Summary
Developmental fate decisions are dictated by master transcription factors (TFs) that interact with cis-regulatory elements to direct transcriptional programs. Certain malignant tumors may also depend on cellular hierarchies reminiscent of normal development but superimposed on underlying genetic aberrations. In glioblastoma (GBM), a subset of stem-like tumor-propagating cells (TPCs) appears to drive tumor progression and underlie therapeutic resistance, yet remain poorly understood. Here, we identify a core set of neurodevelopmental TFs (POU3F2, SOX2, SALL2, OLIG2) essential for GBM propagation. These TFs coordinately bind and activate TPC-specific regulatory elements, and are sufficient to fully reprogram differentiated GBM cells to ‘induced’ TPCs, recapitulating the epigenetic landscape and phenotype of native TPCs. We reconstruct a network model that highlights critical interactions and identifies novel therapeutic targets for eliminating TPCs. Our study establishes the epigenetic basis of a developmental hierarchy in GBM, provides detailed insight into underlying gene regulatory programs, and suggests attendant therapeutic strategies.
Keywords: cis-regulatory elements, enhancers, chromatin, epigenetic states, glioblastoma, stem cells, cellular hierarchy, cellular reprogramming, cancer
Introduction
In mammalian development, stem and progenitor cells differentiate hierarchically to give rise to germ layers, lineages and specialized cell types. These cell fate decisions are dictated and sustained by master regulator transcription factors (TFs), chromatin regulators and associated cellular networks. It is now well established that developmental decisions can be overridden by artificial induction of combinations of ‘core’ TFs that yield induced pluripotent stem (iPS) cells or direct lineage conversion (Hanna et al., 2010; Morris and Daley, 2013; Orkin and Hochedlinger, 2011; Takahashi and Yamanaka, 2006; Vierbuchen and Wernig, 2011). These TFs bind and activate cis-regulatory elements that modulate transcription, and thereby direct cell type-specific gene expression programs (Lee and Young, 2013).
Increasing evidence suggests that certain malignant tumors also depend on a cellular hierarchy, with privileged sub-populations driving tumor propagation and growth. Moreover, many TFs that direct developmental decisions can also function as oncogenes by promoting the re-acquisition of developmental programs required for tumorigenesis (Suva et al., 2013). For example, the pluripotency and neurodevelopmental factor Sox2 is an essential driver of stem-like populations in multiple malignancies. Studies of leukemia pioneered the concept that triggering cellular differentiation can abolish certain malignant programs and override genetic alterations (Ito et al., 2008; Wang and Dick, 2005). Similarly, iPS reprogramming experiments have shown that artificially changing cancer cell identity profoundly alters their properties (Stricker et al., 2013). These findings suggest that epigenetic circuits superimposed upon genetic mutations determine key features of cancer cells.
The extent to which unidirectional differentiation hierarchies underlie tumor heterogeneity remains controversial (Visvader and Lindeman, 2012). For example, recent studies indicate that stem-like cells in breast cancer and melanoma exist in dynamic equilibrium with phenotypically distinct populations incapable of tumor propagation (Chaffer et al., 2013; Roesch et al., 2010). Alternatively, there is evidence supporting more classical hierarchies in other cancers, particularly in leukemias (Wang and Dick, 2005). In GBM models, reversibility seems to depend on the differentiation stimulus and time of exposure. Short-term exposure of GBM stem-like cells to BMP4 is sufficient to abolish their tumor-propagating potential, consistent with unidirectional differentiation (Piccirillo et al., 2006). Serum-triggered differentiation appears to proceed more gradually; short-term exposure can be reversed (Lee et al., 2006; Natsume et al., 2013), while longer-term exposure fully abolishes tumor-propagating potential (Janiszewska et al., 2012; Lee et al., 2006; Wakimoto et al., 2009). A better understanding of the molecular underpinnings that distinguish stem-like cancer cells and control plasticity within tumors is a critical goal with broad implications for diagnosis and therapy.
GBM is the most common malignant brain tumor in adults and remains incurable despite aggressive treatment (Jansen et al., 2010). Genome sequencing and transcriptional profiling studies have highlighted a large number of genetic events and identified multiple biologically relevant GBM subtypes, representing a major challenge for targeted therapy (Sturm et al., 2012; Verhaak et al., 2010). There is strong evidence that differentiation status significantly impacts GBM cell properties, with stem-like cells likely driving tumor propagation and therapeutic resistance (Bao et al., 2006; Chen et al., 2012). Although putative stem-like populations in GBM can be enriched using cell surface markers such as CD133 (Singh et al., 2004), SSEA-1 (Son et al., 2009), CD44 (Anido et al., 2010), and integrin alpha 6 (Lathia et al., 2010), the consistency of the various markers and the extent to which genetic heterogeneity contributes to observed phenotypic differences remains controversial. A TF code for GBM stem-like cells, analogous to those identified in iPS reprogramming and direct lineage conversion experiments, could thus provide critical insights into the epigenetic circuitry underlying GBM pathogenesis.
Here we combine functional genomics and cellular reprogramming to reconstruct the transcriptional circuitry that governs the developmental hierarchy in human GBM. By comparing the epigenetic landscapes of stem-like GBM cells against their differentiated counterparts, we identify four core TFs – POU3F2 (BRN2), SOX2, SALL2 and OLIG2 – whose induction is sufficient to reprogram differentiated GBM into stem-like cells capable of in vivo tumor propagation. We use this TF code to identify candidate tumor propagating cells in primary GBM tumors. Genome-wide binding maps and transcriptional profiles identify key regulatory targets of the core TFs, including the RCOR2/LSD1 histone demethylase complex. RCOR2 can substitute for OLIG2 in the reprogramming cocktail and, moreover, stem-like GBM cells are highly sensitive to LSD1 suppression, thus validating the regulatory model. Our findings demonstrate a cellular hierarchy in GBM, provide detailed insight into its transcriptional and epigenetic basis, and propose therapeutic strategies for eliminating stem-like tumor propagating cells in human GBM.
Results
TF activity and cis-regulatory elements distinguish GBM TPCs
To identify distinguishing features of stem-like GBM cells, we expanded matched pairs of GBM cultures derived from three different human tumors either as stem-like tumor-propagating cells (TPCs) grown in serum-free, spherogenic culture, or as differentiated glioblastoma cells (DGCs) grown as adherent monolayers in serum. The alternate culture conditions confer GBM cells with distinct functional properties, the key of which is their in vivo tumor-propagating potential in orthotopic xenotransplantation limiting dilution assays (Figure 1A and S1) (Chudnovsky et al., 2014; Janiszewska et al., 2012; Lee et al., 2006). This functional difference is accompanied by differences in expression of stem cell (CD133, SSEA-1), astroglial (GFAP), neuronal (beta III tubulin, MAP-2) and oligodendroglial (GalC) markers (Figure 1B, C and S1), consistent with a modulation of the stemness-differentiation axis by serum. Orthotopic xenotransplantation of as few as 50 GBM TPCs leads to formation of tumors that recapitulate major histologic features of GBM (Figure 1D), while as many as 100,000 DGCs fail to initiate tumor. Importantly, although the stem-like TPCs are able to differentiate and expand as monolayers when exposed to serum, DGCs will not expand in serum-free conditions, suggesting that the differentiated state is epigenetically stable. These functional and phenotypic properties suggest that a transcriptional hierarchy predicated on distinct epigenetic circuits is critical for the tumor-propagating potential of GBM cells.
Figure 1. Epigenetic landscapes distinguish functionally distinct GBM models.
(A) GBM cells (MGG8) grown as gliomaspheres in serum-free conditions propagate tumor in vivo while serum-differentiated cells fail to do so. (B) Flow cytometry of MGG8 TPCs shows positivity for the GBM stemlike markers SSEA-1 and CD133, while serum-differentiated cells do not. (C) Cells grow in serum as adherent monolayers and express the differentiation markers GFAP (astroglial), beta III tubulin (neuronal), MAP-2 (neuronal) and GalC (oligodendroglial). (D) Xenografted tumors from MGG8 TPCs (left) are invasive, crossing the corpus callosum (boxed region), infiltrating along white matter tracks (arrowhead). At high magnification, the cells are atypical and mitotic figures are evident (arrow). Xenografted tumors from MGG4 TPCs (right) are more circumscribed but also infiltrate adjacent parenchyma (boxed region, arrowhead). At high magnification areas of necrosis (*) and mitotic figures (arrow) are readily identified. LV: lateral ventricle. (E) TPC-specific, DGC-specific and shared regulatory elements. Shared elements tend to be located proximal to promoters, while the vast majority of TPC- and DGC-specific elements are distal. Motif analyses predict binding sites for TF families within each set of sites. See also Supplemental FigureS1.
To acquire an epigenetic fingerprint of the respective GBM models, we surveyed cis-regulatory elements in three matched pairs of TPCs and DGCs established from three human tumors (Materials and Methods). We specifically mapped histone H3 lysine 27 acetylation (H3K27ac), which marks promoters and enhancers that are ‘active’ in a given cell state (Bulger and Groudine, 2011; Creyghton et al., 2010; Ernst et al., 2011; Hon et al., 2009; Rada-Iglesias et al., 2011; Visel et al., 2009). Unsupervised clustering indicates that the TPCs share similar regulatory element patterning, but are distinct from the DGCs, which are also consistent across the patient-derived samples (Figure S1). This suggests that regulatory element activity in our model correlates more closely with phenotypic state than patient- or tumor-specific genetic background.
To identify TFs that might direct these alternative cell states, we collated sets of TPC-specific, DGC-specific and shared regulatory elements, and searched the underlying DNA sequences for over-represented motifs. TPC-specific elements are strongly enriched for motifs recognized by helix-loop-helix (HLH) and Sry-related HMG box (SOX) family TFs (Figure 1E), while DGC-specific elements are instead enriched for AP1/JUN motifs, consistent with a serum-induced differentiation program (Zhu et al., 2013). We complemented these motif inferences with RNA-Seq expression data and promoter H3K27ac signals for TF genes to identify candidate regulators of the TPC state. This analysis yielded a set of 19 TFs with significantly higher expression in TPCs (Figure 2A–C). Although we previously identified a set of 90 TFs active in GBM stem-like cells (Rheinbay et al., 2013), this more restrictive set is limited to TFs that are specifically active in TPCs and thus candidates for directing their epigenetic state. Notably, 10 of the 19 TFs are HLH or SOX family members, whose cognate motifs were identified in our unbiased analysis of TPC-specific regulatory elements.
Figure 2. Candidate regulators for the specification of alternate epigenetic states in GBM.
(A) A set of 19 TPC-specific TFs is identified based on RNA-Seq expression and promoter H3K27ac signals in TPCs and DGCs. TF family is indicated at right. (B) Western blots confirm exclusive protein expression in TPCs for selected TFs. Lower panel indicates tubulin loading control. (C) ChIP-Seq tracks show H3K27ac signals for loci encoding TPC-specific TFs OLIG1, OLIG2 and SOX2, or (D) the differentiation factor BMP4 in the respective GBM models. TPC-specific TF loci are enriched for TPC-specific regulatory elements.
Derivation of a core TF set sufficient to induce a TPC phenotype
Among the 19 TPC-specific TFs, SOX2, OLIG2 and ASCL1 have been shown to be necessary for spherogenicity and tumor-propagating potential of stem-like GBM cells (Gangemi et al., 2009; Mehta et al., 2011; Rheinbay et al., 2013). However, the hypothesized GBM developmental hierarchy raises the possibility that certain combinations of TFs might be sufficient to reprogram DGCs into TPCs, thus overriding an epigenetic state transition that is irreversible in our model. Notably, several TPC-specific TFs are components of cocktails that have been used to convert fibroblasts into neurons (Pang et al., 2011) or neural stem cells (Lujan et al., 2012). We therefore considered whether these principles of cellular reprogramming could be applied to inter-convert epigenetic states in GBM.
To test the capacity of individual TFs or TF combinations to reprogram GBM cells, we cloned all 19 TPC-specific TFs and ectopically expressed them in DGCs. We then monitored single-cell sphere formation in serum-free conditions, surface marker induction and tumor-propagation by orthotopic xenotransplantation into severe combined immunodeficient (SCID) mice. We first introduced each TF individually. Of the 19 TFs, only SOX1, SOX2 and POU3F2 modestly enhanced spherogenesis, with POU3F2 in particular yielding ~3% sphere formation (compared to ~0% for empty vector and >10% for native TPCs; Figure 3A). These TFs also stimulated weak induction of the stem-cell marker CD133 (Figure 3B). However, orthotopic xenotransplantation of as many of 100,000 DGCs expressing SOX1, SOX2 or POU3F2 failed to initiate tumors in mice (Table S2).
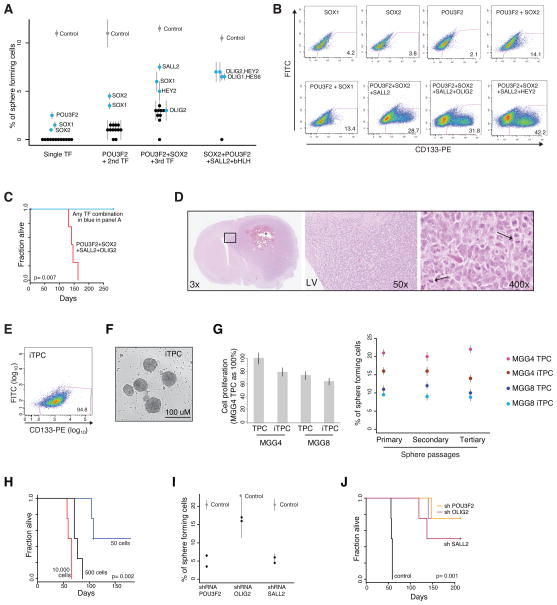
Figure 3. A core TF network for tumor-propagating GBM cells.
(A) Data points indicate percentage of single-cell DGCs capable of forming spheres in serum-free conditions. Each of the 19 TFs in Figure 2A was tested alone (first column, ‘single TF’), in combination with POU3F2 (second column) or in combination with POU3F2 and SOX2 (third column). HLH family TFs were also tested in combination with POU3F2, SOX2 and SALL2 (fourth column), based on an enrichment of HLH motifs in regulatory elements that failed to activate in 3TF-induced DGCs. TF combinations that enhanced in vitro spherogenicity (blue) were selected for in vivo testing. (B) Flow cytometry profiles show expression of the stem cell marker CD133 for DGCs induced by the single, double, triple and quadruple TF combinations with the highest in vitro sphere-forming potential. (C) For TF combinations with in vitro spherogenic potential (blue in panel 3A), 100,000 cells were injected in the brain parenchyma (n=4 mice per TF combination). Survival curve is shown for this in vivo tumor-propagation assay. Only the quadruple TF combination POU3F2+SOX2+SALL2+OLIG2 initiated tumors in mice. (D) Tumor histopathology shows characteristic features of glioblastoma, including necrotic areas (*) and crossing of corpus callosum (boxed area). At high magnification cells show atypical features and mitotic figures are evident (arrows). LV: lateral ventricle. (E) Secondary TPC sphere cultures (“iTPC”) derived from xenotransplant tumors express the stem-cell marker CD133. (F) Contrast field image of iTPC spheres. (G) Left: bar graph shows iTPC and TPC proliferation rates measure by BrdU incorporation. Right: data points indicate percentage of single cells capable of serial sphere formation in three consecutive passages in serum-free conditions. Self-renewal properties and proliferation of iTPCs are comparable to corresponding TPCs. (H) Orthotopic serial xenotransplantation in limiting dilution shows that as few as 50 MGG8 iTPC are sufficient to initiate tumors. (I) Data points indicate in vitro sphere formation of MGG4 TPCs infected with lentivirus containing shRNA for POU3F2, OLIG2 or SALL2, compared to control (two hairpins per TF). (J) Survival curve depicts in vivo tumor propagating potential of MGG4 TPCs infected with POU3F2 shRNA, SALL2 shRNA, OLIG2 shRNA or control shRNA. See also Supplemental FiguresS2–S4.
Reasoning that successful GBM reprogramming might require multiple TFs, we next co-infected DGCs with POU3F2 in combination with each of the other 18 TPC-specific TFs. We found that co-infection of POU3F2 with SOX1 or SOX2 significantly increased in vitro sphere-forming potential and CD133 expression (Figure 3A, B). However, neither 2TF combination nor the SOX1+SOX2 combination initiated tumors in vivo (Table S2). We thus resumed stepwise reconstruction experiments by adding a third TF to the most effective pair (POU3F2+SOX2). Although the addition of SALL2, SOX1, HEY2 or OLIG2 improved our in vitro results, none of these 3TF combinations were sufficient to initiate tumors in vivo (Figure 3A–C).
Failure to achieve complete reprogramming with these TF combinations led us to consider whether TF induction effectively activates TPC-specific regulatory elements, as would be expected in a successful reprogramming experiment. To test this, we mapped H3K27ac-marked regulatory elements in DGCs infected with POU3F2 alone, with the top 2TF combination (POU3F2+SOX2), or with the top 3TF combination (POU3F2+SOX2+SALL2). Each population gained TPC-specific elements and lost DGC-specific elements, with the 3TF combination inducing the most prevalent changes (Figure S2). Yet despite their spherogenic potential and CD133 expression, DGCs expressing the 3TF combination failed to induce a large number of TPC-specific elements. Examination of the subset of TPC-specific regulatory elements that remain silent in these partially reprogrammed cells revealed a strong enrichment for HLH motifs (Figure S2), suggesting that complete reprogramming might require an additional HLH TF.
We therefore supplemented the 3TF combination (POU3F2+SOX2+SALL2) with each HLH factor in the TPC-specific TF set, namely OLIG1, OLIG2, HEY2, HES6 and ASCL1. Although none of these additions significantly enhanced in vitro assay performance, combined induction of POU3F2+SOX2+SALL2+OLIG2 yielded cells capable of tumor initiation in 100% of animals (Figure 3A–C). This 4TF cocktail appears highly specific as four TF combinations with any of the other HLH factors failed to initiate tumors. Moreover, replacement of SOX2 with SOX1 or omission of any single component from the 4TF set yielded cells without tumor initiating properties (Table S2).
Tumors initiated by ‘induced’ TPCs (iTPCs) expressing the four TFs show classical features of high-grade gliomas, including necrosis, atypical cytonuclear features and high mitotic index (Figure 3D). Secondary sphere cultures derived from these tumors express high levels of CD133 and display proliferation and self-renewal properties in serial sphere formation assays, similar to their corresponding TPC lines (Figure 3E–G) (Barrett et al., 2012; Chen et al., 2010). Similarly, serial xenotransplantation of these secondary cultures into SCID mice in limiting dilutions indicates that as few as 50 iTPC cells initiate tumors in 50% of animals, while 500 cells confer tumor initiation in 100% of recipients (Figure 3H). Thus, we have identified a TF cocktail sufficient to reprogram serum-derived differentiated GBM cells into stem-like GBM cells capable of unlimited self-renewal and tumor propagation.
To evaluate the generality of the TF cocktail, we tested its ability to reprogram other DGC models. First, we confirmed that the core TFs were capable of reprogramming a second serum-derived DGC line from a different patient with different genetic aberrations (Figure 3G, 4A). Second, we tested the effects of the TFs in an alternative differentiation model, in which TPCs are differentiated in serum-free conditions by addition of BMP4 (Piccirillo et al., 2006). This treatment caused the cells to adhere and down-regulate the core TFs and CD133 over a 72 hours period. Re-induction of the core TFs in these differentiated GBM cells re-established spherogenic potential and CD133 expression over a one-week period (Figure S3). These data suggest that the core TF circuitry plays a general role in modulating the GBM differentiation axis. Since the specific GBM models investigated here conform to the proneural subtype (Figure S1), further study will be needed to evaluate the role of these TFs in other GBM subtypes (Verhaak et al., 2010).
Figure 4. Core TFs reprogram the epigenetic landscape of DGCs.
(A) Left: heatmap depicts H3K27ac signals for TPC-specific, DGC-specific or shared regulatory elements defined in Figure 1E. Relative to control vector infected DGCs, iTPCs gain H3K27ac over TPC-specific elements and lose H3K27ac over DGC-specific elements, consistent with genome-wide reprogramming of the epigenetic landscape. Right: pie charts show fraction of regulatory elements (dark cyan) in each set with H3K27ac in iTPC. (B) RNA-Seq expression and promoter H3K27ac levels at promoter are shown for TPC-specific TFs defined in Figure 2A (NES: Nestin). (C) Hierarchical clustering of MGG8 DGCs, TPCs and replicate iTPCs (iTPC1/2) by H3K27ac ChIP-Seq signal. (D) RNA-Seq tracks show that core TF mRNAs in iTPCs include 3′UTRs (shaded in gray). This indicates the endogenous loci are reactivated in iTPCs as the exogenous vectors lack 3′UTRs. (E) H3K27ac signal tracks for loci encoding core TFs show that endogenous regulatory elements (highlighted with grey shading) are reactivated in iTPCs. (F) Serum-induced differentiation leads iTPCs to convert to an adherent phenotype, up-regulate differentiation markers GFAP, beta III tubulin, MAP-2, GalC and (G) to lose CD133 expression. (H) Western blots confirm serum-induced differentiation of iTPCs leads to down-regulation of core TFs. Lower panels: tubulin loading control. These data indicate that the core TFs can reprogram DGCs into stem-like GBM cells, which have an epigenetic landscape similar to TPCs that is sustained by endogenous regulatory programs. See also Supplemental Figures S2.
Core TFs fully reprogram the epigenetic state of induced TPCs
To examine the extent to which the four core TFs reprogram the epigenetic state of GBM cells, we surveyed regulatory element activity and TF expression in secondary iTPC sphere cultures. Consistent with their tumor-propagating ability, iTPCs gain H3K27ac at most TPC-specific elements and lose H3K27ac at the majority of DGC-specific elements (Figure 4A). Furthermore, 18/19 TPC-specific TFs are up-regulated in the iTPCs, and most acquire K27ac at their promoter, indicating that their epigenetic landscape closely resembles TPCs (Figure 4B, C). In contrast, DGCs expressing three TFs fail to reset most TPC-specific and DGC-specific regulatory elements (Figure S2). Thus, all four core TFs are required to reprogram the epigenetic landscape of GBM cells, consistent with their requirement for the functional TPC phenotype.
We also considered the mechanistic basis for the sustained phenotype of iTPCs. Several lines of evidence suggest that the four core TFs are expressed from their endogenous loci in the iTPCs, while the exogenously introduced expression vectors are silenced. The endogenous TF genes contain 3′UTRs that distinguish them from the exogenous versions, which lack UTRs. RNA-Seq profiles confirm endogenous transcripts with 3′UTRs for POU3F2, SOX2, SALL2 and OLIG2 in iTPCs, but reveal little or no expression of the exogenous transcripts (Figure 4D). The endogenous TF loci also gain H3K27ac at putative regulatory elements, consistent with their reactivation (Figure 4E). Finally, iTPCs markedly reduce expression of all four TFs and readily differentiate upon exposure to serum (Figure 4F–H), as is indicative of endogenous regulation. These data suggest that induction of the core TFs triggers an epigenetic state transition that is subsequently maintained by endogenous regulatory programs.
Core TFs coordinately expressed in a subset of GBM cells from primary human tumors
To investigate the clinical relevance of our findings, we asked whether the core TFs and corresponding regulatory elements are active in primary human GBM tumors. First we sought to identify individual cells within GBM tumors that co-express all four core factors, postulating that these could represent candidate stem-like TPCs. We performed four-color immunofluorescence and five-color flow cytometry on freshly resected tumors using antibodies against POU3F2, SOX2, SALL2, OLIG2 and CD133. We found that SOX2 identifies the largest set of GBM cells, while SALL2 and POU3F2 have more restricted expression. Image analysis and flow cytometry both identified a small subset of cells in primary tumors (~2–7%) that coordinately express all four TFs (Figure 5A, B). Remarkably, more than 50% of the 4TF-positive cells also express CD133, a striking enrichment over 4TF-negative cells, which almost entirely lack this stem cell marker (Figure 5B). Finally, we also mapped H3K27ac genome-wide in several freshly resected GBM tumors. This bulk analysis revealed significant enrichment for ~50% of TPC-specific regulatory elements, suggesting that they are also active in primary tumor cells (Figure 5C). Collectively, these data suggest that the core TFs, regulatory elements and circuits defined in our TPC model are active in a subset of primary GBM cells that express the stem cell marker CD133 and may underlie tumor propagation.
Figure 5. All four core TFs are coordinately expressed in a subset of primary GBM cells with stem-like markers.
(A) Quadruple immunofluorescence for core TFs in three human GBM samples shows co-expression in a subset of cells; shown at right are the fractions of SOX2+ cells that express each other individual TF or all four TFs in each tumor. (B) Flow cytometry analysis from acutely resected GBM tumors. A majority of cells positive for the four core TFs express the stem-cell marker CD133. Enrichment is significantly greater than for SOX2-expressing cells. (C) Heatmap shows H3K27ac signal from three freshly resected GBM tumors for regulatory elements defined in Figure 1E. Right: pie-charts show fraction of regulatory elements (dark cyan) in each set with H3K27ac. TPC-specific elements show significant enrichment, consistent with a TPC-like regulatory program in a subset of cells.
Essential roles for core TFs and their regulatory targets in GBM TPCs
The identification of TPC-like cells in primary GBM tumors prompted us to investigate the regulatory functions and interactions of the core TFs, reasoning that this might suggest new therapeutic targets or strategies. First, we confirmed that all four TFs are essential for in vitro and in vivo TPC phenotypes. Prior studies had established SOX2 and OLIG2 as essential regulators in this context (Gangemi et al., 2009; Mehta et al., 2011). By performing shRNA-mediated knock-down in TPCs, we showed that POU3F2 and SALL2 are also required for sphere formation in vitro and tumor-propagation in vivo (Figure 3I, J and S4).
To identify direct regulatory targets, we next mapped the binding sites of POU3F2, SOX2, SALL2 and OLIG2 in TPCs using ChIP-Seq with specific antibodies for each factor (Figure 6A and S5). All four TFs preferentially associate with TPC-specific regulatory elements, and there is significant overlap among their binding sites (Figure 6B and S5). As expected, POU3F2, SOX2 and OLIG2 binding sites are enriched for the cognate motifs. However, SALL2 sites are primarily enriched for SOX motifs (Figure 6C), raising the possibility that SALL2 is recruited as a complex. Consistently, co-immunoprecipitation experiments confirmed a direct interaction between SALL2 and SOX2 (Figure S5). Notably, single TF inductions in DGCs indicate that POU3F2 and SOX2 are each capable of activating subsets of TPC-specific elements. In contrast, neither OLIG2 nor SALL2 is able to significantly alter the regulatory landscape of DGCs in isolation (Figure S2). These results suggest that the core TFs cooperatively engage TPC-specific regulatory elements to activate gene expression programs required for GBM propagation.
Figure 6. TF network reconstruction and targeting.
(A) ChIP-Seq signal for core TFs profiled in TPCs (MGG8) shows preferential binding at TPC-specific regulatory elements. (B) Pie charts indicate proportion of TF binding sites that coincide with the indicated sets of regulatory elements. (C) Sequence motifs identified in TF ChIP-Seq peaks. With the exception of SALL2 (see text and Figure S5), motifs correspond to the expected class of TFs, further validating ChIP-Seq experiments. (D) Model for core TF regulatory interactions reconstructed from binding profiles and expression data (see text and methods). Other TFs defined in figure 2A (green) and chromatin regulators (red) are highlighted. (E) Signal tracks depict core TF binding over TPC-specific regulatory elements within loci containing the corresponding TF genes. See also Supplemental Figure S5.
To comprehensively identify functional targets of the core TFs, we collated a list of genes within 50 kb of a bound regulatory element and examined their expression by RNA-Seq in TPCs and DGCs. We identified 325 differentially expressed genes with proximal H3K27ac-marked elements bound by one or more core TFs. These putative direct targets include all four core TF genes and 12 of the 19 TPC-specific TF genes (Figure 6D, E and Table S3), consistent with a role for reciprocal TF interactions in maintaining the TPC regulatory program.
Co-repressor subunit RCOR2 can replace OLIG2 in reprogramming cocktail
We next focused on target genes of the core TFs that are active in TPCs and iTPCs, but not in partially reprogrammed 3TF DGCs, reasoning that these might represent critical nodes for the stem-like GBM cells (see Materials and Methods; Table S4). One nuclear factor satisfying these criteria is the ASCL1 TF, which we previously found to be an essential regulator of Wnt signaling in TPCs (Rheinbay et al., 2013). A second is RCOR2, a co-repressor with essential functions in embryonic stem cells (Yang et al., 2011). RCOR2 resides in a complex with PHF21B and the histone methyltransferase LSD1, both of which were also identified as putative core TF targets (Shi et al., 2005; Yang et al., 2011). LSD1, PHF21B and RCOR2 are differentially expressed in TPC and DGC, with the latter undetectable at mRNA and protein levels in DGCs (Figure 7A, B). We also confirmed a robust physical interaction between RCOR2 and LSD1 in TPCs (Figure 7C).
Figure 7. The LSD1-RCOR2 chromatin complex is essential for GBM TPCs.
(A) Plots depict LSD1 and RCOR2 RNA-Seq expression values for TPCs and DGCs. (B) Western blot for RCOR2 (MGG8 TPC and DGC lysates) confirms exclusive expression in TPC. (C) Western blot for LSD1 on RCOR2 immunoprecipitate indicates co-association between the two proteins in TPCs. (D) Signal tracks depict TF binding and H3K27ac enrichment in the RCOR2 locus. OLIG2 binds a TPC-specific regulatory element in the locus. (E) Survival curve of mice injected with DGCs induced with the combination of POU3F2+SOX2+SALL2+RCOR2 indicates that RCOR2 can substitute for OLIG2 in the cocktail. (F) Coronal section of a xenografted GBM tumor (dashed line) established from iTPCs reprogrammed with the POU3F2+SOX2+SALL2+RCOR2 combination. (G) Representative images of TPCs and DGCs infected with LSD1 shRNA show reduced viability specifically in the TPCs. (H) Bar graphs depict percent viability for MGG4 TPCs or DGCs infected with control shRNA or two different LSD1 shRNAs. LSD1-depletion causes decreased viability in TPCs and has effect on DGCs. (I) Data points indicate in vitro sphere formation of MGG4 TPCs infected with lentivirus shRNA for LSD1 (two hairpins), compared to control in three serial passages. (J) Graph depicts percent viability for TPCs and DGCs (MGG4 and MGG8) and primary astrocytes (NHA) exposed to increasing doses of the synthetic LSD1 inhibitor S2101. A representative image of TPCs exposed to 20uM S2101 for 96 hours is shown below. (K) Survival curve depicts in vivo tumor propagating potential of MGG4 TPCs infected with LSD1 shRNA (two hairpins) or control shRNA. These data suggest that the RCOR2/LSD1 complex is essential for stem-like TPCs, and thus represents a candidate therapeutic target for eliminating this aggressive GBM sub-population. See also Supplemental Figure S4.
Prior studies have suggested that RCOR2 is predominantly expressed in embryonic stem cells, where it plays a role in sustaining pluripotency. Although RCOR2 has not been implicated in GBM, we hypothesized that it might play a key role in initiation and maintenance of TPCs. Since RCOR2 is downstream of OLIG2 in our network, we asked whether it could substitute for OLIG2 in the reprogramming cocktail. We repeated the DGC reprogramming and found that DGCs expressing POU3F2, SOX2, SALL2 and RCOR2 could initiate tumor in 100% of cases, indicating that RCOR2 can replace OLIG2 and establishing it as a key effector of the TPC regulatory program (Figure 7D–F).
Having established a critical role for RCOR2, we next asked whether LSD1, an enzymatic subunit of the RCOR2 complex, might also be essential. We performed shRNA knock-down of LSD1 in TPCs and DGCs, confirming >80% reduction in LSD1 mRNA levels in both cases (Figure S3). Although the proliferation and viability of DGCs were unaffected by the knockdown, the TPC phenotype was profoundly altered, with marked reductions in cell survival and near complete loss of self-renewal in serial sphere formation assays (Figure 7G–I). LSD1 knockdown also caused TPCs to lose their capacity to initiate tumors in vivo (Figure 7K). We also treated TPCs, DGCs and normal human astrocytes with increasing concentrations of the synthetic LSD1 inhibitor S2101 (Mimasu et al., 2010). TPCs lose viability in the presence of 20uM inhibitor, while the DGCs and astrocytes are unaffected (Figure 7J). These findings identify the RCOR2/LSD1 histone demethylase complex as a candidate therapeutic target in stem-like tumor propagating cells in human GBM.
Discussion
Hierarchies of cellular differentiation and the associated epigenetic mechanisms – long the domain of developmental biology – are increasingly appreciated to play critical roles in cancer. Pioneering work in leukemia led to the identification of stem-like cells with high tumor propagating potential that give rise to differentiated progeny bearing identical genetic mutations, knowledge that led to the successful application of differentiation therapy (Ito et al., 2008; Wang and Dick, 2005). Recent studies have established analogous hierarchies in certain solid tumors, including glioblastoma, and thus point to the importance of understanding the epigenetic identities and susceptibilities of such aggressive subpopulations (Binda et al., 2012; Day et al., 2013; Friedmann-Morvinski et al., 2012; Kim et al., 2013; Piccirillo et al., 2006; Singh et al., 2004; Son et al., 2009; Suva et al., 2013).
Here, we identified epigenetic determinants that distinguish an established model of stem-like TPCs in GBM from their differentiated progeny, which normally are unable to reacquire stem-like properties. TFs, regulatory elements and interactions critical for the TPC state were predicted by integrating chromatin maps, RNA expression and TF binding profiles. Four core TFs were found to be sufficient to reprogram differentiated GBM cells into iTPCs that faithfully recapitulate in vitro and in vivo properties of TPCs established directly from human tumors. While SOX2 and OLIG2 have been previously described in GBM, SALL2 and POU3F2 are novel transcriptional regulators in the context of this disease.
All four factors co-bind large numbers of distal regulatory elements with specific activity in TPCs. SOX2 and POU3F2 can each partially reprogram the epigenetic landscape of DGCs on their own, consistent with their partial ability to induce spherogenic growth, and their established roles in direct conversion to neural lineages (Lodato et al., 2013; Lujan et al., 2012; Pang et al., 2011). Furthermore, SALL2 and SOX2 collaborate through a direct physical interaction that has not been previously described. The pervasive regulatory interactions of these neurodevelopmental factors and their efficacy in GBM reprogramming suggest that the malignant hierarchy maintains key features of normal developmental processes.
A limitation of our study is that in vitro TPC and DGC models do not fully recapitulate the diversity of cellular states within primary human GBMs. Nor does our work establish whether retrograde dedifferentiation can actually occur in primary tumors. Nonetheless, the clinical relevance of our findings is supported by (i) the identification of stem-like cells that coordinately express all four factors in primary GBM tumors; (ii) confirmation that large numbers of TPC-specific regulatory elements are active in primary tumors; and (iii) the requirement of all four factors for in vivo tumorigenicity in xenotransplanted mice. Given their demonstrated functionality, the core TFs may have specific advantages for enriching aggressive cellular subsets relative to conventional surface markers that have been defined empirically and remain controversial. This may be most apt in IDH1 wild-type proneural tumors, which account for roughly a quarter of human GBMs and are best represented by our cellular models.
In considering therapeutic opportunities presented by the TPC regulatory program, we focused on potential targets downstream of the core TF network. We identified the RCOR2-LSD1 complex as a key effector of the TPC regulatory program. RCOR2 can substitute for OLIG2 in the reprogramming cocktail, while LSD1 suppression triggered cell death exclusively in TPCs. This vulnerability is consistent with an established requirement for LSD1 in MLL-AF9 leukemia stem cells (Harris et al., 2012). Thus, dissection of the epigenetic circuitry governing a malignant hierarchy may guide therapeutic strategies for targeting cancer stem cells.
In conclusion, we have elucidated epigenetic fingerprints of key subpopulations within the cellular hierarchy of GBM, identified core TFs that direct the hierarchy and control tumor-propagating potential, and established a histone demethylase complex as a candidate therapeutic target in stem-like tumor cells. Although the specific tumor model characterized here conforms to a unidirectional hierarchy, our reprogramming experiment suggests a mechanism by which bidirectional plasticity may occur in certain clinical contexts. Further studies will be needed to assess translational opportunities in GBM and to evaluate the relative merits of hierarchical and plasticity-based cellular models across the diverse spectrum of human malignancies.
Experimental Procedures
Cell Culture
Surgically removed GBM specimens were collected at Massachusetts General Hospital with approval by the Institutional Review Board (IRB protocol 2005-P-001609/16). Tissue was processed as previously described (Rheinbay et al., 2013). See also supplemental methods.
Flow cytometry and Immunofluorescence
CD133/1-PE or CD133/2-APC (Miltenyi Biotec) and SSEA-1-FITC (BD Biosciences) antibodies were used according to manufacturer’s instructions. For TF staining in primary tumors, single cell suspensions were depleted for CD45-positive cells using a MACS separator (Miltenyi Biotec). Antibodies to SOX2 (R&D Systems), POU3F2 (Epitomics), SALL2 (Bethyl) and OLIG2 (R&D Systems) were directly conjugated to fluorophores using Alexa Fluor Conjugation Kits (Invitrogen) or DyLight conjugation kits (Pierce). The CD45-negative fraction was stained with CD133-PE or CD133-APC prior to fixation and permeabilization according to manufacturer’s protocol using the Transcription Factor Buffer Set (BD Pharmingen). Single color controls for all fluorophores were used for compensation. Flow cytometric analysis was conducted with an LSR II flow cytometer (BD Biosciences) and analysis was performed with FlowJo software (Treestar). See also supplemental methods.
ChIP-Seq Assay and 3′end RNA-Seq
ChIP-seq assays were carried out on approximately 1 × 106 cells per histone modification and 107 cells per transcription factor, following the procedures outlined in (Ku et al., 2008; Mikkelsen et al., 2007). For primary GBM, cells were dissociated into single cell suspension followed by depletion for CD45+ inflammatory infiltrate as outlined in previous methods. Immunoprecipitation was performed using antibodies against H3K27ac (Active Motif), POU3F2 (Epitomics), SOX2 (R&D), SALL2 (Bethyl), and OLIG2 (R&D). ChIP DNA samples were made into libraries for sequencing on the Illumina HiSeq 2000 or 2500 following standard procedures. See supplemental methods for ChIP-seq data processing and regulatory network reconstruction. ChIP-Seq data are available for viewing at http://www.broadinstitute.org/epigenomics/dataportal/clonePortals/Suva_Cell_2014.html. For 3′end RNA-seq, total RNA was isolated from cells using the RNeasy Kit (Qiagen). We used 2 ug of total RNA to fragment and polyA isolate the 3′ends of mRNAs. Illumina sequencing libraries were constructed and subjected to high-throughput sequencing. A processing pipeline incorporating Scripture (http://www.broadinstitute.org/software/scripture/) was used to reconstruct the transcriptome and calculate gene expression values as previously described (Mendenhall et al., 2013; Yoon and Brem, 2010). Data accompanying this paper is available through GEO under GSE54792. See also supplemental methods.
Generation of H3K27ac consensus sets
H3K27ac sites shared between 4, 6, 8 TPCs and corresponding DGCs were defined as those that were present in each of the six ChIP-Seq experiments. TPC-specific sites were required to be present in all three TPC lines and not in any of the DGC lines, and accordingly, DGC-specific sites were required to be present in all DGC but not in any of the TPC lines. For heatmaps, H3K27ac or TF signal in a 10kb region for each site was obtained. Total signal was thresholded at the 95th (H3K27ac) or 99th (TFs) percentile and scaled to values between 0 and 1. See also supplemental methods for H3K27ac-based cell type clustering.
Generation of TF list for experimental testing and motif analyses
TFs from the “CSC” and “stem-cell” sets from Rheinbay et al, 2013 were included in the testing set. TFs were then filtered for fold difference between TPCs and DGC, and only those at least 1.5-fold overexpressed in TPC relative to DGC were kept for further analysis. See also supplemental methods.
Knockdown and overexpression experiments
For knockdown experiments, the following lentiviral shRNA set from Thermoscientific were used: POU3F2 (RMM4532-NM_005604), OLIG2 (RHS4531-NM_005806), SALL2 (RHS4531-NM_005407), LSD1 (RHS4531-EG23028). Lentiviruses were produced as previously described (Rheinbay et al., 2013). GBM TPC were selected using 2ug/ml puromycin for 5 days. GBM non-TPC were selected using 1ug/ml puromycin for 5 days. After selection, RNA was extracted (Qiagen RNeasy kit) following manufacturer’s instructions. See also supplemental methods.
Single-cell sphere-formation assay and BrdU
For each condition (shRNA of TFs in GBM TPC or cDNA overexpression in DGC), single cells were plated in 150ul of serum-free medium in a 96 well plate. Sphere number/96 well plate was assessed after 2 weeks. The mean and standard deviation of 2 biological replicates was calculated. In serial sphere forming assays, the same procedure was repeated for 2 additional passages. BrdU assays were performed following manufacturer’s recommendations (Roche).
Chemical inhibition of LSD1
TPCs, DGCs and normal human astrocytes were plated 24 hours prior to addition of the LSD1 inhibitor S2101 (Millipore/Calbiochem). The untreated controls for each cell type received DMSO as vehicle. Dilution series ranged from 0–100 μM. Media and inhibitor were refreshed every 96 hours for a 14-day duration. Percent viability was determined by Trypan blue staining.
Tumorigenicity study
Intracranial injections were performed with a stereotactic apparatus (Kopf Instruments) at coordinates 2.2mm lateral relative to Bregma point and 2.5mm deep from dura mater. Four severe combined immunodeficient (SCID) mice (NCI Frederick) were used per condition. For cDNA overexpression experiments, 100,000 cells were used per mouse, unless otherwise specified. For shRNA experiments, 5000 TPC cells per mouse were injected. Kaplan-Meier curves and statistical significance (log-rank test) were calculated with the R survival package (R, 2008). Animal experiments were approved by the Institutional Animals Care and Use Committee (IACUC) at Massachusetts General Hospital.
Supplementary Material
Significance.
In glioblastoma, a subset of stem-like cells drive tumor propagation and resistance to existing therapies, but remain poorly understood. Suvà et al integrate chromatin profiling and cellular reprogramming to identify a minimal core of neurodevelopmental transcription factors that is sufficient to generate stem-like cells in glioblastoma. The corresponding regulatory network identifies tumor dependencies and suggests alternative approaches to eradicate tumor propagating cells.
Highlights.
distinct epigenetic state enables glioblastoma cells to propagate tumors in vivo
four TFs reprogram differentiated glioblastoma cells into tumor-propagating cells
these four TFs coordinately expressed in stem-like cells in primary human tumors
LSD1 histone demethylase identified as therapeutic target in tumor-propagating cells
Acknowledgments
We thank Timothy Durham, Noam Shoresh, Mia Caplan Uziel and Jing Gao for computational assistance and Charles Epstein, Meital Hatan and the Broad Institute Genome Sequencing Platform for help with data production. We thank Leslie Gaffney and Bang Wong for graphical work, David Dombkowski for flow cytometry, James Kim for histology sections, Erica Shefler, Dave Gennert and John Trombetta for support, Pinky Bautista and Yukako Yagi for slide scanning. We thank Rahul Satija, Leah Escalante, Brian Liau, Richard Koche, Russel Ryan for fruitful discussions and Stephen Elledge for the pINDUCER vectors. M.L.S is supported by Oncosuisse grant BIL-KFS-02590-02-2010 and a Medic Foundation grant. M.N.R is supported by awards from the Burroughs Wellcome Fund and HHMI. This research was supported by funds from the Howard Hughes Medical Institute, the Starr Cancer Consortium, the Burroughs Wellcome Fund, the Harvard Stem Cell Institute and the Klarman Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, Prieto-Sanchez RM, Barba I, Martinez-Saez E, Prudkin L, et al. TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Barrett LE, Granot Z, Coker C, Iavarone A, Hambardzumyan D, Holland EC, Nam HS, Benezra R. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer cell. 2012;21:11–24. doi: 10.1016/j.ccr.2011.11.025. [DOI] [PubMed] [Google Scholar]
- Binda E, Visioli A, Giani F, Lamorte G, Copetti M, Pitter KL, Huse JT, Cajola L, Zanetti N, DiMeco F, et al. The EphA2 receptor drives self-renewal and tumorigenicity in stem-like tumor-propagating cells from human glioblastomas. Cancer cell. 2012;22:765–780. doi: 10.1016/j.ccr.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D’Alessio AC, Young RA, Weinberg RA. Poised Chromatin at the ZEB1 Promoter Enables Breast Cancer Cell Plasticity and Enhances Tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Chudnovsky Y, Kim D, Zheng S, Whyte WA, Bansal M, Bray MA, Gopal S, Theisen MA, Bilodeau S, Thiru P, et al. ZFHX4 Interacts with the NuRD Core Member CHD4 and Regulates the Glioblastoma Tumor-Initiating Cell State. Cell reports. 2014 doi: 10.1016/j.celrep.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day BW, Stringer BW, Al-Ejeh F, Ting MJ, Wilson J, Ensbey KS, Jamieson PR, Bruce ZC, Lim YC, Offenhauser C, et al. EphA3 maintains tumorigenicity and is a therapeutic target in glioblastoma multiforme. Cancer cell. 2013;23:238–248. doi: 10.1016/j.ccr.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell (United States) 2010:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Human molecular genetics. 2009;18:R195–201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, Radovanovic I, Rheinbay E, Provero P, Stamenkovic I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes & development. 2012;26:1926–1944. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet neurology (England) 2010:717–726. doi: 10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al. Phosphorylation of EZH2 Activates STAT3 Signaling via STAT3 Methylation and Promotes Tumorigenicity of Glioblastoma Stem-like Cells. Cancer cell. 2013 doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS genetics. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, Wu Q, Vasanji A, McLendon RE, Hjelmeland AB, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell stem cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, Jaenisch R, Boyer LA. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS genetics. 2013;9:e1003288. doi: 10.1371/journal.pgen.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Huillard E, Kesari S, Maire CL, Golebiowski D, Harrington EP, Alberta JA, Kane MF, Theisen M, Ligon KL, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer cell. 2011;19:359–371. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nature biotechnology. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry. 2010;49:6494–6503. doi: 10.1021/bi100299r. [DOI] [PubMed] [Google Scholar]
- Morris SA, Daley GQ. A blueprint for engineering cell fate: current technologies to reprogram cell identity. Cell research. 2013;23:33–48. doi: 10.1038/cr.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume A, Ito M, Katsushima K, Ohka F, Hatanaka A, Shinjo K, Sato S, Takahashi S, Ishikawa Y, Takeuchi I, et al. Chromatin regulator PRC2 is a key regulator of epigenetic plasticity in glioblastoma. Cancer research. 2013;73:4559–4570. doi: 10.1158/0008-5472.CAN-13-0109. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinbay E, Suva ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL, Rivera MN, et al. An Aberrant Transcription Factor Network Essential for Wnt Signaling and Stem Cell Maintenance in Glioblastoma. Cell reports. 2013;3:1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Molecular cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell stem cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker SH, Feber A, Engstrom PG, Caren H, Kurian KM, Takashima Y, Watts C, Way M, Dirks P, Bertone P, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes & development. 2013;27:654–669. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nature biotechnology (United States) 2011:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell stem cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Wakimoto H, Kesari S, Farrell CJ, Curry WT, Jr, Zaupa C, Aghi M, Kuroda T, Stemmer-Rachamimov A, Shah K, Liu TC, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer research. 2009;69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends in cell biology. 2005;15:494–501. doi: 10.1016/j.tcb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Yang P, Wang Y, Chen J, Li H, Kang L, Zhang Y, Chen S, Zhu B, Gao S. RCOR2 is a subunit of the LSD1 complex that regulates ESC property and substitutes for SOX2 in reprogramming somatic cells to pluripotency. Stem cells. 2011;29:791–801. doi: 10.1002/stem.634. [DOI] [PubMed] [Google Scholar]
- Yoon OK, Brem RB. Noncanonical transcript forms in yeast and their regulation during environmental stress. Rna. 2010;16:1256–1267. doi: 10.1261/rna.2038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.