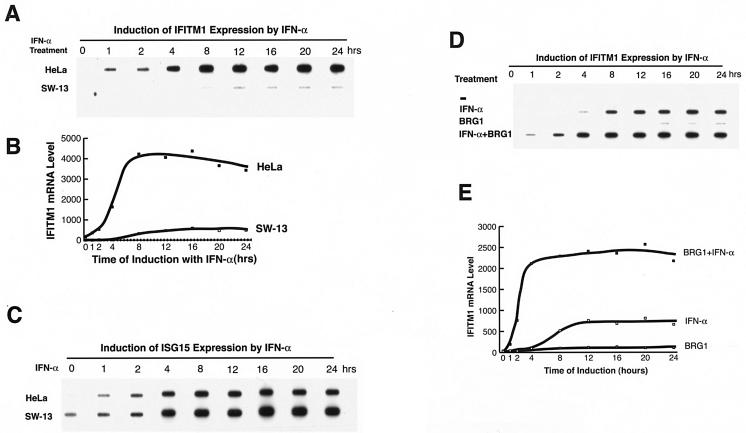
To confirm that the BAF complex prepares the chromatin structure of the IFITM1 promoter for rapid induction by IFN-α, we examined the expression of the IFITM1 gene in HeLa cells as well as in SW-13 cells, which do not have an active BAF complex due to the absence of BRG1 and hBRM subunits (10, 27). Transient expression of BRG1 reconstitutes the active BAF complex in SW-13 cells (45). IFITM1 was induced to high levels in HeLa cells by treatment with IFN-α, with maximal induction occurring between 4 and 8 h (Fig. 4A and B). In contrast, the levels of induction of IFITM1 mRNA by IFN-α were much lower in SW-13 cells, and the induction did not reach its plateau until approximately 12 h. However, the kinetics and levels of induction of another IFN target gene, ISG15, in HeLa cells and SW-13 cells were similar (Fig. 4C), suggesting that the deficient induction of IFITM1 in SW-13 cells was not caused by a defect in IFN signaling.
FIG. 4.
Expression of BRG1 results in more rapid kinetics and higher levels of the IFITM1 gene induction in SW-13 cells in response to IFN-α. (A) Analysis of IFITM1 mRNA expression induced by treatment with IFN-α. Total RNAs extracted from HeLa cells and SW-13 cells treated with IFN-α were reverse transcribed, amplified by PCR with IFITM1 primers, slot blotted onto nylon membrane, and detected by hybridization with a 32P-labeled IFITM1 cDNA probe. (B) The slot blot was quantified by PhosphorImager analysis and plotted after normalization to β actin signals. (C) The same samples were amplified with ISG15 primers and analyzed as described above. (D) SW-13 cells were transfected with pBJ5 or pBJ5-BRG1 for 24 h, followed by treatment with 500 U of IFN-α/ml for various times. The total RNAs were analyzed as described for panel A. (E) Quantification of the samples in panel D by PhosphorImager analysis.
To determine whether the slower kinetics and lower levels of induction of the IFITM1 gene in SW-13 cells was caused by the absence of the BRG1 protein, we transiently expressed BRG1 in SW-13 cells before the treatment with IFN-α. As shown in Fig. 4D, the expression of BRG1 alone induced low levels of IFITM1 expression and IFN-α treatment of BRG1-transfected cells resulted in synergistic activation of the gene. Induction with IFN-α alone reached a plateau at 12 h, but IFN-α treatment in the presence of BRG1 induced the expression of the gene at much higher levels, with an earlier plateau (at approximately 4 h) (Fig. 4E). The expression of BRG1 in SW-13 cells did not alter the induction of the ISG15 gene by IFN-α (data not shown). The expression of the ATPase-dead form of BRG1 did not have a significant effect on the induction of IFITM1 (data not shown), suggesting that the chromatin-remodeling activity of the BAF complex is required for rapid and high-level induction of this gene by IFN-α.
BRG1 increases the accessibility of the IFITM1 promoter.
In order to confirm the mechanism by which BRG1 facilitates the induction of the IFITM1 gene in response to IFN-α by remodeling its chromatin structure, we examined restriction enzyme accessibility at the IFITM1 promoter in the absence and presence of BRG1. SW-13 cells were transiently transfected with a BRG1 expression vector for 24 h, followed by stimulation with IFN-α. Nuclei were isolated and subjected to brief digestion with AvaII or HgiAI. Following complete digestion of the purified DNA with BclI, the cleavage sites were detected by linker ligation-mediated PCR as described above. Stimulation of SW-13 cells with IFN-α for 2 h slightly increased the accessibility of the promoter to AvaII (Fig. 5A and B, compare lanes 1 and 3) and HgiAI (Fig. 5C and D, lanes 1 to 3). Interestingly, IFN-α signaling in the presence of BRG1 dramatically increased the accessibility of the promoter to the restriction enzymes (Fig. 5A, compare lanes 1 and 4, and C, lanes 4 and 5). BRG1 alone without IFN-α stimulation also significantly increased accessibility (Fig. 5A, compare lanes 1 and 2, and C, compare lanes 1 and 4), suggesting that the recruitment of BRG1 to the IFITM1 promoter does not require IFN-α stimulation and that the BAF complex constitutively remodels the chromatin structure at the IFITM1 promoter to a more “open” conformation that is more accessible to transcription activators and RNA polymerase machinery.
Constitutive association of BRG1 with the IFITM1 promoter allows rapid recruitment of ISGF3 complex and RNA polymerase II.
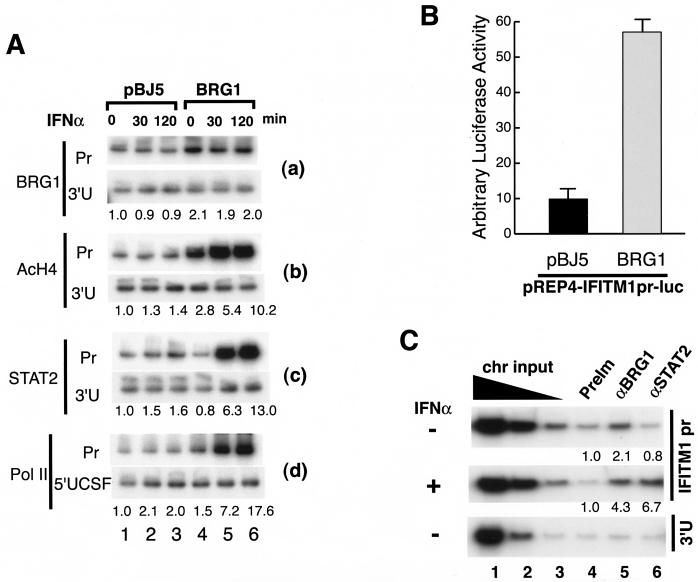
Using ChIP assays, we determined whether the BAF complex binds directly to the IFITM1 promoter (Fig. 6A). Compared to the 3′ untranslated region, the IFITM1 promoter sequence was reproducibly enriched about twofold by the BRG1 antibody from SW-13 cells transiently expressing BRG1 (Fig. 6A, panel a). Interestingly, BRG1 binding was observed even in the absence of IFN-α stimulation (Fig. 6A, compare lanes 1 to 3 and lanes 4 to 6 in panel a), consistent with the persistent open chromatin structure at the IFITM1 promoter in the presence of the active BAF complex. Furthermore, transient expression of BRG1 in SW-13 cells without IFN-α stimulation up-regulated the basal-level expression of the IFITM1 gene, as demonstrated by RT-PCR analysis (Fig. 4D) and by the activity of an IFITM1 luciferase reporter construct (Fig. 6B). The constitutive association of BRG1 with the IFITM1 promoter was not an artifact resulting from the overexpression of BRG1, since the endogenous BRG1 in HeLa cells was also associated with the IFITM1 promoter independent of IFN-α stimulation (Fig. 6C). In contrast, STAT2 association required stimulation with IFN-α (Fig. 6C). No BRG1 or STAT2 binding to the 3′ untranslated region of the IFITM2 gene was detected. Thus, the association of BRG1 with the IFITM1 promoter appears to be constitutive and does not require stimulation with IFN-α.
FIG. 6.
BRG1 is constitutively associated with the IFITM1 promoter. (A) SW-13 cells transfected with pBJ5 or pBJ5-BRG1 for 24 h were treated with 500 U of IFN-α/ml for indicated periods of time. Chromatin lysates were prepared and immunoprecipitated as described in the legend to Fig. 3C with the antibodies indicated on the left side of the panel. After reverse cross-linking, the immunoprecipitated DNA was analyzed by PCR with primers specific to the IFITM1 promoter region (Pr), the 3′ untranslated region (3′U), or the 5′ far-upstream region of the CSF1 gene (5′UCSF). The quantification of the data is shown below each panel. (B) The IFITM1 promoter in the pREP4 reporter vector was cotransfected with pBJ5 or pBJ5-BRG1 into SW-13 cells. Luciferase activity was determined after 24 h of transfection. (C) BRG1 is constitutively associated with the IFITM1 promoter in HeLa cells even without IFN-α stimulation. HeLa cells were stimulated with 500 U of IFN-α/ml for 30 min, followed by chromatin preparation and chromatin immunoprecipitation as described in the legend to Fig. 3C. The purified DNA was analyzed by PCR with the IFITM1 promoter primers (top two panels) or primers for the 3′ untranslated region of the IFITM2 gene (bottom panel). The chromatin (chr) input was diluted 5 times at each step. The preimmune serum (PreIm) and antibodies are indicated above the panels. The quantification of the data after normalization to the preimmune serum is shown below each panel.
Next, we evaluated the levels of histone H4 acetylation at the IFITM1 promoter in the presence and absence of BRG1 and IFN-α. We found that the presence of BRG1 increased the acetylation of histone H4 about threefold at the IFITM1 promoter region compared to the level at the 3′ untranslated region (Fig. 6A, compare lanes 1 and 4 in panel b), consistent with a more open chromatin structure at the promoter region. IFN-α stimulation for 2 h further increased the H4 acetylation at the promoter about 10-fold (Fig. 6A, panel b), resulting in a more open chromatin structure, as demonstrated by the restriction enzyme accessibility data (Fig. 5).
IFN-α signaling activates the ISGF3 complex consisting of STAT1, STAT2, and p48, which relocates to the nucleus and binds to its target promoters (7). As shown in Fig. 6A, panel c, only low levels of STAT2 were associated with the IFITM1 promoter without BRG1, while in the presence of BRG1, IFN-α induced a strong association of STAT2 with the promoter (compare lanes 1 to 3 and lanes 4 to 6). A similar observation was made for p48 (data not shown). As expected, RNA polymerase II binding to the IFITM1 promoter was also enhanced by the presence of BRG1 and stimulation with IFN-α (Fig. 6A, panel d, compare lanes 1 to 3 and lanes 4 to 6). However, significant binding to the 3′ untranslated region (data not shown), but not to the 5′ far-upstream region of CSF1 promoter, was also detected in the presence of BRG1 and IFN-α. The data indicate that significant binding of RNA polymerase II does not occur at the IFITM1 promoter until after IFN-α stimulation. This finding shows that although the chromatin structure of the promoter is already remodeled by the prebound BAF complex, the transcription machinery is not present on the promoter in a poised state. Instead, its assembly is dependent on the cellular signaling cascade. These results show that constitutive binding of the BAF complex facilitated histone H4 acetylation, ISGF3 binding, and the transcription machinery assembly on the IFITM1 promoter upon stimulation with IFN-α.
DISCUSSION
The BAF complex regulates the IFN signaling pathways by multiple mechanisms.
The effectiveness of the cellular antiviral activities is dependent on the rapid transcriptional activation of IFNs and their target genes. Viral infection first activates transcription factors such as IRF3 and NF-κB that bind to their recognition sites in the IFN promoters and activate transcription. Induced IFNs bind to their cell surface receptors and induce the activation of the ISGF3 complex that binds to ISREs and triggers the induction of genes involved in antiviral activities. That the BAF complex has a pivotal role in the expression of not only IFN-inducible genes but also virus-inducible genes is supported by the marked reduction of a series of IFN and poly(I)/poly(C)-inducible genes.
Clearly, the BAF complex mediates cellular antiviral activities by multiple mechanisms. First, this complex is required for maintaining certain basal levels of the IFN target proteins of the antiviral systems, such as Mx1, PKR, OAS3, IFITM1, and ISG20. The existence of the antiviral protein pool at certain levels in the cells may represent the first cellular defense or a barrier against invading pathogens even before the induction of the IFN signaling systems. Indeed, when these proteins were down-regulated by the inhibition of BAF47, the NDV viral yields were significantly higher, suggesting that the innate antiviral activity was impaired (Fig. 2). Second, the BAF complex is required for the induced levels of both the regulators and the target proteins, suggesting that it may play essential roles in IFN-mediated antiviral activities. Indeed, our results show that IFN failed to protect the cells from virus in the absence of BAF47 (Fig. 2). Therefore, the BAF complex controls the antiviral systems at multiple steps in the pathways.
Promoter priming by the BAF complex.
The BAF complex regulates transcription by modifying the chromatin structure. It has been suggested that chromatin-remodeling complexes are recruited to target sites in situ at the time of stimulation (2, 35). In this report, the following observations indicate that the BAF complex regulates the IFN signaling process by a novel mechanism. (i) Reconstitution of the active BAF complex by transient expression of BRG1 in SW-13 cells enhanced the chromatin accessibility of the IFITM1 promoter (Fig. 5), increased its levels of histone H4 acetylation (Fig. 6), and up-regulated the basal-level expression of the IFITM1 gene (Fig. 4) in the absence of IFN-α treatment. (ii) Knockdown of the endogenous BAF complex in HeLa cells by expression of the siRNA targeting BAF47 down-regulated the basal-level expression of the IFITM1 gene (Fig. 1) and reduced the levels of histone H4 acetylation and chromatin accessibility of the IFITM1 promoter (Fig. 3), suggesting that the BAF complex may play a role in modifying the chromatin structure of the promoter even without IFN-α stimulation. (iii) Our ChIP results demonstrate that BRG1 is directly bound to the IFITM1 promoter both in the absence and in the presence of IFN-α (Fig. 6). Furthermore, we found that BRG1 also binds constitutively to the promoters of IFITM3, IFITM2, STAT2, Mx1, OAS3, and TLR3 genes (data not shown). Because of the limited sensitivity of the ChIP assays, we could not conclusively determine whether BAF binding to these promoters was enhanced by IFN treatment.
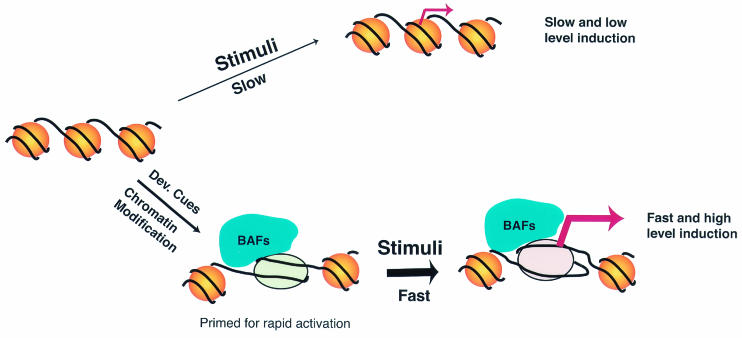
All of these data argue that the binding of the BAF complex does not require IFN-α stimulation, at least in a subset of the IFN target promoters. Therefore, we suggest that in addition to being recruited to some target promoters upon stimulation (2), the BAF complex binds constitutively to a set of IFN-inducible promoters and utilizes ATP-derived energy to maintain the promoters in an open configuration. Without prior chromatin remodeling by the BAF complex, the ISGF3 complex may not efficiently bind the IFITM1 promoter to activate transcription. Following the binding of the ISGF3 complex, histone acetylase such as p300/CBP or GCN5 is recruited to the promoter and results in a more open and stable chromatin structure, which promotes the assembly of the transcription machinery and transcription initiation (5, 29). The chromatin structure at the promoters remodeled by the BAF complex may serve as a marker important for more rapid and higher levels of induction in response to IFN stimulation, as illustrated in Fig. 7. Such a marker provides a mechanism to trigger rapid cellular antiviral responses upon infection of the host.
FIG. 7.
The BAF complex primes the chromatin structure of IFN target promoters for more rapid kinetics and higher levels of induction in response to stimuli.
There are two not-mutually-exclusive potential mechanisms for the constitutive association of BRG1 with the target promoters. One is the constitutive basal-level acetylation of histones that may serve as the docking sites for the BAF complex (9, 15). The other is the possible recruitment of the BAF complex by unidentified transcription factors that bind constitutively to the promoters and are required for the basal-level expression of the genes. This hypothesis is currently under investigation.
Acknowledgments
We thank Tian Chi, John Kelly, Warren Leonard, and Carl Wu for critical reading of the manuscript. K.Z. thanks Warren Leonard for support.
This work was supported by intramural grants to the National Heart, Lung, and Blood Institute, National Institutes of Health.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does ′chromatin remodeling' mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal, S., and A. Rao. 1998. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9:765-775. [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature 383:344-347. [DOI] [PubMed] [Google Scholar]
- 6.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 7.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 8.de La Serna, I. L., K. A. Carlson, and A. N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187-190. [DOI] [PubMed] [Google Scholar]
- 9.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 10.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 11.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 12.Geng, F., Y. Cao, and B. C. Laurent. 2001. Essential roles of Snf5p in Snf-Swi chromatin remodeling in vivo. Mol. Cell. Biol. 21:4311-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidi, C. J., A. T. Sands, B. P. Zambrowicz, T. K. Turner, D. A. Demers, W. Webster, T. W. Smith, A. N. Imbalzano, and S. N. Jones. 2001. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21:3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 15.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 16.Huang, M., F. Qian, Y. Hu, C. Ang, Z. Li, and Z. Wen. 2002. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat. Cell Biol. 4:774-781. [DOI] [PubMed] [Google Scholar]
- 17.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481-485. [DOI] [PubMed] [Google Scholar]
- 18.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 19.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, T., K. Nakayama, J. Penninger, M. Kitagawa, H. Harada, T. Matsuyama, N. Tanaka, R. Kamijo, J. Vilcek, T. W. Mak, et al. 1994. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science 264:1921-1924. [DOI] [PubMed] [Google Scholar]
- 21.Klochendler-Yeivin, A., L. Fiette, J. Barra, C. Muchardt, C. Babinet, and M. Yaniv. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krebs, J. E., M. H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonard, W. J., and J. J. O'Shea. 1998. Jaks and STATs: biological implications. Annu. Rev. Immunol. 16:293-322. [DOI] [PubMed] [Google Scholar]
- 24.Liu, H., H. Kang, R. Liu, X. Chen, and K. Zhao. 2002. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 22:6471-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, R., H. Liu, X. Chen, M. Kirby, P. O. Brown, and K. Zhao. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309-318. [DOI] [PubMed] [Google Scholar]
- 26.Muchardt, C., and M. Yaniv. 1999. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J. Mol. Biol. 293:187-198. [DOI] [PubMed] [Google Scholar]
- 27.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246:780-786. [DOI] [PubMed] [Google Scholar]
- 29.Paulson, M., C. Press, E. Smith, N. Tanese, and D. E. Levy. 2002. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 4:140-147. [DOI] [PubMed] [Google Scholar]
- 30.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 31.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 32.Reinke, H., P. D. Gregory, and W. Horz. 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell 7:529-538. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, C. W., S. A. Galusha, M. E. McMenamin, C. D. Fletcher, and S. H. Orkin. 2000. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. USA 97:13796-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 35.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 36.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 37.Sudarsanam, P., and F. Winston. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 38.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varga-Weisz, P. 2001. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 20:3076-3085. [DOI] [PubMed] [Google Scholar]
- 40.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 41.Wan, M., K. Zhao, S. S. Lee, and U. Francke. 2001. MECP2 truncating mutations cause histone H4 hyperacetylation in Rett syndrome. Hum. Mol. Genet. 10:1085-1092. [DOI] [PubMed] [Google Scholar]
- 42.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 43.Weinmann, A. S., S. E. Plevy, and S. T. Smale. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665-675. [DOI] [PubMed] [Google Scholar]
- 44.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25:619-623. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, K., W. Wang, O. J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G. R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625-636. [DOI] [PubMed] [Google Scholar]