Abstract
Since the invention of the first designer receptors exclusively activated by designer drugs (DREADDs), these engineered G protein-coupled receptors (GPCRs) have been widely applied in investigations of biological processes and behaviors. DREADD technology has emerged as a powerful tool with great potential for drug discovery and development. DREADDs can facilitate the identification of druggable targets and enable researchers to explore the activities of novel drugs against both known and orphan GPCRs. Here, we discuss how DREADDs can be used as novel tools for drug discovery and development.
Keywords: GPCR, DREADD, RASSL, drug discovery, drug development
The GPCR superfamily represents one of the major targets in drug discovery, mainly because of their crucial roles as modulators of nearly every known physiological and pathological process. Not surprisingly, GPCRs represent the canonical targets of at least 30% of the top 200 best-selling drugs in the USA. Indeed, seven out of 39 US Food and Drug Administration (FDA)-approved drugs in 2012 target GPCRs directly or indirectly [1]. According to the report by Global Industry Analysts, the global market for GPCRs is projected to reach US$120.5 billion by 2017 [2]. In addition, abnormal GPCR signaling and/or regulation is associated with the etiology of many disorders [3–5]. Several tools have recently been developed and are now used for drug discovery and development targeting GPCRs. For example, high-throughput (or content) screening (HTS/HCS) is commonly used in automated and robotized processes for the discovery of new ligands and/or modulators of GPCRs [3,6]. Additionally, ligand-based and structure-based methods for in silico screening and ligand design continue to be refined as essential tools for GPCR drug discovery [7–10].
However, the complexity of GPCR signaling and pharmacology needs to be considered when implementing GPCR-based drug screening. To establish effective screening methods and to prioritize druggable targets, it is essential that researchers understand the signaling pathway(s) relevant to a specific physiological and pathophysiological function. If information regarding disease-associated signaling pathways is unclear, multiple assays should be considered [3,11]. By contrast, if more information on the signal transduction of a GPCR target is available, fewer assays might be necessary, and the chance for success in drug discovery is likely to be higher (Figure 1.).
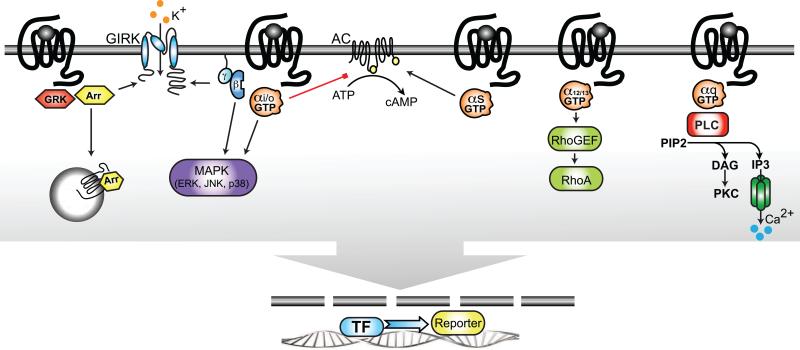
Figure 1.
Receptor-mediated G protein activation. G protein-coupled receptors (GPCRs) are activated by a variety of external stimuli. Upon receptor activation, the G protein exchanges GDP for GTP, causing the dissociation of the GTP-bound ! and !/! subunits and triggering diverse signaling cascades (Gs, Gq, and Gi). Phosphorylation by G protein receptor kinases (GRKs) promotes translocation of the !-arrestins to bind to the receptor and promote internalization. Receptors coupled to different heterotrimeric G protein subtypes can utilize different scaffolds to activate the small G protein/mitogen-activated protein kinase (MAPK) cascade, which can have either stimulatory or inhibitory consequences for the downstream pathways. Owing to these varieties of signaling pathways, information about the signal transduction of GPCR targets is important for deciding which assays should be used to increase chances for success in drug discovery.
The identification of potential therapeutic drugs was based on the concept that one ligand interacted with one receptor, leading to a single cellular output in classical receptor pharmacology [12]. However, there is now abundant evidence leading to a paradigm shift in receptor pharmacology in which one ligand interacts with multiple receptors, each of which can have multiple intracellular mechanisms of action [12–16]. When a GPCR ligand preferentially activates one pathway, it is said to be functionally selective [12] or to have biased signaling [17].
Why DREADDs represent a new approach for deconstructing GPCR actions
DREADDs represent a synthetic biology technology whereby designer GPCRs are created that can be activated solely by inert and extrinsic ligands, have minimal constitutive activity and cannot be activated any known endogenous ligands. In this respect, DREADDs represent second-generation receptors activated solely by synthetic ligands (RASSLs) [18].
Pioneering work in the generation of engineered GPCRs arose from the discovery that the β2-adrenergic receptor mutant D113S was refractory to its endogenous ligand norepinephrine, but acquired measurable but low activity toward synthetic catechol-ethers and esters, normally found to be inert at this receptor [19,20]. These innovative findings were a breakthrough for the initial proof-of-concept that GPCR signaling pathways could be differentially activated by synthetic ligands. Some years later, the Conklin group refined this concept and were able to reduce it to practice by developing the first RASSLs, using κ-opioid receptors as a template. [18,21]. RASSLs have been adopted by many labs and their use has yielded new insights into several physiological and pathological processes (reviewed in [22]). However, RASSLs have not been uniformly adopted because of considerably constitutive activity, the pharmacological activities of the ligands used and pathological phenotypes associated with overexpression [23–25].
The next generation of RASSLs, which we christened DREADDs, was created in the Roth lab in 2007 [26] and was designed to overcome some of the inherent disadvantages of the original RASSLs. DREADDs were developed by the directed molecular evolution of mutant muscarinic acetylcholine receptors (mAChR3) in yeast for activity toward the otherwise pharmacologically inert ligand clozapine N-oxide (CNO) [26–28]. Subsequently, we and others further developed a family of mutant muscarinic receptors for the three G protein-dependent signaling pathways (Gi, Gs, and Gq), which are activated only by CNO (in the nM range), but not by acetylcholine [26,29,30].
How can one use DREADDs for the study of functional selectivity in drug discovery and development? Briefly, as mentioned elsewhere [31], because GPCRs have modified properties that are crucial for regulating individual signal transduction in in vitro systems, DREADDs with different pharmacological profiled are engineered. These specialized DREADDs are then expressed in either targeted cell populations or disease-associated cells [30]. If a particular DREADD in a particular cellular population is efficacious in reversing the disease phenotype, then that cell population can be profiled (e.g., by RNA-seq) to find its druggable targets. These druggable targets can then be expressed in cultured cells and small molecules created to modulate these targets to reproduce the signaling state created by the DREADD. Small drug-like molecules that successfully recapitulate the DREADD-induced signaling could be then tested in animal models of efficacy. We discuss these aspects in detail below.
Functional selectivity 1L GPCR allosterism
One of the most frequently encountered pitfalls during drug discovery is the occurrence of undesirable off-target effects leading to the abandonment of drugs because of safety issues [32]. The identification and design of molecules with high specificity and selectivity remains a challenging endeavor and this is especially true for GPCRs, which can display high levels of cross-reactivity within other members of the family at their orthosteric binding sites. Several drugs show undesirable off-target effects, resulting in cytotoxicity and/or potentially lethal adverse effects. A pertinent example is that of unanticipated agonist activity at the 5-HT2B serotonin receptor leading to valvular heart disease induced by a variety of drugs, including appetite suppressants (e.g., fenfluramine [33]), antiparkinsonian drugs (e.g., cabergoline and pergolide [34]) and some ergot derivatives [4,11]. Thus, for example, because of the highly conserved orthosteric binding sites among aminergic GPCRs, drugs that are active at one biogenic amine GPCR frequently have off-target actions at many others [3,7,13,35,36]. To overcome off-target effects on such homologous GPCR members, conventional allosteric drug discovery has been widely touted as an alternative strategy because such drugs target allosteric rather than orthosteric sites, affording high degrees of selectivity [37].
However, with conventional allosteric drug discovery campaigns, the problem arises that GPCRs expressed in different host systems can show conformational differences and signaling bias [38]. DREADD-based tools aimed at the allosteric modulation of the binding of endogenous ligands could provide a useful strategy because DREADDs can be used directly in multiple model systems after the signaling of a particular receptor is designed and characterized in the cultured cell system, followed by introduction into whole organisms. As shown in Figure 2, the first step in this process is to design DREADDs that mediate unique and specific signal transduction pathways in vitro, which is made possible by the specificity of pharmacologically inert ligands (e.g., CNO). This initial approach can provide important information regarding the complexity of GPCRs in relation to their signaling and pharmacology. By analysis of this information, inferences can be made regarding the signaling pathway relevant to a specific physiological function or disease. Then, these DREADD variants could directly be applied to in vivo systems to recapitulate the designed signaling transduction pathways seen in cell culture systems. The results of this approach could result in clinically validated GPCR targets, and would provide at least insights into new therapeutic possibilities and novel profiles of drugs against GPCR targets.
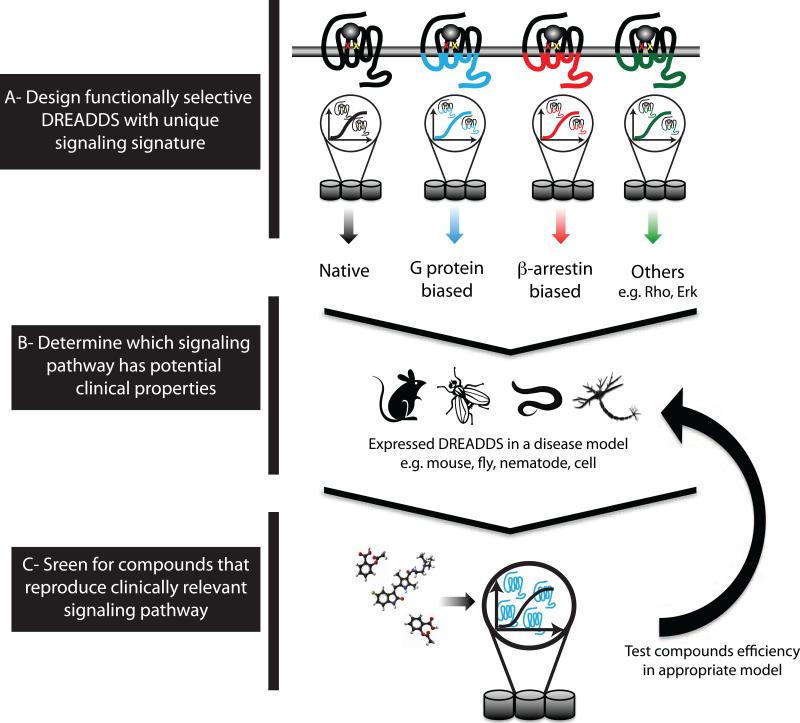
Figure 2.
[LM4] Designer receptors exclusively activated by designer drugs (DREADDs)-based tools aimed at G protein-coupled receptor (GPCR) allosterism. (A) Engineered-functionally selective DREADDs can provide valuable information regarding the complexity of GPCRs in relation to their signaling and pharmacology. (B) After determining which signaling pathway has potential clinical properties, these DREADD variants could directly be applied to in vivo systems. (C) The results of this approach would provide insights into new therapeutic possibilities and novel profiles of drugs against GPCR targets.
Functional selectivity 2
The concept of functional selectivity is based on the observation that the same ligand (agonist, antagonist, etc.) can modulate different signaling pathways through a single GPCR, resulting in different biological and/or physiological responses, depending on which pathway is activated [39,40]. Functional selectivity is well exemplified by the atypical antipsychotic drug aripiprazole, which displays bias for multiple signaling pathways [12,40]. Several β-arrestin-biased ligands were recently reported based on aripiprazole action. A structure–functional–selectivity relationship (SFSR) of the aripiprazole revealed distinct β-arrestin-biased dopamine D2 receptor (D2R) ligands. These are unique D2 β-arrestin-biased agonists that display atypical antipsychotic drug-like activities in vivo [41,42]. Additionally, these studies demonstrated potent antipsychotic-like activity with minimal motoric adverse effects in several in vivo models of antipsychotic drug actions. Thus, arrestin-biased ligands might have unique therapeutic advantages compared with unbiased ligands.
Owing to the phenomenon of functional selectivity, the complete picture of the interactions between a receptor and a ligand can be difficult to assess [3,12,43]. Nevertheless, the concept of functional selectivity can illuminate drug discovery strategies in terms of finding more efficacious drugs with fewer adverse effects [17]. For instance, the study of TRV120027 provides insight into how functional selectivity can be applied to drug discovery. TRV120027 acts as a novel β-arrestin-biased ligand at the angiotensin II type I receptor (AT1R) and antagonizes G protein coupling. These properties make TRV120027 a good candidate for the treatment of acute decompensated heart failure [44]. Given that many ligands can modulate different downstream signaling pathways through GPCRs, one novel approach to identify pathways that are useful for the discovery of biased agonists would be to use DREADDs, or to combine DREADD technology with HTS campaigns. DREADDs enable the testing of ligands on a continuous scale of properties, and can be engineered so that the strength of a particular signaling property is tightly regulated. Instead of searching ligand-based bias, these engineered DREADDs can mediate signal transduction in only one way: G protein-biased or β-arrestin-biased signal transduction. To regulate one of these signaling contexts, on the one hand, DREADDs can be genetically modified to prevent them from interacting with endogenous β-arrestins (e.g., deletion of the β-arrestin interacting domain in DREADDs), resulting in G protein-biased signaling. On the other hand, using chemically induced proximity (CIP) or chemically induced dimerization (CID), DREADDs could be created to modulate selectively only β-arrestin-biased signaling pathways. These features would help to not only understand the function of GPCRs in systemic engagement, but also discover and develop novel drugs featuring better therapeutic efficacy and minimized adverse effects. Through these concepts and technologies, drug screening has already been largely shifted from a single functional assay [e.g., changes in cAMP level or calcium flux in Fluorescence Imaging Plate Reader (FLIPR™)] to multiple end points that facilitate a more complete understanding of GPCRs in relation to their signaling and pathophysiology (Figure 3). Thus, DREADDs open up a new method of modifying GPCR signaling in complex ways.
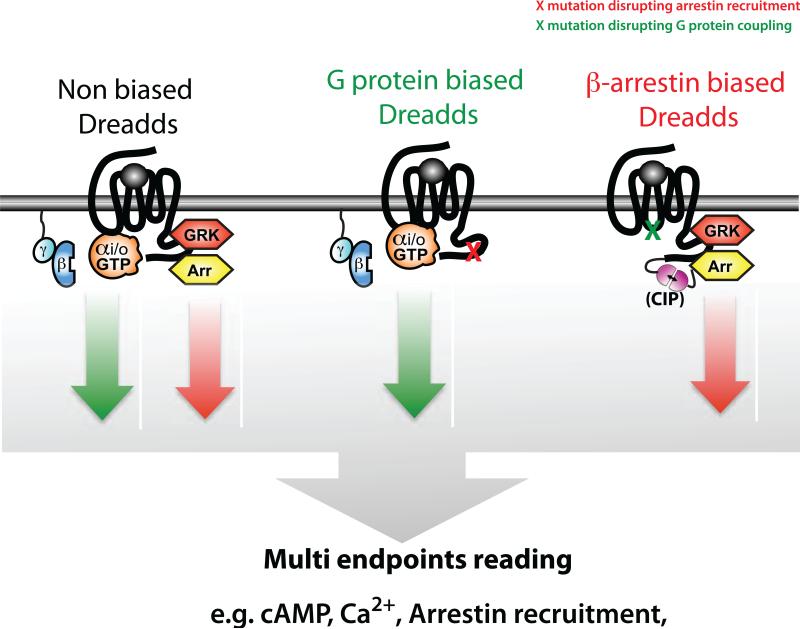
Figure 3.
[LM5] Designer receptors exclusively activated by designer drugs (DREADDs)-dependent biased signal transduction. Nonbiased receptors coupled to different heterotrimeric G protein subtypes mediate different signal transductions. However, engineered DREADDs, either G protein biased or !-arrestin biased, could be used to gain a more complete understanding of G protein-coupled receptors (GPCRs) in relation to their signaling and pathophysiology. These concepts and technologies would help not only to understand the function of GPCRs in systemic engagement, but also to discover and develop novel drugs featuring better therapeutic efficacy and minimized adverse effects. Abbreviation: CIP, chemically induced proximity.
Concluding remarks
Numerous efforts have been made to identify more efficacious and safer drugs targeting GPCRs. However, as knowledge of GPCRs and their signaling cascades and/or functions evolves, more useful and/or effective screening methods are necessary, including the combination of existing screening methods. By enabling exquisite fine-tuning of signaling pathways and, thus, of screening methods, DREADDs can be novel and useful tools in the discovery and development of drugs targeting GPCRs. It seems likely that DREADDs will provide new generations of drugs against GPCRs with unique targeted therapeutic uses, including ‘designer’ drugs, such as allosteric regulators, inverse agonists, and drugs targeting hetero-oligomeric receptor complexes. The combination of DREADDs with new screening technologies will enable the identification of novel drugs against both known and orphan GPCRs. Additionally, DREADD technology is likely to aid in the discovery of allosteric modulators, and might also be useful in addressing target tissue pharmacology early in the discovery cascade, with different experimental readouts capturing different states of the receptor.
Highlights.
DREADD is the acronym forDesigner Receptors Exclusively Activated by Designer Drugs
Available DREADDs are activated by the inert ligand clozapine-N-oxide
DREADDs allow for exquisite fine-tuning of signaling pathways
DREADDs are novel and useful tools in the discovery and development of drugs targeting GPCRs
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Teaser: By allowing exquisite fine-tuning of signaling pathways and thus, of screening methods, DREADDs can be novel and useful tools in the discovery and development of drugs.
References
- 1.Jarvis LM. New drug approvals hit 16-year high in 2012. Chem. Eng. News. 2013;91:15–17. [Google Scholar]
- 2.Analysts GI. G-Protein Coupled Receptors (GPCRS) – A Global Market Report. Global Industry Analysts; 2012. [Google Scholar]
- 3.Allen JA, Roth BL. Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu. Rev. Pharmacol. Toxicol. 2011;51:117–144. doi: 10.1146/annurev-pharmtox-010510-100553. [DOI] [PubMed] [Google Scholar]
- 4.Berger M, et al. The expanded biology of serotonin. Annu. Rev. Med. 2009;60:355–366. doi: 10.1146/annurev.med.60.042307.110802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth BL. Irving Page Lecture: 5-HT(2A) serotonin receptor biology: interacting proteins, kinases and paradoxical regulation. Neuropharmacology. 2011;61:348–354. doi: 10.1016/j.neuropharm.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.May LT, et al. Allosteric modulation of G protein-coupled receptors. Annu. Rev. Pharmacol. Toxicol. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- 7.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlsson J, et al. Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat. Chem. Biol. 2011;7:769–778. doi: 10.1038/nchembio.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besnard J, et al. Automated design of ligands to polypharmacological profiles. Nature. 2012;492:215–220. doi: 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin H, et al. A pharmacological organization of G protein-coupled receptors. Nat. Methods. 2013;10:140–146. doi: 10.1038/nmeth.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang XP, et al. Parallel functional activity profiling reveals valvulopathogens are potent 5-hydroxytryptamine(2B) receptor agonists: implications for drug safety assessment. Mol. Pharmacol. 2009;76:710–722. doi: 10.1124/mol.109.058057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urban JD, et al. Functional selectivity and classical concepts of quantitative pharmacology. J. Pharmacol. Exp. Ther. 2007;320:1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 13.Roth BL, et al. Screening the receptorome to discover the molecular targets for plant-derived psychoactive compounds: a novel approach for CNS drug discovery. Pharmacol. Ther. 2004;102:99–110. doi: 10.1016/j.pharmthera.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Kenakin T. New concepts in drug discovery: collateral efficacy and permissive antagonism. Nat. Rev. Drug Discov. 2005;4:919–927. doi: 10.1038/nrd1875. [DOI] [PubMed] [Google Scholar]
- 15.Kenakin T. Functional selectivity in GPCR modulator screening. Comb. Chem. High Throughput Screen. 2008;11:337–343. doi: 10.2174/138620708784534824. [DOI] [PubMed] [Google Scholar]
- 16.Kenakin TP. Biased signalling and allosteric machines: new vistas and challenges for drug discovery. Br. J. Pharmacol. 2012;165:1659–1669. doi: 10.1111/j.1476-5381.2011.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Violin JD, Lefkowitz RJ. Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol. Sci. 2007;28:416–422. doi: 10.1016/j.tips.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Coward P, et al. Controlling signaling with a specifically designed Gi-coupled receptor. Proc. Natl. Acad. Sci. U. S. A. 1998;95:352–357. doi: 10.1073/pnas.95.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strader CD, et al. A single amino acid substitution in the beta-adrenergic receptor promotes partial agonist activity from antagonists. J. Biol. Chem. 1989;264:16470–16477. [PubMed] [Google Scholar]
- 20.Strader CD, et al. Allele-specific activation of genetically engineered receptors. J. Biol. Chem. 1991;266:5–8. [PubMed] [Google Scholar]
- 21.Redfern CH, et al. Conditional expression and signaling of a specifically designed Gi-coupled receptor in transgenic mice. Nat. Biotechnol. 1999;17:165–169. doi: 10.1038/6165. [DOI] [PubMed] [Google Scholar]
- 22.Conklin BR, et al. Engineering GPCR signaling pathways with RASSLs. Nat. Methods. 2008;5:673–678. doi: 10.1038/nmeth.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sweger EJ, et al. Development of hydrocephalus in mice expressing the G(i)-coupled GPCR Ro1 RASSL receptor in astrocytes. J. Neurosci. 2007;27:2309–2317. doi: 10.1523/JNEUROSCI.4565-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng J, et al. Conditional expression of a Gi-coupled receptor in osteoblasts results in trabecular osteopenia. Endocrinology. 2008;149:1329–1337. doi: 10.1210/en.2007-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redfern CH, et al. Conditional expression of a Gi-coupled receptor causes ventricular conduction delay and a lethal cardiomyopathy. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4826–4831. doi: 10.1073/pnas.97.9.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armbruster BN, et al. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols CD, Roth BL. Engineered G-protein coupled receptors are powerful tools to investigate biological processes and behaviors. Front. Mol. Neurosci. 2009;2:16. doi: 10.3389/neuro.02.016.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong S, et al. Directed molecular evolution of DREADDs: a generic approach to creating next-generation RASSLs. Nat. Protoc. 2010;5:561–573. doi: 10.1038/nprot.2009.239. [DOI] [PubMed] [Google Scholar]
- 29.Alexander GM, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guettier JM, et al. A chemical-genetic approach to study G protein regulation of beta cell function in vivo. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19197–19202. doi: 10.1073/pnas.0906593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farrell MS, Roth BL. Pharmacosynthetics: reimagining the pharmacogenetic approach. Brain Res. 2012 doi: 10.1016/j.brainres.2012.09.043. http://dx.doi.org/10.1016/j.brainres. [DOI] [PMC free article] [PubMed]
- 32.Arrowsmith J. Trial watch: phase III and submission failures: 2007–2010. Nat. Rev. Drug Discov. 2011;10:87. doi: 10.1038/nrd3375. [DOI] [PubMed] [Google Scholar]
- 33.Rothman RB, et al. Evidence for possible involvement of 5-HT(2B) receptors in the cardiac valvulopathy associated with fenfluramine and other serotonergic medications. Circulation. 2000;102:2836–2841. doi: 10.1161/01.cir.102.23.2836. [DOI] [PubMed] [Google Scholar]
- 34.Roth BL. Drugs and valvular heart disease. N. Engl. J. Med. 2007;356:6–9. doi: 10.1056/NEJMp068265. [DOI] [PubMed] [Google Scholar]
- 35.Wacker D, et al. Structural features for functional selectivity at serotonin receptors. Science. 2013 doi: 10.1126/science.1232808. (in press)[LM2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, et al. Structural basis for molecular recognition at serotonin receptors. Science. 2013 doi: 10.1126/science.1232807. (in press)[LM3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christopoulos A. Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nat. Rev. Drug Discov. 2002;1:198–210. doi: 10.1038/nrd746. [DOI] [PubMed] [Google Scholar]
- 38.Digby GJ, et al. Novel allosteric agonists of M1 muscarinic acetylcholine receptors induce brain region-specific responses that correspond with behavioral effects in animal models. J. Neurosci. 2012;32:8532–8544. doi: 10.1523/JNEUROSCI.0337-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth BL, Chuang DM. Multiple mechanisms of serotonergic signal transduction. Life Sci. 1987;41:1051–1064. doi: 10.1016/0024-3205(87)90621-7. [DOI] [PubMed] [Google Scholar]
- 40.Urban JD, et al. Aripiprazole has functionally selective actions at dopamine D2 receptor-mediated signaling pathways. Neuropsychopharmacology. 2007;32:67–77. doi: 10.1038/sj.npp.1301071. [DOI] [PubMed] [Google Scholar]
- 41.Allen JA, et al. Discovery of beta-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18488–18493. doi: 10.1073/pnas.1104807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, et al. Structure-functional selectivity relationship studies of beta-arrestin-biased dopamine D(2) receptor agonists. J. Med. Chem. 2012;55:7141–7153. doi: 10.1021/jm300603y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mailman RB. GPCR functional selectivity has therapeutic impact. Trends Pharmacol. Sci. 2007;28:390–396. doi: 10.1016/j.tips.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boerrigter G, et al. TRV120027, a novel beta-arrestin biased ligand at the angiotensin II type I receptor, unloads the heart and maintains renal function when added to furosemide in experimental heart failure. Circ. Heart Fail. 2012;5:627–634. doi: 10.1161/CIRCHEARTFAILURE.112.969220. [DOI] [PubMed] [Google Scholar]