Abstract
Posttranscriptional maturation of the 3′ end of eukaryotic pre-mRNAs occurs as a three-step pathway involving site-specific cleavage, polymerization of a poly(A) tail, and trimming of the newly synthesized tail to its mature length. While most of the factors essential for catalyzing these reactions have been identified, those that regulate them remain to be characterized. Previously, we demonstrated that the yeast protein Pbp1p associates with poly(A)-binding protein (Pab1p) and controls the extent of mRNA polyadenylation. To further elucidate the function of Pbp1p, we conducted a two-hybrid screen to identify factors with which it interacts. Five genes encoding putative Pbp1p-interacting proteins were identified, including (i) FIR1/PIP1 and UFD1/PIP3, genes encoding factors previously implicated in mRNA 3′-end processing; (ii) PBP1 itself, confirming directed two-hybrid results and suggesting that Pbp1p can multimerize; (iii) DIG1, encoding a mitogen-activated protein kinase-associated protein; and (iv) PBP4 (YDL053C), a previously uncharacterized gene. In vitro polyadenylation reactions utilizing extracts derived from fir1Δ and pbp1Δ cells and from cells lacking the Fir1p interactor, Ref2p, demonstrated that Pbp1p, Fir1p, and Ref2p are all required for the formation of a normal-length poly(A) tail on precleaved CYC1 pre-mRNA. Kinetic analyses of the respective polyadenylation reactions indicated that Pbp1p is a negative regulator of poly(A) nuclease (PAN) activity and that Fir1p and Ref2p are, respectively, a positive regulator and a negative regulator of poly(A) synthesis. We suggest a model in which these three factors and Ufd1p are part of a regulatory complex that exploits Pab1p to link cleavage and polyadenylation factors of CFIA and CFIB (cleavage factors IA and IB) to the polyadenylation factors of CPF (cleavage and polyadenylation factor).
Most eukaryotic mRNAs contain a 3′ poly(A) tail, an appendage synthesized in a posttranscriptional processing reaction that involves site-specific cleavage, polyadenylation of the upstream cleavage product, and mRNA-specific trimming of the newly synthesized poly(A) tail. These reactions take place in a large complex that must interact with components of the transcription and splicing machineries, recognize several distinct sequence elements near the 3′ end of a transcript, cleave endonucleolytically with high precision, add and trim a fairly homogenous length of adenylate residues to different templates, and then remodel the resulting mRNP for efficient passage to and through the nuclear pore (47). In yeast, the complex and its supplementary factors include poly(A)-binding protein (Pab1p), cleavage and polyadenylation factor IA (CFIA), cleavage and polyadenylation factor IB (CFIB), cleavage and polyadenylation factor (CPF), and poly(A) nuclease (PAN) (47). CFIA is comprised of four subunits, Rna14p, Rna15p, Pcf11p, and Clp1p; CFIB is only a single polypeptide, Hrp1p/Nab4p; CPF contains numerous subunits, including Yhh1p, Ydh1p, Ysh1p, Mpe1p, Ref2p, Pta1p, Pfs2p, Pti1p, Swd2p, Fip1p, Glc7p, Yth1p, Ssu72p, Yor179cp, Ydl094cp, and, of course, poly(A) polymerase [Pap1p] (30, 47). PAN encompasses Pan2p, Pan3p, and Pbp1p (7, 9).
The core sequence requirements for 3′-end formation in yeast include the polyadenylation-cleavage site (PyAn), an “efficiency element” (UAUAUA, or repeats thereof) located a variable number of nucleotides 5′ to the poly(A) site, and a “positioning element” (AAUAAA, AAAAAA, and related sequences) located approximately 20 nucleotides (nt) 5′ to the poly(A) site. Unlike their mammalian counterparts, the yeast elements tend to be degenerate and redundant, a fact that explains the minimal effects on gene expression of some deletions that encompass normal 3′-processing signals (28, 47).
Cleavage and polyadenylation can be reproduced in vitro and appear to be coupled mechanistically, but such coupling is not required (i.e., precleaved substrates can be polyadenylated). While a detailed mechanistic understanding of the roles of the different factors remains to be established, current models for yeast polyadenylation (28, 30, 45, 47) suggest that the CPF complex recognizes the poly(A) site via interactions of Yhh1p, Ydh1p, and Yth1p with sequences surrounding the site of cleavage (3, 15, 16, 30) and that Hrp1p binds to the efficiency element while its ability to bind Rna14p acts to recruit CFIA to the positioning element (10, 20, 29). Subsequent to cleavage (the subunit that catalyzes the cleavage reaction is unknown), the Fip1p subunit of CPF recruits poly(A) polymerase and poly(A) synthesis on the newly created 3′ end ensues (22, 35). The extent of polyadenylation is regulated at least in part by Pab1p action on Pap1p and by its recruitment of PAN (8, 22, 32, 43, 48).
PAN is a yeast enzyme that trims newly added poly(A) tails after their default polyadenylation to lengths of 70 to 90 A's. Poly(A) trimming by PAN is mRNA specific, and analyses of three different mRNAs indicate that such trimmed tails have lengths ranging from 55 to 71 A's (8). PAN is a Pab1p-dependent 3′-to-5′ poly(A) exoribonuclease comprised of two subunits, Pan2p and Pan3p, that respectively, appear to have catalytic and regulatory roles (7, 9, 31). Pan3p, the positive regulator of PAN activity, interacts with Pab1p, thus providing substrate specificity for this nuclease (32a). Additional 3′-processing regulatory activities of Pab1p, as well as Pap1p, have been suggested by their two-hybrid interactions. For example, Pbp1p (Pab1p-binding protein 1) was identified in a two-hybrid screen using the carboxy terminus of Pab1p as bait (32). In the absence of Pbp1p, 3′ termini of pre-mRNAs are properly cleaved but lack full-length poly(A) tails (32). Since Pbp1p may also interact with Pan2p, it has been suggested that its role in poly(A) tail maturation is realized by negative regulation of PAN (32).
In an effort to further clarify the role of Pbp1p in poly(A) tail synthesis and maturation, we have carried out a two-hybrid screen using the PBP1 gene as bait. Among the Pbp1p-interacting proteins that were identified in this screen were Fir1p and Ufd1p, factors that have been previously shown to interact with Pap1p (13). Functional analyses of extracts prepared from pbp1Δ and fir1Δ cells and from cells lacking the Fir1p interactor, Ref2p, all demonstrate an inability to polyadenylate pre-mRNAs to normal lengths. Kinetic analyses of the aberrant polyadenylation carried out in vitro by all three extracts distinguish three types of defects. Collectively, these studies lend credence to the hypothesis that Pbp1p is a negative regulator of PAN activity and that Fir1p and Ref2p are, respectively, a positive regulator and a negative regulator of poly(A) synthesis.
MATERIALS AND METHODS
General methods.
Preparation of standard yeast media and methods of cell culture were as described previously (36). Transformations of yeast cells for library screens and creation of gene disruptions were done by the high-efficiency method (18); all other yeast transformations utilized the rapid method (42). DNA manipulations employed standard techniques (40). PCR amplifications were performed with Taq DNA polymerase (46) and confirmed, where appropriate, with DNA sequencing according to the method of Sanger et al. (41) or by PCR sequencing by the Nucleic Acid Facility at the University of Massachusetts Medical School. Plasmid DNAs were propagated in Escherichia coli strain DH5α. The new gene name, PBP4 (Pab1p-binding protein 4), was reserved for locus YDL053C in accordance with the naming guidelines of the Saccharomyces Genome Database (http://www.yeastgenome.org/).
Oligonucleotides.
The oligonucleotides used in this study were prepared by Operon, Inc., and are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence 5′→3′ |
|---|---|
| CYC1-NdeI1 | GTATTAAGAACGTTATTTATATTTCATATGTTTCTTTTTTTTCTGTACAAACGCG |
| CYC1-NdeI2 | CGCGTTTGTACAGAAAAAAAAGAAACATATGAAATATAAATAACGTTCTTAATAC |
| FIR1-5 | ATCATACCCGTAGTCCTCGCTGC |
| FIR1-3 | CATGTGCCAAGGATGATCTTTGG |
| FIR1-D5-U | TAGCACCCGCACACCGCAGCGGCCTGTGATATTCCGCATTGGAAATTAATGTGGCTGTGGTTTCAGGGTCC |
| FIR1-D3-U | CGATGAGCTGGTCAAATCTGTTTACCAGAACAATTTGTAGGGTTGACAAACGCATGAAATCCTTCATTTGC |
| PBP2-5 | CAAACTTATCCATAGCCACATCC |
| PAN2-5 | TAACGTACTATGTGGGCATAGAGG |
| PAN2-3 | GAAGTCTAATTCTGGCGTGTGATGAC |
| PAN2-D5-L | CTAGAACACAATTGCTATACTGAGTTTCTGAATGGTGAATGTATTCGCACAGAATCAAATTCGATGACTGG |
| PAN2-D3-L | GTGCGCTGGTGGCTCTTGAGATTACGTGAAAGGCACTGCACCATATAACTTTTTGTGTGGTGCCCTCCTCC |
| PAN3-5 | TACATATCAGACCTTTACAGGGTAACC |
| PAN3-3 | GATGTGGCGAACAAGGGATGAATCGC |
| PAN3-D5-L | CGATGATAGGTAAGCCTAAAAGACTAGTCAGACGTATCTTACGCCCGCACAGAATCAAATTCGATGACTGG |
| PAN3-D3-L | GTATAATTAATATATATGTGTGTATCTACATTATTTGGTGTTGTTTAACTTTTTGTGTGGTGCCCTCCTCC |
| PBP2-3 | AAAGAGAAATAATTCCGCGTACG |
| PBP2-D5-L | AGCGCGGCATTAAATAATCTTTCTGTAATACTCTTTAGCTCAATTCGCACAGAATCAAATTCGATGACTGG |
| PBP2-D3-L | TCTGTATTTTTATTTTCTATGTGTTTTTATTGACTAGCAGTATATTAACTTTTTGTGTGGTGCCCTCCTCC |
| PBP4-5 | TGTTCACTGTTTAATATCTATCC |
| PBP4-3 | CGGAAAGGCTAAGTGAACTATGC |
| PBP4-D5-U | TTATTACTTATTTACGATACAATTTTCCCTTTAATCTAGTACGAATTAATGTGGCTGTGGTTTCAGGGTCC |
| PBP4-D3-U | TGTTTGACTCCTTTTTGCGTTATGAAACGTGATGCTTCGATATTCACAAACGCATGAAATCCTTCATTTGC |
| REF2-5 | GTGTGAGTAAGTGAGGTTAAAGC |
| REF2-3 | GTGCCAGTAGCATCGTCATGTCC |
| REF2-D5-L | AAAGAAACGATACGATAAGTAAAGACACTGTGGAAGAATTGAACACGCACAGAATCAAATTCGATGACTGG |
| REF2-D3-L | AAATGAGTATATATACTACATGTTTATGTATCAGCATGTCATAGCTAACTTTTTGTGTGGTGCCCTCCTCC |
Yeast strains.
The yeast strains used in this study and their sources are listed in Table 2. Strains yDM372, yDM374, yDM376, yDM378, yME40, and yME43 were constructed by PCR-based gene deletion as described previously (6). For deletion of PBP2, REF2, PAN2, and PAN3, oligonucleotide pairs PBP2-D5-L/PBP2-D3-L, REF2-D5-L/REF2-D3-L, PAN2-D5-L/PAN2-D3-L, and PAN3-D5-L/PAN3-D3-L were respectively used to amplify the LEU2 marker from pJJ252 (27). For deletion of FIR1 and PBP4, oligonucleotide pairs FIR1-D5-U/FIR1-D3-U, and PBP4-D5-U/PBP4-D3-U were respectively used to amplify the URA3 marker from the genomic DNA of a URA3 wild-type yeast strain. PCR products to be used for gene deletion were electrophoresed on agarose gels, and the full-length products were recovered using a QIAquick gel extraction kit (Qiagen, Inc.). The recovered products were transformed into yeast strain yAS306, and transformants were selected on appropriate media. Genomic DNA was isolated from cultures started from individual colonies, and the presence of the disruption was confirmed by PCR using the gene-specific primers FIR1-5/FIR1-3 (for fir1Δ), PBP2-5/PBP2-3 (for pbp2Δ), PBP4-5/PBP4-3 (for pbp4Δ), REF2-5/REF2-3 (for ref2Δ), PAN2-5/PAN2-3 (for pan2Δ), and PAN3-5/PAN3-3 (for pan3Δ). Construction of the fir1Δ/ref2Δ strain, yDM428, was achieved by PCR disruption of REF2 in the fir1Δ strain, yDM376, as described above. The pan2Δ/pbp1Δ and pan3Δ/pbp1Δ strains yDM432 and yDM434, respectively, were generated by integration of a pbp1::TRP1 fragment isolated from pDM114 at the wild-type PBP1 locus in pan2Δ and pan3Δ strains yME40 and YME43, respectively.
TABLE 2.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| L40 (yDM61) | MATaade2 his3Δ200 leu2-3, 112 trp1-901 LYS::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ gal4 gal80 | S. Hollenberg |
| AMR70 (yDM62) | MATαade2 his3Δ200 leu2-3, 112 trp1-901 LYS::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ gal4 gal80 | S. Hollenberg |
| yAS306 (yDM117) | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 | A. Sachs |
| yDM146 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 pbp1::LEU2 | 32 |
| yDM372 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 pbp2::LEU2 | This study |
| yDM374 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 ref2::LEU2 | This study |
| yDM376 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 fir1::URA3 | This study |
| yDM428 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 fir1::URA3 ref2::LEU2 | This study |
| yDM378 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 pbp4::URA3 | This study |
| yME40 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 pan2::LEU2 | This study |
| yME43 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 pan3::LEU2 | This study |
| yDM432 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 pan2::LEU2 pbp1::TRP1 | This study |
| yDM434 | MATaade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 pan3::LEU2 pbp1::TRP1 | This study |
Plasmid construction.
Plasmid pDM335 was created to allow for the generation of in vitro-transcribed “precleaved” CYC1 precursor RNA substrates for in vitro polyadenylation reactions. Plasmid pGEM4-CYC1 (1) was subjected to site-directed mutagenesis with oligonucleotide primers CYC1-NdeI1 and CYC1-NdeI2 using a Stratagene Quik Change kit as directed by the manufacturer. The resulting construct was sequenced and subjected to restriction analysis to confirm the introduction of an NdeI restriction site. Runoff in vitro transcription reactions using the resulting plasmid and T7 RNA polymerase produce a 183-nucleotide precursor RNA ending at the normal 3′-end cleavage site of the CYC1 pre-mRNA.
Plasmid pDM114, used to generate pbp1::TRP1 strains, was constructed as follows. Initially, a 0.7-kb ClaI/BamHI fragment of PBP1 was inserted into the pBluescript II SK+ phagemid forming plasmid pDM98. This plasmid was then digested with EcoRV, and a SmaI/PvuII fragment from pJJ281 (27) was inserted to create pDM114. A SacI/ClaI fragment from pDM114 was used in yeast transformations to make the pbp1::TRP1 strains.
Two-hybrid screening.
Yeast strain L40 (23) (Table 2) harboring the lexA(DB)-PBP1 FL plasmid (pDM125) was transformed with GAL4(AD)-fused yeast genomic DNA libraries (25) (generously provided by Philip James and Elizabeth Craig, University of Wisconsin Medical School, Madison) and plated on synthetic complete (SC) medium without Leu, Trp, or His. Transformants were screened as described previously (32). The extent of protein interaction was assessed by monitoring the level of HIS3 reporter activity using 3-aminotriazole (3-AT) as a competitive inhibitor of His3p (i.e., imidazole glycerol-phosphate dehydratase). The amount of 3-AT on which cells can grow correlates with the level of HIS3 expression, thus indicating the relative strength of protein-protein interaction. This was assayed by growing colonies from each transformation in SC −Leu, −Trp broth, serially diluting them in water, and applying them as spots to SC −Leu, −Trp and SC −Leu, −Trp, −His plates containing 0, 5, 10, 20, 40, 60, 80, or 100 mM 3-AT.
In vitro 3′-end processing assays.
Whole-cell yeast extracts were prepared from stationary-phase cells (A600 of ∼4.0) as described previously (32). CYC1 precleaved precursors were generated for in vitro polyadenylation assays by using MEGAscript T7 transcription kits (Ambion, Inc.) to produce runoff transcripts of NdeI-cut plasmid pDM335. All reactions were performed at 25°C with 30 μg of extract in a final volume of 25 μl. The resulting products were analyzed on 6% polyacrylamide-7 M urea gels and visualized by autoradiography. Poly(A) tail lengths were measured against molecular weight markers using densitometry and are reported as the peak amount of polyadenylation in a given reaction. The data shown are representative of multiple independent determinations.
RESULTS
Identification of factors that interact with Pbp1p.
Using an in-frame lexA fusion to the entire PBP1 gene [PBP1(1-722)] (Fig. 1A) as bait, a two-hybrid screen (4, 5) was conducted to identify factors that interact with yeast Pbp1p. To assay interaction, we monitored the activity of two reporters, (lexAop)4-HIS3 and (lexAop)8-lacZ. We screened approximately 3 × 106 transformants and identified 64 clones that both grew in the absence of histidine and demonstrated significant β-galactosidase activity. These clones passed several tests for specificity, including the failure to activate transcription of the reporter genes in the absence of the lexA-PBP1(1-722) construct, a lack of interaction with the lexA(DB) vector alone, and a lack of interaction with lexA(DB) fused to the lamin, MOT1, MTF1, and PAF1 genes or the PAB1 P-H gene fragment (32). After sequencing the inserts from plasmids isolated in the screen, a total of five genes encoding putative Pbp1p-interacting proteins were identified (Table 3). These included one previously uncharacterized gene (designated PBP4) and four known genes, FIR1/PIP1, UFD1/PIP3, DIG1, and PBP1 itself.
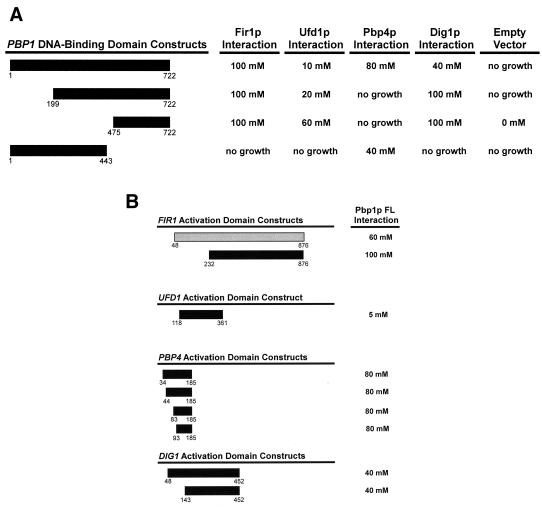
FIG. 1.
Mapping protein-protein interaction domains in Pbp1p and its interacting partners. Protein fragments used in two-hybrid analyses are denoted as bars, with numbers indicating the span of amino acids encoded by each construct. Proteins that interacted activated transcription of the HIS3 gene, producing resistance to the competitive inhibitor 3-AT. The results are expressed as the highest concentration of 3-AT (on plates of SC medium lacking His, Leu, and Trp) that still allowed substantial cellular growth; “no growth” indicates that cells could grow in the presence of histidine, but were unable to grow on medium lacking histidine (SC −Leu, −Trp, −His). (A) N-terminal and C-terminal truncations of Pbp1p were used to map the sites of interaction with factors identified in the two-hybrid screen. Each PBP1 fragment was tested against FIR1(281-925), UFD1-FL (full length), PBP4(93-185), DIG1(48-452), and empty GAL4(AD) vector. The PBP1(1-722) allele is identical to that previously designated PBP1-FL (32). (B) Fragments of interacting proteins identified in the two-hybrid screen with full-length Pbp1p.
TABLE 3.
Genes encoding putative Pbp1p-interacting proteins
| Gene | Homology or function | Mol mass (kDa) | Gene disruption | No. of times isolated |
|---|---|---|---|---|
| FIR1/PIP1 | mRNA 3′-end processing | 99 | Nonessential | 7 |
| UFD1/PIP3 | mRNA 3′-end processing | 40 | Essential | 1 |
| PBP1 | mRNA 3′-end processing | 79 | Nonessential | 6 |
| PBP4 | Unknown | 20 | Nonessential | 15 |
| DIG1 | MAP kinase-associated protein | 49 | Nonessential | 8 |
Initial characterization of Pbp1p-interacting factors.
A summary of some basic properties of the Pbp1p interactors identified in our screen and the genes that encode them is presented in Table 3. Two of these factors, Fir1p and Ufd1p, have previously been implicated in mRNA 3′-end processing. FIR1 has been shown to have two-hybrid interactions with the processing factor Ref2p (38), as well as with poly(A) polymerase (Pap1p) (13). The GAL4(AD)-FIR1 fusion proteins we identified promoted very strong interactions with Pbp1p, as indicated by the ability of the Fir1p/Pbp1p complex to mediate resistance up to 100 mM 3-AT (Fig. 1). FIR1 fragments were recovered seven times and included two different N-terminal fusions (Table 3 and Fig. 1B). Ufd1p was also identified as a Pap1p-interacting protein in the same analysis that identified Fir1p (13). Independently, an allele of UFD1 was isolated in a genetic screen for mutants that promoted the stabilization of a short-lived ubiquitinated β-galactosidase reporter (26). Although this allele stabilized the reporter protein, it failed to alter its posttranslational ubiquitination. Therefore, the exact role of UFD1 in the ubiquitin-mediated proteolytic pathway remains unclear. Our analysis identified a single clone of UFD1 that fused amino acids 118 to 361 to the GAL4(AD) (Fig. 1B). This fusion protein interacted weakly with full-length Pbp1p, mediating resistance to only 5 mM 3-AT (Fig. 1B). A subsequent analysis, expressing a full-length UFD1 clone, increased Ufd1p-Pbp1p interaction marginally, mediating resistance to 10 mM 3-AT (Fig. 1A).
PBP4 (YDL053C) was the only gene encoding a Pbp1p interactor that had not been characterized previously. Fifteen PBP4 clones were isolated (Table 3) and encompassed four unique N-terminal fusions to GAL4(AD) (Fig. 1B). In each instance, the Pbp1p-Pbp4p interaction mediated resistance to 80 mM 3-AT, suggesting that these two proteins can associate tightly. PBP4 is predicted to encode a 20-kDa polypeptide that has no significant homology to other known proteins. This gene is not required for cell viability, and its deletion did not result in any defect in growth rate (Table 3) (data not shown). This result is consistent with those obtained in the systematic analysis of all Saccharomyces cerevisiae deletion alleles (17).
Our previous studies had shown that Pbp1p is capable of self-interaction (32), and we thus anticipated the isolation of PBP1 fragments in this screen. This prediction was borne out with the isolation of six PBP1(339-722) clones that promoted resistance to 20 mM 3-AT (data not shown). This observation was consistent with data from a deletion analysis in which we demonstrated that a lexA-PBP1 fusion protein including Pbp1p amino acids 357 to 722 was also resistant to 20 mM 3-AT (32a). Surprisingly, the interacting fragments identified here did not, however, include a fragment approximating PBP1(199-722), the smallest PBP1 fragment that our deletion analyses showed was capable of multimerization and high-level (80 mM) 3-AT resistance (32a).
DIG1, a gene required for regulation of the mating and filamentous growth responses, was unanticipated as a potential PBP1 interactor. This gene encodes one of two highly homologous proteins that interact with the Fus3p and Kss1p mitogen-activated protein (MAP) kinases to repress the Ste12p transcriptional activator (12, 44). Eight DIG1 clones were isolated (Table 3), from which we identified two unique N-terminal fusions to GAL4(AD) (Fig. 1B). In each instance, Pbp1p/Dig1p interaction mediated resistance to 40 mM 3-AT. The lack of independent evidence suggesting a role for Dig1p in mRNA 3′-end processing led us to minimize further studies of this protein.
Mapping domains of protein-protein interactions.
Directed two-hybrid analyses with PBP1 fragments were utilized to identify the Pbp1p domains involved in specific protein-protein interactions (Fig. 1A). Three of the four factors identified in our screen were shown to interact with the Pbp1p C terminus. For Fir1p, the Pbp1p(475-722) fragment was necessary and sufficient to promote interaction. For Ufd1p and Dig1p, the strength of interaction increased when the Pbp1p N terminus was removed. For example, the enhancement of Pbp1p-Dig1p interaction promoted by deletion of Pbp1p amino acids 1 to 199 increased 3-AT resistance of the relevant clones from 40 mM to 100 mM (Fig. 1A). Similarly, deletion of Pbp1p amino acids 1 to 199 marginally strengthened association of Pbp1p and Ufd1p, but further truncation (of amino acids 1 to 475) enhanced interaction markedly, resulting in an increase in 3-AT resistance from 10 mM to 60 mM (Fig. 1A). Both of these sets of interaction patterns suggest that the Pbp1p N-terminal domain may fold in such a way as to inhibit or regulate a subset of this protein's interactions. The association of Pbp1p with Pbp4p was unique in that it was the only interaction requiring the Pbp1p N terminus. This was manifested by the ability of Pbp1p(1-443) to interact with Pbp4p, while Pbp1p(199-722) and Pbp1p(475-722) were unable to bind the same factor (Fig. 1A).
The isolation of multiple fragments of some genes encoding Pbp1p interactors allowed for partial mapping of the respective interacting domains on these proteins. Since the original screen was conducted with genomic DNA libraries, the results are biased to the identification of genes encoding factors whose interaction with Pbp1p does not require the N terminus of the protein. Fir1p, Ufd1p, and Dig1p all utilize domains within the C-terminal two-thirds of the respective proteins for interaction with Pbp1p (Fig. 1B). Pbp4p, which is composed of only 185 amino acids, requires only its C-terminal half for interaction with Pbp1p, thus delineating a relatively small binding site of 93 amino acids (Fig. 1B).
Pbp1p-interacting factors regulate the extent of polyadenylation.
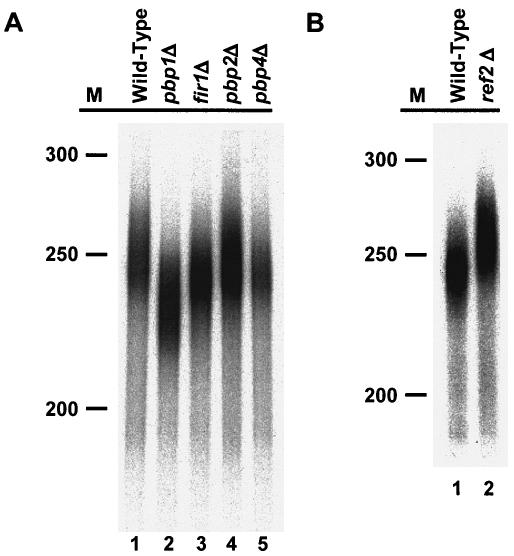
Two of the Pbp1p interactors we identified appear to play a role in mediating proper mRNA 3′-end formation. Deletion of FIR1 causes less efficient use of cryptic 3′-end cleavage sites in vivo, while loss of Ufd1p results in a substantial decrease in the amount of polyadenylation in vitro (13, 38). Since our earlier studies demonstrated that loss of Pbp1p diminishes the extent of polyadenylation in vitro (32), we sought to determine whether the nonessential factors identified in our two-hybrid screens with Pbp1p and Pab1p (32) behave similarly. To that end, we prepared multiple independent 3′-end processing extracts from wild-type, pbp1Δ, fir1Δ, pbp2Δ (32), and pbp4Δ strains and tested their ability to synthesize poly(A) tails in vitro on precleaved substrates. As described previously (32) and as illustrated in Fig. 2A, pbp1Δ extracts routinely synthesized poly(A) tails that were approximately 15 to 30 nt shorter than those synthesized by wild-type extracts (compare lanes 1 and 2). Similarly, extracts from fir1Δ cells consistently generated tails that were 5 to 7 nt shorter than those produced by wild-type extracts (compare lanes 1 and 3). In contrast, extracts from pbp2Δ and pbp4Δ cells polyadenylated the precursor transcript to the same extent as that derived from wild-type cells (compare lanes 1 with lanes 4 and 5).
FIG. 2.
The extent of in vitro polyadenylation is altered in extracts derived from fir1Δ, ref2Δ, and pbp1Δ strains. RNA processing extracts were prepared from the indicated strains and used for in vitro polyadenylation with a precleaved 183-nt CYC1 pre-mRNA. Radiolabeled ATP was used to monitor the reaction products. The reactions were terminated after 1 h, electrophoresed on 6% acrylamide gels, and visualized by autoradiography. M, radiolabeled DNA markers of indicated sizes (nucleotides).
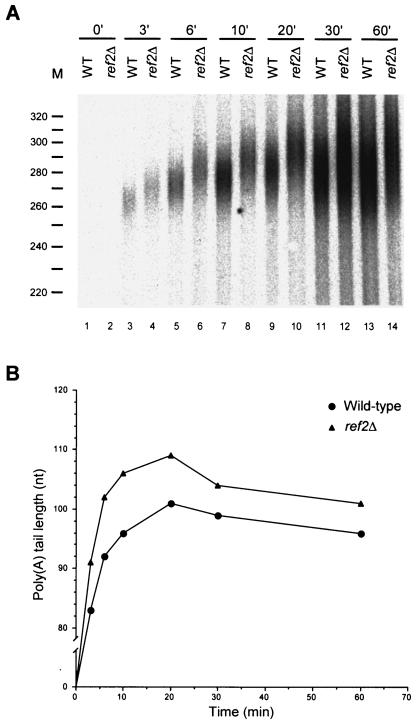
Fir1p was originally identified as a Ref2p-interacting protein (38). Loss of Ref2p appears to reduce the rate of pre-mRNA cleavage (37) and influence cleavage at weak 3′-end cleavage sites (38) but has not been shown to affect polyadenylation per se. Given the effects observed in Pbp1p- and Fir1p-deficient extracts (Fig. 2A), we sought to determine if loss of Ref2p also altered the extent of polyadenylation. We tested several extracts prepared from ref2Δ cells and observed that the poly(A) tails synthesized in those extracts were routinely 5 to 15 nt longer than those synthesized by extracts from wild-type cells (Fig. 2B, compare lanes 1 and 2).
Complementation of defects in poly(A) tail synthesis and maturation.
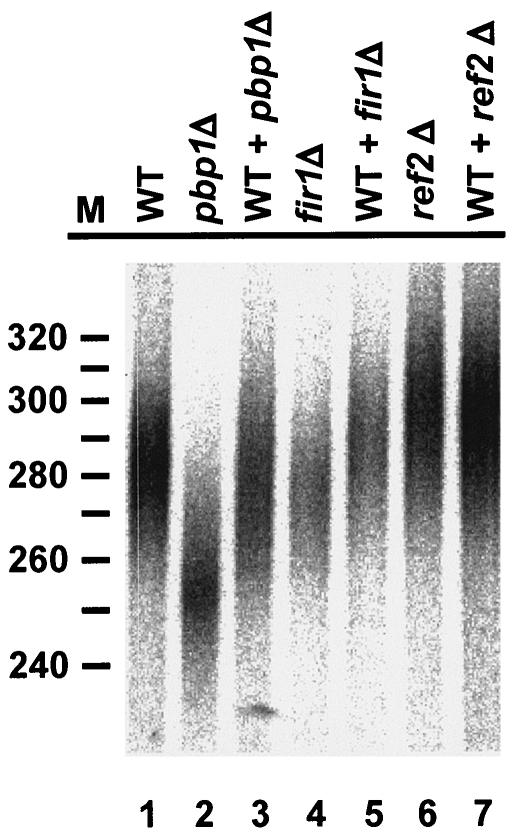
To demonstrate that the changes in poly(A) tail lengths observed in pbp1Δ, fir1Δ, and ref2Δ extracts were specific, we sought to test for biochemical complementation of the respective defects. Since we and others have been unable to purify functional recombinant Pbp1p, Fir1p, and Ref2p, we tested the effects of mixing extracts from the various deletion strains with extract prepared from wild-type cells (Fig. 3). Mixing wild-type extract with either pbp1Δ or fir1Δ extracts reversed the respective defects and restored polyadenylation to near-wild-type levels (Fig. 3, compare lanes 1 and 2 with lane 3 and lanes 1 and 4 with lane 5), but assays containing wild-type and ref2Δ extracts showed only marginal effects (compare lane 1 with lanes 6 and 7). Further experiments, combining extracts from pbp1Δ and ref2Δ cells, fir1Δ and ref2Δ cells, and fir1Δ and pbp1Δ cells, led to full restoration of wild-type poly(A) lengths (data not shown). These results demonstrate that the mutant extracts have specific defects in poly(A) tail synthesis or maturation.
FIG. 3.
Wild-type extracts reverse defects in poly(A) tail synthesis and maturation. RNA processing extracts prepared from pbp1Δ, fir1Δ, and ref2Δ strains were mixed with equal amounts of wild-type extracts for 30 min on ice and then used for in vitro polyadenylation of a precleaved CYC1 pre-mRNA. Reaction conditions and product analyses were identical to those described in the legend to Fig. 2.
Fir1p and Ref2p regulate poly(A) polymerase.
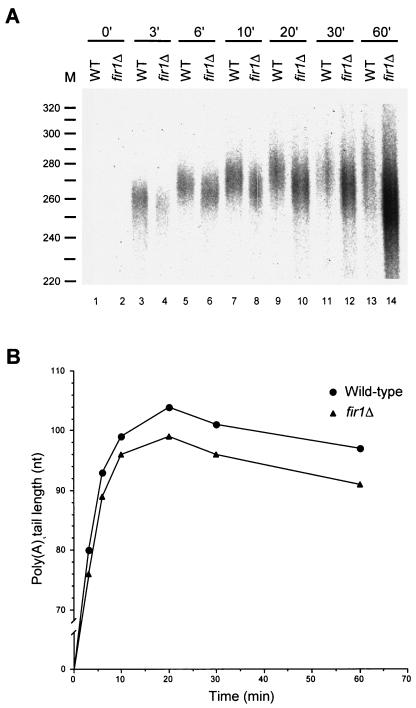
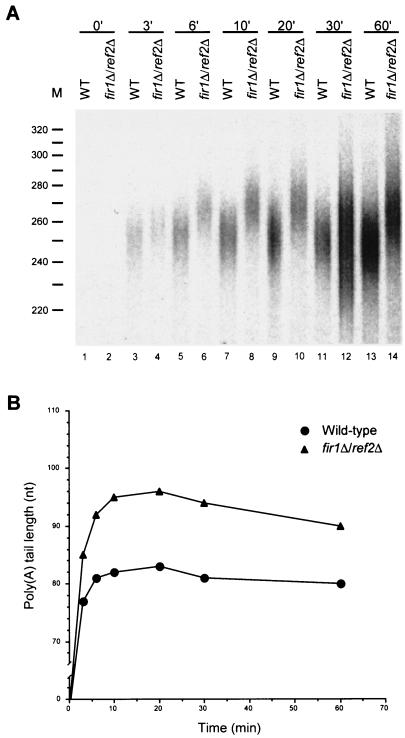
The data of Fig. 2 and 3 show that extracts from fir1Δ and ref2Δ cells fail to polyadenylate pre-mRNAs to normal lengths. To assess whether these effects are mediated at the level of poly(A) addition or trimming, we analyzed the respective time courses of polyadenylation in vitro. If Fir1p or Ref2p is required for poly(A) synthesis, poly(A) tail lengths in extracts lacking these factors should differ from those produced in wild-type extracts throughout the course of the reaction. However, if either of these factors regulates poly(A) trimming, the initial lengths of poly(A) tails produced in wild-type and mutant extracts should be very similar but then change as the rate of poly(A) shortening is affected. The results of this type of experiment are shown in Fig. 4 and 5, where the A panels are autoradiographs of the poly(A) tail length distributions and the B panels are graphic depictions of the peak amount of polyadenylation observed at each time point of the reaction. In these experiments, extracts derived from fir1Δ and ref2Δ cells displayed differences from wild-type extracts in the sizes of poly(A) tails initially added to substrate pre-mRNAs. In fir1Δ extracts, poly(A) tail lengths were initially 3 to 6 nt shorter than those generated by wild-type extracts and this difference was maintained throughout the reaction (Fig. 4). This result suggests that the difference in the extent of polyadenylation observed in fir1Δ extracts results from the loss of Fir1p stimulation of Pap1p activity. In extracts made from ref2Δ cells, the poly(A) tail lengths were initially longer than those produced by wild-type extracts (Fig. 5). This difference continued to increase during the polyadenylation phase of the reaction and peaked after 10 min of incubation. Beyond that point, the reaction switched to a deadenylation phase where the rates of poly(A) removal were roughly equivalent in both the wild-type and ref2Δ extracts. This result suggests that the increase in the extent of polyadenylation observed in the ref2Δ extracts is attributable to a loss of Ref2p inhibition of Pap1p activity. Since fir1Δ and ref2Δ extracts had opposing effects, we examined the effects on polyadenylation activity engendered by the combined loss of both genes. As shown in Fig. 6, extracts prepared from fir1Δ/ref2Δ cells demonstrate that the ref2Δ-like polyadenylation activity is dominant, i.e., poly(A) tails are longer than those produced by wild-type extracts.
FIG. 4.
Fir1p positively regulates poly(A) synthesis. (A) RNA processing extracts were prepared from wild-type and fir1Δ strains and used for in vitro polyadenylation of precleaved CYC1 pre-mRNA. Reactions, and their analysis, were identical to those depicted in Fig. 2, except that samples were taken at various times, as indicated. (B) The peak amount of polyadenylation from each reaction was determined by densitometry and graphed versus the time of incubation.
FIG. 5.
Ref2p negatively regulates poly(A) synthesis. (A) RNA processing extracts were prepared from wild-type and ref2Δ strains and used for in vitro polyadenylation of a precleaved CYC1 pre-mRNA as in Fig. 4. (B) The peak amount of polyadenylation was determined and plotted as in Fig. 4.
FIG. 6.
Extracts of fir1Δ/ref2Δ cells are phenotypically identical to ref2Δ extracts. (A) RNA processing extracts were prepared from wild-type and fir1Δ/ref2Δ strains and used for in vitro polyadenylation of a precleaved CYC1 pre-mRNA as in Fig. 4. (B) The peak amount of polyadenylation was determined and plotted as in Fig. 4.
Pbp1p negatively regulates poly(A) trimming.
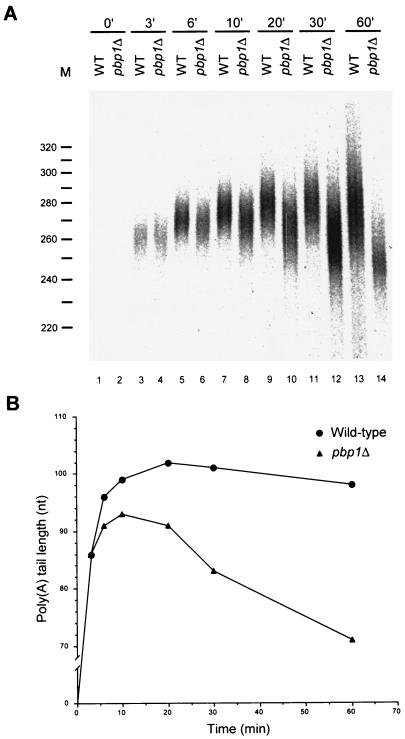
The production of shorter poly(A) tails by Pbp1p-deficient extracts (Fig. 2 and 3) could be attributable to a lack of stimulation of poly(A) synthesis or a failure to negatively regulate poly(A) trimming (e.g., by PAN). As above, to differentiate between these possibilities, we assayed the time course of polyadenylation in extracts derived from wild-type and pbp1Δ cells. The data in Fig. 7 demonstrate that the polyadenylation reaction has at least two phases, an initial poly(A) synthesis phase followed by a phase in which the poly(A) tail is then subjected to shortening. Initially, the products produced by both the wild-type and pbp1Δ extracts are identical in size (Fig. 7A, compare lanes 3 and 4). However, by 6 min of reaction, the poly(A) tails produced by the pbp1Δ extract are smaller than those of the wild-type extract, and this difference becomes more pronounced over time (Fig. 7B). In the wild-type extract, poly(A) tail lengths increase in size for 20 min and then are gradually trimmed. In contrast, the polyadenylation phase of the pbp1Δ extract is over by 10 min of incubation and is followed by poly(A) trimming that occurs at a considerably faster rate than that observed in the wild-type extract. Consistent with the results of earlier experiments (Fig. 2) (32), the poly(A) tails produced by pbp1Δ and wild-type extracts by 60 min of incubation differed by 27 nt (Fig. 7B). This kinetic analysis indicates that pbp1Δ extracts are unable to properly regulate poly(A) maturation. We conclude that Pbp1p is most likely a negative regulator of PAN, with its loss allowing for both premature initiation and a more rapid rate of poly(A) shortening.
FIG. 7.
Pbp1p is a negative regulator of poly(A) trimming. (A) RNA processing extracts were prepared from wild-type and pbp1Δ strains and used for in vitro polyadenylation of precleaved CYC1 pre-mRNA as in Fig. 4. (B) The peak amount of polyadenylation from each reaction was determined and plotted as in Fig. 4.
Pbp1p negatively regulates poly(A) nuclease.
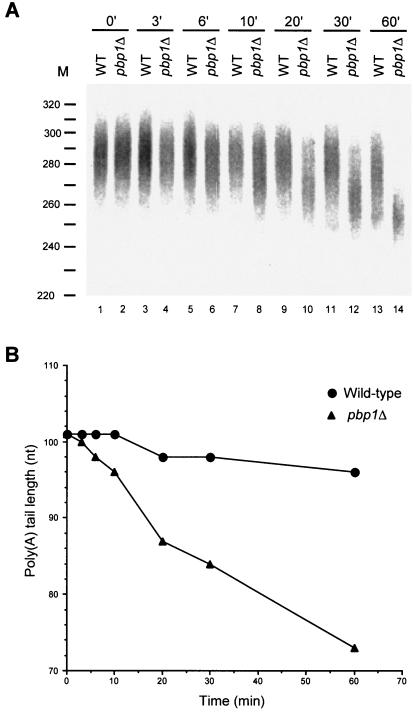
To test whether Pbp1p directly regulates PAN activity, we compared the abilities of wild-type and pbp1Δ extracts to degrade fully adenylated poly(A) tails. Fully adenylated, radiolabeled substrate was prepared by incubating our standard CYC1 precleaved 3′ RNA fragment in wild-type extract for 20 min (Fig. 3 to 7). Purified substrate was then added to wild-type and pbp1Δ extracts, and the kinetics of poly(A) removal was analyzed. Figure 8 shows that poly(A) shortening rates are approximately sevenfold faster in pbp1Δ extracts than in wild-type extracts, implying that the presence of Pbp1p somehow inhibits the activity of a poly(A) nuclease.
FIG. 8.
The absence of Pbp1p accelerates poly(A) shortening. (A) Fully adenylated, radiolabeled substrate was purified after incubating the precleaved CYC1 3′ RNA fragment in wild-type extract for 20 min and then incubated with wild-type and pbp1Δ extracts. Assay conditions were the same as those used for polyadenylation reactions except that labeled and unlabeled ATP were omitted. Samples were taken at various times, as indicated, and analyzed by gel electrophoresis and autoradiography. (B) The peak amount of poly(A) from each reaction was determined and plotted as in Fig. 4.
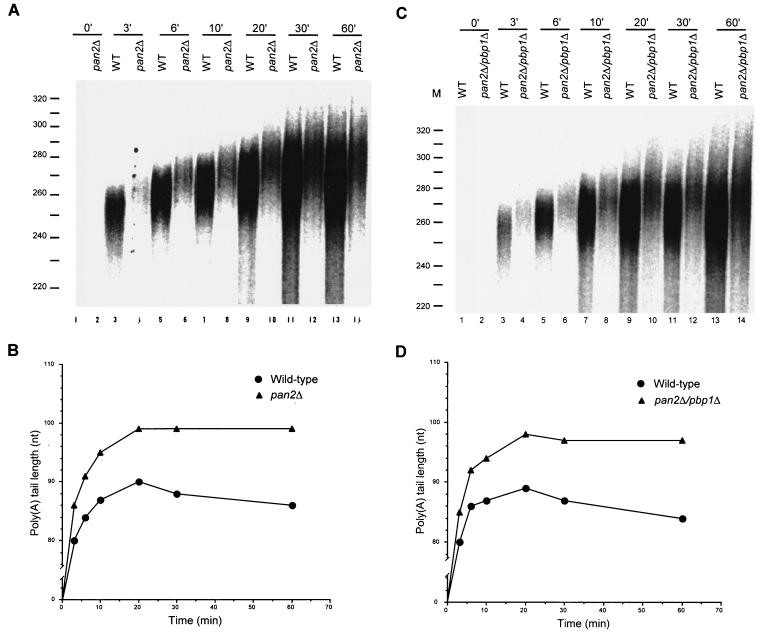
PAN is the most likely candidate for the target of Pbp1p's regulatory activity. To address this possibility definitively, we compared the kinetics of polyadenylation in extracts prepared from wild-type, pan2Δ, and pan2Δ/pbp1Δ cells. As shown in Fig. 9A and B, pan2Δ extracts accumulate significantly longer poly(A) tails than wild-type extracts. Interestingly, this result reflects the combined effect of an early increase in poly(A) synthesis as well as a reduction in the rate of poly(A) trimming. Since extracts of pan2Δ/pbp1Δ and pan2Δ cells exhibit the same characteristics (Fig. 9), we conclude that Pbp1p does indeed regulate PAN. This conclusion is supported by additional experiments demonstrating similar results with extracts prepared from pan3Δ cells and pan3Δ/pbp1Δ cells (data not shown).
FIG. 9.
Pbp1p is a negative regulator of poly(A) nuclease. (A and C) RNA processing extracts were prepared from wild-type, pan2Δ, and pan2Δ/pbp1Δ strains and used for in vitro polyadenylation of precleaved CYC1 pre-mRNA as in Fig. 4. (B and D) The peak amount of polyadenylation from each reaction was determined and plotted as in Fig. 4.
DISCUSSION
The addition of a poly(A) tail to most eukaryotic mRNAs provides a binding site for a major class of posttranscriptional regulatory factors, the poly(A)-binding proteins (11, 19, 33). These highly conserved polypeptides bind poly(A) using one or more RNA recognition motifs and then play key roles in the pathways of gene expression. In the nucleus, poly(A)-binding proteins regulate the ultimate length of the poly(A) tail by inhibiting its polymerization and stimulating its maturation (2, 8, 48). Association with these proteins is also required for the nuclear export of some mRNAs (33). In the cytoplasm, poly(A)-binding proteins facilitate the formation of the “closed loop” structure of the mRNP that is crucial for subsequent activities that promote translation initiation and termination, recycle ribosomes, and stabilize the mRNA (24, 33). Since they lack any apparent catalytic activity, it has been postulated that poly(A)-binding proteins provide a scaffold for the binding of factors that mediate these steps in gene expression while simultaneously preventing the binding of factors that enable the terminal steps of mRNA degradation (33). In higher eukaryotes, these sequential contributions are made by distinct nuclear and cytoplasmic poly(A)-binding proteins, but in yeast, most (but not all) of these functions have been attributed to a single protein, Pab1p (21, 33).
The creation of a binding site for these cis-acting effectors of gene expression, i.e., the process of polyadenylation, is an elaborate mechanism involving a large complex of factors. These factors must interact with components of the transcription and splicing machineries, recognize several distinct sequence elements near the 3′ end of a transcript, cleave endonucleolytically with high precision, add and trim a fairly homogenous length of adenylate residues to different templates, and then remodel the resulting mRNP for efficient passage to and through the nuclear pore (47). In yeast, the factors other than Pab1p responsible for these steps have been identified by a combination of biochemical and genetic methods and include CFIA, CFIB, CPF, and PAN (47). Almost all of the factors present in these complexes have been identified, with most encoding proteins essential for cell viability (30, 47). Although these factors have been categorized by their associated biochemical function, these complexes are interconnected. For example, CPF and CFIA appear to be linked via several interactions, including Fip1p/Rna14p, Psf2p/Rna14p, as well as multiple contacts between Ydh1p, Yhh1p, Ysh1p, Pta1p and Clp1p, Pcf11p, and Rna14p (30, 34, 35).
In this and a prior study (32), we have identified at least three factors (Pbp1p, Ref2p, and Fir1p) that influence the final size of a yeast poly(A) tail synthesized in vitro. While Ref2p has previously been shown to be a copurifying CPF component (14), evidence for the linkage of Pbp1p and Fir1p to the polyadenylation apparatus has depended largely on their genetic interactions (i.e., Fir1p's interactions with Pap1p and Ref2p's and Pbp1p's interactions with Pab1p) (32, 38). By extending the list of Pbp1p's interactors to include Ufd1p, Fir1p, and Pan2p (32a) and by obtaining more extensive evidence of RNA processing defects in extracts of pbp1Δ, ref2Δ, and fir1Δ cells, it now appears likely that Pbp1p, Ref2p, and Fir1p are bona fide regulators of yeast polyadenylation. Fir1p and Ref2p appear to affect poly(A) synthesis directly, since the effects on poly(A) tail lengths engendered by their loss are largely evident at the onset of polyadenylation (Fig. 4 to 6). Initial poly(A) tail lengths were decreased by the loss of Fir1p and increased by the loss of Ref2p, leading us to conclude that these two proteins can either act as respective positive and negative regulators of poly(A) polymerase (Pap1p) or mediate their effects indirectly through other components of CPF. The epistasis of the ref2Δ phenotype over that seen with fir1Δ extracts (Fig. 6) is intriguing in light of the known interactions of Fir1p and Pap1p and suggests that regulatory interactions within CPF are multifaceted.
Whereas Fir1p and Ref2p regulate poly(A) synthesis, several independent observations indicate that Pbp1p negatively regulates PAN during poly(A) tail maturation. The collective evidence includes our recent demonstration of interactions between Pbp1p and Pan2p (32a) and experiments here showing that pbp1Δ extracts have faster rates of poly(A) trimming than wild-type extracts (Fig. 7 and 8) and that this enhanced nucleolytic activity is dependent on functional Pan2p and Pan3p (Fig. 9) (data not shown). Since pbp1Δ extracts also pass prematurely from the synthesis to the maturation phase of the polyadenylation reaction (Fig. 7B), it is possible that Pbp1p may also help control an early step in poly(A) synthesis. However, Pbp1p does not appear to participate in the switch in Pap1p activity from processive to distributive activity (48), so its absence may simply allow for premature PAN recruitment and poly(A) trimming that would normally follow the binding of Pab1p to the elongating poly(A) tail. The extremely large poly(A) lengths observed in vivo in pab1, pan2, and pan3 mutants (9, 39) suggest that the latter step may also play a role in terminating the synthesis reaction.
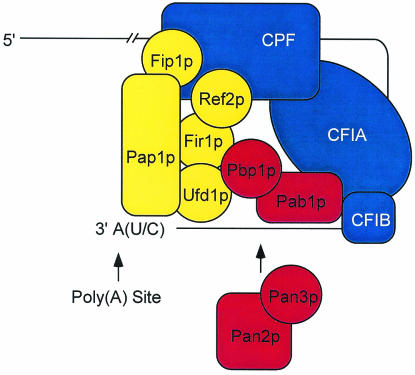
The experiments presented here and elsewhere (32a) define a new set of interactions linking CPF and CFIA that help to coordinate control of poly(A) tail synthesis and maturation. A model that incorporates these new regulators and some of their interactions into the general scheme for polyadenylation in yeast is shown in Fig. 10. It should be noted that many of the interactions portrayed as direct have only been inferred from one experimental approach (e.g., two-hybrid analysis) and may thus be bridged. Moreover, although the model implies the existence of multiple simultaneous interactions, sequential dynamic interactions are more likely. As befits a putative scaffolding protein (33), Pab1p is shown playing a central role, interacting with CFIA and -B, Pbp1p, and Pan3p to orchestrate poly(A) tail maturation (by Pan2p, Pan3p, and Pbp1p) (8) while simultaneously bridging the CFI factors to those associated with Pap1p via its interaction with Pbp1p. This arrangement, which also depicts the principal Pap1p interactors, takes into account the linkage that two of them (Fir1p and Ufd1p) have to Pbp1p as well as the observed positive and negative regulatory effects of Fir1p and Ref2p on polyadenylation. In addition, Fip1p is positioned to carry out its role as a mediator of Pap1p's transition from processive to distributive activity (22, 35, 48) and Ufd1p is included in this model by virtue of its interactions with Pap1p (13) and Pbp1p (Table 1 and Fig. 1). Ufd1p's status as the product of an essential gene precluded the possibility of our testing the consequences of its absence by analysis of cell extracts from a deletion strain, but subsequent analyses using other methodologies will allow more definitive tests of its possible regulatory roles. While the lack of in vitro polyadenylation phenotypes in pbp2Δ and pbp4Δ extracts suggested that Pbp2p and Pbp4p did not have roles in poly(A) tail synthesis or maturation, we cannot exclude the possibility that these factors have a function in pre-mRNA cleavage or mRNA export.
FIG. 10.
Factors associated with poly(A) polymerase (Pap1p) and poly(A)-binding protein (Pab1p) act to regulate poly(A) tail synthesis and maturation. In this model, the protein interactions identified in this study are superimposed on the core factors (blue) known to be required for pre-mRNA 3′ processing. These new interactions are postulated to link CPF to the CFI subunits and to play a role in coordinating polyadenylation factors (yellow) with those involved in poly(A) trimming (red).
Acknowledgments
This work was supported by a grant to A.J. from the National Institutes of Health (GM61096).
We thank Neptune Mizrahi and Claire Moore for plasmids and strains and members of our laboratory for discussions of the experiments and comments on the manuscript.
REFERENCES
- 1.Amrani, N., M. E. Dufour, N. Bonneaud, and F. Lacroute. 1996. Mutations in STS1 suppress the defect in 3′ mRNA processing caused by the rna15-2 mutation in Saccharomyces cerevisiae. Mol. Gen. Genet. 252:552-562. [DOI] [PubMed] [Google Scholar]
- 2.Amrani, N., M. Minet, M. Le Gouar, F. Lacroute, and F. Wyers. 1997. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 17:3694-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barabino, S. M., M. Ohnacker, and W. Keller. 2000. Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 19:3778-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14:920-924. [PubMed] [Google Scholar]
- 5.Bartel, P. L., and S. Fields. 1997. The yeast two-hybrid system. Oxford University Press, New York, N.Y.
- 6.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeck, R., S. Tarun, Jr., M. Rieger, J. A. Deardorff, S. Muller-Auer, and A. B. Sachs. 1996. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 271:432-438. [DOI] [PubMed] [Google Scholar]
- 8.Brown, C. E., and A. B. Sachs. 1998. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 18:6548-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, C. E., S. Z. Tarun, Jr., R. Boeck, and A. B. Sachs. 1996. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5744-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, S., and L. E. Hyman. 1998. A specific RNA-protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res. 26:4965-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coller, J. M., N. K. Gray, and M. P. Wickens. 1998. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 12:3226-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831-2848. [DOI] [PubMed] [Google Scholar]
- 13.del Olmo, M., N. Mizrahi, S. Gross, and C. L. Moore. 1997. The Uba2 and Ufd1 proteins of Saccharomyces cerevisiae interact with poly(A) polymerase and affect the polyadenylation activity of cell extracts. Mol. Gen. Genet. 255:209-218. [DOI] [PubMed] [Google Scholar]
- 14.Dichtl, B., D. Blank, M. Ohnacker, A. Friedlein, D. Roeder, H. Langen, and W. Keller. 2002. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10:1139-1150. [DOI] [PubMed] [Google Scholar]
- 15.Dichtl, B., D. Blank, M. Sadowski, W. Hubner, S. Weiser, and W. Keller. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 21:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dichtl, B., and W. Keller. 2001. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 20:3197-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 18.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 19.Gray, N. K., J. M. Coller, K. S. Dickson, and M. Wickens. 2000. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19:4723-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross, S., and C. L. Moore. 2001. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 21:8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hector, R. E., K. R. Nykamp, S. Dheur, J. T. Anderson, P. J. Non, C. R. Urbinati, S. M. Wilson, L. Minvielle-Sebastia, and M. S. Swanson. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21:1800-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helmling, S., A. Zhelkovsky, and C. L. Moore. 2001. Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Mol. Cell. Biol. 21:2026-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobson, A. 1996. Poly(A) metabolism and translation: the closed loop model, p. 451-480. In J. W. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, E. S., P. C. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442-17456. [DOI] [PubMed] [Google Scholar]
- 27.Jones, J. S., and L. Prakash. 1990. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6:363-366. [DOI] [PubMed] [Google Scholar]
- 28.Keller, W., and L. Minvielle-Sebastia. 1997. A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr. Opin. Cell Biol. 9:329-336. [DOI] [PubMed] [Google Scholar]
- 29.Kessler, M. M., M. F. Henry, E. Shen, J. Zhao, S. Gross, P. A. Silver, and C. L. Moore. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 11:2545-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyburz, A., M. Sadowski, B. Dichtl, and W. Keller. 2003. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res. 31:3936-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowell, J. E., D. Z. Rudner, and A. B. Sachs. 1992. 3′-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 6:2088-2099. [DOI] [PubMed] [Google Scholar]
- 32.Mangus, D. A., N. Amrani, and A. Jacobson. 1998. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 18:7383-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Mangus, D. A., M. C. Evans, N. S. Agrin, M. Smith, P. Gongidi, and A. Jacobson. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol, in press. [DOI] [PMC free article] [PubMed]
- 33.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223.1-223.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohnacker, M., S. M. L. Barabino, P. J. Preker, and W. Keller. 2000. The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 19:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preker, P. J., J. Lingner, L. Minvielle-Sebastia, and W. Keller. 1995. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell 81:379-389. [DOI] [PubMed] [Google Scholar]
- 36.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Russnak, R., K. W. Nehrke, and T. Platt. 1995. REF2 encodes an RNA-binding protein directly involved in yeast mRNA 3′-end formation. Mol. Cell. Biol. 15:1689-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russnak, R., S. Pereira, and T. Platt. 1996. RNA binding analysis of yeast REF2 and its two-hybrid interaction with a new gene product, FIR1. Gene Expr. 6:241-258. [PMC free article] [PubMed] [Google Scholar]
- 39.Sachs, A. B., and R. W. Davis. 1989. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58:857-867. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soni, R., J. P. Carmichael, and J. A. Murray. 1993. Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr. Genet. 24:455-459. [DOI] [PubMed] [Google Scholar]
- 43.Tacahashi, Y., S. Helmling, and C. L. Moore. 2003. Functional dissection of the zinc finger and flanking domains of the Yth1 cleavage/polyadenylation factor. Nucleic Acids Res. 31:1744-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tedford, K., S. Kim, D. Sa, K. Stevens, and M. Tyers. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7:228-238. [DOI] [PubMed] [Google Scholar]
- 45.Wahle, E., and U. Kuhn. 1997. The mechanism of 3′ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog. Nucleic Acid Res. Mol. Biol. 57:41-71. [DOI] [PubMed] [Google Scholar]
- 46.White, T. J., N. Arnheim, and H. A. Erlich. 1989. The polymerase chain reaction. Trends Genet. 5:185-189. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhelkovsky, A., S. Helmling, and C. Moore. 1998. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol. Cell. Biol. 18:5942-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]