Abstract
Receptor tyrosine kinase (RTK) signals regulate the specification of a varied array of tissue types by utilizing distinct modules of proteins to elicit diverse effects. The RSK proteins are part of the RTK signal transduction pathway and are thought to relay these signals by acting downstream of extracellular signal-regulated kinase (ERK). In this study we report the identification of ribosomal S6 kinase 4 (Rsk4) as an inhibitor of RTK signals. Among the RSK proteins, RTK inhibition is specific to RSK4 and, in accordance, is dependent upon a region of the RSK4 protein that is divergent from other RSK family members. We demonstrate that Rsk4 inhibits the transcriptional activation of specific targets of RTK signaling as well as the activation of ERK. Developmentally, Rsk4 is expressed in extraembryonic tissue, where RTK signals are known to have critical roles. Further examination of Rsk4 expression in the extraembryonic tissues demonstrates that its expression is inversely correlated with the presence of activated ERK 1/2. These studies demonstrate a new and divergent function for RSK4 and support a role for RSK proteins in the specification of RTK signals during early mouse development.
Receptor tyrosine kinase (RTK) signals have established roles in a variety of physiological processes, including cellular proliferation and migration, tissue specification, and hormone regulation (reviewed in references 50, 53, and 65). RTK signals are relayed, in part, by the GTPase RAS and a cascade of kinases which include RAF, MEK, and extracellular signal-regulated kinase (ERK) (reviewed in reference 5). Though members of this pathway have been extensively studied, the mechanism by which the activation of these seemingly generic signals can illicit such a diversity of specific functions is still an area of intense investigation.
Studies of the p90 ribosomal S6 kinases (RSKs) indicate that these proteins may aid in the specification of RTK signals. The RSK proteins are intercellular serine/threonine kinases and were among the first identified targets of ERK (39, 64). Their protein structure comprises an ERK binding site as well as two distinct functional kinase domains; the C-terminal kinase domain is important for autophosphorylation, while the N-terminal kinase domain phosphorylates other target substrates (11, 26, 39, 76). In mammals, there are four p90Rsk genes, termed Rsk1 to -4 (RPS6K A1, A3, A2, and A6, respectively) (2, 47, 75, 77). Comparative analyses of RSK1-4 suggest that these proteins may have distinct roles for specifying ERK signals. Biochemical studies of RSK1-3 demonstrate that these proteins have different binding specificities for ERK and, after RTK stimulation, interact with ERK for different lengths of time (55, 76). In addition, RSK1 has limited interaction with identified targets of RSK2 (18), and Rsk1, -2, -3, and -4 genes are expressed in different patterns during late embryonic stages and in adult tissues (2, 42, 75). The biochemical differences among the family members suggest that there may be functional differences among the RSK proteins on a cellular level. Moreover, the differential expression patterns of the Rsk genes suggest distinct roles for the specification of RTK signals during mammalian embryogenesis.
Genetic analysis of mice has shown that many components of the RTK pathway are critical for the formation and patterning of the extraembryonic tissues, the precursors of the placenta (27, 54, 58, 74). Analysis of activated ERK during early mouse development further demonstrates that RTK signals are stimulated in the extraembryonic tissue (21). We wanted to identify molecules that could modify the outcome of RTK signals in the developing mouse embryo. Given the well-established role of RTK signals in the extraembryonic tissue, we decided to use this tissue as a source for identifying molecules that can mediate RTK signals. Unfortunately, the mouse embryo is small and confined to the uterus during the time at which RTK signals in the extraembryonic tissue are most influential. To overcome these technical challenges, we devised a method to identify mediators of RTK signals in the extraembryonic tissue by exploiting the more tractable Xenopus embryo as a screening tool (6, 14). In Xenopus the RTK pathway is necessary for the formation of the mesoderm, a tissue that has been well studied (reviewed in reference 29). Based on the knowledge that the core proteins of RTK signaling are conserved across species and utilized for the formation of different types of tissues, we screened a mouse expression library containing extraembryonic tissue for molecules that, upon overexpression, could alter the fate of Xenopus mesoderm.
In this screen for RTK modulators, we identified ribosomal S6 kinase 4 (RSK4), the fourth and least-studied member of the RSK family (75). Though similar in structure to the other RSK family members, RSK4 has a function that is distinct from that of RSK1 to RSK3. Consistent with this functional divergence, the inhibitory activity of RSK4 for RTK signaling is dependent upon a region that is not conserved in the other RSK proteins. Analysis of RSK4 in the developing mouse embryo demonstrates that Rsk4 is at the right time and place for this functional activity. Rsk4 expression is altered in mice without fibroblast growth factor (FGF) signals, suggesting that RTK signals regulate Rsk4 in vivo. These data provide the first evidence that an RSK protein may play an important role in the regulation of RTK signaling and tissue patterning in the developing mammalian embryo. Moreover, this study provides further support for the notion that though similar, the RSK proteins have diverse and distinct functions in the regulation of RTK signals.
MATERIALS AND METHODS
Construction of the mouse expression library.
Construction and subsequent gridding of the 6.5-day postcoitum (d.p.c.) mouse expression library has been previously described (6, 7, 14).
Construction of DNA plasmids.
The expression library used for the screen was constructed with the pCS105 expression vector (35). The identified Rsk4 gene was subcloned from this vector into pCS107 (6) at SalI/NotI sites. Using a PCR strategy, MYC-tagged clones were constructed by cloning products into the EcoRI site of MTpCS3+, a modified version of pCS2 (54, 56, 70) (for MTpCS3+ sequence, visit http://sitemaker.med.umich.edu/dlturner.vectors). The original Rsk4pCS105 plasmid was used as a template for the MYC-tagged Rsk4 and Δ1-96Rsk4 constructs. Rsk2 pMT2, provided by C. Bjorbaek (24), and Homo sapiens Rsk1 pKH3 and H. sapiens Rsk3 pkH3 (55) were the templates for the Rsk2, -1, and -3 MYC constructs, respectively. Primers with EcoRI sequences were used to amplify the Rsk4 open reading frame (ORF) (forward [F], 5′ CGTCAGAATTCTATGCTGAATTTTAGAAGGACACGCC; reverse [R], 5′ GCGTACGAATTCGGACTGAAGAGCACAAGACTCTTA), Δ1-96Rsk4 (F, 5′ GCGTCAGAATTCTCCTGAAGCGGCGATGCTACCGTT; R, 5′ GCGTACGAATTCGGACTGAAGAGCACAAGACTCTTA), Mus musculus Rsk2 (F, 5′ AGCCTCTAGAAATGCC GCTGGCGCACGTGGCG; R, 5′ GAGCCTCTAGAACAGGGCTGTTGAGGTG). H. sapiens Rsk1 (F, 5′ GCGTCAGGATTCTATGCCGCTCGCCC; R, 5′ GCGTCAGGATTCTCAGG GTGGTGG) and H. sapiens Rsk3 (F, 5′ GCGTCAGGATTCTATGGACCTGAGCATG; R, 5′ GCGTCAGGATTCTCTACAGCCGCGTGG). Protein analysis using the TNT coupled reticulolysate system (Promega, Madison, Wis.) with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis for MYC demonstrated bands at the expected sizes. Δ91-860Rsk4 was constructed from Rsk4 in MTpCS3+. MTpCS3+Rsk4 was cut with XhoI enzyme to excise all but the first 272 bp of the Rsk4 gene. Rsk4 in MTpCS3+ was also used as the template for the KΔQ221Rsk4 construct. QuikChange site-directed mutagenesis (Stratagene, La Jolla, Calif.) was used with the following primer and its reverse complement: F, 5′ GGGCAGCTCTATGCAATGCAGGTGTTAAG AAAAGCTT. Mutations were confirmed by sequence analysis. The full length of the clone was sequenced to ensure that no other mutations were present. In vitro transcription and translation of this clone, followed by SDS-PAGE and Western blot analysis with MYC antibody, demonstrated that a protein at the expected size was formed.
Transcription of RNA.
DNA constructs were linearized, and RNA was transcribed with an mMESSAGE Machine kit (Ambion, Austin, Tex.). The sources of the constructs are given in Table 1.
TABLE 1.
DNA constructs used in this study
| RNAa | Vector | Linearization | Reference |
|---|---|---|---|
| Fgf8 | pCS105 | AscI | |
| Erk sem | pGEMHE | NheI | 51 |
| p21Ras | pSP64 | EcoRI | 71 |
| M.m. Rsk2 | MTpCS3+ | ApaI | This work |
| H.s. Rsk3 | MTpCS3+ | ApaI | This work |
| X.l. FARsk1 | pOTV-LIC | NotI | 28 |
| H.s. Rsk1 | MTpCS3+ | ApaI | This work |
| Noggin | pCS2+ | NotI | 61 |
| Activin | pSP64T | AscI | 68 |
| Smad2 | pCS105 | AscI | 7 |
| Rsk4 | MTpCS3+ | ApaI | This work |
| Δ1-96Rsk4 | MTpCS3+ | ApaI | This work |
| Δ91-860Rsk4 | MTpCS3+ | ApaI | This work |
| KΔQ221Rsk4 | MTpCS3+ | ApaI | This work |
M.m., M. musculus; H.s., H. sapiens; X.l., X. laevis.
Microinjections.
Female frogs were primed for ovulation with human chorionic gonadotropin (19). Embryos were collected into 0.1× MR solution (52), fertilized in vitro, and dejellied with 2.5% cysteine, pH 8.0. Embryos were transferred into 0.3× MR with 2.5% Ficoll for injection. Embryos were injected at the one-cell stage in either the presumptive ectoderm (for explant assays) or the presumptive mesoderm (for in situ analysis). Injection of Xenopus laevis oocytes was completed as previously described (63).
In situ hybridizations.
Plasmids used to generate antisense probes are listed in Table 2.
TABLE 2.
Antisense probes used in this study
| Probe | Vector | Reference |
|---|---|---|
| Xbra | pGEM7 | 60 |
| Sox17β | pSPJC2L | 36 |
| Endodermin | pBS | 57 |
| Goosecoid | pG500 | 13 |
| MyoD | pSP73 | 34 |
| Rsk4Δ91-860 | pCS107 | This work |
X. laevis embryos were developed to stage 10.5 or 12.5 and fixed in MEMFA (52). Single in situ analysis was performed as described previously (30). Double in situ analysis was completed using a modified protocol (22) which has been previously described (14). Detection of β-galactosidase (β-Gal) protein was performed as described previously (62) with the replacement of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside with 6-chloro-3-indolyl-β-d-galactoside (Red-Gal; Research Organics, Cleveland Ohio).
Mouse in situ hybridizations were completed using a modified protocol (10, 72): embryos were dissected from pregnant Swiss Webster mice (Simmonson Lab, Gilroy, Calif.). Embryos were blocked with 2% BM Block (Roche, Indianapolis, Ind.) in 1× MAB buffer (52) and incubated at 4°C with anti-digoxigenin-AP Fab fragments (Roche) at a 1/2,000 dilution. Embryos were developed with BM Purple AP substrate (Roche). The antisense probe for Rsk4 was synthesized by using the Rsk4Δ91-860 construct as a template.
Ectodermal explants and RT-PCR assays.
Embryos were injected into the presumptive ectoderm at the one-cell stage, developed to stage 8-9, and transferred to 0.75× NAM (52) solution for tissue excision. Explants were cultured in 0.75× NAM until stage 10.5. RNA was isolated, and reverse transcription (RT)-PCR was performed as previously described (73). Primers used are listed in Table 3.
TABLE 3.
Primers used in this study
| Gene | Primer sequencea | Reference |
|---|---|---|
| Xbra | F, GGATCGTTATCACCTCTG | 73 |
| R, GTGTAGTCTGTAGCAGCA | ||
| Ef1α | F, CAGATTGGTGCTGGATATGC | 73 |
| R, ACTGCCTTGATGACTCCTAG | ||
| ODC | F, CAGCTAGCTGTGGTGTGG | 1 |
| R, CAACATGGAAACTCACACC | ||
| Sox17β | F, AACTCCCACCAGCAGGCTACTTTG | |
| R, TGTCAATGTCACTCTCCAGATGTCC | ||
| Gsc | F, ACAACTGGAAGCACTGGA | 73 |
| R, TCTTATTCCAGAGGAACC | ||
| Endodermin | F, TATTCTGACTCCTGAAGGTG | 57 |
| R, GAGAACTGCCCATGTGCCTC | ||
| NCAM | F, CACAGTTCCACCAAATGC | 31 |
| R, GGAATCAAGCGGTACAGA | ||
| Sef | F, GTCGAATTCGCCTGCAACGACCAAGTGGC | |
| R, CTGCGAATTCGCCCTCGTGCTTCAGTTTG | ||
| Sprouty2 | F, CATTCCTTGTTTCAGGC | |
| R, GATGGGAGAGTCCTTGG |
F, forward; R, reverse.
Mouse whole-mount antibody analysis.
Mouse whole-mount antibody staining was completed as previously described (21) with polyclonal phospho-p44/42 mitogen-activated protein kinase (MAPK) (Thr202/Tyr204) antibody [Phospho-p44/42MAPK(Thr202/Tyr204)] (Cell Signaling Technology, Beverly, Mass.). Staining was detected using Tyramide signal amplification kit no. 15 (Molecular Probes, Eugene, Oreg.) or diaminobenzidine.
In vitro culture of mouse embryos.
Embryos were dissected and cultured as described previously (21).
Protein assays and immunoblots.
Embryos were injected with RNA at the one-cell stage. Ectoderm was excised at stage 8-9 and allowed to develop until stage 10.5 in 0.75× NAM. Tissue was lysed in NOP buffer (150 mM NaCl, 10 mM Tris [pH 7.5], 1 mM MgCl2, 0.75 mM CaCl2, 2% IGPal) with general protease inhibitors (Sigma, St. Louis, Mo.), 10 mM NaF (Sigma), 10 nM sodium orthovanadate (Sigma), and 5 mM β-glyceraldehyde (Sigma). Lysates were centrifuged at 4°C for 10 min, and the supernatant was removed. Samples were analyzed by SDS-PAGE and Western blotting with the following antibodies: Phospho-p44/42MAPK(Thr202/Tyr204) E10 monoclonal, p42 MAP kinase 3A7 monoclonal, and phospho-Mek1/2 (Ser217/221) (Cell Signaling Technology), anti-rabbit immunoglobulin, horseradish peroxidase (HRP)-linked whole antibody from donkey (Amersham, Piscataway, N.J.), and HRP-labeled polyclonal anti-mouse immunoglobulin (Pharmingen, San Diego, Calif.).
Kinase assays.
One-cell-stage Xenopus embryos were injected with Rsk4, Rsk2, Δ1-96Rsk4, and KΔQ221Rsk4 RNA. Protein lysates were harvested at the gastrula stage and analyzed for kinase activity using the S6 kinase assay (Upstate, Lake Placid, N.Y.). Five experiments were analyzed for P32 transfer. The counts per minute (cpm) of Rsk4, Rsk2, and Δ1-96Rsk4 were significantly different (P < 0.05) from the cpm of the control (uninjected embryo lysates) when analyzed individually with a matched t test. The cpm of KΔQ221Rsk4 were not significantly different from that of the control lysates.
RESULTS
Identification of Rsk4.
To identify molecules that regulate RTK signaling, we performed a functional screen in which pools of RNA from a 6.5-d.p.c. mouse expression cDNA library (14) were injected into the presumptive mesoderm of the one-cell X. laevis embryo. Embryos were cultured to stage 10.5 (midgastrulation) and analyzed for the expression of the T-box transcription factor, Xbrachyury (Xbra) (60), a downstream target of RTK signals and a molecular marker of mesoderm. Using this strategy, a pool of RNA was identified for its ability to disrupt the Xbra expression pattern in the absence of any noticeable cell death. From the pool, a single clone was identified which encoded a 4-kb transcript with a 2.6-kb ORF. The ORF exhibited similarity to the Ribosomal S6 kinase (p90Rsk) family and 89% identity to the human Rsk4 (RPS6KA6) gene (75), indicating that it is the mouse orthologue of human Rsk4.
Rsk4 is the fourth member of the mammalian p90rsk gene family (39). Human Rsk4 was originally identified in a positional cloning study as a candidate for an X-linked mental retardation syndrome, although no mutations within the Rsk4 gene were identified in patients affected with the syndrome (75). To date, the biochemical and cellular characteristics of RSK4 have not been described. The cloning of Rsk4 in a functional screen for Xbra inhibition suggested that Rsk4 could modulate RTK signals.
As seen in Fig. 1, injection of Rsk4 into the marginal zone of the developing embryos results in the disruption of normal Xbra expression (Fig. 1E). Upon further development, these embryos have severe defects, including patent blastopores and deformed neural tubes (Fig. 1F and G). The resulting tailbud-stage embryos are truncated along the anterior-posterior axis (Fig. 1H). These developmental abnormalities are consistent with the disruption of mesoderm and are similar to the phenotypes seen in embryos injected with other RTK inhibitors, such as dominant-negative forms of the FGF receptor and RAF proteins (3, 67).
FIG. 1.
Rsk4 is identified in a functional expression screen. Rsk4 was injected into the marginal zone of a one-cell embryo (E to H). Uninjected embryos were developed under the same conditions (A to D). In situ analysis shows that Rsk4-injected embryos at the gastrula stage (E) display disrupted Xbra expression compared to controls (A). Upon further development, Rsk4-injected embryos have a patent blastopore (F), fail to properly neurulate (G), and are truncated at the tailbud stage (H). The development of uninjected controls is also shown (B to D). The arrows demarcate the blastopore (B and F) and neural tube (C and G).
Xbra inhibition by Rsk4 is local.
To further characterize the effect of Rsk4 on Xbra, one-cell-stage embryos were injected at the margin with both Rsk4 and a lineage tracer, lacz. Embryos were allowed to develop until stage 10.5, fixed, and analyzed for both β-Gal activity and Xbra RNA. While embryos injected with lacz alone showed colocalization of β-Gal activity and Xbra RNA, embryos injected with Rsk4 and lacz showed no cellular colocalization of β-Gal activity and Xbra RNA (Fig. 2B and D). Additionally, in double in situ analysis for Rsk4 and Xbra performed on Rsk4-injected embryos, there was no colocalization of the two RNAs (Fig. 2L and N). These results indicate that Rsk4 acts locally to inhibit Xbra expression.
FIG. 2.
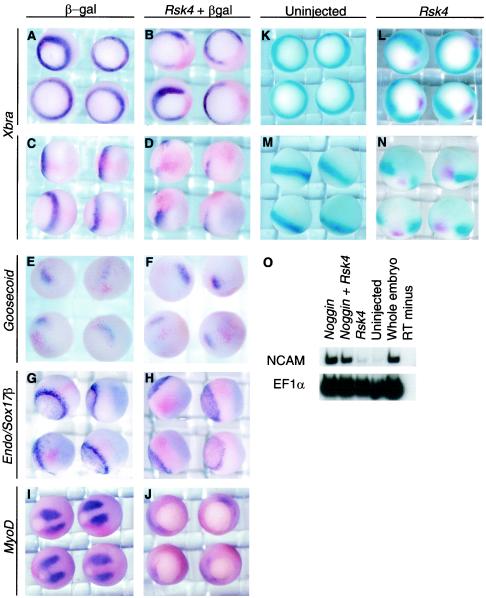
Characterization of embryos injected with Rsk4. (A to J) X. laevis one-cell-stage embryos were injected at the margin with lacZ RNA (A, C, E, G, and I) or both lacz and Rsk4 RNA (B, D, F, H, and J). Embryos were allowed to develop to stage 10.5 (A to H) or stage 12.5 (I and J), stained for β-Gal activity, and then analyzed by in situ hybridization. Xbra expression (A to D) is shown from the vegetal (A and B) and lateral (C and D) view. Goosecoid expression (E and F) is shown from a dorsal-vegetal view. Sox17β and Endodermin expression (G and H) are shown from a dorsal-vegetal view. MyoD expression is shown (I and J). X. laevis was injected into the margin of one-cell-stage embryos with Rsk4 RNA. Double in situ analysis was performed for Xbra (turquoise) and Rsk4 (magenta) (K to N). Uninjected controls are shown (K and M). The presumptive ectoderm tissue of one-cell embryos was injected with Noggin and Noggin with Rsk4. Ectodermal explants were excised and developed in vitro to neurula stage. RNA was isolated, and RT-PCR was performed for NCAM, a neural marker, and EF1α, a loading control (O).
Rsk4 specifically disrupts the formation of mesoderm.
The inhibition of Xbra and the resulting developmental phenotype in Rsk4-injected embryos were consistent with a disruption of RTK signaling. To assess the specificity of this disruption, we studied the role Rsk4 had in regulating markers of other tissues. Rsk4-injected embryos were analyzed at gastrula stages by in situ hybridization with markers for different types of mesoderm (Fig. 2). At stage 10.5, the mesoderm is patterned along the dorsal-ventral axis (reviewed in reference 48). In situ analysis of a dorsally restricted marker, Goosecoid (Gsc) (16), revealed that injection of Rsk4 did not affect the specification of dorsal tissues (Fig. 2F). However, expression of MyoD, a helix-loop-helix transcription factor that marks the more ventral, somitic mesoderm (34), was disrupted in Rsk4-injected embryos at stage 12.5 (Fig. 2J). Analysis of Sox17β and Endodermin, markers of endoderm (36, 57) (both stained in Fig. 2H), demonstrated a slight reduction of expression in Rsk4-injected embryos compared to controls. Endodermin normally stains a subset of mesodermal cells (57), and we suspect that this slight reduction of expression of these markers is a loss of expression in these cells. Rsk4-injected embryos have severe developmental defects and often die prior to neurulation. To investigate the effect of Rsk4 expression on the formation of neural tissue, the naive ectoderm of the one-cell X. laevis embryo was injected with the bone morphogenic protein antagonist and neural inducer Noggin (45, 61, 78) with and without Rsk4 RNA. Injected ectodermal tissue was excised at the blastula stage (herein referred to as ectodermal explants) and cultured in vitro. At the neurula stage, RNA was isolated from the ectodermal explants and analyzed by RT-PCR to detect the neural marker NCAM (37). Results of this experiment showed that Rsk4 does not inhibit the signals that induce neural tissue (Fig. 2O). This in situ analysis led us to conclude that Rsk4 did not affect the expression of all tissue types and therefore was acting on a specific signaling pathway.
Rsk4 disrupts FGF signaling.
The signaling pathways that regulate the specification, as well as the dorsal-ventral patterning, of the germ layers in Xenopus have been well studied. Cursory analysis of the tissue formation of Rsk4-injected embryos allowed for analysis of the effect of Rsk4 overexpression on several pathways, including the FGF and transforming growth factor beta (TGF-β) pathways. Both the FGF and TGF-β signaling pathways are integral to the proper induction of Xbra (reviewed in reference 29). The TGF-β pathway can also induce endoderm and specify dorsal mesoderm (16, 33). These signaling pathways are not independent of each other, since TGF-β signals cannot induce general mesoderm in the absence of the FGF pathway. For example, in the presence of dominant-negative FGF receptor, ectopic TGF-β signals can no longer induce Xbra or MyoD but maintain the ability to induce the dorsal mesoderm marker Gsc (20, 44). The large effect of Rsk4 overexpression on somitic mesoderm, but not dorsal or endodermal tissues, is consistent with a specific effect of Rsk4 on the FGF pathway. To further investigate this possibility, the effect of Rsk4 on the FGF and TGF-β pathways was analyzed in more detail.
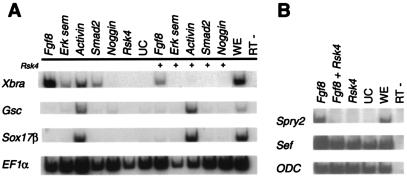
To investigate the effect of Rsk4 on the FGF and TGF-β pathways, an ectodermal explant assay was performed. Activin, encoding a TGF-β ligand, Smad2, encoding a mediator of TGF-β signals, and Fgf8 were injected into the presumptive ectoderm of one-cell embryos, with or without Rsk4. Ectoderm explants were cultured to stage 10.5 and then harvested for RNA. RT-PCR was performed to detect downstream targets of the TGF-β and FGF pathways. As shown in Fig. 3, Gsc, a dorsal mesoderm marker, and Sox17β, an endoderm marker, are induced by Activin. (The level of Smad2 RNA injected was not high enough to induce dorsal or endodermal genes in this experiment.) These inductions are maintained in the presence of Rsk4, indicating that Rsk4 does not directly inhibit the TGF-β pathway (Fig. 3A). Analysis of Xbra demonstrated that the presence of Rsk4 inhibited Fgf8, Activin, and Smad2 from inducing Xbra. Previous studies have shown that if the FGF pathway is abrogated, TGF-β cannot induce Xbra (20, 44). Therefore, this result is consistent with Rsk4 directly disrupting both the FGF pathway and Xbra-inducing capabilities of the TGF-β pathway or the FGF pathway only. We further investigated the effect of Rsk4 on two FGF-specific targets, Sprouty2 and Sef (49, 69). Fgf8 enhanced the expression of both Sprouty2 and Sef. This up-regulation was impeded by the presence of Rsk4 (Fig. 3B), indicating that Rsk4 inhibits FGF signals. Additionally, Rsk4 inhibits the induction of Xbra by both FGF and TGF-β signals. Since the FGF pathway is required for TGFβ induction of Xbra and the injection of Rsk4 does not impede the transcription of other downstream signals of TGF-β, we conclude that Rsk4 is acting specifically on the FGF pathway.
FIG. 3.
Rsk4 specifically blocks the Fgf pathway. (A) Embryos were injected into the presumptive ectoderm tissues with mesoderm inducing molecules with or without Rsk4. The ectodermal explants were excised and cultured to the gastrula stage. RNA was isolated, and RT-PCR was performed for Xbra, Gsc, Sox17β, and EF1α. (B) The presumptive ectodermal tissue of one-cell embryos was injected with Fgf8 with or without Rsk4. Ectodermal explants were excised, and RNA was collected and analyzed by RT-PCR for Sprouty2, Sef, and ODC, a loading control. UC, uninjected control explants; WE, whole embryo; RT-, reverse transcriptase minus.
Rsk4 inhibits the RAS-ERK pathway.
In order to elucidate the molecular mechanism of Rsk4 function, we investigated the effect of Rsk4 on the RAS-ERK pathway, the mediator of FGF signals. The FGF ligands signal through an RTK receptor and the RAS-ERK pathway, a kinase cascade which includes the p21RAS, RAF, MEK, and ERK proteins (reviewed in reference 5). Constitutively active molecules of the FGF receptor, p21RAS, RAF, and ERK, can all induce Xbra (43, 46, 71). To investigate whether Rsk4 affects this pathway, we injected embryos with p21Ras and Drosophila melanogaster Erksem (51), encoding a constitutively active ERK. Both of these molecules are capable of inducing Xbra expression in Xenopus (43, 71). The presence of Rsk4 disrupted the ability of both p21Ras and D. melanogaster Erksem to induce Xbra expression (Fig. 3A and 4A). These results indicate that Rsk4 can block the RAS-ERK pathway and that Rsk4 likely acts directly on or downstream of ERK.
FIG. 4.
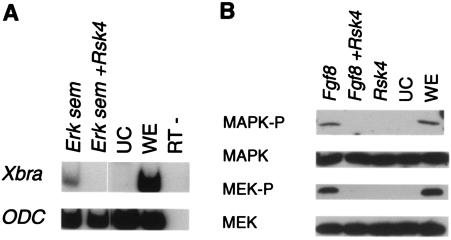
Rsk4 disrupts the RAS-MAPK pathway. (A) The presumptive ectodermal tissue of one-cell embryos was injected with the Erksem or p21Ras, with or without Rsk4. RNA was isolated from injected tissue, and RT-PCR was performed for Xbra and ODC. (B) Fgf8 was injected into the presumptive ectoderm of one-cell embryos with or without Rsk4. Ectodermal explants were excised and allowed to develop to gastrula stage. Proteins were isolated and analyzed by SDS-PAGE and Western blot techniques for p42MAPK and phospho-p44/42MAPK(Thr202/Tyr204). UC, uninjected control explants; WE, whole embryo; RT-, reverse transcriptase minus.
To further investigate the effect of Rsk4 on ERK, we analyzed the phosphorylation state of ERK in the presence of Rsk4. Fgf8 was injected into the presumptive ectoderm of the one-cell-stage embryo, with or without Rsk4 RNA. Ectodermal explants were excised and allowed to develop until gastrulation. As can be seen in Fig. 4B, embryos injected with Fgf8 alone had active ERK as determined by the phosphorylation at Tyr202 and Thr204 (4). However, active ERK was either reduced or absent in embryos injected with Fgf8 and Rsk4. That Rsk4 is able to block the inducing capabilities of D. melanogaster Erksem, and active ERK was not present in Rsk4-injected embryos, suggests that Rsk4 is acting at the level of ERK.
RTK inhibition is specific to Rsk4.
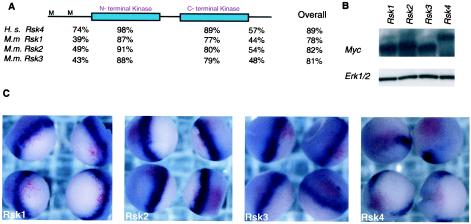
The RSK proteins have high similarity (Fig. 5A), and RSK1, -2, and -3 have been used interchangeably in biochemical assays to illicit similar effects (55, 76). A few studies have suggested a role for RSK proteins in negative feedback inhibition (23, 24, 40); however, RSK1, -2, and -3 have not been shown to directly inhibit RTK signaling. To investigate whether the other Rsk genes were capable of Xbra disruption, H. sapiens Rsk1, M.m. Rsk2, and H. sapiens Rsk3 (2, 77) were injected into the marginal zone of one-cell X. laevis embryos and analyzed by in situ analysis for Xbra expression at the gastrula stage. Xbra expression was not specifically disrupted by any these genes (Fig. 5C). To measure the protein level in the injected embryos, 20 embryos from each batch of injected embryos were removed prior to fixation. Protein was isolated from these embryos and analyzed by SDS and Western blotting for the MYC-tag protein. As can be seen in Fig. 5B, the embryos had similar amounts of protein.
FIG. 5.
The Rsk family. (A) Using DNA Star MegaAlign, the conversed region of RSK4 (97 to 860 amino acids) was aligned to mouse RSK1, RSK2, and RSK3 and human RSK4. The percent identities for the N terminus, N-terminal kinase, C-terminal kinase, and C-terminal domains are listed below the schematized protein structure. M marks the methionine at positions 1 and 97 in the full-length RSK4 protein. The second M is the start of the region of homology of RSK4 to the RSK family. (B) One-cell-stage embryos were coinjected at the marginal zone with lacz and Rsk1, Rsk2, Rsk3, or Rsk4. Embryos were allowed to develop to gastrula stages, at which point 20 embryos from each variable were removed for the isolation of protein and the remaining were fixed. Protein lysates were analyzed by SDS-PAGE and Western blotting with MYC antibody. (C) The fixed embryos were analyzed for β-Gal activity and Xbra expression.
Since the other Rsk homologs do not inhibit Xbra, we hypothesized that perhaps RSK4 was simply a more active isoform than the other RSK proteins. To investigate this, we analyzed the effects of the constitutively active Rsk clone, FARsk1 (28). FARsk1 was designed to mimic the hyperphosphorylated state of active RSK protein and has acidic residues at all of the known phosphorylation sites. Unlike its wild-type counterpart, FARsk1 induces oocyte maturation in the absence of progesterone. FARsk1 was injected into the marginal zone of the embryos and analyzed by in situ hybridization for Xbra. Xbra expression was not disrupted in embryos injected with this molecule (data not shown).
Conversely, we investigated whether Rsk4 could mimic the activity of FARsk1 in maturing X. laevis oocytes. Stage 6 oocytes were injected with Rsk4 RNA, and germinal vesicle breakdown was measured both visually and by ERK activity. Unlike results with FARsk1 (28), the Rsk4-injected oocytes did not mature in the absence of progesterone (data not shown). Since the Rsk genes are not interchangeable in our assays, we conclude that Rsk4 has an intrinsic property that is sufficient for the disruption of the RAS-ERK pathway and is not found in the other Rsk genes.
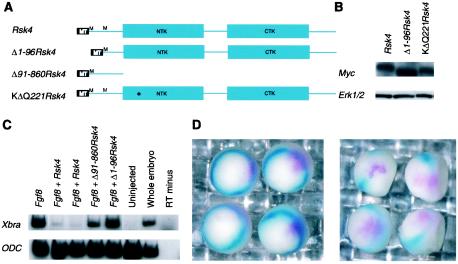
Mutational analysis of Rsk4.
To identify regions of Rsk4 that confer Xbra-inhibiting activity, we deleted regions of the Rsk4 gene that are divergent from the other Rsk genes (Fig. 6A). Members of the p90RSK family have a distinct protein structure, comprised of two functional kinase domains as well as an ERK (ERK1/2) docking site (26, 39, 76). Alignment of the Rsk4 translated ORF with members of the p90RSK family shows that these domains are highly conserved (Fig. 5 and data not shown). However, mouse RSK4 has a 96-amino-acid extension at its N terminus that is not present in the published sequences of mouse RSK1-3 or human RSK4 (2, 41, 75). No known protein motifs are present in this extension. However, it contains a high number of positively charged residues. To test whether this N-terminal extension conferred the mesoderm-inhibiting activity of RSK4, a Δ1-96Rsk4 construct was created. Further analysis of the clone demonstrated that it encodes a MYC-tagged protein of the expected size (Fig. 6B) and it is a functional kinase (see Materials and Methods). Fgf8 RNA was injected into the presumptive ectoderm of one-cell embryos with or without RNA generated from the Δ1-96Rsk4 construct. Ectodermal explants were excised, RNA was generated, and RT-PCR analysis for Xbra was performed. While the full-length Rsk4 was able to block the induction of Xbra by Fgf8, Δ1-96Rsk4 could not (Fig. 6C). To test whether the N-terminal region alone could confer RSK4 activity, an N-terminal clone, Rsk4Δ91-860, was constructed and analyzed. This clone did not impede the induction of Xbra by Fgf8 (Fig. 6C).
FIG. 6.
Rsk4 activity is dependent upon its N terminus and its N-terminal kinase. (A) Schematics of the RSK4 constructs, including Δ1-96Rsk4, Δ91-860Rsk4, and the KΔQ221Rsk4 clones. (B) Rsk4, Δ1-96Rsk4, and the KΔQ221Rsk4 RNAs were injected into one-cell-stage Xenopus embryos. At gastrula stage, protein was isolated and analyzed by SDS-PAGE and Western blotting with MYC antibody. (C) The presumptive ectoderm of a one-cell embryo was injected with Fgf8 and the various Rsk4 constructs. Ectodermal explants were excised, allowed to develop to gastrula stage, and analyzed by RT-PCR for the presence of Xbra. (D) Using site-directed mutagenesis, a K-to-Q transition at amino acid 221 was made in RSK4. RNA from this construct, KΔQ221Rsk4 (left panel), and the original Rsk4 construct (right panel) was injected into one-cell embryos at the marginal zone. Double in situ analysis was performed for detection of Xbra (turquoise) and Rsk4 (magenta). Uninjected, uninjected control explants; RT minus, reverse transcriptase negative.
Since the N-terminus region was necessary but not sufficient for Rsk4 activity, we wanted to investigate the relative importance of the kinase domain in mediating RSK4 activity. Members of the RSK protein family encode a protein with two distinct kinase domains (39). Previous mutational analysis of these kinase domains in other RSK family members has demonstrated differential activities for these domains. The carboxy-terminal kinase is generally required for autophosphorylation, while the N-terminal kinase domain is required for the phosphorylation of other target substrates (11, 26). A lysine-to-glutamine substitution at the predicted ATP binding site in the N-terminal kinase domain of RSK4 was created to remove the putative N-terminal kinase activity. The effect of this construct, K221QRsk4, on Xbra expression was analyzed by in situ hybridization. Rsk4 or K221QRsk4 was injected into the margin of one-cell embryos. The embryos were fixed at stage 10.5 and analyzed by double in situ hybridization. While Rsk4 RNA did not colocalize with Xbra RNA (Fig. 6D, right panel), K221QRsk4 and Xbra staining overlapped (Fig. 6D, left panel), indicating that the N-terminal kinase activity is likely required for Rsk4 activity in our assay. Protein lysates isolated from embryos injected with similar amounts of K221QRsk4 and Rsk4 RNA had similar amounts of protein (Fig. 6B). In assays for kinase activity, lysates from Rsk4-injected embryos had significantly different cpm than those from uninjected embryos, while lysates isolated from embryos injected with K221QRsk4 did not (see Materials and Methods). These results indicate that Rsk4 is likely dependent on its N-terminal kinase activity for its Xbra-inhibiting capability.
Rsk4 expression in the developing mouse gastrula.
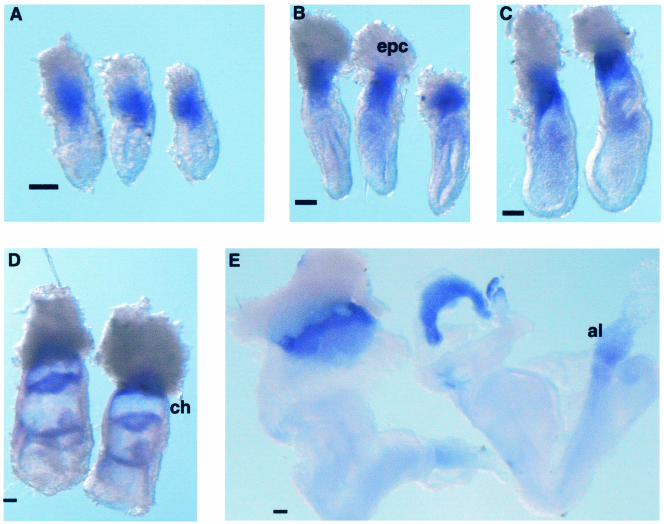
After first identifying and characterizing mouse RSK4 as a negative regulator of the RAS-ERK pathway in X. laevis, we proceeded to investigate the role of RSK4 in the mouse embryo. Since the Rsk4 gene was identified from an expression library derived from 6.5-d.p.c. mouse embryos, we examined the expression pattern of Rsk4 during this developmental time. Whole-mount in situ analysis revealed that Rsk4 localizes to the extraembryonic tissue of the mouse at 6.0 to 8.5 d.p.c. At 6.0 to 7.5 d.p.c., expression is restricted to central extraembryonic tissue: it is not expressed in the ectoplacental cone or the extraembryonic cells most proximal to the embryonic/extra-embryonic junction (Fig. 7A to C). Rsk4 expression is strong in the chorion at 7.5 d.p.c. (Fig. 7D) and persists to 8.5 d.p.c. (Fig. 7E). Expression can also be seen in the allantois (Fig. 7E). The strong expression of Rsk4 in the extraembryonic tissue suggests a possible role in the specification of these tissues.
FIG. 7.
Rsk4 expression in the early developing mouse embryo. (A to E) In situ analysis was performed on 6.0- to 8.5-d.p.c. embryos. At 6.0 (A), 6.5 (B), and 7.0 (C) d.p.c., expression is restricted to the central region of the extraembryonic tissue; it is not expressed in the ectoplacental cone or the extraembryonic cells most proximal to the embryonic/extraembryonic junction. (D and E) Rsk4 expression is strong in the extraembryonic ectoderm of the chorion at 7.5 d.p.c. (D) and persists to 8.5 d.p.c. (E). (E) Expression can also be seen in the allantois. Epc, ectoplacental cone; ch, chorion; al, allantois. Space bar = 100 μm.
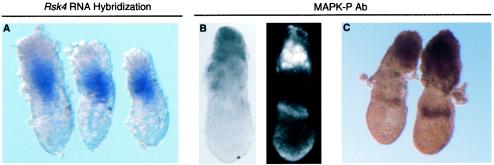
FGF and RAS-ERK signaling are known to have important roles in the specification of the extraembryonic tissue. Mice with targeted mutations in Fgf4, FgfR2, Grb2, or Mek1 exhibit peri-implantation lethality due in part to a lack of trophoblast proliferation (25, 27, 58, 74). Trophoblast stem cells have been derived from the extraembryonic ectoderm at these stages and require FGF-RAS-ERK signaling for maintenance of their stem cell properties (66). Analysis of the temporal and spatial expression of dual phosphorylated ERK1 and -2 (active ERK) in the early developing mouse showed that there is strong ERK signaling in the extraembryonic ectoderm and ectoplacental cone (21). This pattern of ERK is restricted compared to the expression of FgfR2, encoding an RTK receptor, which is located throughout the extraembryonic ectoderm (17). To compare the expression of Rsk4 and active ERK, RNA in situ analysis and whole-mount antibody staining was performed on similarly staged embryos (Fig. 8). The region of Rsk4 expression is proximally and distally flanked by areas of tissue with active ERK. There is dark staining of active ERK in the ectoplacental cone and in a ring of tissue just proximal to the embryonic/extraembryonic junction. The juxtaposed nature of these two expression patterns suggests that Rsk4 may be playing a role in restricting the domain of activated ERK in the extraembryonic tissue.
FIG. 8.
Comparison of Rsk4 and active MAPK expression localization. Embryos at stage 6.25 d.p.c. were analyzed for Rsk4 RNA (A) and active MAPK protein expression (B and C). Whole-mount antibody stain for MAPK was detected by HRP visualization with both a tyramide amplification kit (B) and diaminobenzidine staining (C). The left panel in B is the light view of the same embryo under fluorescence on the right panel.
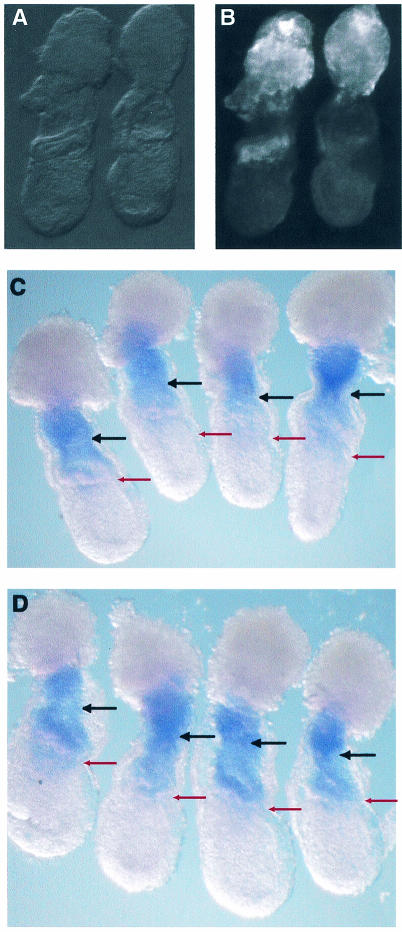
Rsk4 expression is altered in embryos cultured in FGF inhibitor.
The localization of activated ERK just proximal to the embryonic/extraembryonic junction is dependent upon FGF signaling. Embryos cultured in the FGF inhibitor SU5402 lack prominent activated ERK in this ring of tissue (Fig. 9B, right embryo). In contrast, active ERK expression in the proximal ectoplacental cone persists. This indicates that FGF is required for active ERK expression at the embryonic/extraembryonic junction but not in the ectoplacental cone (21). We hypothesized that if Rsk4 were regulated by FGF signaling, Rsk4 expression would be altered in embryos cultured in SU5402. Therefore, we analyzed the expression pattern of Rsk4 in embryos cultured in SU5402. A litter of 6.5-d.p.c. mice was dissected from a pregnant mouse and divided into two experimental treatments. One group was cultured in vitro in media supplemented with SU5402. The other was cultured in media with dimethyl sulfoxide (DMSO), the solvent of SU5402. These two groups were then further divided. Half of each group was examined for Rsk4 expression, and half was examined for the presence of activated ERK. The expression patterns of both Rsk4 and active ERK in the DMSO-cultured embryos were normal. In contrast, the mice cultured in SU5402 lacked active ERK expression in the ring just proximal to the extraembryonic/embryonic junction. Additionally, Rsk4 expression in these embryos was expanded distally (Fig. 9D). The expansion of Rsk4 into regions without active ERK but not into regions where active ERK was still present suggests that the formation of the Rsk4 and active ERK boundary is dependent upon FGF signals.
FIG. 9.
Rsk4 expression is altered in embryos cultured in FGF inhibitor. (A and B) 6.5-d.p.c. embryos were cultured in DMSO (embryos on the left of panels A and B) or SU5402, an FGF inhibitor (embryos on the right of panels A and B). Whole-mount antibody staining was performed for active MAPK. Embryos were visualized by light (A) and fluorescence (B) microscopy. (C and D) Embryos were cultured in DMSO (C) or SU5402 (D) and analyzed by in situ hybridization for Rsk4. Embryos cultured in SU5402 have distal expansion of Rsk4 compared to DMSO controls. Red and black arrows demarcate the embryonic/extraembryonic junction and the distal boundary of normal Rsk4 expression, respectively.
DISCUSSION
RSK4 function is distinct from those of other RSK proteins.
RTK signals are required for a variety of biological processes. The molecules comprising this pathway utilize distinct modules of proteins to render specific cellular responses. We demonstrate that the RSK proteins may be involved in generating subtle variations in different cellular environments. Studies have characterized RSK1 to -3 as downstream mediators of RTK signals (55, 76). In this study, we identified mouse RSK4. In contrast to the other RSK proteins, overexpression of RSK4 does not mediate, but rather inhibits, RTK signals. The ability of RSK4 to inhibit RTK signals is dependent upon the 96 amino acids at the amino-terminal end of the protein. This region is not conserved in the other RSK proteins. These data add to the growing evidence that though the RSK proteins are highly similar and can all be activated with stereotyped signals, subtle differences among them may result in distinct cellular roles (55, 76).
In vivo analysis of Rsk4.
The extraembryonic patterning events that are regulated by RTK signals in the mouse are among the earliest in the developing mammal. Given the genetic and functional support for RTK signals in the extraembryonic tissue, we felt that this tissue was ideal for the identification of RTK modulators, despite some of the technical challenges. To facilitate the study of this small, less accessible tissue, we capitalized on the Xenopus embryo. Using an expression cDNA library from an early developing mouse embryo, we performed a functional screen with Xenopus to identify molecules that regulate and/or specify RTK signals and found Rsk4. Despite the obvious caveats of this nonconventional method, the subsequent demonstration of specific tissue expression of Rsk4 validated our initial studies with the frog embryo. Rsk4 is expressed in the extraembryonic tissue, a tissue in which the regulation of RTK signals is critical for proper development. Moreover, Rsk4 is localized to the central region of the extraembryonic tissue, and its expression is flanked both distally and proximally by the expression of active ERK. The most distal expression of active ERK is a layer of cells that forms a discrete ring (21). Maintenance of such a specific pattern requires a tight regulation of local signals and therefore often involves intricate feedback loops. In the absence of FGF, an RTK ligand, active ERK is lost distally and Rsk4 expression expands into the ring of cells that normally contains active ERK. This indicates that Rsk4 and activated ERK signals have a dynamic interaction in vivo. This interaction may regulate the specification of the distal extraembryonic cells. Though the exact role of these distal cells remains unknown, they are located in an area of active signaling that likely plays a role in the patterning of both embryonic and extraembryonic tissues (reviewed in references 8 and 9). Further studies of Rsk4 in this area may shed more light on the function of these cells, but certainly data from this report provide the first evidence that an RSK may be involved in the modulation of RTK signals in the early developing embryo.
How and where does Rsk4 act?
Results from mutational analysis provide some information about how RSK4 might act. Site-directed mutagenesis of the lysine in the N-terminal kinase domain removed the ability of RSK4 to disrupt RTK signals. This suggests that the inhibition of RTK signals by RSK4 is a result of the phosphorylation of substrate. Candidates for the molecular target of RSK4 kinase activity are plentiful. RSK4 could act by directly phosphorylating and inhibiting a member of the RAS-ERK pathway. Alternatively, it could act indirectly by either activating an inhibitor or repressing expression of downstream transcriptional targets. These possibilities can all be supported by studies of the other RSK isoforms. In a study using rat PC12 epidermal growth factor (EGF)-cultured cells, RSK2 was shown to phosphorylate SOS, a membrane-tethered protein that mediates RTK activities (23). From this result, it was hypothesized that RSK2 could feedback negatively on the EGF pathway, though no further evidence in support of this idea has been put forth. Although hypothesized for RSK2, we do not support this mode of action for RSK4. RSK4 is able to inhibit constitutively active forms of RAS and ERK, which are both downstream of SOS. Therefore, we think it is more likely that RSK4 can inhibit RTK signaling at the level of or downstream of ERK. This model is supported in the literature, where it is shown that RSK isoforms localize to the nucleus upon RTK activation (12). Moreover, RSK2 has been shown to up-regulate Estrogen Receptor α and c-Fos transcription, suggesting a role for the RSK proteins in transcriptional regulation (15, 18, 38).
Deletion of the N terminus of RSK4, a region not conserved in the other RSK proteins, removes the ability of RSK4 to disrupt RTK signaling, indicating that these N-acids are required for RSK4 activity. Since BLOCK analysis (32) did not identify any known protein motifs, there is little indication of the exact role for this region in conferring RSK4 activity. Protein from the Δ1-96Rsk4 construct maintains its ability to phosphorylate S6, suggesting that deletion of the N terminus does not disrupt the kinase ability of RSK4. Though no defined motifs were identified, the N terminus is comprised of many charged residues, suggesting that it could be targeting RSK4 to the nucleus or allowing RSK4 to interact with DNA. Analysis of Sef and Sprouty2, two negative feedback regulators of the FGF-RAS-ERK pathway, demonstrates that these genes are not transcriptionally regulated by RSK4. However, this does not exclude the possibility that RSK4 could be interacting with SEF and SPROUTY2 proteins or that RSK4 is interacting with other nuclear targets.
Our biochemical experiments suggest that RSK4 confers its Xbra inhibition by blocking signaling directly on or downstream of ERK, since RSK4 is able to block D. melanogaster Erksem from inducing its transcription. Consistent with RSK4 acting directly on ERK, tissues injected with Fgf8 and Rsk4 do not have phosphorylated ERK protein. Functional screens such as the one described in this report are inherently prone to the caveats of expressing genes at high, often nonphysiologic levels. Therefore, it is formally possible that RSK4, with its carboxy-terminal ERK binding site, is simply sequestering and inhibiting the activation of ERK to act as a dominant-negative molecule. We think this is unlikely for several reasons. First, the RSK4 putative N-terminal kinase activity is required for Rsk4 activity. Additionally, the other RSK isoforms, in which the C-terminal ERK docking site is conserved (59), do not maintain this function. Finally, mutational analysis demonstrates that without the N terminus of the gene, Rsk4 loses its ability to disrupt the FGF pathway. Since the conserved carboxy-terminal ERK binding site is still present on the Δ1-96Rsk4 clone, its presence cannot be the sole reason for the described Rsk4 function. Thus, while the data are consistent with RSK4 directly disrupting active ERK, the evidence that RSK4 requires its kinase activity and that the amino-terminally truncated RSK4 with an intact ERK binding site is not sufficient for its function suggests a mechanism of action that is more complex than simple sequestration of ERK by RSK4. The identification of the mechanism by which Rsk4 inhibits RTK signals and the identification of other Rsk4 targets will be exciting avenues for investigation. The use of trophoblast stem cells for biochemical assays in combination with RSK4 analysis in genetically perturbed mouse models should aid in these pursuits.
Summary.
As a result of the conservation of the RAS-ERK cascade and the requirement of this pathway in multiple organisms for the specification of different tissue types, we were able to identify Rsk4 in a cross-species screen. Since its function is dependent upon a region of the gene that is divergent from the other Rsk family members, it would have been challenging to identify the full length of this gene by homology only. Among products of the Rsk genes, the ability to inhibit RTK signals is specific to RSK4, adding to the evidence that not all RSK functions are similar. Rsk4 is expressed in the extraembryonic region of the developing gastrula in a pattern that is consistent with its activity. Further investigation of Rsk4 in the contexts of general RAS-ERK signaling and the development of the extraembryonic tissues should add to these fields of investigation.
Acknowledgments
We thank M. Whitman, J. Blenis, J. Farrell, J. Maller, and C. Bjorbaek for their generosity with DNA constructs. We thank J. Hebert, P. Roux, C. Smith, J. Myers, G. Barsh, S. Kim, P. Jackson, K. Knox, E. Ray, D. Ko, E. Chiao, A. Borchers, A. Hufton, and N. Mitiku for their sage scientific advice.
This work was supported by a grant from the National Institutes of Health (ROI HD 41557). Support for A.P.M. was provided by the Medical Scientist Training Program (GM07365).
REFERENCES
- 1.Agius, E., M. Oelgeschlager, O. Wessely, C. Kemp, and E. M. De Robertis. 2000. Endodermal nodal-related signals and mesoderm induction in Xenopus. Development 127:1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcorta, D. A., C. M. Crews, L. J. Sweet, L. Bankston, S. W. Jones, and R. L. Erikson. 1989. Sequence and expression of chicken and mouse rsk: homologs of Xenopus laevis ribosomal S6 kinase. Mol. Cell. Biol. 9:3850-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaya, E., P. A. Stein, T. J. Musci, and M. W. Kirschner. 1993. FGF signalling in the early specification of mesoderm in Xenopus. Development 118:477-487. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, N. G., J. L. Maller, N. K. Tonks, and T. W. Sturgill. 1990. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature 343:651-653. [DOI] [PubMed] [Google Scholar]
- 5.Avruch, J., A. Khokhlatchev, J. M. Kyriakis, Z. Luo, G. Tzivion, D. Vavvas, and X. F. Zhang. 2001. Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog. Horm. Res. 56:127-155. [DOI] [PubMed] [Google Scholar]
- 6.Baker, J. C., R. S. Beddington, and R. M. Harland. 1999. Wnt signaling in Xenopus embryos inhibits bmp4 expression and activates neural development. Genes Dev. 13:3149-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker, J. C., and R. M. Harland. 1996. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes Dev. 10:1880-1889. [DOI] [PubMed] [Google Scholar]
- 8.Beddington, R. S., and E. J. Robertson. 1998. Anterior patterning in mouse. Trends Genet. 14:277-284. [DOI] [PubMed] [Google Scholar]
- 9.Beddington, R. S., and E. J. Robertson. 1999. Axis development and early asymmetry in mammals. Cell 96:195-209. [DOI] [PubMed] [Google Scholar]
- 10.Belo, J. A., T. Bouwmeester, L. Leyns, N. Kertesz, M. Gallo, M. Follettie, and E. M. De Robertis. 1997. Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech. Dev. 68:45-57. [DOI] [PubMed] [Google Scholar]
- 11.Bjorbaek, C., Y. Zhao, and D. E. Moller. 1995. Divergent functional roles for p90rsk kinase domains. J. Biol. Chem. 270:18848-18852. [DOI] [PubMed] [Google Scholar]
- 12.Blenis, J., J. Chung, E. Erikson, D. A. Alcorta, and R. L. Erikson. 1991. Distinct mechanisms for the activation of the RSK kinases/MAP2 kinase/pp90rsk and pp70-S6 kinase signaling systems are indicated by inhibition of protein synthesis. Cell Growth Differ. 2:279-285. [PubMed] [Google Scholar]
- 13.Blumberg, B., C. V. Wright, E. M. De Robertis, and K. W. Cho. 1991. Organizer-specific homeobox genes in Xenopus laevis embryos. Science 253:194-196. [DOI] [PubMed] [Google Scholar]
- 14.Borchers, A. G., A. L. Hufton, A. G. Eldridge, P. K. Jackson, R. M. Harland, and J. C. Baker. 2002. The E3 ubiquitin ligase GREUL1 anteriorizes ectoderm during Xenopus development. Dev. Biol. 251:395-408. [DOI] [PubMed] [Google Scholar]
- 15.Chen, R. H., P. C. Juo, T. Curran, and J. Blenis. 1996. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene 12:1493-1502. [PubMed] [Google Scholar]
- 16.Cho, K. W., B. Blumberg, H. Steinbeisser, and E. M. De Robertis. 1991. Molecular nature of Spemann's organizer: the role of the Xenopus homeobox gene goosecoid. Cell 67:1111-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciruna, B. G., and J. Rossant. 1999. Expression of the T-box gene Eomesodermin during early mouse development. Mech. Dev. 81:199-203. [DOI] [PubMed] [Google Scholar]
- 18.Clark, D. E., C. E. Poteet-Smith, J. A. Smith, and D. A. Lannigan. 2001. Rsk2 allosterically activates estrogen receptor alpha by docking to the hormone-binding domain. EMBO J. 20:3484-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Condie, B. G., and R. M. Harland. 1987. Posterior expression of a homeobox gene in early Xenopus embryos. Development 101:93-105. [PubMed] [Google Scholar]
- 20.Cornell, R. A., and D. Kimelman. 1994. Activin-mediated mesoderm induction requires FGF. Development 120:453-462. [DOI] [PubMed] [Google Scholar]
- 21.Corson, L. B., Y. Yamanaka, K. M. Lai, and J. Rossant. 2003. Spatial and temporal patterns of ERK signaling during mouse embryogensis. Development 130:4527-4537. [DOI] [PubMed] [Google Scholar]
- 22.Doniach, T., and T. J. Musci. 1995. Induction of anteroposterior neural pattern in Xenopus: evidence for a quantitative mechanism. Mech. Dev. 53:403-413. [DOI] [PubMed] [Google Scholar]
- 23.Douville, E., and J. Downward. 1997. EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene 15:373-383. [DOI] [PubMed] [Google Scholar]
- 24.Dufresne, S. D., C. Bjorbaek, K. El-Haschimi, Y. Zhao, W. G. Aschenbach, D. E. Moller, and L. J. Goodyear. 2001. Altered extracellular signal-regulated kinase signaling and glycogen metabolism in skeletal muscle from p90 ribosomal S6 kinase 2 knockout mice. Mol. Cell. Biol. 21:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldman, B., W. Poueymirou, V. E. Papaioannou, T. M. DeChiara, and M. Goldfarb. 1995. Requirement of FGF-4 for postimplantation mouse development. Science 267:246-249. [DOI] [PubMed] [Google Scholar]
- 26.Fisher, T. L., and J. Blenis. 1996. Evidence for two catalytically active kinase domains in pp90rsk. Mol. Cell. Biol. 16:1212-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giroux, S., M. Tremblay, D. Bernard, J. F. Cardin-Girard, S. Aubry, L. Larouche, S. Rousseau, J. Huot, J. Landry, L. Jeannotte, and J. Charron. 1999. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr. Biol. 9:369-372. [DOI] [PubMed] [Google Scholar]
- 28.Gross, S. D., A. L. Lewellyn, and J. L. Maller. 2001. A constitutively active form of the protein kinase p90Rsk1 is sufficient to trigger the G2/M transition in Xenopus oocytes. J. Biol. Chem. 276:46099-46103. [DOI] [PubMed] [Google Scholar]
- 29.Harland, R., and J. Gerhart. 1997. Formation and function of Spemann's organizer. Annu. Rev. Cell Dev. Biol. 13:611-667. [DOI] [PubMed] [Google Scholar]
- 30.Harland, R. M. 1991. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36:685-695. [DOI] [PubMed] [Google Scholar]
- 31.Hemmati-Brivanlou, A., and D. A. Melton. 1994. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell 77:273-281. [DOI] [PubMed] [Google Scholar]
- 32.Henikoff, S., and J. G. Henikoff. 1994. Protein family classification based on searching a database of blocks. Genomics 19:97-107. [DOI] [PubMed] [Google Scholar]
- 33.Henry, G. L., I. H. Brivanlou, D. S. Kessler, A. Hemmati-Brivanlou, and D. A. Melton. 1996. TGF-beta signals and a pattern in Xenopus laevis endodermal development. Development 122:1007-1015. [DOI] [PubMed] [Google Scholar]
- 34.Hopwood, N. D., A. Pluck, and J. B. Gurdon. 1989. MyoD expression in the forming somites is an early response to mesoderm induction in Xenopus embryos. EMBO J. 8:3409-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu, D. R., A. N. Economides, X. Wang, P. M. Eimon, and R. M. Harland. 1998. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell 1:673-683. [DOI] [PubMed] [Google Scholar]
- 36.Hudson, C., D. Clements, R. V. Friday, D. Stott, and H. R. Woodland. 1997. Xsox17α and -β mediate endoderm formation in Xenopus. Cell 91:397-405. [DOI] [PubMed] [Google Scholar]
- 37.Jacobson, M., and U. Rutishauser. 1986. Induction of neural cell adhesion molecule (NCAM) in Xenopus embryos. Dev. Biol. 116:524-531. [DOI] [PubMed] [Google Scholar]
- 38.Joel, P. B., J. Smith, T. W. Sturgill, T. L. Fisher, J. Blenis, and D. A. Lannigan. 1998. pp90rsk1 regulates estrogen receptor-mediated transcription through phosphorylation of Ser-167. Mol. Cell. Biol. 18:1978-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones, S. W., E. Erikson, J. Blenis, J. L. Maller, and R. L. Erikson. 1988. A Xenopus ribosomal protein S6 kinase has two apparent kinase domains that are each similar to distinct protein kinases. Proc. Natl. Acad. Sci. USA 85:3377-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinuya, M., K. Takishima, and G. Mamiya. 2000. Detection of kinases that phosphorylate 14-3-3 binding sites of Raf-1 using in situ gel kinase assay. Biol. Pharm. Bull. 23:1158-1162. [DOI] [PubMed] [Google Scholar]
- 41.Kispert, A., R. J. Stoger, M. Caparros, and B. G. Herrmann. 1999. The mouse Rsk3 gene maps to the Leh66 elements carrying the t-complex responder Tcr. Mamm. Genome 10:794-802. [DOI] [PubMed] [Google Scholar]
- 42.Kohn, M., H. Hameister, M. Vogel, and H. Kehrer-Sawatzki. 2003. Expression pattern of the Rsk2, Rsk4 and Pdk1 genes during murine embryogenesis. Gene Expr. Patterns 3:173-177. [DOI] [PubMed] [Google Scholar]
- 43.LaBonne, C., B. Burke, and M. Whitman. 1995. Role of MAP kinase in mesoderm induction and axial patterning during Xenopus development. Development 121:1475-1486. [DOI] [PubMed] [Google Scholar]
- 44.LaBonne, C., and M. Whitman. 1994. Mesoderm induction by activin requires FGF-mediated intracellular signals. Development 120:463-472. [DOI] [PubMed] [Google Scholar]
- 45.Lamb, T. M., A. K. Knecht, W. C. Smith, S. E. Stachel, A. N. Economides, N. Stahl, G. D. Yancopolous, and R. M. Harland. 1993. Neural induction by the secreted polypeptide noggin. Science 262:713-718. [DOI] [PubMed] [Google Scholar]
- 46.MacNicol, A. M., A. J. Muslin, and L. T. Williams. 1993. Raf-1 kinase is essential for early Xenopus development and mediates the induction of mesoderm by FGF. Cell 73:571-583. [DOI] [PubMed] [Google Scholar]
- 47.Moller, D. E., C. H. Xia, W. Tang, A. X. Zhu, and M. Jakubowski. 1994. Human rsk isoforms: cloning and characterization of tissue-specific expression. Am. J. Physiol. 266:C351-C359. [DOI] [PubMed] [Google Scholar]
- 48.Munoz-Sanjuan, I., and A. H.-Brivanlou. 2001. Early posterior/ventral fate specification in the vertebrate embryo. Dev. Biol. 237:1-17. [DOI] [PubMed] [Google Scholar]
- 49.Nutt, S. L., K. S. Dingwell, C. E. Holt, and E. Amaya. 2001. Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. 15:1152-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nystrom, F. H., and M. J. Quon. 1999. Insulin signalling: metabolic pathways and mechanisms for specificity. Cell Signal 11:563-574. [DOI] [PubMed] [Google Scholar]
- 51.Oellers, N., and E. Hafen. 1996. Biochemical characterization of rolledSem, an activated form of Drosophila mitogen-activated protein kinase. J. Biol. Chem. 271:24939-24944. [DOI] [PubMed] [Google Scholar]
- 52.Peng, H. B. 1991. Xenopus laevis: practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol. 36:657-662. [PubMed] [Google Scholar]
- 53.Porter, A. C., and R. R. Vaillancourt. 1998. Tyrosine kinase receptor-activated signal transduction pathways which lead to oncogenesis. Oncogene 17:1343-1352. [DOI] [PubMed] [Google Scholar]
- 54.Rossant, J., and J. C. Cross. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2:538-548. [DOI] [PubMed] [Google Scholar]
- 55.Roux, P. P., S. A. Richards, and J. Blenis. 2003. Phosphorylation of p90 ribosomal S6 kinase (RSK) regulates extracellular signal-regulated kinase docking and RSK activity. Mol. Cell. Biol. 23:4796-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rupp, R. A., L. Snider, and H. Weintraub. 1994. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8:1311-1323. [DOI] [PubMed] [Google Scholar]
- 57.Sasai, Y., B. Lu, S. Piccolo, and E. M. De Robertis. 1996. Endoderm induction by the organizer-secreted factors chordin and noggin in Xenopus animal caps. EMBO J. 15:4547-4555. [PMC free article] [PubMed] [Google Scholar]
- 58.Saxton, T. M., A. M. Cheng, S. H. Ong, Y. Lu, R. Sakai, J. C. Cross, and T. Pawson. 2001. Gene dosage-dependent functions for phosphotyrosine-Grb2 signaling during mammalian tissue morphogenesis. Curr. Biol. 11:662-670. [DOI] [PubMed] [Google Scholar]
- 59.Smith, J. A., C. E. Poteet-Smith, K. Malarkey, and T. W. Sturgill. 1999. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J. Biol. Chem. 274:2893-2898. [DOI] [PubMed] [Google Scholar]
- 60.Smith, J. C., B. M. Price, J. B. Green, D. Weigel, and B. G. Herrmann. 1991. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell 67:79-87. [DOI] [PubMed] [Google Scholar]
- 61.Smith, W. C., and R. M. Harland. 1992. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell 70:829-840. [DOI] [PubMed] [Google Scholar]
- 62.Smith, W. C., and R. M. Harland. 1991. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell 67:753-765. [DOI] [PubMed] [Google Scholar]
- 63.Sohaskey, M. L., and J. E. Ferrell, Jr. 2002. Activation of p42 mitogen-activated protein kinase (MAPK), but not c-Jun NH(2)-terminal kinase, induces phosphorylation and stabilization of MAPK phosphatase XCL100 in Xenopus oocytes. Mol. Biol. Cell 13:454-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sturgill, T. W., L. B. Ray, E. Erikson, and J. L. Maller. 1988. Insulin-stimulated MAP-2 kinase phosphorylates and activates ribosomal protein S6 kinase II. Nature 334:715-718. [DOI] [PubMed] [Google Scholar]
- 65.Tan, P. B., and S. K. Kim. 1999. Signaling specificity: the RTK/RAS/MAP kinase pathway in metazoans. Trends Genet. 15:145-149. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka, S., T. Kunath, A. K. Hadjantonakis, A. Nagy, and J. Rossant. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072-2075. [DOI] [PubMed] [Google Scholar]
- 67.Tang, T. L., R. M. Freeman, Jr., A. M. O'Reilly, B. G. Neel, and S. Y. Sokol. 1995. The SH2-containing protein-tyrosine phosphatase SH-PTP2 is required upstream of MAP kinase for early Xenopus development. Cell 80:473-483. [DOI] [PubMed] [Google Scholar]
- 68.Thomsen, G., T. Woolf, M. Whitman, S. Sokol, J. Vaughan, W. Vale, and D. A. Melton. 1990. Activins are expressed early in Xenopus embryogenesis and can induce axial mesoderm and anterior structures. Cell 63:485-493. [DOI] [PubMed] [Google Scholar]
- 69.Tsang, M., R. Friesel, T. Kudoh, and I. B. Dawid. 2002. Identification of Sef, a novel modulator of FGF signalling. Nat. Cell Biol. 4:165-169. [DOI] [PubMed] [Google Scholar]
- 70.Turner, D. L., and H. Weintraub. 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8:1434-1447. [DOI] [PubMed] [Google Scholar]
- 71.Whitman, M., and D. A. Melton. 1992. Involvement of p21ras in Xenopus mesoderm induction. Nature 357:252-254. [DOI] [PubMed] [Google Scholar]
- 72.Wilkinson, D. 1992. In situ hybridisation: a practical approach. IRL Press, Oxford University, Oxford, United Kingdom.
- 73.Wilson, P. A., and D. A. Melton. 1994. Mesodermal patterning by an inducer gradient depends on secondary cell-cell communication. Curr. Biol. 4:676-686. [DOI] [PubMed] [Google Scholar]
- 74.Xu, X., M. Weinstein, C. Li, M. Naski, R. I. Cohen, D. M. Ornitz, P. Leder, and C. Deng. 1998. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development 125:753-765. [DOI] [PubMed] [Google Scholar]
- 75.Yntema, H. G., B. van den Helm, J. Kissing, G. van Duijnhoven, F. Poppelaars, J. Chelly, C. Moraine, J. P. Fryns, B. C. Hamel, H. Heilbronner, H. J. Pander, H. G. Brunner, H. H. Ropers, F. P. Cremers, and H. van Bokhoven. 1999. A novel ribosomal S6-kinase (RSK4; RPS6KA6) is commonly deleted in patients with complex X-linked mental retardation. Genomics 62:332-343. [DOI] [PubMed] [Google Scholar]
- 76.Zhao, Y., C. Bjorbaek, and D. E. Moller. 1996. Regulation and interaction of pp90(rsk) isoforms with mitogen-activated protein kinases. J. Biol. Chem. 271:29773-29779. [DOI] [PubMed] [Google Scholar]
- 77.Zhao, Y., C. Bjorbaek, S. Weremowicz, C. C. Morton, and D. E. Moller. 1995. RSK3 encodes a novel pp90rsk isoform with a unique N-terminal sequence: growth factor-stimulated kinase function and nuclear translocation. Mol. Cell. Biol. 15:4353-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zimmerman, L. B., J. M. De Jesus-Escobar, and R. M. Harland. 1996. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 86:599-606. [DOI] [PubMed] [Google Scholar]