Abstract
Background
Radioresistance in human tumors has been linked in part to a subset of cells termed cancer stem cells (CSCs). The prominin 1 (CD133) cell surface protein is proposed to be a marker enriching for CSCs. We explore the importance of DNA repair in contributing to radioresistance in CD133+ lung cancer cells.
Materials and Methods
A549 and H1299 lung cancer cell lines were used. Sorted CD133+ cells were exposed to either single 4Gy or 8Gy doses and clonogenic survival measured. γ-H2AX immunofluorescence and quantitative real time PCR was performed on sorted CD133+ cells both in the absence of IR and after two single 4Gy doses. Lentiviral shRNA was used to silence repair genes.
Results
A549 but not H1299 cells expand their CD133+ population after single 4Gy exposure, and isolated A549 CD133+ cells demonstrate IR resistance. This resistance corresponded with enhanced repair of DNA double strand breaks (DSBs) and upregulated expression of DSB repair genes in A549 cells. Prior IR exposure of two single 4Gy doses resulted in acquired DNA repair upregulation and improved repair proficiency in both A549 and H1299. Finally Exo1 and Rad51 silencing in A549 cells abrogated the CD133+ IR expansion phenotype and induced IR sensitivity in sorted CD133+ cells.
Conclusions
CD133 identifies a population of cells within specific tumor types containing altered expression of DNA repair genes that are inducible upon exposure to chemotherapy. This altered gene expression contributes to enhanced DSB resolution and the radioresistance phenotype of these cells. We also identify DNA repair genes which may serve as promising therapeutic targets to confer radiosensitivity to CSCs.
Keywords: Cancer stem cells, CD133, Radiation resistance
Introduction
The emergence of cancer stem cells (CSCs) in recent years as drivers of tumor maintenance and chemoresistance has resulted in new approaches to studying the biology of cancer. CSCs have been implicated in a multitude of human malignancies including tumors of the brain, breast, lung, colon, pancreas, and others [1–5]. While they are identified using cell surface markers, CSCs are phenotypically distinct from the bulk of a tumor by their ability to reconstitute tumors using a small number of cells. CSCs have been associated with an increased metastatic potential and chemoresistance in multiple human cancers and thus may be a significant contributor to tumor relapse and treatment failure [6, 7].
One such marker believed to enrich for a CSC population is Prominin 1 (CD133). CD133 is a transmembrane glycoprotein with still unknown function, although it is found in embryonic stem cells, normal adult stem cells and circulating endothelial progenitors. CD133 has three known isoforms (CD133-1, CD133-2, CD133-3) which are expressed on different tissue types, and loss of CD133 has been correlated with an increase in terminal differentiation[8, 9]. CD133 expression has been associated with chemoresistance and increased metastatic potential in multiple human cancers although the mechanistic relationship between Prominin 1 and these phenotypes is still unknown.
Many factors are thought to contribute to the chemoresistance observed in cancer stem cells including an upregulated capacity for DNA repair. Bao et. al demonstrated that human glioma stem cells become enriched after radiation therapy in primary tumors. Sorted CD133+ cells were shown to exhibit an activated capacity for DNA repair, including basal phosphorylation of the checkpoint proteins Chk1 and Chk2. Inhibition of Chk1 with a selective inhibitor resulted in subsequent IR sensitivity suggesting that DNA repair proteins may provide promising therapeutic targets for CSCs[10]. While groups have demonstrated an increased DNA repair capacity in CD133+ cells including in medulloblastoma cells[11] and prostate cancer cell lines [12] others have shown that CD133+ cells do not equate to increased radiation resistance nor to an enhanced DNA repair capacity [13, 14] and thus the impact of CD133 on radioresistance and enhanced DNA repair may be tumor type specific.
In human lung cancer cells, CD133 has also been shown to enrich for a cancer stem cell like population[3, 15]. Indeed Shien et. al (2012) examined the expression of CD133 in 50 patients with non-small cell lung cancer and found that those patients with CD133− positive samples had significantly worse 5-year survival rates than those that were CD133− negative[16]. Iida et. al have proposed that hypoxic conditions promote CD133 expression in lung cancer cell lines via Oct4 and Sox2 activity[17] and groups including Bertolini et. al have shown that in lung cancer cell lines CD133+ cells are enriched in response to cisplatin therapy[18]. Liu et. al have recently proposed that this resistance is due to activation of the notch pathway and that sensitivity can be induced via notch inhibition[19]. However the role that DNA repair plays in contributing to IR resistance in CD133+ lung cancer cells has yet to be fully elucidated.
In this manuscript we demonstrate the role that DNA repair capacity plays in the ionizing radiation (IR) response of CD133 populations in two human lung cancer cells lines. We compare CD133+ IR resistance in A549 cells against H1299 cells and show that CD133 contributes to radiation resistance in only A549 cells in part due to increased capacity to resolve DNA double strand breaks (DSBs). We additionally examined the DNA repair capacity and mRNA gene expression levels of multiple DNA repair genes in CD133+ vs. CD133− cells and found that only in A549 cells did the CD133+ population exhibit a higher basal expression of these genes. We treated both cell lines with multiple exposures of ionizing radiation and after a recovery period found that CD133+ cells demonstrated an acquired IR resistance in both cell lines, and that in H1299 cells this corresponded with an induction of DNA repair expression. Finally we used lentiviral mediated shRNA silencing of two consistently upregulated CD133+ repair genes (Rad51 and Exo1) in A549 cells and showed that this inhibition abrogated CD133 expansion post IR and also conferred radiation sensitivity in CD133+ cells. This work demonstrates that CD133 identifies a population of cells with altered expression of DNA repair genes that are induced by exposure to radiation therapy. This altered expression appears to contribute to radiation resistance of these cells, thus identification of critical repair elements in CD133+ cells may yield promising therapeutic targets.
Methods
Cell lines
Human non small cell lung cancer cell lines A549 and H1299 acquired from ATCC were used in these studies. H1299 cells contain a homozygous partial deletion of p53 which results in loss of p53 protein while A549 cells are WT for p53 but contain a KRAS mutation.
Flow cytometry
For CD133 IR expansion studies A549 and H1299 cells were treated with 4Gy IR and at multiple time points cells were trypsinized, incubated with CD133 antibody (293C3-Miltenyi Biotec), and flow cytometry performed on a BD LSRII instrument to measure the percent of PE+ cells.
CD133 Separation
Separation of CD133+ cells from CD133− cells was performed using the CD133 microbead kit (Miltenyi Biotec) and MACs separation columns (MS columns-Miltenyi). Separation was confirmed via flow cytometry using the CD133 antibody on sorted cells.
Clonogenic Survival Assays
After CD133 separation, 100 cells were plated in 3 cm2 plates and treated with an IR dose range. 10–14 days post treatment the plates were stained with crystal violet for colony counting. Only colonies containing >50 cells were counted. Student’s t-tests were performed to determine significance.
Immunostaining
Cells were grown on coverslips and 24 hours post plating were treated with 4Gy of ionizing radiation. At the indicated time points, cells were fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, blocked with 5% NGS, and incubated at 1:500 for one hour in primary phospho-histone H2AX antibody (Millipore-05-636). Cells were washed three times in PBS and incubated at 1:500 for 45 minutes with Alexa Fluor 488. Coverslips were washed and mounted onto coverslips with Dapi fluoromount G (southern biotech). Images were acquired with a fluorescent microscope (Nikon) with a 40× objective and analyzed in AIM Image Browser.
Real-time PCR
Separated CD133 cells were assessed for gene expression via realtime PCR. RNA was extracted using the Trizol method from CD133− and CD133+ cells and cDNA synthesis was performed (Superscript III First Strand Kit-Invitrogen). Real-time PCR was performed using validated primers (Applied Biosystems). Student’s t-tests were performed to determine significance.
Lentiviral Gene Silencing
Rad51 and Exo1 were silenced via shRNA transduction with validated clones (Sigma). Lentiviral particles were synthesized via HEK293 cells and target cells were infected, selected for with puromycin, and clones were assessed for verification of gene silencing.
Results
CD133+ cell enrichment post IR treatment
Lung cancer cells have previously been shown to expand their CD133 population following chemotherapy[18]. Here we wanted to assess the effect of ionizing radiation on this CD133 expansion phenotype. A549 and H1299 cells were treated with 4Gy IR and the percent of CD133+ cells in culture at multiple timepoints (24, 48, 72 hrs post treatment) was assessed via flow cytometry. We show in A549 cells by 48 hours the percentage of CD133+ cells had more than doubled compared to untreated, and expanded approximately five-fold by 72 hours. In H1299 cells the expansion was not significant at any time point measured (Supplement 1).
Radiosensitivity and DNA repair capacity of separated CD133+ cells
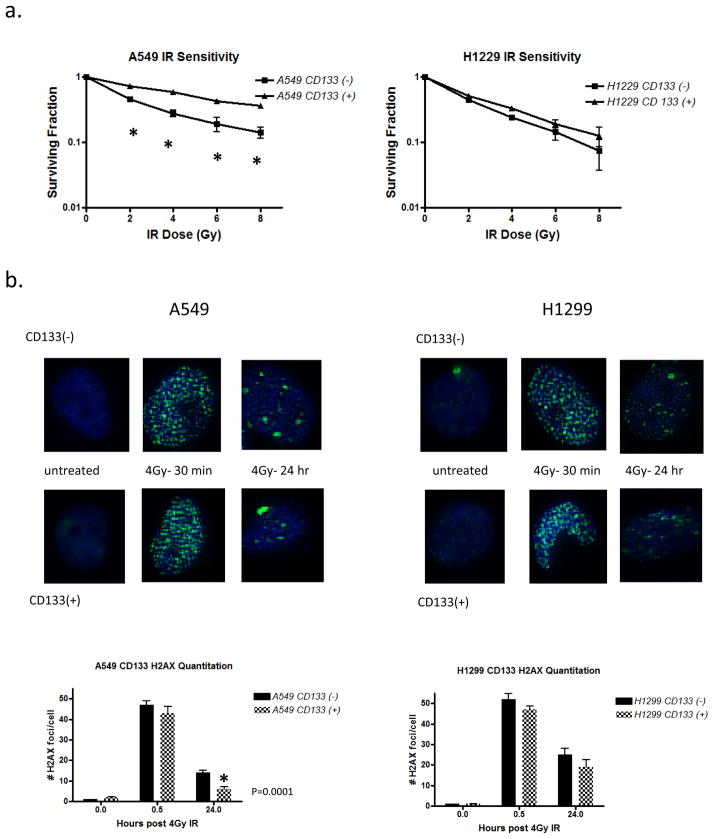
To study whether CD133 contributes to radiation resistance in these cell lines we performed magnetic separation of CD133+ from CD133− cells (Miltenyi) and subjected these sorted cells to an IR dose range of 2Gy-8Gy. The purity of magnetically sorted cells was confirmed via flow cytometry (Supplement 2) Cell viability was assessed 10–14 days post treatment via clonogenic survival. In A549 cells separated CD133+ cells conferred approximately a 2-fold increase in radiation resistance, while H1299 cells demonstrated an insignificant difference in IR sensitivity (Figure 1a). To assess the role of DNA repair proficiency in the radiation response data we quantified γ-H2AX immunofluorescence staining as a measure of DNA double strand breaks on CD133− and CD133+ cells from A549 and H1299 cells at multiple time points following irradiation. We found that in A549 cells exclusively the CD133+ cells were able to completely resolve DSBs by 24 hours post treatment while the CD13− cells retained persistent H2AX foci. In H1299 cells both CD133− and CD133+ populations retained some persistent DSBs by 24 hours (Figure 1b). This data corroborates previous suggestions that CD133 serves as a marker for radioresistance only in specific cell and tumor types.
Figure 1. The CD133 marker promotes IR resistance in A549 cells but not H1229 cells.
(a) CD133+ cells were magnetically separated from CD133− cells and IR sensitivity was measured via clonogenic survival on the two populations. Cells were treated with an IR dose range of 2–8Gy and permitted to grow for 10–14 days prior to survival analysis. Unpaired t-test performed with two-tailed p value shown, error bars = SEM. (b) Representative images of γ-H2AX immunostaining. 50 cells per treatment were analyzed for foci formation and the % positive was plotted.
DNA repair gene expression analysis in CD133+ cells
To confirm that the increased ability of A549 CD133+ cells to resolve H2AX foci was due to an enhanced capacity for DNA repair we performed real time PCR analysis on separated CD133+ cells in A549 and H1299 cells. We hypothesized that the level of IR resistance observed in Figure 1 would correlate with basal gene expression levels of DNA repair genes.
We examined the expression of genes implicated in multiple DNA repair pathways, but a particular focus was given to DNA double strand break (DSB) repair pathways due to the dominant type of IR induced DNA lesions. Double strand breaks are repaired by two conserved pathways in mammalian cells, homologous recombination (HR) and non-homologous end joining (NHEJ). HR has higher fidelity but requires a homologous sister chromatid for repair thus can only be utilized in the S and G2 phase of the cell cycle[20, 21].
In addition to DSB repair genes BRCA1, Exo1, Mre11, Rad51, Ku70 and DNA PKC we included representative genes critical for the DNA checkpoint response (Chk1), crosslink repair (Fan1), and mismatch repair (Msh2). Gene expression analysis demonstrated that A549 cells contained basal upregulation of all genes examined in the CD133+ population. On the other hand in H1299 cells we observed no significant increase in any of the genes analyzed (Figure 2), similar to Figure 1 in which only A549 CD133+ cells demonstrated IR resistance and an increased capacity for DSB repair. This expression data was confirmed using western blotting (Supplement 3).
Figure 2. CD133+ cells display basally upregulated DNA repair genes in A549 cells only.
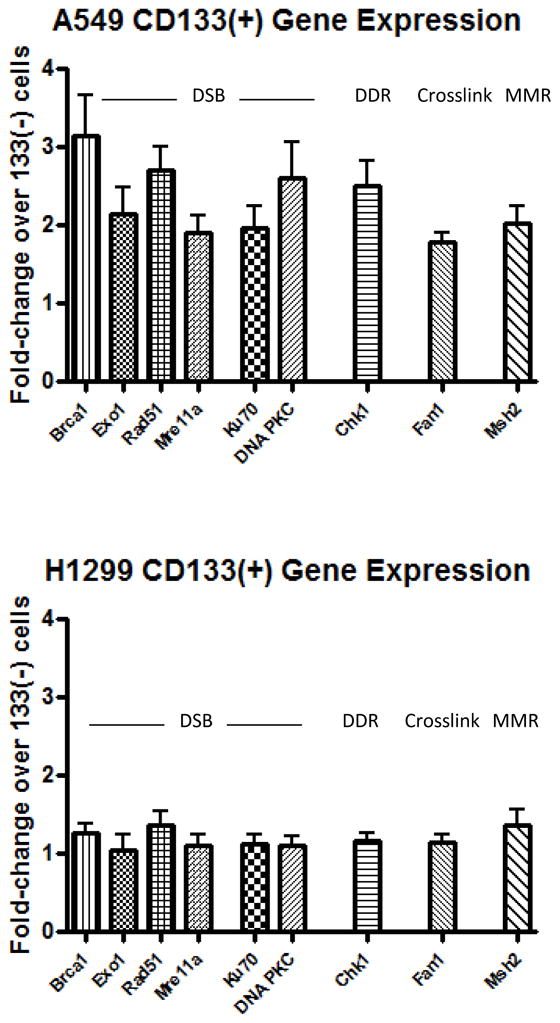
RNA was isolated from magnetically separated CD133− and CD133+ cells in A549 (A) and H1229 (B) cell lines and real time PCR analysis was performed examining multiple DNA repair genes. Error bars= SEM.
Contribution of IR exposure to DNA repair induction in CD133+ cells
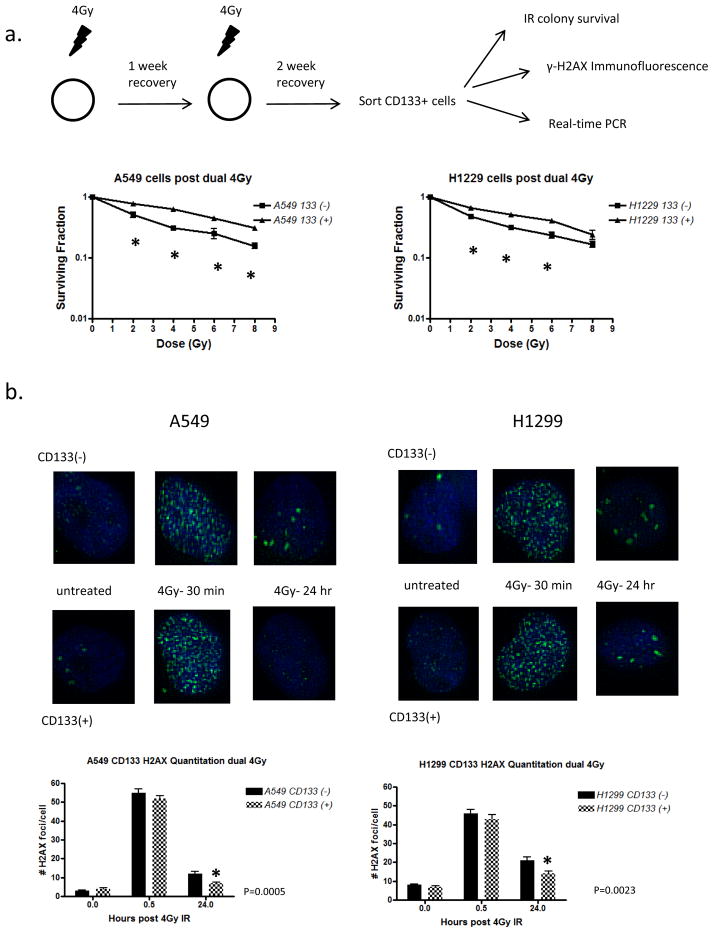
While DNA repair capacity did not appear to be critical for the CD133+ IR sensitivity of H1299 cells, we asked whether prior exposure to IR would result in an activation of CD133+ cells and enhance their capacity for repair. This would help define whether IR in physiological settings contribute to an acquired CD133+ radioresistance.
We irradiated A549 and H1299 cells with 4Gy IR and after a 1 week recovery period, exposed them again to 4Gy IR. After another 2 week recovery period these cells were harvested and CD133+ cells separated. IR sensitivity, persistence of γ-H2AX, and DNA repair gene expression was measured in CD133+ vs. CD133− cells. In A549 cells we again found that CD133+ cells displayed increased radioresistance over CD133− cells at each dose measured, however we also observed that H1299 CD133+ cells exhibited a significant increase in IR resistance at the 2Gy, 4Gy, and 6Gy doses, with a trend toward increased resistance at 8Gy (Figure 3a). To determine whether an increase in DNA repair capacity contributed to this IR resistance we again performed γ-H2AX staining on sorted CD133+ and CD133− cells and found that both A549 and H1299 CD133+ cells were able to resolve DSBs by 24 hours post treatment (Figure 3b). Gene expression analysis confirmed this data as H1299 cells demonstrated a moderate increase in expression of all genes analyzed (Figure 4). The DNA repair expression and IR sensitivity changes of CD133+ H1299 cells after IR recovery suggests that prior IR exposure has a repair gene-inducing effect on CD133+ cells and results in a promotion of radiation resistance.
Figure 3. CD133 contributes to radioresistance in cells surviving multiple IR doses in both A549 and H1229 cells via upregulation of DNA repair genes.
(a) A549 and H1229 cells were treated as diagrammed. Sorted cells were treated with an IR dose range and RNA was extracted for real time PCR analysis. Error bars = SEM. (b) Representative images of γ-H2AX immunostaining. 25 cells per treatment were analyzed.
Figure 4.
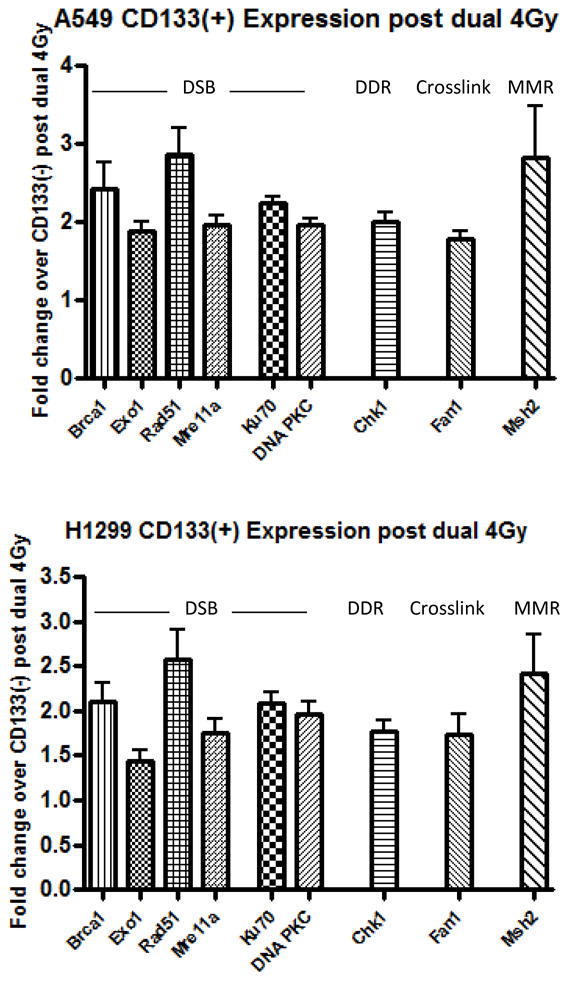
CD133+ cells display upregulated DNA repair genes in both A549 and H1299 following IR exposure RNA was isolated from magnetically separated CD133− and CD133+ cells in A549 (A) and H1229 (B) cell lines and real time PCR analysis was performed examining multiple DNA repair genes. Error bars= SEM.
Exo1 and Rad51 silencing in A549 cells and effects on CD133+ cells
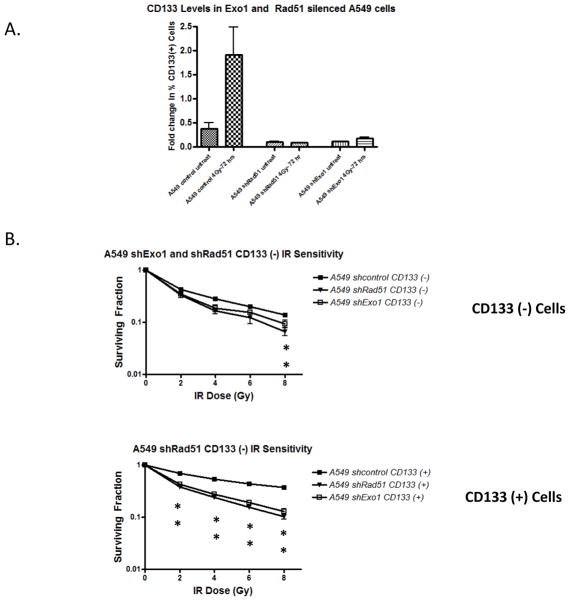
After determining that DNA repair contributes to CD133+ radioresistance in A549 cells, we were interested in studying whether silencing critical repair proteins would confer radiation sensitivity on these cells. For this study we silenced Rad51 and Exonuclease1 (Exo1) due to their observed upregulation in CD133+ cells and the fact that homologous recombination has been proposed to be vital to CSCs in their response to chemotherapy. We sought to determine the subsequent effect on CD133+ IR induced expansion as well as CD133+ IR sensitivity. Rad51 is a critical HR protein that controls strand invasion during homologous mediated repair. Rad51 foci form rapidly after DSB induction and loss of Rad51 is associated with severe HR defects[22, 23]. Additionally Rad51 has been proposed to be an effective therapeutic target in multiple cancer models[24, 25]. Exo1 is another critical HR protein although its precise role in the pathway is still being elucidated. Exo1 has been implicated in the early end resection step of HR and its loss is associated not only with defective HR but also with impaired checkpoint activation[26, 27].
We silenced these genes using lentiviral shRNA (Supplement 4) and examined IR expansion and CD133+ IR sensitivity. While A549 cells transduced with a scrambled shRNA expanded significantly 72 hours post IR, silencing of both Exo1 and Rad51 resulted in complete loss of the expansion. In addition separated CD133+ cells from both A549 shExo1 and shRad51 display increased radiation sensitivity compared to the control cells at each dose tested, while CD133− cells were only more sensitive than control at the 8Gy dose (Figure 5). This data suggests that the upregulation of DNA repair observed in A549 cells does contribute directly to the IR resistance, and thus inhibition of appropriate repair proteins may be an effective way to target CD133+ cells.
Figure 5. Rad51 and Exo1 silencing in A549 cells abrogates the IR expansion phenotype and induces CD133 IR sensitivity.
(A) Exo1 and Rad51 were silenced via lentiviral transduction in A549 cells. Mixed populations were irradiated with 4Gy IR and the percent CD133+ cells 72 hours post IR were measured.
(B) CD133+ cells were magnetically separated from CD133− cells and IR sensitivity was measured via clonogenic survival. Cells were treated with either 4Gy or 8Gy and permitted to grow for 10–14 days prior to survival analysis.
Discussion
This work provides insight into the role of DNA repair in promoting radiation resistance in CD133+ lung cancer cells. We showed that CD133 selected for a radioresistant population in A549 but not H1299 cells and that this correlated with their altered basal expression of important DNA repair genes as well as enhanced DSB repair capacity. Additionally we showed that CD133+ cells surviving multiple rounds of IR treatment induce genes associated with an increased capacity for DNA repair. In H1299 cells the previously sensitive CD133+ population acquired radiation resistance which was linked to enhanced DNA repair capacity in these cells. Finally we demonstrated that silencing critical DNA repair genes abrogated the expansion of A549 cells after IR treatment and also induced IR sensitivity in CD133+ cells. Our work begins to characterize an IR induced enrichment and expansion of CD133+ cells caused in part by upregulation of DNA repair capacity, and also suggests that inhibition of DNA repair proteins may provide a promising therapeutic direction. Since CSCs appear in part responsible to post treatment rebound and relapse of many solid tumors, and since we have observed that CSCs rely on DNA repair, targeting these pathways may impact tumor regrowth.
The case for targeted approaches to CSC therapy is becoming stronger. As discussed earlier Bao et. al and others have demonstrated in glioblastomas that CD133+ cells are more radioresistant than CD133− and that Chk1 specific inhibitors can specifically target these cells. Work in other cancer models has suggested that this may also be true for a plethora of diseases. Mihatsch et. al (2011) used ALDH1 as a marker for cancer stem cells in human lung cancer (A549) and breast cancer (SK-BR-3) and found that ALDH1+ cells displayed increased radioresistance and increased DNA repair capacity, and that PI3K inhibition induced significant radiation sensitivity in these cells[28]. Other groups including the previously mentioned Liu et. al demonstrated that CD133+ cells in H460 and H661 lung adenocarcinomas were also resistant to cisplatin and that pre-treatment with γ-secretase inhibitors or Notch1 shRNA resulted in increased drug sensitivity. Mizugaki et. al (2013) analyzed primary samples of human patients with non-small cell lung cancers and found that high CD133 expression correlated with late pathological stages of the disease and that high expression was correlated with an unfavorable prognosis[29], suggesting that the in vitro data correlates with what is observed in the human disease. Similar to Roppolo et. al (2009) and Dittfield et. al (2009) we show that CD133+ resistance to therapy may be tumor type specific, as A549 but not H1299 CD133+ cells correlated with increased radiation resistance, but our work goes in detail to demonstrate that increased DNA repair capacity, and in particular a reliance on specific DNA repair genes may be a major contributing factor for observed radiation resistance.
Our work identified Rad51 as a gene of interest contributing to CD133 radiation resistance. Rad51 has been proposed as a suitable therapeutic target in multiple cancer types due to its critical role in the strand invasion step of HR. Ko et. al (2008) silenced Rad51 in human lung cancer lines A549 and H1650 and found that it increased sensitivity to both cisplatin and mitomycin C[25]. Others such as Tsai et. al (2010) also examined Rad51 silencing in human lung cancer cells and found they were able to induce sensitivity of previously drug resistant cells to gemcitabine[30]. Quiao et. al (2005) additionally demonstrate in human non-small cell lung cancer patient samples that high Rad51 expression correlated with a worse patient prognosis. Our data shows that Rad51 silencing in human lung cancer cells induces radiation sensitivity due to the reliance of CD133+ cells on this gene for DSB repair, and thus may be a suitable target for all CSCs demonstrating increased DNA reliance.
Exo1 on the other hand is a very interesting potential therapeutic target. Exo1 has been implicated in the end resection step of HR and thus is essential for activation of not only homologous recombination but also cell cycle checkpoints. While it has yet to be targeted for cancer therapy studies we believe that its ability to confer IR sensitivity to CD133+ cells, as well as its possible disruption of the checkpoint activation also makes it a very desirable target for cancer therapy. We identified additional DNA repair factors, notably BRCA1 and DNA-PKcs which were significantly upregulated basally in A549 CD133+ cells and could also serve as promising CSC specific therapeutic targets. While their targeting has been proposed in multiple cancer models, their efficacy in inducing radiosensitivity in CSCs has largely yet to be explored [31–33].
Multiple factors are believed to contribute to the radioresistance observed in cancer stem cells. In addition to the DNA repair aspect that we have discussed in this manuscript, CSCs are believed to rely on a hypoxic niche environment which has been shown to result in therapy resistance and metastatic potential. Groups attempting to disrupt this niche via inhibition of the hypoxia inducible factors have had success in conferring sensitivity in glioblastoma models[34]. A point of interest for our work is that groups have demonstrated that hypoxic conditions can result in decreased expression of homologous recombination proteins including Rad51. Chan et. al (2008) demonstrate this in H1299 cells cultured chronically at 0.2% O2[35], and Bindra et. al (2007) use MCF-7 cells cultured at 0.5% 02 to show that Rad51 is repressed under hypoxic conditions[36]. How these studies relate to CSCs is not currently well understood. Chan et. al demonstrate that the hypoxic cells with decreased HR capacity were also hypersensitive to MMC and cisplatin, which contradicts what has been shown in other CSC models. Thus the role of hypoxia on HR functionality in CD133+ and other CSC cells needs to be further examined, as it will be interesting to determine the hypoxic response in CD133+ vs. CD133− cells.
CSCs are also believed to maintain an environment with very low levels of reactive oxygen species (ROS) via upregulation of ROS scavengers to minimize potential genomic damage. Efforts to inhibit scavengers such as glutathione are also underway[37]. Other hypotheses such as activation of signaling pathways and drug efflux pumps have also been proposed[38, 39]. Thus effective CSC specific therapies will likely involve some combination of drugs targeting these multiple mechanisms, but our work begins to demonstrate the importance of DNA repair as a mediator of IR resistance in human lung cancer cells and establishes potential targets to combat this resistance.
Supplementary Material
Supplement 1: CD133+ cells expand following ionizing radiation in human lung cancer cell lines.
A549 and H1299 human lung cancer cell lines were treated with 4Gy ionizing radiation and the percent positive CD133 cells were measured using cytometric analysis every 24 hours post treatment.
Supplemental 2: Purity of magnetically sorted CD133 cells
Cells were sorted via magnetic separation (Miltenyi). Sorted cells were stained with PE tagged CD133 antibody (293C-Miltenyi) and analyzed via flow cytometry.
Supplemental 3: Confirmation of gene expression data using western blotting
CD133+ cells were magnetically separated from A549 and H1299 cells. Western blot analysis was performed on cell lysates from each population probing for DNA repair proteins Exo1, Ku70, Rad51 and Msh2 to demonstrate basal protein levels in CD133− and CD133+ cells.
Supplemental 4: Confirmation of lentiviral knockdown of Exo1 and Rad51 in A549 cells
A549 cells were transduced with lentiviral shRNA specific to Exo1, Rad51, or scrambled control (Sigma). After puromycin selection cells were probed for Exo1 and Rad51 using western blot analysis to determine the levels of protein knockdown.
Acknowledgments
This work was supported by the Cytometry & Imaging Microscopy Core Facility of the Case Comprehensive Cancer Center (P30CA43703) and the Radiation Resources Core Facility of the Case Comprehensive Cancer Center (P30 CA43703). It was also supported by National Institutes of health grants R01AG024916 and R01CA063193 and molecular therapeutics grant 5T32GM008803-10.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Al-Hajj M, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104(24):10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eramo A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15(3):504–14. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 4.Li C, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 5.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 6.Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26(17):2839–45. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Y, Ramena G, Elble RC. The role of cancer stem cells in relapse of solid tumors. Front Biosci (Elite Ed) 4:1528–41. doi: 10.2741/e478. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Li SY. Research progression of CD133 as a marker of cancer stem cells. Chin J Cancer. 29(3):243–7. doi: 10.5732/cjc.009.10587. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, et al. CD133, Stem Cells, and Cancer Stem Cells: Myth or Reality? Curr Colorectal Cancer Rep. 7(4):253–259. doi: 10.1007/s11888-011-0106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 11.Blazek ER, Foutch JL, Maki G. Daoy medulloblastoma cells that express CD133 are radioresistant relative to CD133− cells, and the CD133+ sector is enlarged by hypoxia. Int J Radiat Oncol Biol Phys. 2007;67(1):1–5. doi: 10.1016/j.ijrobp.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. Chk1 knockdown confers radiosensitization in prostate cancer stem cells. Oncol Rep. 28(6):2247–54. doi: 10.3892/or.2012.2068. [DOI] [PubMed] [Google Scholar]
- 13.Ropolo M, et al. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Mol Cancer Res. 2009;7(3):383–92. doi: 10.1158/1541-7786.MCR-08-0409. [DOI] [PubMed] [Google Scholar]
- 14.Dittfeld C, et al. CD133 expression is not selective for tumor-initiating or radioresistant cell populations in the CRC cell lines HCT-116. Radiother Oncol. 2009;92(3):353–61. doi: 10.1016/j.radonc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 15.Tirino V, et al. The role of CD133 in the identification and characterisation of tumour-initiating cells in non-small-cell lung cancer. Eur J Cardiothorac Surg. 2009;36(3):446–53. doi: 10.1016/j.ejcts.2009.03.063. [DOI] [PubMed] [Google Scholar]
- 16.Shien K, et al. Prognostic impact of cancer stem cell-related markers in non-small cell lung cancer patients treated with induction chemoradiotherapy. Lung Cancer. 77(1):162–7. doi: 10.1016/j.lungcan.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Iida H, et al. Hypoxia induces CD133 expression in human lung cancer cells by up-regulation of OCT3/4 and SOX2. Int J Oncol. 40(1):71–9. doi: 10.3892/ijo.2011.1207. [DOI] [PubMed] [Google Scholar]
- 18.Bertolini G, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106(38):16281–6. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu YP, et al. Cisplatin selects for multidrug-resistant CD133+ cells in lung adenocarcinoma by activating Notch signaling. Cancer Res. doi: 10.1158/0008-5472.CAN-12-1733. [DOI] [PubMed] [Google Scholar]
- 20.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27(3):247–54. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 21.Sancar A, et al. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 22.Krejci L, et al. Homologous recombination and its regulation. Nucleic Acids Res. 40(13):5795–818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schild D, Wiese C. Overexpression of RAD51 suppresses recombination defects: a possible mechanism to reverse genomic instability. Nucleic Acids Res. 38(4):1061–70. doi: 10.1093/nar/gkp1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hine CM, Seluanov A, Gorbunova V. Use of the Rad51 promoter for targeted anti-cancer therapy. Proc Natl Acad Sci U S A. 2008;105(52):20810–5. doi: 10.1073/pnas.0807990106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko JC, et al. Involvement of Rad51 in cytotoxicity induced by epidermal growth factor receptor inhibitor (gefitinib, IressaR) and chemotherapeutic agents in human lung cancer cells. Carcinogenesis. 2008;29(7):1448–58. doi: 10.1093/carcin/bgn130. [DOI] [PubMed] [Google Scholar]
- 26.Tomimatsu N, et al. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst) 11(4):441–8. doi: 10.1016/j.dnarep.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia V, et al. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 479(7372):241–4. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mihatsch J, et al. Selection of radioresistant tumor cells and presence of ALDH1 activity in vitro. Radiother Oncol. 99(3):300–6. doi: 10.1016/j.radonc.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Mizugaki H, et al. CD133 expression: a potential prognostic marker for non-small cell lung cancers. Int J Clin Oncol. doi: 10.1007/s10147-013-0541-x. [DOI] [PubMed] [Google Scholar]
- 30.Tsai MS, et al. Down-regulation of Rad51 expression overcomes drug resistance to gemcitabine in human non-small-cell lung cancer cells. J Pharmacol Exp Ther. 335(3):830–40. doi: 10.1124/jpet.110.173146. [DOI] [PubMed] [Google Scholar]
- 31.Davidson D, et al. Small Molecules, Inhibitors of DNA-PK, Targeting DNA Repair, and Beyond. Front Pharmacol. 4:5. doi: 10.3389/fphar.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark-Knowles KV, O’Brien AM, Weberpals JI. BRCA1 as a Therapeutic Target in Sporadic Epithelial Ovarian Cancer. J Oncol. 2010:891059. doi: 10.1155/2010/891059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, et al. Expression of DNA-PKcs and BRCA1 as prognostic indicators in nasopharyngeal carcinoma following intensity-modulated radiation therapy. Oncol Lett. 5(4):1199–1204. doi: 10.3892/ol.2013.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan N, et al. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res. 2008;68(2):605–14. doi: 10.1158/0008-5472.CAN-07-5472. [DOI] [PubMed] [Google Scholar]
- 36.Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007;26(14):2048–57. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- 37.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8(7):579–91. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 38.Moitra K, Lou H, Dean M. Multidrug efflux pumps and cancer stem cells: insights into multidrug resistance and therapeutic development. Clin Pharmacol Ther. 89(4):491–502. doi: 10.1038/clpt.2011.14. [DOI] [PubMed] [Google Scholar]
- 39.Sartelet H, et al. CD133 expression is associated with poor outcome in neuroblastoma via chemoresistance mediated by the AKT pathway. Histopathology. 60(7):1144–55. doi: 10.1111/j.1365-2559.2012.04191.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 1: CD133+ cells expand following ionizing radiation in human lung cancer cell lines.
A549 and H1299 human lung cancer cell lines were treated with 4Gy ionizing radiation and the percent positive CD133 cells were measured using cytometric analysis every 24 hours post treatment.
Supplemental 2: Purity of magnetically sorted CD133 cells
Cells were sorted via magnetic separation (Miltenyi). Sorted cells were stained with PE tagged CD133 antibody (293C-Miltenyi) and analyzed via flow cytometry.
Supplemental 3: Confirmation of gene expression data using western blotting
CD133+ cells were magnetically separated from A549 and H1299 cells. Western blot analysis was performed on cell lysates from each population probing for DNA repair proteins Exo1, Ku70, Rad51 and Msh2 to demonstrate basal protein levels in CD133− and CD133+ cells.
Supplemental 4: Confirmation of lentiviral knockdown of Exo1 and Rad51 in A549 cells
A549 cells were transduced with lentiviral shRNA specific to Exo1, Rad51, or scrambled control (Sigma). After puromycin selection cells were probed for Exo1 and Rad51 using western blot analysis to determine the levels of protein knockdown.