Abstract
Although the molecular mechanisms by which host cells defend themselves against viral infection have been studied in great depth, and countermeasures viruses employ to suppress such defensive responses have been widely documented, relatively little attention has been devoted toward elucidating how such interactions between virus and host are resolved over multiple rounds of infection. Here we describe the design, synthesis, and validation of a dual-color fluorescent reporter system to study how viral infections spread through a host cell monolayer and how the cellular innate immune system mounts an antiviral response. We employed recombinant, red fluorescent protein (RFP) expressing mutants of a prototypical RNA virus, vesicular stomatitis virus (VSV) to enable identification and tracking of infected cells. Further, we generated stable reporter cells that use green fluorescent protein (GFP) to report on the expression of IFIT2, an interferon stimulated gene (ISG) involved in the interference of viral protein translation, and a marker of antiviral defense activation. The presence of the fluorescent protein reporters had minimal effects on the normal behavior of the cells or viruses. Moreover, expression of the virus and cell reporters correlated with the kinetics of viral replication and activation of an anti-viral response, respectively. This two-color system enabled us to track and quantify in live cells how viral replication and activation of host defensive responses play out over multiple rounds of infection. Initial study of propagating infections demonstrated that antiviral activation over multiple rounds was critical for slowing and ultimately halting the spread of infection.
INTRODUCTION
Shortly after a virus enters a cell and initiates replication, pathogen recognition receptors (PRRs) of the host cell detect the presence of viral nucleic acids and proteins and trigger signaling cascades that activate antiviral defenses. A competition ensues, pitting the ability of the virus to replicate and suppress the host-cell response against the ability of its host cell to launch defenses to inhibit the viral replication, and induce signaling to warn neighboring cells. Decades of research have elucidated many diverse molecular mechanisms of the virus-host arms race within an infected cell; however, less attention has been paid to how the release of virus and signals from an infected cell affects the subsequent spread of infection.
PRRs such as the toll-like receptors and RNA helicases are responsible for the initial detection of protein or nucleic acid species produced by the virus. For infection by RNA viruses, it is thought to be primarily RNA helicases such as RIG-I or MDA5 that detect aberrant RNA intermediates such as double-stranded RNA (dsRNA) or uncapped, unencapsidated forms of single-stranded RNA (ssRNA) (reviewed in Gerlier and Lyles, 2011; Kawai and Akira, 2006; Randall and Goodbourn, 2008). These sensors then set off signaling cascades that lead to the production of type-I interferons (IFNs) and other secreted cytokines. These signaling molecules can then feedback through the type I interferon receptor (IFNAR) on the infected cell or stimulate further gene expression in neighboring cells. Such interferon stimulated genes (ISGs) can have additional signaling roles or direct the degradation of viral RNA, prevent translation of viral proteins, or carry out other antiviral activities.
To effectively replicate in the face of potentially diverse antiviral responses, viruses have evolved a wide range of strategies to either suppress or evade these defenses (Andrejeva et al., 2004; García-Sastre and Biron, 2006; Unterstab et al., 2005). In the species used in this work, vesicular stomatitis virus (VSV), its matrix(M) protein translocates to nuclear pores and prevents the export of the host mRNAs through a Rae1 dependent interaction with the nucleoporin Nup98. This interaction largely suppresses the activation of host defensive response genes (Petersen et al., 2000; Rajani et al., 2012; Stojdl et al., 2003). Here, we employ a VSV mutant that carries a methionine-to-arginine point mutation in its M protein (M51R) that largely abolishes this function and is a useful tool for probing the ability of the host to respond in the absence of viral suppression (Ahmed et al., 2003; Rajani et al., 2012).
The transcriptional activity of type I IFNs and ISGs have been probed by promoter-reporter constructs, originally with luciferase or CAT reporters (Bluyssen et al., 1994; Wang et al., 2000), but more recently with fluorescent protein readouts (Martínez-Sobrido et al., 2006; Nguyen et al., 2009). Such fluorescent reporters have allowed for the discrimination of individual cell activity and demonstrated a spatially heterogeneous response of individual cells to viral stimuli. An IFN-β-GFP reporter cell line was used to show that in response to parainfluenza infection only a small minority of cells are able to activate their IFN-β gene, but that this minority is sufficient to produce enough secreted cytokine to activate ISGs in the majority of neighboring cells (Chen et al., 2010).
The engineering of recombinant virus strains to express fluorescent proteins provides a facile method to identify infected cells and track the spread of infection in cells and tissues (Duprex et al., 1999; Duprex et al., 2000; Jöns and Mettenleiter, 1997; Lam et al., 2006; Payne et al., 2006). For example, we employed a recombinant strain of human cytomegalovirus (HCMV), engineered to express a GFP-fusion with immediate early (IE2) protein of HCMV, to track the spread of focal infections in monolayers of normal human dermal fibroblasts (Lam et al., 2006). In a 45-day culture the expression of viral proteins in the cell monolayer corresponded with their temporal order; specifically, the viral immediate early protein (IE2) was detected at larger radii of the expanding infection than the late expressed viral glycoprotein B, which was detected by immunolabeling. Further, the directional spread of virus, detectable by the GFP-IE2 fusion, correlated with the alignment of host-cell clusters, made visible by cytoskeletal staining. This study highlighted how the spatial patterns of viral gene expression across a spreading infection can reflect local virus-cell interactions. In later work recombinant strains of adenovirus engineered to express GFP during infection have been used to study how initial cell infections influence subsequent spatial patterns of spread within cell monolayers (Hofacre et al., 2012; Wodarz et al., 2012; Yakimovich et al., 2012).
Antiviral cytokine signaling from infected to uninfected cells can establish an antiviral state in uninfected cells and thereby prepare them to resist infection. Therefore, for many viruses, including VSV, the ability to effectively suppress the host innate immune response is critical not only for initial replication, but also for permitting spread to neighboring susceptible cells. In this work, we investigated how different strains of VSV either suppress or activate innate immunity, and thereby impact the ability of the VSV infection to spread, using a 2D cell culture monolayer model. To provide a read-out on the activities of both the virus and the host, we developed a dual-color fluorescent reporter system that uses red fluorescent protein (RFP) to indicate viral activity by cloning an RFP gene into the VSV genome, and green fluorescent protein (GFP) to report on host innate immune activity through a promoter-reporter construct for the well-characterized antiviral gene IFIT2.
Previous work showed that host innate immunity is involved in inhibiting the spread of VSV infection in cell monolayers (Duca et al., 2001; Haseltine et al., 2008; Lam et al., 2005). Here we engineer a direct spatial read-out on not only the spread of infection but also the local activity of the host innate immune response. We adapted and combined two fluorescent reporters, recombinant virus strains and stably integrated transcriptional reporter cells, to create this system. We generated these novel biological tools, compared them to their non-fluorescent progenitors, and validated that the fluorescent signal was correlated with the associated biological function. We then used these tools in a 2D model of infection spread to show that the native capacity of VSV to suppress its host innate immunity is critical for its ability to spread.
MATERIALS AND METHODS
Cells lines and media
All cell lines were cultured at 37°C and 5%CO2. Baby hamster kidney (BHK) cells used in rescuing new VSV strains and performing plaque assays were a generous gift of Dr. Isabel Novella. They were cultured in MEM media supplemented with 10%FBS and 5% GlutaMax. Human prostate cancer (PC3) cells were received from ATCC and cultured in RPMI 1640 media supplemented with 10% FBS. 293TN cells used in packaging lentiviral particles were received from System Biosciences and were cultured in DMEM 10% FBS. Opti-MEM serum-free media was used to dilute all DNA and liposome reagents for transfection.
Reverse genetics and rescue of infectious virus
Plasmids pBS-N, pBS-P, pBS-L, and pVSVFL(+) (Lawson et al., 1995), for the production of VSV N, P, and L proteins and antigenomic VSV RNA under a T7 promoter were generously provided by Dr. Valery Grdzelishvili. The well-known methionine to arginine substitution at the 51st position in the M protein was introduced via oligonucleotide-directed mutagenesis and inserted into pVSVFL(+) digested with XbaI and NheI (Fermentas) to create pVSV-M51R(+). The DsRed-Express-DR (DsRed-Ex), DsRed2, or ZsGreen1-DR (ZsGreen) (Clontech cat #: 632423, 632404, 632428, respectively) gene was inserted into the fifth genomic position of both pVSVFL(+) and pVSV-M51R(+) with In-Fusion (Clontech) after digestion with NheI. To eliminate subcloning, the primers used to amplify the fluorescent protein genes contained an overlap with the target vector, an additional VSV transcription unit (Das et al., 2006), and a sequence complementary to the gene of interest (Supplemental table 1).
For all VSV plasmids, infectious virus was rescued via published techniques(Lawson et al., 1995; Whelan et al., 1995) Briefly, BHK cells at 36°C were infected with T7 expressing vaccinia virus (VVT7) (Fuerst et al., 1986) and co-transfected with pBS-N, pBS-P, pBS-L, and the VSV strain-specific plasmid via Lipofectamine (Invitrogen). The rescued VSV was separated from VVT7 via filtration with a 0.22µm Millex GV filter unit (Millipore, Billerica, MA), plaque purified, and amplified on BHK cells into a master stock. The presence of the desired fluorescent protein or M51R mutation was confirmed in the plasmids and master viral stocks by Sanger sequencing.
Generation of IFIT2 reporter cells
The promoter region of human IFIT2 (also known as ISG54), 412 nt upstream of the transcription start site was synthesized by IDT DNA with flanking XhoI and BamHI sites and delivered in the pIDTSMART-Kan plasmid. The promoter region was excised by double digestion with XhoI and BamHI (Fermentas) and ligated into pZsGreen (Clontech 632428), and transformed into DH5α cells (Invitrogen). The resulting IFIT2 promoter-ZsGreen construct was subsequently sub-cloned into the lentiviral vector Duet011 (Addgene) by PCR amplifying the region of interest (Phusion polymerase- NEB) with overlapping tails homologous for the PacI and XhoI restriction sites in Duet011. The region between the two cut sites in Duet011 was removed by restriction digest (Fermentas) and the IFIT2-ZsGreen amplified region was integrated with the Clontech In Fusion ligase. The resulting Duet011-IFIT2-ZsGreen construct was transformed into Clontech Stellar Competent cells and plasmid purified with Qiagen Plasmid Midi kit. Lentiviral particles were packaged on 293TN cells (System Biosciences) by co-transfection of Duet011-IFIT2-ZsGreen with psPAX2, and pMD2.G packaging plasmids provided by Addgene (molar ratio approx.: 1:2:2) using Qiagen Superfect reagent (1 µgDNA : 3 µl Superfect). After overnight incubation, media containing liposomes was removed and replaced. Forty-eight hours later, supernatants containing lentiviral particles were harvested and minimally purified by low speed centrifugation to remove cell debris. Lentiviral containing supernatants were mixed 50:50 with fresh growth media supplemented with 0.8 µg/ml polybrene to transduce target cells. Twenty-four hours after transduction media was removed the target cells were passaged into hygromycin containing media. Initial testing of reporter responsiveness was carried out by stimulating the transduced cells by transfecting poly(I:C) and adding IFN-alpha. After growth in selective media for about two weeks, monoclonal colonies were created by limiting dilution. Monoclonal lines were tested for responsiveness to infection with VSV-M51R and the most responsive line was used in subsequent experiments.
Virus replication assays
Cells were infected at the MOI stated, calculated by titering each strain by plaque assay on BHK cells and counting the target cell type by hemocytometer. A minimal volume of virus inoculum was added to the cells and allowed to absorb for 1hr at 37°C. The inoculum was then removed, cells were washed 1X with DPBS and the appropriate media type supplemented with 2% FBS was added to the cells. At various timepoints post infection the supernatant was removed from the well and stored at −80°C. The supernatants were titered on BHK cells regardless of the cell line used to produce the virus. Titers were determined by plaque assay (Zhu et al., 2009), virus was serially diluted and then adsorbed to cells in 6-well plates for 1 hr at 37°C. After removing inoculum and washing with DPBS, cells were overlaid with semi-solid 2% FBS media containing 0.6% agar noble. Plates were incubated for 18–20 hrs, agar was removed, cells were fixed with a solution of 4% PFA and 5% sucrose in PBS for 20 min, washed with PBS and stained with crystal violet. Plaques were counted with two replicate wells for each timepoint, and PFU/cell was calculated.
Flow cytometry
PC3 IFIT2-ZsGreen cells were infected at the MOI indicated with either the VSV-rWT-DsRed-Ex or VSV-M51R-DsRed-Ex strains. 20 hours post infection cells were harvested by trypsination and centrifugation, fixed with PFA, and washed with PBS. They were then blocked with blocking buffer (3% BSA, 0.05% triton X-100 in PBS) for 20 min and incubated with the primary α-VSV-G antibody, (mouse mAb KeraFast EB0010) at 1:1000 dilution, for 2 hrs at RT. Primary antibody was removed, cells were washed, reblocked, and then incubated in the secondary Ab (goat α-mouse AlexaFluor647- Jackson ImmunoResearch) at 1:500 for 1 hr. Cells were washed and resuspended in RPMI 2% FBS for FACS analysis. Cells were analyzed on a BD LSR II equipped with a 488nm laser to detect ZsGreen, a 561nm laser to detect DsRed, and a 633nm laser to detect AF647. Cells were first gated on FSC/SSC to discriminate between whole cells and debris. A minimum of 10,000 single cells per sample were analyzed. FlowJo was used to determine gates and percentages.
Live cell microscopy
All timelapse microscopy experiments were performed on an Nikon TE Eclipse 300 fitted with an outer warming chamber set to 37°C (InVivo Scientific) and a stage-top incubator chamber (Pathology Devices) set to 37°C, 5%CO2, and 75% relative humidity. Fluorescence illumination was provided by a Chroma PhotoFluor and controlled with a Lambda 10-2. Stage automation was carried out with a Prior ProScanII and all automated imaging was controlled with MetaMorph v.7.7.8
RT-qPCR
PC3 IFIT2-ZsGreen reporter cells were infected with VSV-M51R-DsRed-Ex at MOI 50, staggered at regular intervals over 24 hrs. Cells were harvested, total cellular RNA was isolated using a Qiagen RNeasy vacuum isolation kit. Total RNA was reverse transcribed to cDNA using Promega GoScript RT system with random hexamer primers. Quantitative PCR was performed using BioRad SsoFast SYBR Green Supermix on a BioRad CFX96 with primers specific for the coding region of ZsGreen, IFIT2, or β-actin. Ct values were normalized to β-actin and expressed as fold over mock-infected cells.
Western Blotting
Total cell lysates were harvested by lysing cells in RIPA buffer (Sigma R0278) with 1µl/ml protease inhibitor (Sigma P8340). Protein concentration was determined by BCA assay (Thermo #23227) and samples were diluted to equal concentration with PBS. Samples were run on 10% SDS-PAGE gel (Amresco NextGel 10% - M256) in a BioRad Critterion cassette, and transferred to PVDF membrane (Bio-Rad 162-0238). Membranes were blocked in TBS-T 5% BSA for 1 hr and blotted in primary antibody O/N at 4°C. (B-actin Sigma-A5441, 1:2000 in TBS-T 5%BSA; Mx1 Abgent-AM2061, 1:2000 in TBS-T 5%BSA). Detection was with LiCor NIR secondary Abs, diluted 1:5000 in TBS-T 5%BSA, incubated 1 hr at RT and scanned with a LiCor Odyssey and analyzed with ImageStudio2.0.
Spreading infection model
PC3 IFIT2-ZsGreen cell monolayers were infected with either VSV-WT-DsRed-Ex or M51R-DsRed-Ex at MOI ~0.0001 (plaque forming concentration). Virus inoculum was adsorbed for 1 hr at 37°C 5% CO2, removed, and cells were washed 1X with DPBS. Cells were overlaid with RPMI 2% FBS supplemented with 0.6% agar to prevent fluid flows and Hoestch 33342 (Anaspec #83218) diluted 1:20,000 as a live-cell nuclear stain. The monolayers were then imaged using live-cell microscopy described above. Approximately 150mm2 per condition were imaged every 4 hours for 48 hrs post infection.
RESULTS
Synthesis of a DsRed reporter of viral replication
To track infections in live cells we designed a new DsRed-expressing variant of VSV. Previous reports had demonstrated that fluorescent proteins could be incorporated into the VSV genome without major disruption of normal viral function and had established methods for this reverse genetic engineering (Dalton and Rose, 2001; Whelan et al., 1995; Wollmann et al., 2010). DsRed-Ex was inserted into the genome of both rWT VSV as well as the M51R mutant, which has been demonstrated to be deficient in its ability to inhibit host innate immunity (Petersen et al., 2000; Stojdl et al., 2003) (Figure 1A). We first compared the kinetics of virus production of the rWT and M51R-DsRed-Ex strains to their non-fluorescent progenitors. BHK (MOI 10) and PC3 (MOI 50) cells were infected with each of the four strains and supernatant samples were collected at various timepoints post-infection. On BHK cells the growth kinetics and final titer of all 4 strains were similar (data not shown), indicating that introduction of fluorescent protein had minimal impact on the overall ability of the virus to replicate. Similarly, on PC3 cells both the rWT and M51R-DsRed-Ex strains grew as least as well as their non-fluorescent parent strains (Figure 2).
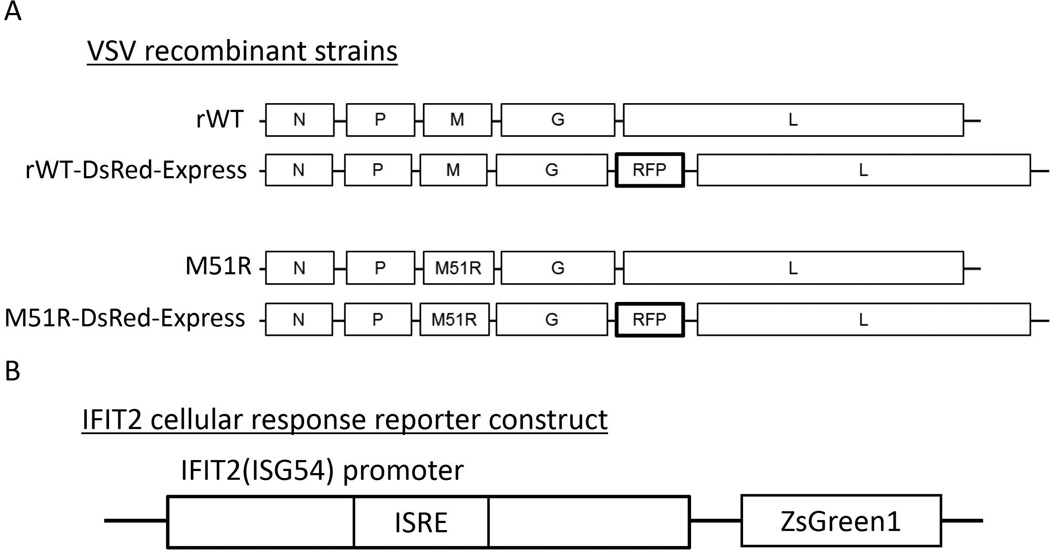
Figure 1. Schematic of recombinant constructs used in this work.
(A) We created recombinant forms of both VSV-WT and the M51R mutant containing a point mutation in the matrix protein that abolishes the ability to inhibit host innate immune response, using a reverse genetics system. We further altered these strains by inserting DsRed-Express or other fluorescent protein in the 5th position of the genome. (B) We also created an IFIT2 reporter construct by inserting the IFIT2 promoter containing the ISRE site into a ZsGreen1 expression plasmid. We sub-cloned this construct into a lentiviral vector and transduced onto PC3 cells to stably integrate this reporter transgene.
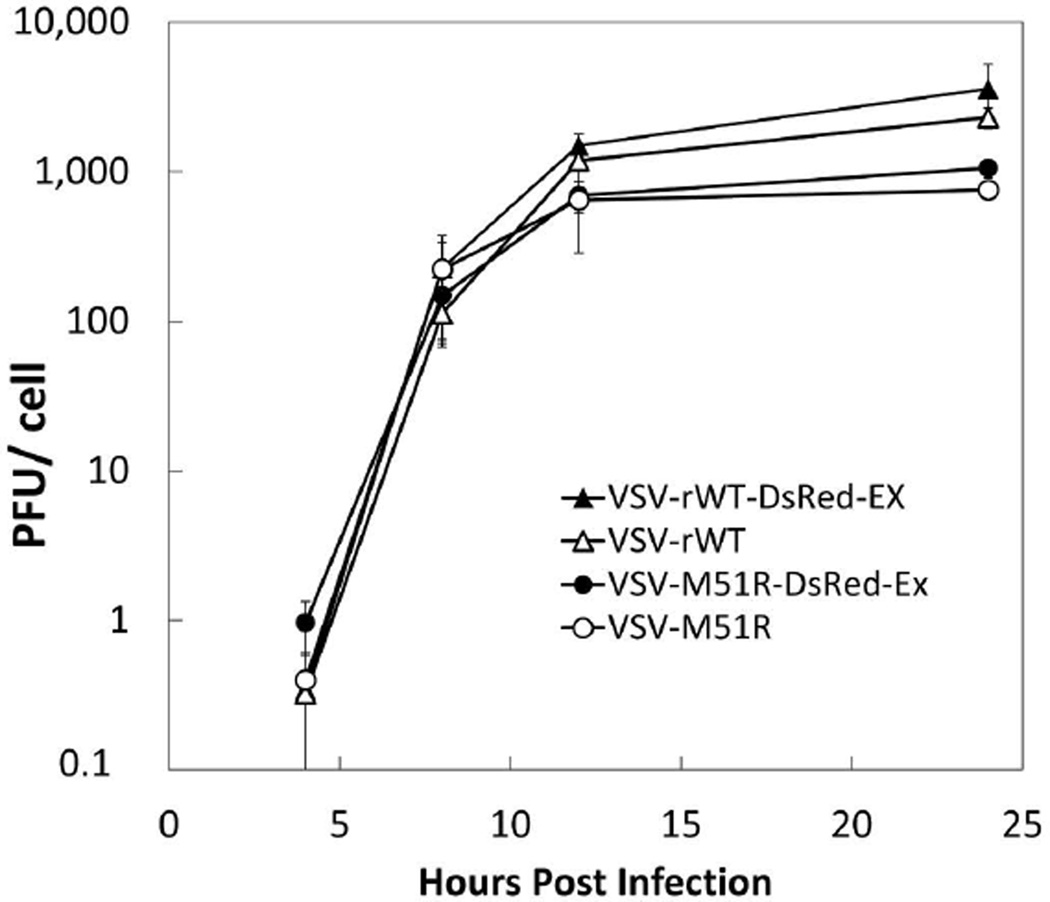
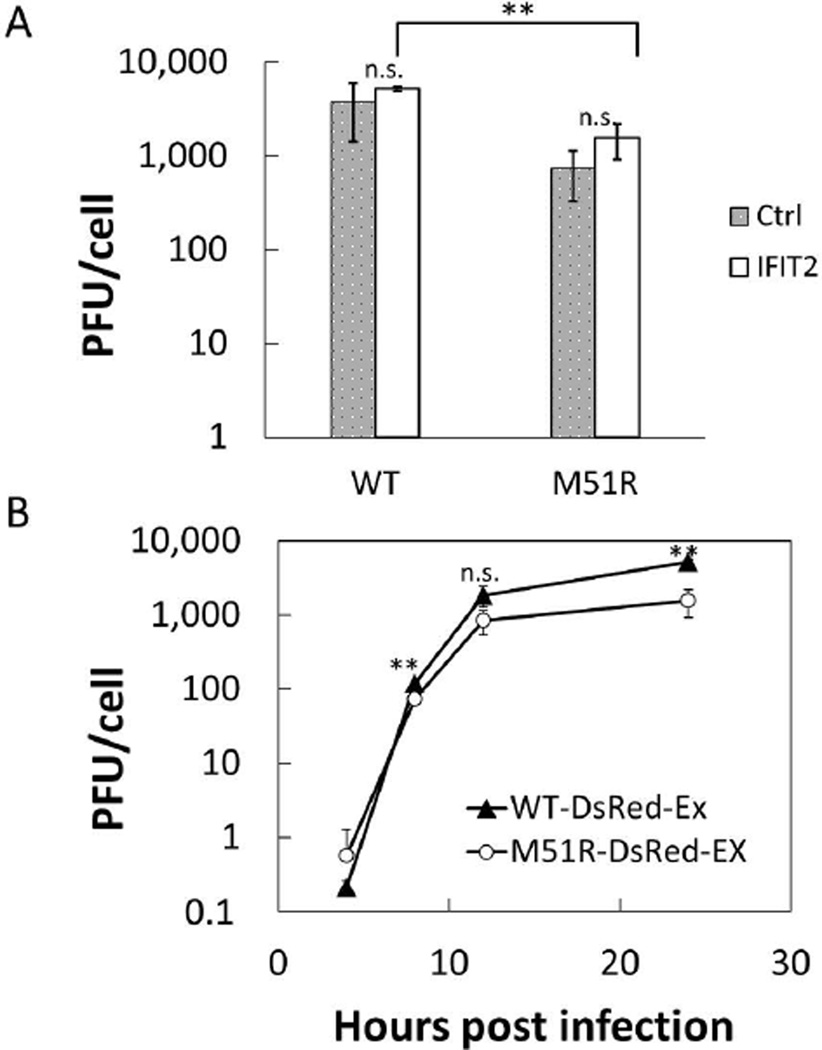
Figure 2. Insertion of fluorescent protein into VSV genome does not attenuate replication.
PC-3 IFIT2-ZsGreen1 reporter cells were infected at an MOI of 50 with either the VSV-WT-DsRed-Express (filled triangles), VSV-M51R-DsRed-Express (filled circles), or the non-fluorescent parent of each strain (open triangles, open circles, respectively). Supernatants were harvested at 4, 8, 12, 24 HPI and titered for infectious virus by plaque assay. Each data point represents the mean of 3 biological replicates with the error bars representing 1 standard deviation. We found that the fluorescent strains replicated on the target reporter cells similarly to their non-fluorescent parent strains.
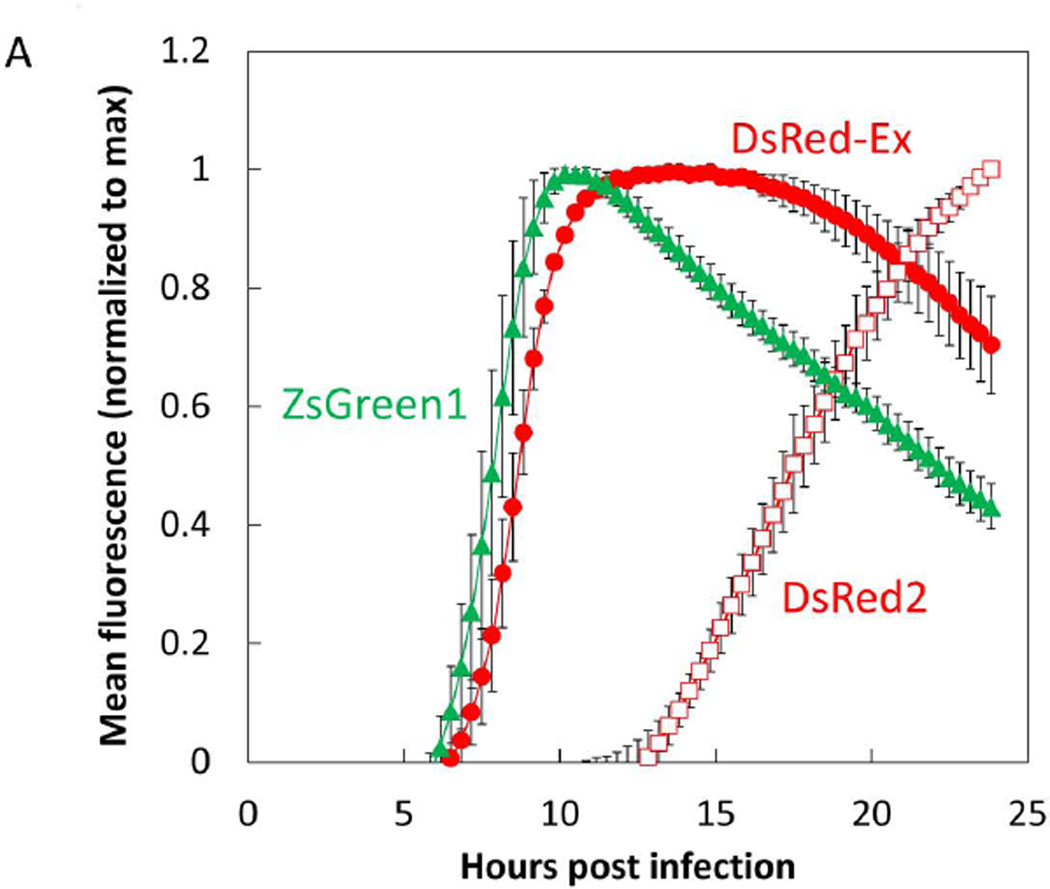
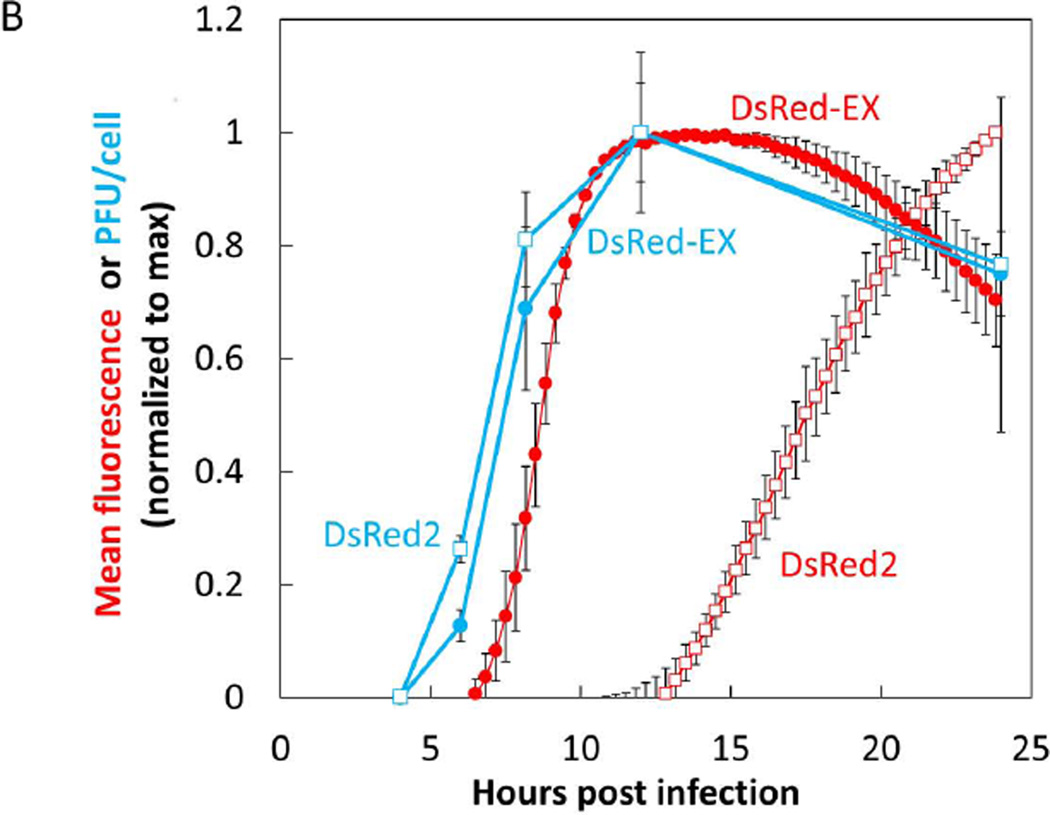
In addition to the DsRed-Ex, we also cloned the red fluorescent protein DsRed2 and the green fluorescent protein ZsGreen into the 5th position of the VSV genome and were successful in rescuing the M51R mutant form of all these fluorescent strains. We then infected BHK cells at an MOI of 10 with the VSV-M51R variants of DsRed-Ex, DsRed2, and ZsGreen. Using timelapse live-cell microscopy we determined the earliest time post infection at which a fluorescent signal could be detected above background and the timepoint at which we reached maximum signal (Figure 3). These infections were imaged in parallel with supernatant sampling which were titered for infectious viral counts by plaque assay. Both the DsRed-Ex and ZsGreen strains produced detectable fluorescence at approximately 6 hours post infection and reached a maximal level between 10–12 hrs post infection, while the DsRed2 fluorescence was delayed six hours relative to the other two strains (Figure 3A). Comparing the fluorescence kinetics to the rates of virus production, we detected initial virus release at about 4–5 hours post infection, while virus production leveled out between 10–12 HPI for all three strains (DsRed-Ex and DsRed2 shown in Figure 3B). The temporal correlation of the fluorescence and virus production for the DsRed-Ex strain indicated that it would be a suitable candidate to use as our read-out of viral replication in further studies, and highlighted the need to select and validate a fluorescent protein read-out. Additionally, the similarity of fluorescence kinetics for DsRed-Ex and ZsGreen when expressed from the identical viral genomes indicated that these two fluorescent proteins have very similar translation and maturation kinetics. Therefore differences in detectable expression between these two proteins should reflect other biological factors, indicating their suitability for red/green pairing to study their relative kinetics of expression.
Figure 3. Fluorescence kinetics of DsRed-Ex are similar to ZsGreen kinetics and correlated with virus production.
(A) BHK cells were infected at an MOI of 10 with the VSV-DsRed-Express, VSV-DsRed2, or VSV-ZsGreen1- strain. Infected monolayers were imaged by live-cell microscopy every 20 min with 4 fields per well at 10X magnification. Total field intensity was quantified and normalized to the number of cells in the field imaged (cell number determined by counting nuclei). Background intensity calculated from mock infected wells was subtracted, the total field intensity was normalized to the maximum value of each well, and the normalized value of 3 replicate wells was averaged. Data points represent the mean of 3 replicate wells with error bars of 1 standard deviation. DsRed-Ex fluorescence (red filled circle) was found to have similar kinetic parameters (earliest time detectible, time to max level) as ZsGreen1 fluorescence (green filled triangle). Conversely DsRed2 fluorescence (red open square) was delayed 5–6 hrs relative to the other two strains.
(B) BHK cells were infected at an MOI of 10 with either the VSV-DsRed-Express or VSV-DsRed2 strain. Infected monolayers were imaged by live-cell microscopy every 20 min and mean field intensity was determined and expressed as described in 3A. Supernatant samples from parallel wells were harvested at regular intervals and titered by plaque assay with 3 biological replicates at each timepoint. Virus titers at each timepoint were averaged and both mean values and 1 standard deviation were expressed as normalized to the maximum timepoint (12 HPI for both strains). DsRed-Ex fluorescence (red filled circle) was found to have similar kinetic parameters (earliest time detectible, time to max level) as DsRed-Ex virus production (blue filled circle). Conversely DsRed2 fluorescence (red open square) was delayed ~9 hrs relative to virus production (blue open square).
Virus production is similar on both progenitor and IFIT2 reporter cells
To create a promoter-reporter construct for the host innate response (Figure 1B) we selected IFIT2, a gene with a well-established role as an interferon stimulated gene involved in antiviral defense, that has also previously been used in transcriptional reporters (Nguyen et al., 2009). While IFIT2 does respond to IFN stimulation, it also has been shown to be activated by Sendai virus without requiring IFN signaling through direct IRF-3 activation (Elco et al., 2005). Most importantly for this study, IFIT2 has been demonstrated to have a key role in protection against VSV infection spread in mouse models (Fensterl et al., 2012). Using current NCBI consensus human IFIT2 sequences we synthesized the ~400 nt region upstream of the transcription start site (IDT DNA), including the conserved ISRE transcription factor binding site and cloned into a promoterless pZsGreen (Clontech) vector. Due to low levels of transfection by liposome-based methods we opted to use a lentiviral approach to achieve stable gene integration. The lentiviral constructs were transduced onto target human prostate cancer (PC3) cells in the presence of polybrene. PC3 cells were selected as they have been demonstrated to be more resistant to VSV infection than most immortalized cell lines, reflecting their intact and robust innate immune response (Ahmed et al., 2004; Carey et al., 2008). The transduced cells were grown in the presence of selective media for 2 weeks and then monoclonal populations were created by limiting dilution. Twelve healthy monoclonal lines were then tested for responsiveness by infection with VSV-M51R and analyzed by flow cytometry. The highest responding line (>90%) was selected for further study.
Virus production for the PC3-IFIT2 reporter cells was compared to their non-reporter progenitor cells 24 hours post infection (Figure 4A). We found that there was no significant difference in virus replication between the two cell types for either the rWT or M51R DsRed-Ex strains. We did observe a significant difference between the rWT and M51R strains in 24 h endpoint titers when grown on the IFIT2-reporter cells. For earlier time points production by the rWT and M51R mutant were more similar (Figure 4B), consistent with a previous report where infection of PC3 cells with these two strains produced similar levels of virus (Ahmed et al., 2004).
Figure 4. VSV production on PC3 IFIT2-ZsGreen reporter cells.
(A) Replication of both the rWT-DsRed-Ex and M51R-DsRed-Ex at MOI 50 was compared between the PC3 IFIT2-ZsGreen reporter cells and their PC3 progenitors. Virus production at 24 HPI was determined by plaque assay for 3 biological replicates. No significant difference was found for either WT or M51R between the two cells types (n.s. p> 0.1). However, we did observe a small but significant difference in viral production between the WT and M51R strains on the IFIT2 reporter cells (** p< 0.001, Student’s t-test)
(B) Replication kinetics of the VSV-WT-DsRed-Ex (filled triangles) and VSV-M51R-DsRed-Ex (filled circles) at MOI 50 on the PC-3 IFIT2-ZsGreen reporter cells. WT replication was higher than M51R at 8 and 24 HPI, but not at 4 and 12 HPI (** p<0.002, Student’s t-test).
Fluorescent signals are sensitive readouts of biological activity
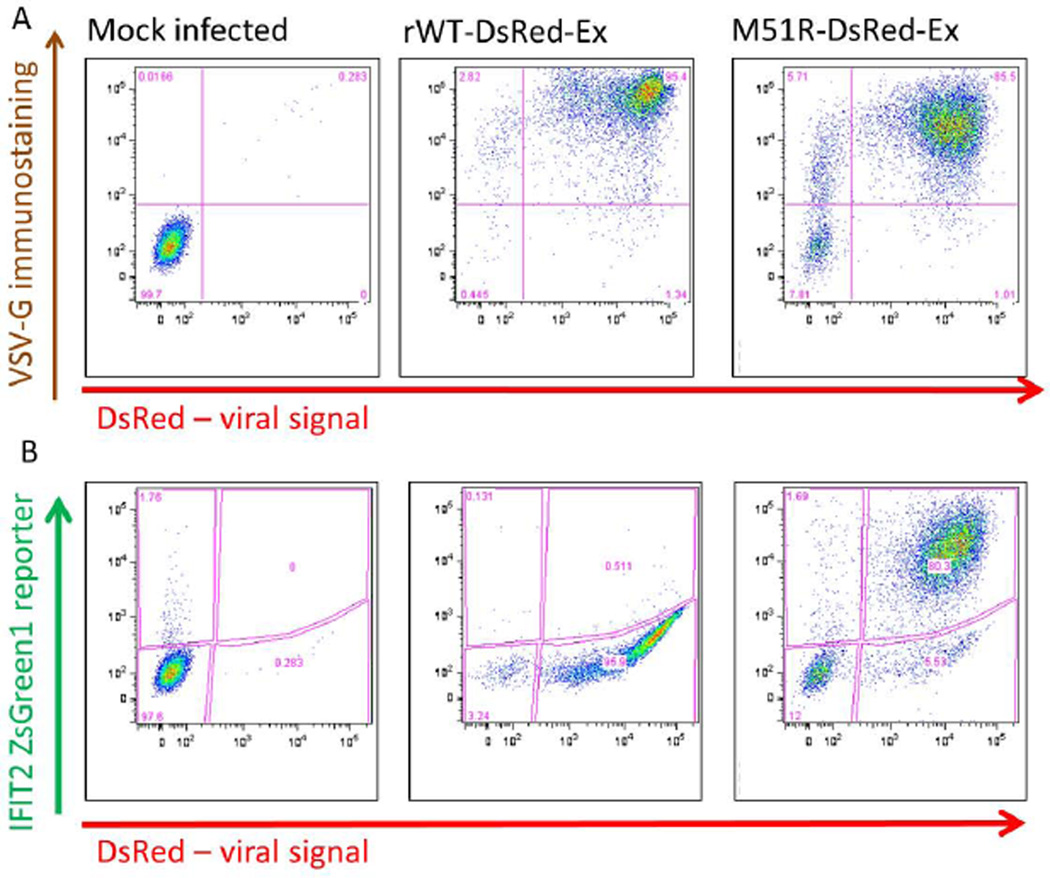
The selected PC3 IFIT2-ZsGreen reporter line and the VSV rWT and M51R-DsRed-Ex strains were first characterized by flow cytometry (Figure 5). Reporter cells were infected with MOI 100 of either strain and harvested 20 hours post infection. Cells were fixed with PFA and then immunostained with an antibody recognizing the external epitope of VSV-G protein. For both strains the majority of cells (>85%) were positive for both VSV-G and DsRed expression (Figure 5A). The gating percentages along with the shape of the populations suggest that infected cells produce external VSV-G to levels detectable by immunostaining earlier than they express detectable internal DsRed. While this indicates that the DsRed may be a slight lagging indicator of viral protein expression, the two signals are still well correlated, supporting use of the DsRed signal as a read-out for virus production.
Figure 5. Flow cytometry results confirm both viral and IFIT2 reporter function.
PC-3 IFIT2-ZsGreen1 reporter cells were infected with VSV-WT-DsRed-Ex or VSV-M51R-DsRed-Ex at MOI 50. 20 HPI cells were harvested and immunostained for VSV-G protein on the cell surface, and analyzed by flow cytometry. In both strains DsRed expression was found to be correlated with VSV-G protein although in the M51R infection, a small population of cells (~5%) were G positive but not DsRed positive, suggesting it is a somewhat less sensitive indicator of infection. The results also demonstrate that while the WT infection suppresses the activation of IFIT2, the M51R strongly induces its activity.
The unique characteristic of the VSV-M51R mutant’s inability to inhibit host innate immune responses were used to investigate the function of the IFIT2-ZsGreen reporter (Figure 5B). The vast majority of cells infected at MOI 100 with the rWT strain expressed viral DsRed, while only a minor fraction (<1%) were positive for ZsGreen, suggesting that the WT virus is highly effective in suppressing host innate responses. Conversely, infection with the M51R mutant produced a similar although slightly lower level of DsRed activity, but a high percentage of cells positive (>80%) for the IFIT2 reporter. This demonstrates that the reporter is highly sensitive to internal viral replication in the absence of suppression by VSV M protein.
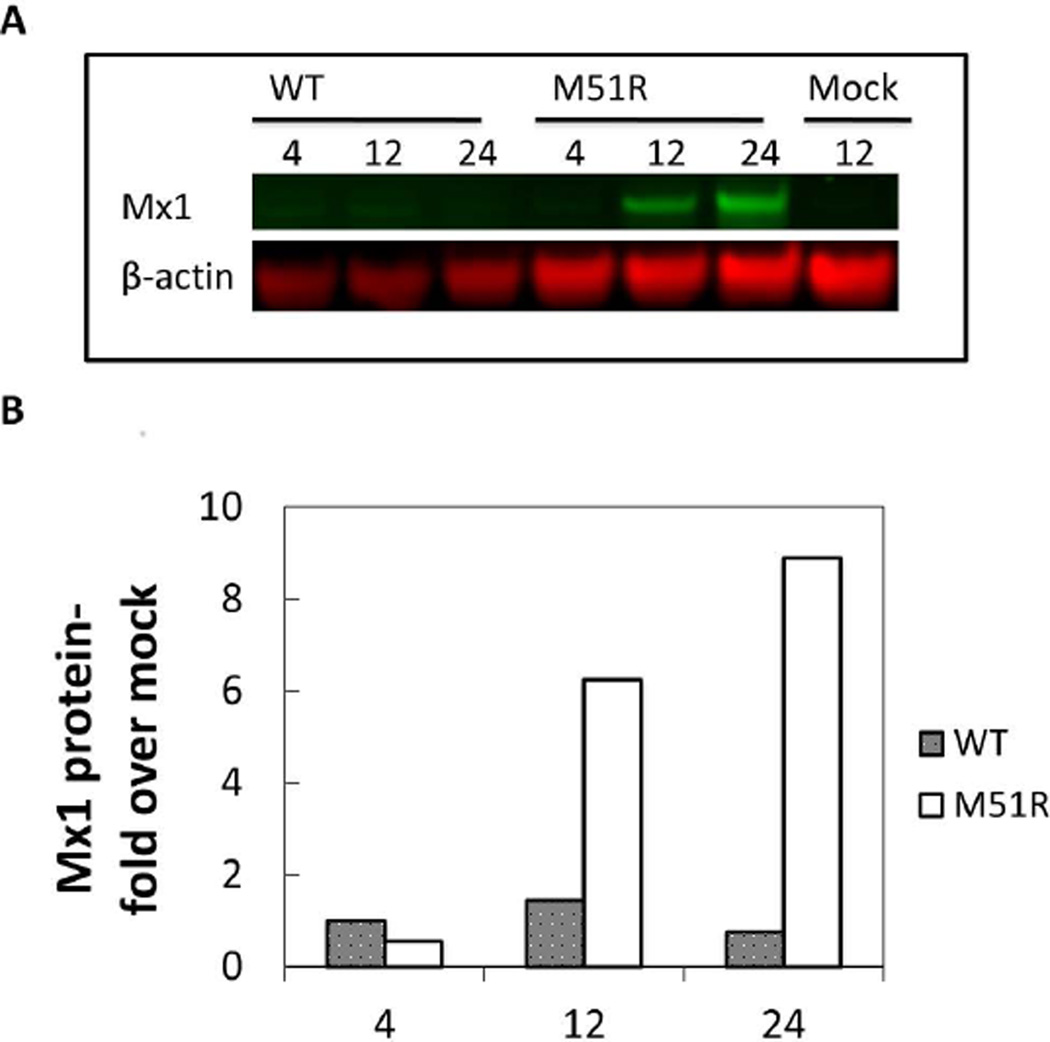
To corroborate that the IFIT2-ZsGreen reporter was accurately reporting on normal IFIT2 activity we used RT-qPCR to show that both the ZsGreen and IFIT2 mRNA were expressed with similar kinetics in response to VSV-M51R infection (Supp. Figure 1). At the protein level we lacked a specific antibody to IFIT2, so we instead tested the activity of a related interferon stimulated antiviral gene, Mx1, which we could readily detect by western blot (Figure 6). While WT-DsRed-Ex did not induce detectable Mx1 expression, infection with the M51R mutant strongly induced Mx1 protein on timescales that were similar to IFIT2-ZsGreen reporter expression.
Figure 6. Pattern of antiviral protein production is similar to IFIT2-ZsGreen reporter.
PC-3 IFIT2-ZsGreen1 reporter cells were infected with VSV-WT-DsRed-Ex or VSV-M51R-DsRed-Ex at MOI 50. (A) At 4, 12, 24 HPI total cell lysates were harvested and Western blotted for Mx1 protein expression. Two biological replicates were performed at each timepoint (representative blot shown).
(B). Band intensities were determined using Image Studio(LiCor) software. Each Mx1 band was normalized to β-actin band for that sample, replicates were averaged and expressed as fold over mock infected cells (harvested at 12 hrs). Results demonstrate that M51R strongly induced Mx1 expression while WT largely suppressed its activity.
Dual-color fluorescent reporters suggest role for innate immune inhibition of VSV infection spread
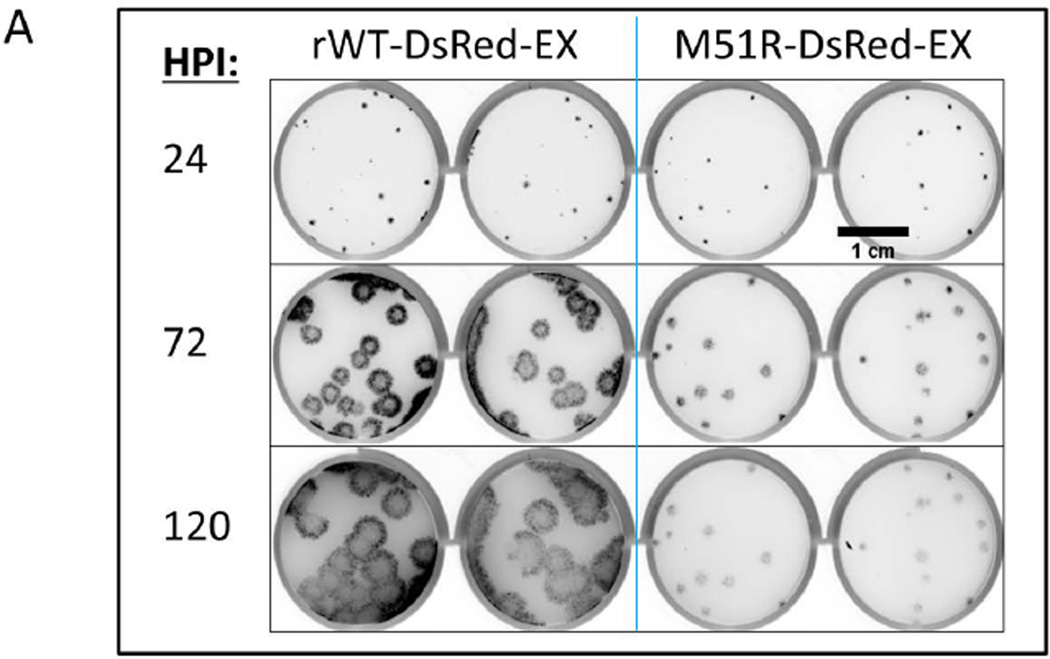
We initiated multiple isolated infections (MOI ~0.0001) on PC3 IFIT2-ZsGreen reporter cell monolayers and constrained their spread with a semi-solid agar overlay. We investigated the spread phenotypes of both the WT and M51R DsRed-Ex viruses using a fluorescent plate scanner (GE Typhoon 9000) to gauge overall plaque size (Figure 7A). An infected plate with a low density of plaques was imaged every 24 hours for 5 days. While WT and mutant plaques were similar in size after 24 hours, by 72 hours the WT infection was clearly spreading faster than the mutant M51R. WT continued to spread out to 120 hours post infection while the mutant essentially stopped at 72 hours.
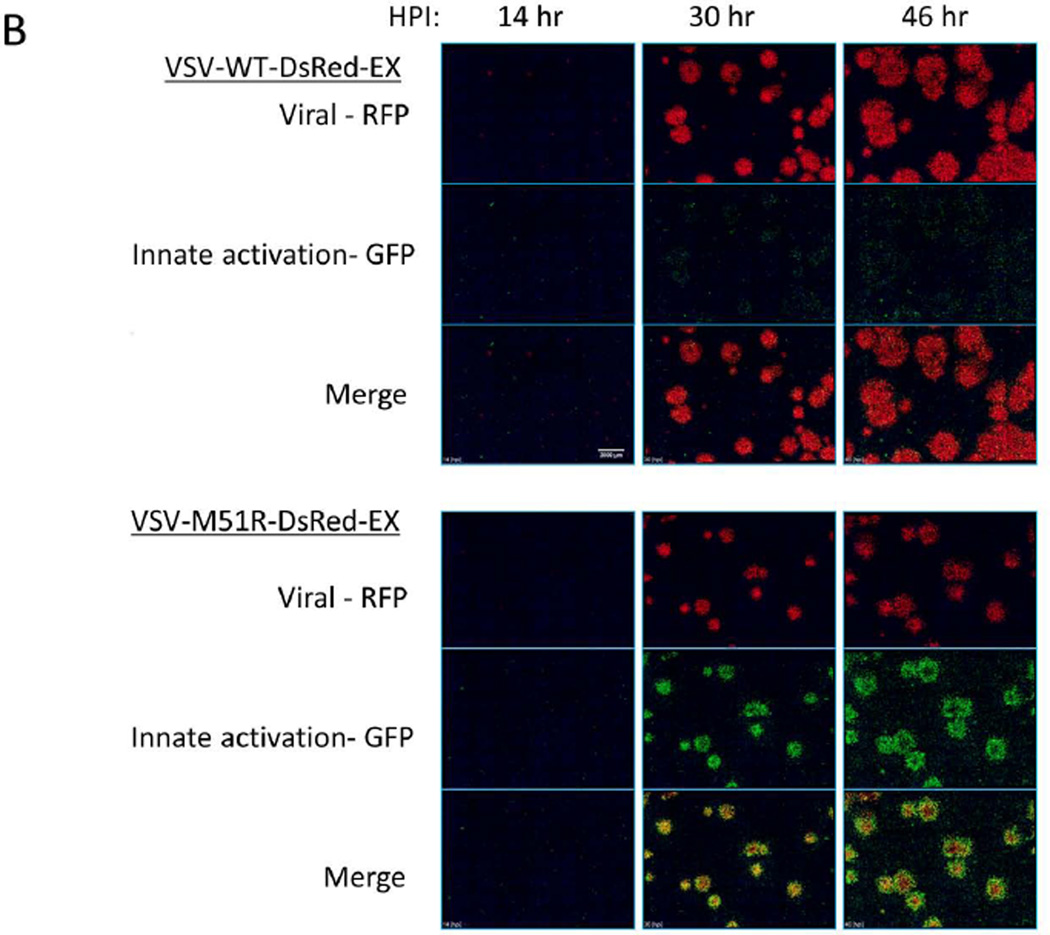
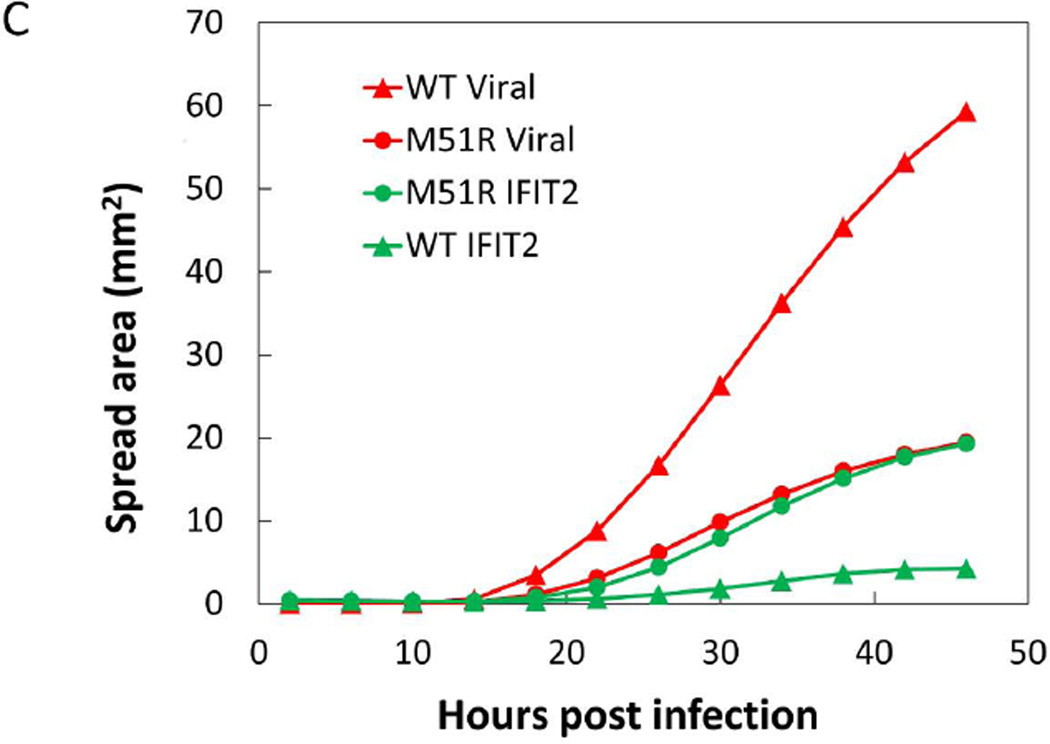
Figure 7. VSV spreading infection model suggests role of IFIT2 in limiting viral propagation.
(A) PC3 IFIT2-ZsGreen reporter cells were seeded into a 12 well plate and infected at a low (plaque forming) MOI and overlaid with a semi-solid media containing 0.6% agar. Every 24 hrs post infection the plate was removed from the incubator and scanned for RFP on a fluorescent plate scanner (GE Typhoon 9000), and then returned to continue incubation. At 24 HPI the WT and mutant plaques had grown to similar size, however by 72 HPI the WT plaques were considerably larger and continued to spread out to 120 HPI, while the M51R infections did not spread after 72 HPI.
(B) WT and M51R plaque infections were tracked using an automated fluorescent microscope fitted with a stage-top incubator chamber. Each images shown is a montage of a 7×7 array taken with a 4× magnification objective. Images underwent flat field correction and log transformation to enable visualization of both very dim and bright cells in the same image. Single channel images (red – viral RFP; green – IFIT2 reporter) display relative size and intensity of the two signals at timepoints shown, while the merged images show the location of the infected (red), IFIT2 active (green) and both infected and activated (yellow – orange) cells relative to each other. In general the WT infection produced strong infection spread and little IFIT2 activation. Conversely, in the M51R infection most cells were initially both infected and activated (see 30 hr), however, by later timepoints a ring of activated cells (green) had developed around the periphery of most plaques along with a dimmer more diffuse activation of cells distal to the infected regions. Scale bar is 2000 µm.
(C) Area of both infection and IFIT2 activation spread was determined by analysis of single channel images. Using ImageJ software, images were thresholded to identify positive pixels and total area above threshold was determined for each time-point.
We next investigated the activity of the IFIT2 reporter in the spreading infection context. Using a live-cell stage-top chamber and automated microscopy we continuously observed the infection spread until approximately 48 hours post infection (Figure 7B). Similar to the high MOI infections, the WT virus largely suppressed the activity of the IFIT2 reporter and the resulting expansion of the infection spread area was relatively rapid (Figure 7C). Conversely, infections with the M51R virus initially activated the IFIT2 reporter only in infected cells, but at later timepoints uninfected cells, beyond the infected area, were also activated. The higher IFIT2 activity correlated with a much smaller extent of infection spread, and by later timepoints it appeared that the rate of spread was beginning to slow; however, environmental conditions in the stage-top chamber prevented observation beyond 48 hrs.
DISCUSSION
We sought here to develop a dual-color system that would report not only viral replication and activation of the host cellular anti-viral response, but also the relative timing of these processes. By inserting ZsGreen (GFP) and DsRed-Ex (RFP) into the identical viral backbone and observing the fluorescence expression of both, we demonstrated that these two fluorescent proteins have similar translation and maturation kinetics, making them a suitable pair for development of the dual-color system. Further, to functionally test the host reporter we exploited the well-characterized inability of the VSV M51R mutant to inhibit host antiviral responses. Using both fluorescence microscopy and flow cytometry we found that high MOI infection with rWT virus largely suppressed activation of the IFIT2-GFP reporter while M51R strongly induced, or failed to suppress, IFIT2 activation. To link the GFP reporter function with activity of the native gene IFIT2, we found that expression of both mRNAs was activated at similar times to similar levels. We further sought to correlate reporter expression with levels of IFIT2 protein; however, in the absence of an IFIT2-specific antibody we employed as a surrogate the related gene Mx1. Mx1 produced the same on/off behavior in response to either M51R or WT, respectively, as did the IFIT2 reporter. Based on the indiscriminate mechanism of WT VSV innate immune inhibition, it is plausible that these two related genes are reacting to the infection in a similar manner, and that the activity of the reporter is indicative of the broader antiviral response.
While we observed a robust difference of the IFIT2 reporter in response to either WT or M51R VSV in a high MOI infection, we found relatively small differences in the resulting virus produced. This suggests that during an initial round of infection the antiviral response does not greatly impact the ability of VSV to replicate, consistent with previous reports (Ahmed et al., 2004). However, given that in vivo infections are more typically multi-round processes, we investigated the impact of the antiviral response in our multi-round plaque-spread model. Based on the previous high MOI experiments we were confident that the both the RFP and GFP reporters were valid indicators of their respective biological functions. In spreading plaque infections, we found that both WT and M51R strains initially spread at similar rates, but the M51R infections eventually slowed to a stop while the WT infections continued to spread. Both strains produced a general low level of IFIT2 activation beyond 30 HPI, but infection with M51R also produced a more robust host response, particularly in the region immediately adjacent to and ahead of the spreading infected region. It remains an open question whether this slowing of the M51R infection spread is due to activation by cytokine paracrine signaling, direct interaction with the virus, or some combination of these factors. Some reports have indicated that cytokine signaling through the Jak/Stat pathway is required for IFIT2 activation (Terenzi et al., 2007; Wacher et al., 2007), while others have shown that PRR recognition and IRF-3 can directly activate IFIT2 (Elco et al., 2005); ongoing investigations aim to discriminate which of these two pathways is critical for the inhibition of VSV infection spread. In summary, our results here highlight how the dynamics of a multi-round spreading infection, illuminated by reporters of viral replication and a host anti-viral response, reflect a layering of virus-cell interactions that is undetectable or obscured by more traditional single-round bulk measures.
Supplementary Material
PC-3 IFIT2-ZsGreen1 reporter cells were infected by M51R-DsRed-Ex at MOI 50. Total cellular RNA was harvested at regular intervals, reverse transcribed to cDNA and equivalent amounts of either IFIT2 (black circles) or ZsGreen (open circles) mRNA were determined by qPCR and are presented as fold over mock infection. We found that in response to M51R infection both IFIT2 and its ZsGreen1 reporter were expressed with similar kinetics and to similar levels.
ACKNOWLEDGEMENTS
We thank Dr. Valery Grdzelishvili for providing reagents and training for the creation of recombinant VSV strains, and Dr. Richard Randall for helpful discussions on the generation of stable reporter cell lines. We are grateful for support from the NIH (R01AI091646), an NSF GRFP fellowship (DGE-1256259) to A.S., and an NHGRI training grant to the Genomic Sciences Training Program (5T32HG002760) to A.B.
Footnotes
The authors declare that no conflicts of interest exist.
References
- Ahmed M, Cramer SD, Lyles DS. Sensitivity of prostate tumors to wild type and M protein mutant vesicular stomatitis viruses. Virology. 2004;330:34–49. doi: 10.1016/j.virol.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Ahmed M, McKenzie MO, Puckett S, Hojnacki M, Poliquin L, Lyles DS. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. Journal of Virology. 2003;77:4646–4657. doi: 10.1128/JVI.77.8.4646-4657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluyssen HAR, Vlietstra RJ, Made AVANDER, Trapman J. The interferon-stimulated gene 54 K promoter contains two adjacent functional interferon-stimulated response elements of different strength, which act synergistically for maximal interferon-a inducibility. EurJBiochem. 1994;220:395–402. doi: 10.1111/j.1432-1033.1994.tb18636.x. [DOI] [PubMed] [Google Scholar]
- Carey BL, Ahmed M, Puckett S, Lyles DS. Early steps of the virus replication cycle are inhibited in prostate cancer cells resistant to oncolytic vesicular stomatitis virus. Journal of Virology. 2008;82:12104–12115. doi: 10.1128/JVI.01508-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Short JaL, Young DF, Killip MJ, Schneider M, Goodbourn S, Randall RE. Heterocellular induction of interferon by negative-sense RNA viruses. Virology. 2010;407:247–255. doi: 10.1016/j.virol.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton KP, Rose JK. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology. 2001;279:414–421. doi: 10.1006/viro.2000.0736. http://www.ncbi.nlm.nih.gov/pubmed/11162797. [DOI] [PubMed] [Google Scholar]
- Das SC, Nayak D, Zhou Y, Pattnaik AK. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. Journal of Virology. 2006;80:6368–6377. doi: 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca Ka, Lam V, Keren I, Endler EE, Letchworth GJ, Novella IS, Yin J. Quantifying viral propagation in vitro: toward a method for characterization of complex phenotypes. Biotechnology Progress. 2001;17:1156–1165. doi: 10.1021/bp010115m. [DOI] [PubMed] [Google Scholar]
- Duprex WP, McQuaid S, Hangartner L, Billeter Ma, Rima BK. Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. Journal of Virology. 1999;73:9568–9575. doi: 10.1128/jvi.73.11.9568-9575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprex WP, McQuaid S, Roscic-Mrkic B, Cattaneo R, McCallister C, Rima BK. In vitro and in vivo infection of neural cells by a recombinant measles virus expressing enhanced green fluorescent protein. Journal of Virology. 2000;74:7972–7979. doi: 10.1128/jvi.74.17.7972-7979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elco CP, Guenther JM, Bryan RG, Sen GC. Analysis of Genes Induced by Sendai Virus Infection of Mutant Cell Lines Reveals Essential Roles of Interferon Regulatory Factor 3, NF-κB, and Interferon but Not Toll-Like Receptor 3. Journal of Virology. 2005;79:3920–3929. doi: 10.1128/JVI.79.7.3920-3929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathogens. 2012;8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sastre A, Biron C a. Type 1 interferons and the virus-host relationship: a lesson in détente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Gerlier D, Lyles DS. Interplay between Innate Immunity and Negative-Strand RNA Viruses: towards a Rational Model. Microbiology and Molecular Biology Reviews. 2011;75:468–490. doi: 10.1128/MMBR.00007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine EL, Lam V, Yin J, Rawlings JB. Image-guided modeling of virus growth and spread. Bulletin of Mathematical Biology. 2008;70:1730–1748. doi: 10.1007/s11538-008-9316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacre A, Wodarz D, Komarova NL, Fan H. Early infection and spread of a conditionally replicating adenovirus under conditions of plaque formation. Virology. 2012;423:89–96. doi: 10.1016/j.virol.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Jöns A, Mettenleiter TC. Green fluorescent protein expressed by recombinant pseudorabies virus as an in vivo marker for viral replication. Journal of Virological Methods. 1997;66:283–292. doi: 10.1016/s0166-0934(97)00065-7. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nature Immunology. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Lam V, Boehme KW, Compton T, Yin J. Spatial Patterns of Protein Expression in Focal Infections of Human Cytomegalovirus. Biotechnology and Bioengineering. 2006;93:1–11. doi: 10.1002/bit.20786. [DOI] [PubMed] [Google Scholar]
- Lam V, Duca Ka, Yin J. Arrested spread of vesicular stomatitis virus infections in vitro depends on interferon-mediated antiviral activity. Biotechnology and Bioengineering. 2005;90:793–804. doi: 10.1002/bit.20467. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Sobrido L, Zúñiga EI, Rosario D, García-Sastre A, de la Torre JC. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. Journal of Virology. 2006;80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DN, Kim P, Martínez-Sobrido L, Beitzel B, García-Sastre A, Langer R, Anderson DG. A novel high-throughput cell-based method for integrated quantification of type I interferons and in vitro screening of immunostimulatory RNA drug delivery. Biotechnology and Bioengineering. 2009;103:664–675. doi: 10.1002/bit.22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AF, Binduga-Gajewska I, Kauffman EB, Kramer LD. Quantitation of flaviviruses by fluorescent focus assay. Journal of Virological Methods. 2006;134:183–189. doi: 10.1016/j.jviromet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Petersen JM, Her LS, Varvel V, Lund E, Dahlberg JE. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Molecular and Cellular Biology. 2000;20:8590–8601. doi: 10.1128/mcb.20.22.8590-8601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajani KR, Pettit Kneller EL, McKenzie MO, Horita Da, Chou JW, Lyles DS. Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription. PLoS Pathogens. 2012;8:e1002929. doi: 10.1371/journal.ppat.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. The Journal of General Virology. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Stojdl DF, Lichty BD, TenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Terenzi F, White C, Pal S, Williams BRG, Sen GC. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. Journal of Virology. 2007;81:8656–8665. doi: 10.1128/JVI.00322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterstab G, Ludwig S, Anton A, Planz O, Dauber B, Krappmann D, Heins G, Ehrhardt C, Wolff T. Viral targeting of the interferon-{beta}-inducing Traf family member-associated NF-{kappa}B activator (TANK)-binding kinase-1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13640–13645. doi: 10.1073/pnas.0502883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacher C, Müller M, Hofer MJ, Getts DR, Zabaras R, Ousman SS, Terenzi F, Sen GC, King NJC, Campbell IL. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. Journal of Virology. 2007;81:860–871. doi: 10.1128/JVI.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li M, Zheng H, Muster T, Palese P, Beg aa, García-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. Journal of Virology. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Ball LA, Barr JN, Wertz GT. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz D, Hofacre A, Lau JW, Sun Z, Fan H, Komarova NL. Complex spatial dynamics of oncolytic viruses in vitro: mathematical and experimental approaches. PLoS Computational Biology. 2012;8:e1002547. doi: 10.1371/journal.pcbi.1002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann G, Rogulin V, Simon I, Rose JK, van den Pol AN. Some attenuated variants of vesicular stomatitis virus show enhanced oncolytic activity against human glioblastoma cells relative to normal brain cells. Journal of Virology. 2010;84:1563–1573. doi: 10.1128/JVI.02040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimovich A, Gumpert H, Burckhardt CJ, Lütschg Va, Jurgeit A, Sbalzarini IF, Greber UF. Cell-free transmission of human adenovirus by passive mass transfer in cell culture simulated in a computer model. Journal of Virology. 2012;86:10123–10137. doi: 10.1128/JVI.01102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yongky A, Yin J. Growth of an RNA virus in single cells reveals a broad fitness distribution. Virology. 2009;385:39–46. doi: 10.1016/j.virol.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PC-3 IFIT2-ZsGreen1 reporter cells were infected by M51R-DsRed-Ex at MOI 50. Total cellular RNA was harvested at regular intervals, reverse transcribed to cDNA and equivalent amounts of either IFIT2 (black circles) or ZsGreen (open circles) mRNA were determined by qPCR and are presented as fold over mock infection. We found that in response to M51R infection both IFIT2 and its ZsGreen1 reporter were expressed with similar kinetics and to similar levels.