Abstract
The Sox-2 gene is expressed in embryonic stem (ES) cells and neural stem cells. Two transcription enhancer regions, Sox-2 regulatory region 1 (SRR1) and SRR2, were described previously based on their activities in ES cells. Here, we demonstrate that these regulatory regions also exert their activities in neural stem cells. Moreover, our data reveal that, as in ES cells, both SRR1 and SRR2 show their activities rather specifically in multipotent neural stem or progenitor cells but cease to function in differentiated cells, such as postmitotic neurons. Systematic deletion and mutation analyses showed that the same or at least overlapping DNA elements of SRR2 are involved in its activity in both ES and neural stem or progenitor cells. Thus, SRR2 is the first example of an enhancer in which a single regulatory core sequence is involved in multipotent-state-specific expression in two different stem cells, i.e., ES and neural stem cells.
Stem cells have been identified in various organs, including hematopoietic tissue and the nervous system, and play a central role in tissue generation during development (for details, see references 22, 49, and 53). These stem cells are also present in adult animals, where they participate in tissue repair and the homeostasis of each tissue. Stem cells share the properties of self-renewal and the ability to generate at least one (but usually more) differentiated cell types, suggesting the presence of common genetic programs to maintain these unique biological properties of stem cells. To date, an increasing amount of data from global transcriptional profiling analyses have piled up (10, 15, 19, 36, 45, 50), and these analyses have led to the identification of a number of genes which are commonly expressed in more than two different types of stem cells. Indeed, some, such as the integrin alpha 6 and polycystic kidney disease 2 genes, have been shown to be expressed in all of the three best-characterized types of stem cells, i.e., embryonic stem (ES) cells, neural stem cells, and hematopoietic stem cells (15, 36). However, it is not known whether the expression of these genes is supported by a single regulatory enhancer which operates in different types of stem cells or whether such expression merely reflects the combined actions of multiple different regulatory regions in which individual regulatory enhancers function only in specific stem cells. Recently, Cairns et al. (7) demonstrated that a portion of the first intron of the c-kit gene supports its expression in both hematopoietic and germ cell lineages. However, because the enhancer region was not finely mapped, it is not known at present whether a single regulatory sequence indeed participates in gene expression in these two different cell lineages.
Sox-2, a transcriptional factor bearing a high-mobility-group box, is one of the examples expressed in more than two different types of stem cells (4, 57). In fact, zygotically transcribed Sox-2 mRNA is detected in the inner cell mass, epiblasts, and germ cells in early mouse embryos. Sox-2 is also expressed in trophoblast stem cells, which correspond to the stem cells of extraembryonic ectodermal tissues (4). In these cell lineages, Sox-2 expression is restricted to cells with stem cell characteristics and no longer expressed in cells with restricted developmental potential. Besides being expressed in early mouse embryos, Sox-2 is also expressed in the developing central nervous system (6, 12, 20, 34, 51, 52, 57). Sox-2 expression is first detected uniformly in the neural plate, in which most of the cells are multipotent. However, once the columnar epithelium of the neural plate acquires a more complex stratum structure, the expression becomes restricted to the germinal layer, where multipotent neural stem cells are enriched. Moreover, Graham et al. (11) recently demonstrated that the signaling of members of the SoxB1 transcription factor group, which includes the Sox-2 protein, is sufficient to maintain the panneural properties of neural progenitor cells.
Recent microarray analyses have pointed out significant similarities between ES cells and neural stem cells at the transcriptional level (36). Therefore, it is assumed that the ways of supporting stem cell-specific gene expression in these two distinct types of stem cells are intimately related and that at least some of the genes whose expressions overlap in ES and neural stem cells are regulated by common enhancers which function in these two distinct stem cells. We pursued the possibility that the Sox-2 gene possesses such a regulatory region(s).
The two regulatory regions termed Sox-2 regulatory region 1 (SRR1) and SRR2, which were identified based on their activities in pluripotent ES cells, were reported previously (48). Moreover, it was shown that SRR2 has a regulatory core sequence comprising octamer and Sox-2 binding sequences and that SRR2 exhibits its activity by recruiting the Oct-3/4-Sox-2 or Oct-6-Sox-2 complex to it in ES cells. In this report, we demonstrate that both SRR1 and SRR2 also exert their activities in neural stem cells. We also show that SRR2 utilizes the same core sequence to support its multipotent-state-specific enhancer activity in both ES and neural stem cells.
MATERIALS AND METHODS
Plasmid constructions.
For constructing β-geo reporter plasmids (see Fig. 2), the splice acceptor portion of the engrailed gene and internal ribosome entry site (IRES) region were removed from pGT1.81IresBgeo (27) and either SRR1 or SRR2 was subcloned together with a herpes simplex virus thymidine kinase (tk) promoter (positions −109 to +51) (21). SRR1 and SRR2 encompass the regions from positions −3937 to −3487 and +3641 to +4023 of Sox-2 genomic DNA, respectively, in which the transcription start site is considered position +1 (see reference 54). The location of SRR1 has been narrowed down to this short region (451 bp) (S.M. and A.O., unpublished data) from previous work (48). SRR1 was recovered by PCR, while the SRR2 portion was obtained from cloned genomic DNA.
FIG. 2.
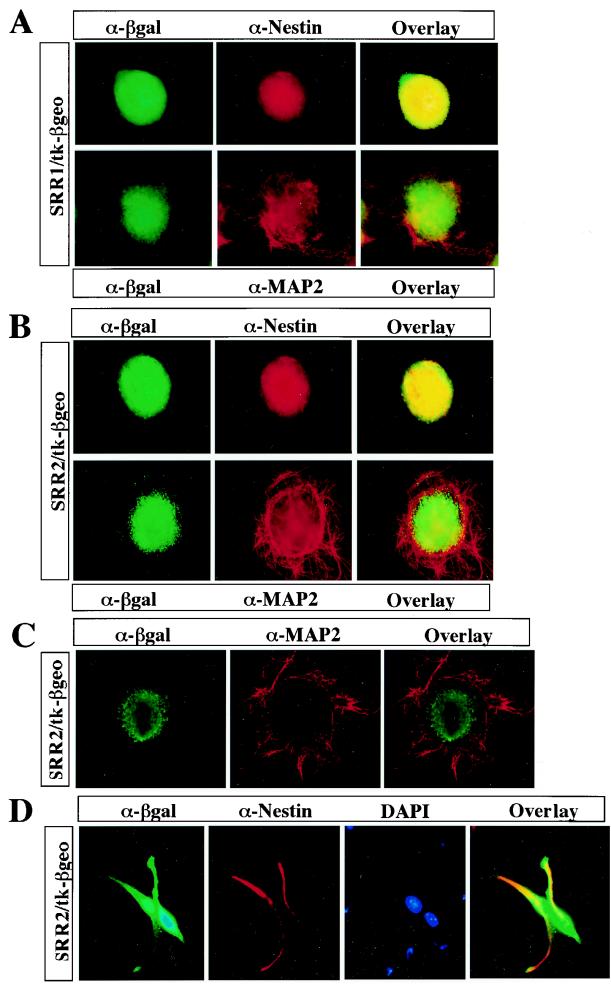
Function of SRR1 and SRR2 in ES cells subjected to neural differentiation. ES cells carrying SRR1 (A) or the SRR2/tk-β-geo transgene (B) were subjected to neural differentiation using retinoic acid as described by Bain et al. (5). These cells were then cultured on coverslips coated with PDL and laminin for 2 days. After fixation, these cells were immunostained with a combination of two stains, either nestin-β-Gal or MAP2-β-Gal. (C) The ES cells carrying the SRR2/tk-β-geo transgene were subjected to neural differentiation and immunostained with MAP2-β-Gal as described for panel B. These cells were then inspected with a confocal microscope. (D) The ES cell-derived neural cells bearing the SRR2/tk-β geo transgene were dissociated to the single-cell level, transferred to coverslips coated with PDL and laminin, and immunostained with nestin-β-Gal. α, antibody.
For constructing the ptk-Venus reporter plasmids shown in Fig. 3A, the second intron enhancer of the rat nestin gene (+1162 to +1798) (17) was amplified by PCR. As for SRR1 and SRR2, the same DNA fragments used to obtain the results shown in Fig. 2 were used. These regulatory regions were subcloned into the SalI/NcoI site of Venus/pCS2 (28), which carries the Venus reporter gene together with the tk promoter with the aid of linkers. Venus is a modified form of enhanced yellow fluorescent protein bearing a unique set of amino acid substitutions (F46L, F64L, M153T, V163A, and S175G) and exhibits enhanced fluorescence because of these mutations (28). For constructing EF1-Venus, the EF1 promoter region was recovered from the pEF-BOS vector (26) and subcloned into the SalI/NcoI site of Venus/pCS2. The internal deletion and nucleotide substitution mutants of SRR2 shown in Fig. 7A were all obtained by PCR-based procedures in which mutOct and mutSox carried the same mutations of triple-point mutants 19 and 16, respectively, which were described previously (48). For constructing the 4×CORE plasmid, the SRR2 core sequence, 5′-GGCAGCCATTGTGATGCA TATGGATTA-3′, was multimerized to four copies according to the method of Nishimoto et al. (30) and subcloned into the SalI/NcoI site of the Venus reporter together with the tk promoter.
FIG. 3.
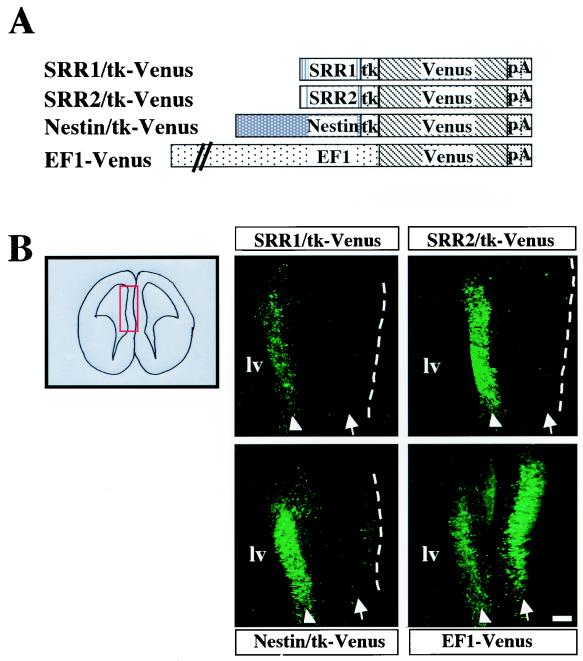
Characterization of SRR1 and SRR2 activities in the developing brain by in vivo electroporation. (A) Reporter plasmids used for in utero electroporation. pA represents the poly(A) signal from simian virus 40. (B) SRR1 and SRR2 support rather specific reporter gene expression in the ventricular zone of the developing brain. The reporter plasmids were electroporated into E13.5 mouse brain. The red rectangle represents the brain portion in which the reporter plasmids were introduced. After 4 days, brains were recovered from fetuses and sliced with a vibratome, and reporter gene expression was inspected. The dorsal portion is to the top, while the medial portion is to the right. Arrowheads and arrows indicate the ventricular zone and cortical plate of the brain, respectively, while dotted lines correspond to the outer surface of the brain. lv, lateral ventricle. The white bar corresponds to 100 μm.
FIG. 7.
The same core sequence of SRR2 is involved in its enhancer activities in ES cells and neural stem or progenitor cells. (A) Schematic representation of wild-type (WT) and various types of mutants of SRR2. “del.” followed by a number indicates a mutant regulatory region with a deletion. The Sox- and Oct-like sequences are indicated as filled and open circles, respectively. The open and filled boxes represent noncoding and coding regions of the Sox-2 gene, respectively. Numbers represent the positions where the adenine nucleotide of the transcription initiation codon is set to +1. These DNA fragments were subcloned into the ptk-Venus reporter plasmid. (B) The Oct- and Sox-like elements of SRR2 are required for its enhancer activity in neural stem cells. The Venus reporter plasmids shown in panel A were introduced into E13.5 mouse brain together with an internal control DsRed reporter gene, which is connected to the chicken β-actin promoter. After 48 h, Venus and DsRed reporter gene expression was inspected. The orientation of sectioned brains was the same as that in Fig. 3B. (C) All of the POU III class octamer factors expressed in brain show the potential to augment SRR2 activity together with Sox-2. COS cells were transfected with 0.07 μg of octamer factor expression vectors and with increasing amounts of Sox-2 vector, as indicated, as well as tk-Luc reporter plasmid (0.7 μg) bearing SRR2. An internal control luciferase gene (0.7 μg) of Renilla reniformis were also transfected. The transcriptional level was estimated as described in Materials and Methods. Data were obtained from five independent experiments with comparable results.
For constructing the puro-Venus reporter gene, coding regions of these two protein were fused with the aid of a linker and subcloned into the plasmid bearing the polyadenylation sequence from the PGK gene (S.M. and H.N., unpublished data). Subsequently, various regulatory regions used to obtain the results shown in Fig. 4A and B were individually subcloned into the vector.
FIG. 4.
SRR1 and SRR2 do not function in trophoblast and hematopoietic stem cells. (A) Functional analyses of SRR1 and SRR2 in ES cells and trophoblast stem cell-like cells. The puro-Venus reporter genes bearing the indicated regulatory regions were introduced to ZHBTc4 ES cells (32), and puromycin-resistant transformants were obtained. These ES cells were then converted to cells with trophoblast stem cell characteristics as described by Niwa et al. (32). Typical examples of stable ES cell transformants and cells converted to trophoblast stem cell-like cells were shown under fluorescence microscopy. The latter cells were also counterstained with DAPI (4′,6′diamidino-2-phenylindole). Arrows indicate ES cell-derived trophoblast stem cell-like cells with a regular epithelial cell morphology (44). TS,trophoblast stem cell-like cells. (B) Expression level of puro-Venus protein in ES cells and trophoblast stem cell-like cells. Whole-cell extracts were prepared from various ES cells and trophoblast stem cell-like cells, and Western blot analyses were performed using anti-GFP antibody, which recognizes the Venus portion of the fusion protein. (C) SRR1 and SRR2 fail to display their enhancer activities in hematopoietic stem cells. The hematopoietic stem cell-enriched CD34− KSL cell population from mouse bone marrow was prepared according to the method of Osawa et al. (33) and transduced with the lentiviruses shown in Fig. 1A. After 48 h, the production of Venus reporter protein was inspected.
All expression vectors of octamer factors and the Sox-2 protein were constructed by subcloning cDNAs carrying entire coding regions of these proteins into the EcoRI site of the pCAG vector (31), while construction of the SRR2/tk-luciferase reporter gene was described previously (48).
ES cell culture and transfection.
E14 ES cells were cultured as described previously (29). The β-geo reporter plasmid bearing SRR1 or SRR2 was introduced into ES cells by electroporation according to the method of Thomas and Capecchi (46). After selection with G418, the drug-resistant clones were picked and expanded for subsequent analyses.
In vitro differentiation of ES cells.
Neural differentiation of ES cells was performed essentially as described by Bain et al. (5). Briefly, E14 ES cells were cultured as usual on a feeder layer with leukemia inhibitory factor-supplemented medium. Subsequently, embryoid bodies were generated by culturing cells in bacterial-grade dishes. These embryoid bodies were maintained as a suspension culture for 8 days, and cells were exposed to all-trans-retinoic acids (0.5 μM) for the last 4 days. Subsequently, cell aggregates were plated onto tissue culture dishes precoated with poly-d-lysine (PDL) and laminin and cultured for another 2 days before being subjected to immunohistochemical analyses. Establishment and culture of trophoblast stem cells by cultivating ZHTc4 ES cells with FGF-4, heparin, and tetracycline were done as described by Niwa et al. (32).
Neurosphere culture.
Neurospheres were generated from an embryonic day 13.5 (E13.5) or E17.5 mouse forebrain according to the method of Reynolds et al. (38), with slight modifications utilizing B-27 supplement (Invitrogen), 20 ng of epidermal growth factor (Becton Dickinson) per ml, and 20 ng of basic fibroblast growth factor (Roche) per ml in place of a defined hormone mix and salt mixture.
Generation of lentivirus vectors and infection of neurosphere and hematopoietic stem cells.
For constructing self-inactivating vector plasmids used to obtain the results shown in Fig. 1 and 4C, pCS-CDF-CG-PRE, a modified form of pCS-CG-PRE (43), was used as starting material. pCS-CDF-CG-PRE carries a polypurine tract which increases the efficiency of infection (H. Miyoshi, unpublished data). First, the cytomegalovirus promoter and green fluorescent protein (GFP) cDNA portions were removed from the plasmid, and Venus reporter cassettes connected to one of the regulatory regions shown in Fig. 1 were then subcloned. For the UTF1 regulatory region, the genomic DNA region from +991 to +2041 (29) was used, while for all other regulatory regions, the same portions used for constructing Venus reporter plasmids were used. The production of pseudotyped human immunodeficiency virus type 1-based lentivirus possessing vesicular stomatitis virus G protein (VSV-G) was carried out according to the method of Miyoshi et al. (25) in which the VSV-G protein allows the virus to infect mammalian cells in general (for details, see references 24 and 25). The vector titers were determined as described by Tahara-Hanaoka et al. (43).
FIG. 1.
SRR1 and SRR2 show enhancer activities equivalent to that of the nestin regulatory region in neurospheres. (A) Schematic representation of the self-inactivating vector (pCS-CDF-CG-PRE) and Venus reporter genes bearing distinct regulatory regions. The exact coordinate of each regulatory enhancer in the original gene locus is described in Materials and Methods. Abbreviations used are as follows: CMV RU5, the 5′ long terminal repeat of the human immunodeficiency virus in which the U3 enhancer-promoter region is replaced by the cytomegalovirus promoter; 5′SD, splice donor site; RRE, rev responsive element; 3′SA, splice acceptor site; cPPT, central polypurine tract; CTS, central termination sequence; PRE, woodchuck hepatitis virus posttranscriptional regulatory element; and delURU5, 3′ long terminal repeat lacking the enhancer-promoter portion of U3. (B) Functional analyses of SRR1 and SRR2 in forebrain-derived neurospheres. At 72 h postinfection with lentiviruses, reporter gene expression in neurosphere cells was examined under a fluorescence microscope as well as in bright fields (Bf). (C) Both SRR1 and SRR2 show the enhancer activities rather specifically in neural stem or progenitor cell populations. Neurosphere cells were infected with lentiviruses as described for panel B and maintained in the multipotent state or induced to differentiate with FBS. These cells were then dissociated to the single-cell level, and Venus expression was quantitated on a FACSCalibur device. The green portion and the orange line correspond to data obtained from a neural stem or progenitor cell population and differentiation-induced cells, respectively. The numbers in vertical and horizontal axes represent cell counts and Venus fluorescence intensity, respectively.
For infecting lentivirus, neurospheres were dissociated and immediately infected with the virus at a multiplicity of infection of 1.0. Under this condition, about 70% of neurosphere cells were infected. These cells were cultured for another 3 days in serum-free medium containing appropriate growth factors so that the cells could maintain a multipotent state as described above or in medium containing 10% fetal bovine serum (FBS).
Murine hematopoietic stem cells with a CD34−/low c-Kit+ Sca-1+ Lin− phenotype (CD34− KSL) were prepared from bone marrow cells according to the method of Osawa et al. (33). Briefly, bone marrow cells were obtained from the tibias and femurs of C57BL/6 mice. These bone marrow cells were overlaid with sodium metrizoate, and low-density cells were harvested. Lineage-positive cells were removed from these cells by utilizing biotinylated antilineage markers (Mac1, Gr-1, B220, CD4, CD8, and TER119), and CD34−/low c-Kit+ Sca-1+ cells were sorted by fluorescence-activated cell sorting with a Vantage SE (Becton Dickinson). These hematopoietic stem cell-enriched cells were transduced with lentivirus bearing the Venus reporter gene at a multiplicity of infection of 300 as described by Tahara-Hanaoka et al. (43). Under these conditions, about 50% of CD34− KSL cells were infected.
In utero electroporation.
Gene transfer into the developing mouse brains and subsequent analyses were done as described by Saito and Nakatsuji (40). ICR strain mice were used for the analyses. Data shown in Fig. 7B were obtained with coinjection of one of the Venus reporter plasmids shown in Fig. 7A with an internal control DsRed reporter plasmid in which DsRed expression is supported by the chicken β-actin promoter (31). In all cases, 3 μl of solution containing 0.1 pmol of DNA was injected.
Immunostaining.
Indirect immunocytochemistry was carried out with cells that had been cultured on coverslips (for in vitro-differentiated ES cells) or in slide chambers (for clonally grown neurospheres) coated with PDL and laminin. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature. For immunohistochemical analyses, brains were recovered from 17.5-day-postcoitum (d.p.c.) embryos in which the SRR2/tk-Venus reporter had been introduced at 13.5 d.p.c. by in utero electroporation and embedded in OCT materials after fixation with 4% paraformaldehyde. The frozen sections (thickness, 20 μm) of fetal brains were incubated with anti-nestin, MAP2, or phosphohistone H3 antibody together with anti-GFP antibody. Immunostaining was performed as described by Saba et al. (39) with appropriate Alexa Fluor dye-conjugated secondary antibodies from Molecular Probes.
Antibodies.
The following antibodies were used for immunostaining analyses: anti-Sox-2 (rabbit immunoglobulin G [IgG]; CHEMICON), anti-phosphohistone H3 (clone 6G3; Cell Signaling Technology), anti-MAP2 (clone HM-2; Sigma), anti-glial fibrillary acidic protein (anti-GFAP) (rabbit IgG; Sigma), O4 (clone 81; CHEMICON), antinestin (clone Rat401; BD PharMingen), anti-GFP (rabbit antiserum; MBL), and anti-β-galactosidase (anti-β-Gal) (rabbit IgG; Cappel). These antibodies were diluted appropriately according to the suppliers' recommendations.
Luciferase assay.
COS cells in 60-mm-diameter dishes were transfected by lipofection using lipofectamine 2000 (Invitrogen) with the amounts of reporter and expression vectors indicated in the legend to Fig. 7. The total amount of DNA was adjusted to 8 μg with pUC18. After 48 h of transfection, transcription levels were determined by the dual-luciferase system according to the instructions of the manufacturer (Promega).
RESULTS
SRR1 and SRR2 exert their activities in forebrain-derived neurosphere cells.
The Sox-2 gene is expressed in various types of stem cells, including ES cells and neural stem cells (4, 35, 56, 57). Since two regulatory regions, SRR1 and SRR2, which are involved in the expression of the Sox-2 gene in pluripotent ES cells, have already been identified (48), we examined whether these regulatory regions also work in multipotent neural stem cells. We first produced lentiviruses carrying the Venus reporter gene, which encodes a fluorescent protein that exhibits enhanced fluorescence (28), and one of the various regulatory regions shown in Fig. 1A. We then used these viruses to infect cells from neurosphere colonies which had been generated from the forebrains of 13.5-d.p.c. mouse embryos. Culture continued for another 72 h. As expected, the EF1 promoter and nestin enhancer, the best-characterized neural stem cell- or progenitor cell-specific enhancer (17), were active and drove the transcription of the Venus reporter gene in the majority of neurosphere cells (Fig. 1B). More importantly, a substantial number of cells also became Venus positive when these cells were infected with viruses bearing SRR1 or SRR2, whereas the tk promoter alone or the ES cell-specific regulatory element of the UTF1 gene (29) did not show any obvious effect. To evaluate the strength of these regulatory regions on reporter gene expression, these neurosphere cells were dissociated and their fluorescent intensity was quantitated by fluorescence-activated cell sorting analysis. These analyses revealed that SRR1 and SRR2 display activities equivalent to that of the nestin enhancer (Fig. 1C). From these results, we concluded that both SRR1 and SRR2 are able to function in neural stem cell-enriched neural cells. We also confirmed that these regulatory regions, like the nestin enhancer, do not have a prominent effect on reporter gene expression when cells were cultured under differentiation-inducing conditions (Fig. 1C), indicating that both SRR1 and SRR2 display their activities rather specifically in multipotent neural stem and progenitor cell populations.
SRR1 and SRR2 direct β-geo reporter gene expression in nestin-positive cells derived from ES cells.
To further characterize the specificities of SRR1 and SRR2 activities in neural cells, we took advantage of their activities in pluripotent ES cells. That is, we made two tk promoter/β-geo reporter constructs in which either SRR1 or SRR2 is connected to the promoter. These plasmids were introduced into ES cells by electroporation, and stable transformants were obtained. These cells were subjected to neural differentiation in which cells are cultured as embryoid bodies for 4 days in the absence of retinoic acids and then cultured for another 4 days in the presence of retinoic acids (see Materials and Methods). It should be noted that ES cell-specific enhancers, such as that of the UTF1 gene, lost their entire activity during this procedure and that the activities of both SRR1 and SRR2 also profoundly diminished within the first 4 days of this procedure (data not shown). We then examined whether SRR1 and SRR2, whose activities decreased in the beginning of the process, were reactivated when complete neural differentiation procedures were applied to the cells. We performed immunological staining procedures with these differentiation-induced cells to compare the expression profiles of β-Gal directed by SRR1 or SRR2 and endogenous neural-lineage cell markers. As shown in Fig. 2A, we found that the expression of β-Gal in cells in which SRR1 directed β-Gal protein production significantly overlapped that in cells possessing nestin, but the protein was not detected in cells with MAP2, one of the markers for postmitotic neurons. We obtained essentially the same results with SRR2 (Fig. 2B). Moreover, as shown in Fig. 2C, we confirmed the completely mutually exclusive patterns of expression of β-Gal directed by SRR2 and MAP2 with the aid of confocal microscopy. To determine the extent of coexpression of the β-Gal and nestin proteins more precisely, cells bearing the SRR2/β-Gal transgene were disaggregated after induction of neural differentiation and then immunostained. A representative example is shown in Fig. 2D. We found that about 96% of β-Gal-positive cells were also positive for nestin. Likewise, 94% of β-Gal-positive cells were positive for nestin when cells bearing the SRR1/β-Gal transgene were used (data not shown). Thus, these results also indicate that both SRR1 and SRR2 exert their functions rather specifically in neural stem and progenitor cells but not in postmitotic neurons.
SRR1 and SRR2 display their enhancer activities rather specifically in cells located in the ventricular zone of the developing brain.
Next, we examined whether SRR1 and SRR2 are able to function in developing brains of mouse embryos. We employed in utero electroporation, by which DNAs injected into the lateral or third ventricle of the mouse embryo brain are efficiently delivered into brain cells (40). We introduced the Venus reporter plasmids shown in Fig. 3A into the brains of 13.5-d.p.c. mice and maintained them in the uterus. Four days after electroporation, the brains were recovered, sliced with a vibratome, and viewed under a fluorescence microscope. By this method, DNA is delivered exclusively into cells facing the ventricle, which are mainly neural stem or progenitor cells. While neural stem cells make cell division in the ventricular zone, some of their descendants, postmitotic neurons, migrate into the cortical plate. As expected, many cells in both the cortical plate and the ventricular zone were labeled by the transfection of EF1-Venus, in which process Venus expression is driven by the generally active EF1 promoter. In contrast, the transfection of SRR2/tk-Venus labeled mainly the ventricular zone and only a limited number of cells in the cortical plate, indicating that the transcriptional stimulating activity of SRR2 in neural stem or progenitor cells declined abruptly when cells differentiated and migrated to the cortical plate portion. This assumption was supported by the fact that Nestin/tk-Venus also gave essentially the same Venus expression pattern. SRR1 also supported ventricular-zone-specific Venus expression, albeit rather weakly compared with what occurred with SRR2 and the nestin enhancer. As expected, cells with the tk promoter alone and those with the UTF1 enhancer could not produce appreciable amounts of Venus-positive cells in these assays (data not shown).
SRR1 and SRR2 do not exert their enhancer activities in trophoblast and hematopoietic stem cells.
Sox-2 expression is not restricted to ES cells and neural stem cells; it is also expressed in trophoblast stem cells (4, 55). Moreover, it has been shown that the protein is required for the development of trophoblastic stem cell-derived extraembryonic ectodermal tissues (4). Therefore, we examined whether SRR1 and SRR2 can also function in the trophoblast stem cells. To address this issue, we used a genetically manipulated ES cell line, ZHBTc4, in which the Oct-3/4 gene shows tetracycline-regulated expression. This ES cell line can be easily converted to cells with trophoblast stem cell characteristics by culturing in the presence of FGF-4, heparin, and tetracycline but in the absence of leukemia inhibitory factor (32, 44). The SRR1, SRR2, UTF1, and EF1 regulatory regions were individually connected to the puro-Venus reporter gene, which encodes a fusion protein of Venus and puromycin-detoxifying enzyme (see Materials and Methods). Subsequently, these reporter genes were introduced to the ES cells by lipofection, and stable puromycin-resistant transformants were obtained based on these enhancer activities in pluripotent ES cells. The activities of these regulatory regions in the ES cells were evident not only by the puromycin-resistant phenotype but also from the fluorescence of the fusion protein (Fig. 4A). However, when these cells were converted to trophoblast stem cell-like cells, fluorescence from the fusion protein was concomitantly extinguished in all cases except in the cells bearing the EF1-puro-Venus reporter gene. We also examined reporter gene expression by Western blot analysis using an anti-GFP polyclonal antibody which recognizes Venus. Consistent with the data shown in Fig. 4A, none of the regulatory regions except for the EF1 promoter were able to contribute to the production of a detectable amount of the fusion protein in these trophoblast stem cell-like cells, although all of these regulatory regions were able to support reporter gene expression in ES cells (Fig. 4B). It should be noted that a high level of endogenous Sox-2 gene expression is detected in these cells (data not shown). Thus, these results postulate that both SRR1 and SRR2 fail to support gene expression in trophoblast stem cells and that Sox-2 expression in these cells is supported by a distinct regulatory region(s).
We next examined the possible function of SRR1 and SRR2 in hematopoietic stem cells. Although the Sox-2 gene appears not to be expressed in hematopoietic stem cells (36), there is the possibility that these isolated regulatory regions somehow function in these cells. For this purpose, we transduced the reporter genes into CD34− KSL cells, which are a highly enriched cell population for hematopoietic stem cells (33), using the same set of VSV-G pseudotype lentiviruses used to obtain the results shown in Fig. 1. Only the EF1 promoter exerted its activity in these hematopoietic cells, whereas all of other regulatory regions did not show a detectable effect on the expression of the reporter gene in these cells (Fig. 4C). Thus, together with the fact that both SRR1 and SRR2 fail to function in trophoblast stem cells, these regulatory regions do not promiscuously show their transcription-stimulating activities in stem cells.
Immunohistochemical analyses of cells in which SRR2 functions in the developing brain.
For subsequent analyses, we decided to concentrate on SRR2 in the developing brain because we have already systematically analyzed the molecular basis of SRR2 activity in ES cells (48). That is, we believe that similar systematic analyses of the region in neural stem and progenitor cells may allow us to compare for ES and neural stem cells the molecular bases of SRR2-mediated transcriptional stimulating activity.
Based on these considerations, we first examined molecular aspects of the cells in which SRR2 functions in the developing brain. After electroporation with the SRR2/tk-Venus reporter plasmid, brain slices were immunostained for Venus protein together with antibody for nestin or MAP2 protein. We also examined the endogenous Sox-2 protein by the same procedure. As shown in Fig. 5A, endogenous Sox-2 was present rather predominantly in the ventricular zone of the 17.5-d.p.c. embryonic brain. Similarly, SRR2 directed Venus reporter expression rather specifically in the ventricular zone (Fig. 5B). Anti-nestin antibody revealed that the expression of Venus and the expression of nestin overlapped in the ventricular zone (Fig. 5B to D; magnified views are shown in panels E to G). To determine this coexpression conclusively, cells were recovered from developing brains after electroporation of the reporter plasmid. These cells were then transferred to coverslips and immunostained for Venus and nestin. Representative data are shown in Fig. 5H to N. These analyses revealed that about 92% of Venus-positive cells were nestin positive. We assume that cells which were positive for nestin but negative for Venus in most cases represent cells that were not transfected with the reporter gene. We also compared the levels of expression of Venus and MAP2 and found that the expressions of these proteins were mutually exclusive (Fig. 5O to Q). Analyses of the expression of Venus and phosphohistone H3, a specific marker for mitotic cells revealed that some of the Venus-positive cells were also positive for phosphohistone H3 (Fig. 5R to T). Again, cells which were positive for phosphohistone H3 but negative for Venus were assumed to be nontransfected cells. In any event, from these results, we conclude that, as in in vitro-cultured cells, SRR2 exerts its function mainly in rapidly proliferating nestin-positive neural stem or progenitor cells and not in MAP2-positive postmitotic neurons in the developing brain.
FIG. 5.
SRR2 exerts its enhancer activity in neural stem or progenitor cells but not in postmitotic neurons in developing brains. Sections from normal E17.5 mouse brain (A) or brains in which the SRR2/tk-Venus reporter gene had been introduced by in vivo electroporation (B to G and O to T) were immunostained. (H to N) Cells were recovered from brains after incorporation of the reporter plasmid by in utero electroporation, transferred to a coverslip, and immunostained. The antibodies (α) used are indicated at the top of each panel. (E to G) Magnified views of panels B to D. The orientation of the brain sections is the same as that shown in Fig. 3B. White bars in panels B, E, O, and R correspond to 100 μm. Arrows in panel T indicate cells which are doubly positive for GFP and phosphorylated histone H3. lv, lateral ventricle.
From the homogeneous expression profile of the Venus reporter gene in the ventricular zone of the embryonic brain, it is obvious that SRR2 exerts its activity in neural progenitor cells. However, none of the above-described analyses unequivocally demonstrate that SRR2 functions in multipotent neural stem cells. Thus, we performed a clonogenic analysis to examine whether SRR2 is also active in neural stem cells. By this method, neurospheres were generated from fetal brains in which the SRR2/tk-Venus reporter gene had been introduced by in vivo electroporation and plated at clonal density. Then, a spherical colony generated from a single Venus-positive cell shown in Fig. 6A was recovered. By following the same protocol, we also prepared neurospheres from healthy developing brains in which no DNA had been introduced and compared the self-renewal activities of neurospheres derived from nonselected control cells and those derived from a single cell in which SRR2 functioned. As shown in Fig. 6B, these analyses revealed that the cells in which SRR2 functioned showed activity comparable to that of control neurosphere cells in terms of their ability to produce secondary neurospheres. We next examined multipotent properties of the cell in which SRR2 functioned. After trypsinization and expansion of neurospheres derived from a single Venus-positive cell, immunostaining analyses were performed after induction of differentiation with FBS. As shown in Fig. 6C to E, these cells generated both MAP2-positive neurons and GFAP-positive astrocytes upon differentiation. Figure 6F to H show that neurosphere cells derived from the same single cell generated O4-positive oligodendrocytes as well as GFAP-positive astrocytes. Thus, these results confirmed that the cells in which SRR2 functioned gave rise to all three different neural lineages when cells were induced to differentiate. From these results, we conclude that at least a portion of cells showing SRR2-dependent Venus expression in the developing brain are multipotent neural stem cells.
FIG. 6.
Cells in which SRR2 functions were converted to all three different neural lineages (neuron, astrocyte, and oligodendrocyte) upon differentiation. (A) Preparation of neurospheres generated from a single cell in which SRR2 functions. Neurospheres prepared from SRR2/tk-Venus-transfected brains were plated at clonal density with serum-free medium containing basic fibroblast growth factor and epidermal growth factor for 7 days, and neurospheres derived from single Venus-positive cells were selected. (B) Self-renewal activity of cells in which SRR2 functions. Neurospheres of similar size (about 0.2 mm in diameter) derived from a single Venus-positive cell were individually dissociated and recultured. The number of generated colonies was counted under a microscope. The data were obtained from 16 independent colonies. The neurospheres derived from nontransfected brain were used as a control. (C to H) Multipotent properties of cells in which SRR2 functions. The expanded neurospheres derived from a single cell in which SRR2 functions were dissociated, split into two wells of a slide chamber coated with PDL and laminin, and cultured in medium containing 1% FBS. After 7 days, cells were fixed and subjected to immunostaining procedures. One of the chambers was stained with anti-GFAP and anti-MAP2 antibodies (C to E), while the other was stained with anti-GFAP antibody and O4 antibody (F to H). Venus protein is not present in the differentiation-induced cells at a detectable level (data not shown), and the green fluorescent color in panels C and F is exclusively due to Alexa Fluor 488 dye-conjugated secondary antibody bound to cells via the GFAP-anti-GFAP antibody complex. α, antibody.
The same core sequence of SRR2 is involved in gene expression in ES cells and neural stem or progenitor cells.
To localize the SRR2 core sequence involved in gene expression in neural stem or progenitor cells, we made a series of internal-deletion mutants of SRR2 connected to the Venus reporter gene (Fig. 7A). Subsequently, these plasmids were individually introduced into the 13.5-d.p.c. mouse brains by in utero electroporation. After 48 h, brains were recovered and Venus expression was examined as described for Fig. 3B. These analyses revealed that the transcription-stimulating activity of SRR2 was profoundly impaired in the del.9 and del.10 mutants but that all of the other mutants showed activities equivalent to that of wild-type SRR2 (Fig. 7B). It should be noted that deleted regions of these mutants encompass the octamer and Sox-2 binding site-like sequences which have been shown to play a critical role in the enhancer activity of SRR2 in ES cells (48). To examine the possible involvement of the octamer and Sox-2 binding site-like sequences in SRR2 in the neural stem or progenitor cell population, we made two different nucleotide substitution mutants, mutOct and mutSox, in which octamer and Sox-2 site-like sequences were impaired so as not to serve as octamer factor and Sox binding sites, respectively (see Materials and Methods). The transcription-stimulating activities of these SRR2 mutants were analyzed in developing brains as described above. These analyses revealed that both of these mutants failed to exhibit significant activities in this system, indicating that, as in the ES cells, both the octamer and Sox-2 binding site-like sequences play a pivotal role in supporting the enhancer activities of SRR2 in developing brains. Moreover, these octamer and Sox-2 binding site-like sequences, when multimerized to four copies, were sufficient to support ventricular-zone-restricted reporter gene expression in developing brains.
We have previously demonstrated that the Oct-3/4-Sox-2 complex makes a major contribution to SRR2 activity in ES cells (48). Because the same or at least overlapping regulatory sequences are involved in SRR2 activity in neural stem or progenitor cells, it is possible to assume that a similar protein complex(es) supports SRR2 activity in the brain. However, Oct-3/4 protein is not present in the brain, although Sox-2 is present in neural stem or progenitor cells. Therefore, it is conceivable that certain other octamer factors present in the developing brain may contribute to SRR2 activities in this tissue. We note from the literature (1, 9, 13, 16, 23, 41, 42) and microarray analyses (N. Masuyama, S. Miyagi, A. Okuda, and Y. Gotoh, unpublished data) that four different octamer factors, i.e., Brn-1, Brn-2, Brn-4, and Oct-6, in addition to the widely expressed Oct-1 factor, are present in neural stem or progenitor cells. Moreover, our analyses revealed that, except for Oct-1, all of these factors were able to bind to the SRR2 sequence together with Sox-2 in gel shift DNA binding assays (data not shown). Therefore, we examined the potential involvement of these octamer factors and the Sox-2 protein in SRR2 activity. To this end, we introduced a luciferase reporter gene bearing wild-type SRR2 together with expression vectors of octamer factors and Sox-2 by transient transfection into COS cells which were devoid of endogenous Sox-2 protein. As shown in Fig. 7C, increasing the amount of the Sox-2 expression vector boosted the level of transcription especially when a certain amount of octamer factor expression vectors were cotransfected. Indeed, except for Oct-1, all other octamer factors present in the brain showed activities equivalent to or even higher than that of Oct-3/4. The observed transcriptional activation is SRR2 dependent, because no elevation of transcriptional level was detected when the experiments were done with the reporter gene which lacks SRR2 (data not shown). Thus, these results indicate that all of these octamer factors except for Oct-1 have the potential to contribute to SRR2 activity in the developing brain, although we cannot eliminate the possibility that a novel neural stem or progenitor cell-specific octamer factor or another type of transcription factor(s) that contributes to SRR2 activity in the brain remains to be identified.
DISCUSSION
The Sox-2 gene is known to be expressed in neural stem or progenitor cells as well as in ES cells (4, 6, 56, 57). In the present study, we have demonstrated that two Sox-2 regulatory regions, SRR1 and SRR2, which were previously identified based on their activities in pluripotent ES cells, also function in neural stem or progenitor cell populations. There is a precedent for a regulatory region which supports gene expression in different tissues through the same DNA core element. Indeed, it has been demonstrated that the mafK gene encoding one of the small Maf proteins possesses an enhancer that functions in both cardiac muscle and hematopoietic cells and that distinct sets of GATA transcription factors are involved in each tissue to activate the enhancer (18). Thus, the transcriptional regulatory mechanism described in the present study is not restricted to stem cells but may operate in many aspects of development.
About the regulatory element involved in neural Sox-2 expression, Zappone et al. (57) have previously reported the identification of the specific region acting as an enhancer in the developing telencephalon. Now we know that SRR1 activity in ES cells and the telencephalon-specific enhancer activity which Zappone et al. identified are defined by a single regulatory region, i.e., SRR1. Moreover, as with SRR2, SRR1 exerts its activity in ES cells and neural stem or progenitor cells in a similar manner by utilizing the common core sequence in which the octamer-like sequence also plays a central role (S. Nicolis, personal communication).
The activity of SRR2 in the developing brain has been examined for the first time in this study. We found that SRR2 was able to function in neural stem or progenitor cell population by experiments using neurospheres and also by neural differentiation of ES cells. The in utero electroporation analyses further corroborate and extend the results obtained with the in vitro culture systems. Indeed, these analyses clearly demonstrate that SRR2 is able to display its activity in neural stem or progenitor cells in the developing brains of mouse embryos. This system also allowed us to perform clonogenic analyses for demonstrating that at least a portion of cells in which SRR2 functions in the developing brain are multipotent neural stem cells. Zappone et al. (57) had previously shown that deletion of a DNA region carrying SRR1 resulted in the loss of the expression of the β-geo reporter gene, which was integrated into the Sox-2 gene locus by homologous recombination in the telencephalic portion of developing brain. These results may cast doubt on the function of SRR2 in this portion of brain. However, this doubt should be banished. Because of the vector design, Zappone et al. deleted SRR2 as well as SRR1 during integration of the reporter gene in the Sox-2 locus. This happened because SRR2, which is located in close proximity to the coding region of the gene, was not identified when they had done the analyses. Therefore, the loss of reporter gene expression in the telencephalon which Zappone et al. demonstrated is the consequence of the loss of both SRR1 and SRR2. Moreover, it should be noted that transgenic analyses revealed that, like SRR1, SRR2 functioned as a telencephalon-specific regulatory region (S.M. and A.O., unpublished data). Thus, we assume that both of them play at least a certain role in supporting the high level of Sox-2 gene expression in the developing brain, although we do not know at present whether either one of them plays a more prominent role than the other.
Sox-2 is not the sole protein which is expressed in both ES and neural stem cells. Recent microarray technology revealed the significant similarity between these stem cells at the transcriptional level (36), although significant genetic dissimilarities have also been documented (8). One prominent characteristic shared by ES cells and neural stem cells is the ability to propagate in tissue culture systems without significantly losing multipotent properties, although neural stem cells would not grow indefinitely in vitro (22, 53). On the other hand, in the case of most other types of somatic stem cells, significant spontaneous differentiation occurs during the expansion of cells in vitro, and hematopoietic stem cells are one of the typical examples (3, 37). Therefore, it is possible to speculate that similar genetic regulatory networks operating in ES cells and neural stem cells are involved in sustaining the common biological property of these stem cells. Alternatively, as discussed by Ramalho-Santos et al. (36), this global overlap in expressed genes is simply due to the result of conversion of embryonic ectodermal cells to neural cells by a default mechanism (14). In any event, it is tempting to speculate that these commonly expressed genes also possess regulatory regions which are similar to SRR1 or SRR2 in sequences and/or element organizations.
Detailed analyses for characterizing the SRR2 core sequence revealed that the same or at least overlapping sequences were involved in its activity in ES cells and in the developing brain. For ES cells, the fact that SRR2 activity is mostly defined by the Oct-3/4-Sox-2 complex has previously been demonstrated (48). Although Sox-2 is present in the developing brain, Oct-3/4 is essentially not present in this tissue. Therefore, we assume that similar protein complexes, such as the Brn-1-Sox-2 complex, contribute to SRR2 activities in the developing brain. Indeed, we have demonstrated that POU III class octamer factors present in brain, such as Brn-1 and Brn-2, show the potential to augment SRR2 activity together with that of Sox-2 in heterologous cells. We think that this Sox-2-mediated gene regulation with the aid of certain octamer factors in neural stem cells is particularly interesting. In fact, a large amount of data underscores the importance of the Oct-3/4-Sox-2 complex for maintaining the pluripotent state of ES cells by controlling the expression of many genes in ES cells (2, 4, 29, 47, 48, 56). Therefore, the presence of transcriptional regulation by similar protein complexes in neural stem cells indicates that these complexes also play a crucial role in preventing neural stem or progenitor cells from differentiating into postmitotic cells. It is noteworthy that essentially the same set of octamer factors are also shown to be involved in supporting the expression of genes for nestin and brain fatty acid binding protein, which are, like Sox-2, preferentially expressed in neural stem or progenitor cells in the brain (17). Thus, these results indicate that the requirement of these octamer factors is the general mechanism for restrictive gene expression in neural stem or progenitor cell populations. However, these octamer factors should not be thought sufficient to produce a stem or progenitor cell-specific expression profile in the developing brain; rather, they require some other factors' functions since these octamer factors by themselves are not exclusively expressed in neural stem or progenitor cells but are also expressed in subsets of differentiated cells (for details, see references 23 and 42). In this context, we assume that the specific activity of SRR2 in the stem or progenitor population is specified mainly by the Sox-2 protein by itself.
As described above, we have characterized the regulatory elements which support specific gene expression in these two distinct stem cells. Moreover, we have demonstrated that, at least for SRR2, transcription is activated by a common core sequence in ES and neural stem or progenitor cells. We hope that the data presented here may lead to the unraveling of the broader aspect of a common regulatory network which defines the nature of the stem cell state of ES cells and neural stem cells and possibly stem cells in general.
Acknowledgments
We are indebted to Atsushi Miyawaki, Hiroyuki Miyoshi, and Yoshinobu Sugitani for providing the Venus/pCS2, pCS-CDF-CG-PRE, and Brn-1, -2, and -4 vectors, respectively. We also thank Shinji Sasaki and Shinji Hirotsune for technical advice about immunohistochemical analyses and helpful discussion.
This work was supported in part by the Ministry of Education, Science, Sports, and Culture and was especially supported by a Ministry grant to Saitama Medical School Research Center for Genomic Medicine. This work was also performed as a part of the Rational Evolutionary Design of Advanced Biomolecules (REDS) Project, Saitama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, supported by the Japan Science and Technology Agency.
REFERENCES
- 1.Alvarez-Bolado, G., M. G. Rosenfeld, and L. W. Swanson. 1995. Model of forebrain regionalization based on spatiotemporal patterns of POU-III homeobox gene expression, birthdates, and morphological features. J. Comp. Neurol. 355:237-295. [DOI] [PubMed] [Google Scholar]
- 2.Ambrosetti, D. C., C. Basilico, and L. Dailey. 1997. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interaction facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 17:6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonchuk, J., G. Sauvageau, and R. K. Humphries. 2002. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109:39-45. [DOI] [PubMed] [Google Scholar]
- 4.Avilion, A. A., S. K. Nicolis, L. H. Pevny, L. Perez, N. Vivian, and R. Lovell-Badge. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17:126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bain, G., D. Kittchens, M. Yao, J. E. Huettner, and D. I. Gottlied. 1995. Embryonic stem cells express neuronal properties in vitro. Dev. Biol. 168:342-357. [DOI] [PubMed] [Google Scholar]
- 6.Cai, J., Y. Wu, T. Mirua, J. L. Pierce, M. T. Lucero, K. H. Albertine, G. J. Spangrude, and M. S. Rao. 2002. Properties of a fetal multipotent neural stem cell (NEP cell). Dev. Biol. 251:221-240. [DOI] [PubMed] [Google Scholar]
- 7.Cairns, L. A., E. Moroni, E. Levantini, A. Giorgetti, F. G. Klinger, M. C. Magli, B. Giglioni, and S. Ottolenghi. 2003. Kit regulatory elements required for expression in developing hematopoietic and germ cell lineages. Blood 102:3954-3962. [DOI] [PubMed] [Google Scholar]
- 8.D'Amour, K. A., and F. H. Gage. 2003. Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc. Natl. Acad. Sci. USA 100:11866-11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frantz, G. D., A. P. Bohner, R. M. Akers, and S. K. McConnell. 1994. Regulation of the POU domain gene SCIP during cerebral cortical development. J. Neurosci. 14:472-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geschwind, D. H., J. Ou, M. C. Easteday, J. D. Dougherty, R. L. Jackson, Z. Chen, H. Antoine, A. Terskikh, I. L. Weissman, and S. F. Nelson. 2001. A genetic analysis of neural progenitor differentiation. Neuron 29:325-339. [DOI] [PubMed] [Google Scholar]
- 11.Graham, V., J. Khudyakov, P. Ellis, and L. Pevny. 2003. SOX2 functions to maintain neural progenitor identity. Neuron 39:749-765. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi, S., P. Lewis, L. Pevny, and A. P. McMahon. 2002. Efficient gene modulation in mouse epiblast using a Sox2 Cre transgenic mouse strain. Gene Expr. Patterns 2:93-97. [DOI] [PubMed] [Google Scholar]
- 13.He, X., M. N. Treacy, D. M. Simmons, H. A. Ingraham, L. W. Swanson, and M. G. Rosenfeld. 1989. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature 340:35-41. [DOI] [PubMed] [Google Scholar]
- 14.Hemmati-Brivanlou, A., and D. Melton. 1997. Vertebrate embryonic cells will become nerve cells unless told otherwise. Cell 88:13-17. [DOI] [PubMed] [Google Scholar]
- 15.Ivanova, N. B., J. T. Dimos, C. Schaniel, J. A. Hackney, K. A. Moore, and I. R. Lemischka. 2002. A stem cell “molecular signature.” Science 298:601-604. [DOI] [PubMed] [Google Scholar]
- 16.Jaegle, M., W. Mandemakers, L. Broos, R. Zwart, A. Karis, P. Visser, F. Grosveld, and D. Meijer. 1996. The POU factor Oct-6 and Schwann cell differentiation. Science 273:507-510. [DOI] [PubMed] [Google Scholar]
- 17.Josephson, R., T. Muller, J. Pickel, S. Okabe, K. Reynolds, P. A. Turner, A. Zimmer, and R. D. McKay. 1998. POU transcription factors control expression of CNS stem cell-specific genes. Development 125:3087-3100. [DOI] [PubMed] [Google Scholar]
- 18.Katsuoka, F., H. Motohashi, K. Onodera, N. Suwabe, J. D. Engel, and M. Yamamoto. 2000. One enhancer mediates mafk transcriptional activation in both hematopoietic and cardiac muscle cells. EMBO J. 19:2980-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko, M. S. H., J. Kichen, X. Wang, T. A. Treat, X. Wang, A. Hasegawa, T. Sun, M. J. Grahovac, G. J. Kargul, M. K. Lim, Y. Cui, Y. Sano, T. Tanaka, Y. Liang, S. Mason, P. D. Paonessa, A. D. Sauls, G. E. DePalma, R. Sharara, L. B. Rowe, J. Eppig, C. Morrell, and H. Doi. 2000. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development 127:1737-1749. [DOI] [PubMed] [Google Scholar]
- 20.Li, M., L. Pevny, R. Lovell-Badge, and A. Smith. 1998. Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr. Biol. 8:971-974. [DOI] [PubMed] [Google Scholar]
- 21.Luckow, B., and G. Schutz. 1987. CAT constructs with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 15:5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshak, D. R., R. L. Gardner, and D. Gottlieb. 2001. Stem cell biology. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.McEvilly, R. J., M. O. de Diaz, M. D. Schonemann, F. Hooshmand, and M. G. Rosenfeld. 2002. Transcriptional regulation of cortical neuron migration by POU domain factors. Science 295:1528-1532. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi, H., U. Blomer, M. Takahashi, F. H. Gage, and I. M. Verma. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi, H., K. A. Smith, D. E. Mosier, I. M. Verma, and B. E. Torbett. 1999. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science 283:682-686. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima, S., and S. Nagata. 1990. pEF-BOS: a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mountford, P., B. Zevnik, A. Duwel, J. Nicholis, M. Li, C. Dani, M. Robertson, I. Chambers, and A. Smith. 1994. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. USA 91:4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90. [DOI] [PubMed] [Google Scholar]
- 29.Nishimoto, M., A. Fukushima, A. Okuda, and M. Muramatsu. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19:5453-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimoto, M., S. Miyagi, T. Katayanagi, M. Tomioka, M. Murmatsu, and A. Okuda. 2003. The embryonic Octamer factor 3/4 displays distinct DNA binding specificity from those of other Octamer factors. Biochem. Biophys. Res. Commun. 302:581-586. [DOI] [PubMed] [Google Scholar]
- 31.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 32.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 33.Osawa, M., K. Hanada, H. Hamada, and H. Nakauchi. 1996. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273:242-245. [DOI] [PubMed] [Google Scholar]
- 34.Pevny, L., and M. S. Rao. 2003. The stem-cell menagerie. Trends Neurosci. 26:351-359. [DOI] [PubMed] [Google Scholar]
- 35.Pevny, L. H., and R. Lovell-Badge. 1997. Sox genes find their feet. Curr. Opin. Genet. Dev. 7:338-344. [DOI] [PubMed] [Google Scholar]
- 36.Ramalho-Santos, M., S. Yoon, Y. Matsuzaki, R. C. Mulligan, and D. A. Melton. 2002. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298:597-600. [DOI] [PubMed] [Google Scholar]
- 37.Reya, T., A. W. Duncan, L. Ailles, J. Domen, D. C. Scherer, K. Willert, L. Hintz, R. Nusse, and I. L. Weissman. 2003. A role for Wnt signalling in self-renewal of hematopoietic stem cells. Nature 423:409-414. [DOI] [PubMed] [Google Scholar]
- 38.Reynolds, B. A., W. Tetzlaff, and S. Weiss. 1992. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J. Neurosci. 12:4565-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saba, R., N. Nakatsuji, and T. Saito. 2003. Mammalian BarH1 confers commissural neuron identity on dorsal cells in the spinal cord. J. Neuorsci. 23:1987-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito, T., and N. Nakatsuji. 2001. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 240:237-246. [DOI] [PubMed] [Google Scholar]
- 41.Schonemann, M. D., A. K. Ryan, R. J. McEvilly, S. M. O'Connell, C. A. Arias, K. A. Kalla, P. Li, P. E. Sawchenko, and M. G. Rosenfeld. 1995. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 9:3122-3135. [DOI] [PubMed] [Google Scholar]
- 42.Sugitani, Y., S. Nakai, O. Minowa, M. Nishi, K. Jishage, H. Kawano, K. Mori, M. Ogawa, and T. Noda. 2002. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 16:1760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tahara-Hanaoka, S., K. Sudo, H. Ema, H. Miyoshi, and H. Nakauchi. 2002. Lentiviral vector-mediated transduction of murine CD34− hematopoietic stem cells. Exp. Hematol. 30:11-17. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka, S., T. Kunath, A. K. Hadjantonakis, A. Nagy, and J. Rossant. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072-2075. [DOI] [PubMed] [Google Scholar]
- 45.Terskikh, A. V., M. C. Easterday, L. Li, L. Hood, H. I. Kornblum, D. H. Geschwind, and I. L. Weissman. 2001. From hematopoiesis to neuropoiesis: evidence of overlapping genetic programs. Proc. Natl. Acad. Sci. USA 96:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas, K. R., and M. R. Capecchi. 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51:503-512. [DOI] [PubMed] [Google Scholar]
- 47.Tokunaga, Y., E. Kaiho, M. Maruyama, K. Takahashi, K. Mitsui, M. Maeda, H. Niwa, and S. Yamanaka. 2003. Fbx15 is a novel target of Oct-3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 23:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomioka, M., M. Nishimoto, S. Miyagi, T. Katayanagi, N. Fukui, H. Niwa, M. Muramatsu, and A. Okuda. 2002. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 30:3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai, R. Y., R. Kittappa, and R. D. McKay. 2002. Plasticity, niches, and the use of stem cells. Dev. Cell 6:707-712. [DOI] [PubMed] [Google Scholar]
- 50.Tsai, R. Y., and R. D. McKay. 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16:2991-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchikawa, M., Y. Kamachi, and H. Kondoh. 1999. Two distinct subgroups of group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech. Dev. 84:103-120. [DOI] [PubMed] [Google Scholar]
- 52.Uchikawa, M., Y. Ishida, T. Takemoto, Y. Kamachi, and H. Kondoh. 2003. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4:509-519. [DOI] [PubMed] [Google Scholar]
- 53.Weissman, I. L., D. J. Anderson, and F. H. Gage. 2001. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiation. Annu. Rev. Cell Dev. Biol. 17:387-403. [DOI] [PubMed] [Google Scholar]
- 54.Wiebe, M. S., P. J. Wilder, D. Kelly, and A. Rizzino. 2000. Isolation, characterization, and differential expression of the murine Sox-2 promoter. Gene 246:383-393. [DOI] [PubMed] [Google Scholar]
- 55.Wood, H., and V. Episkopou. 1999. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 86:197-201. [DOI] [PubMed] [Google Scholar]
- 56.Yuan, H., N. Corbi, C. Basilico, and L. Dailey. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9:2635-2645. [DOI] [PubMed] [Google Scholar]
- 57.Zappone, M. V., R. Galli, R. Catena, N. Meani, S. De Biasi, E. Mattei, C. Tiveron, A. L Vescovi, R. Lovell-Badge, S. Ottolenghi, and S. K. Nicolis. 2000. Sox2 regulatory sequences direct expression of a β-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development 127:2367-2382. [DOI] [PubMed] [Google Scholar]