Abstract
HIRA-like (Hir) proteins are evolutionarily conserved and are implicated in the assembly of repressive chromatin. In Saccharomyces cerevisiae, Hir proteins contribute to the function of centromeres. However, S. cerevisiae has point centromeres that are structurally different from the complex centromeres of metazoans. In contrast, Schizosaccharomyces pombe has complex centromeres whose domain structure is conserved with that of human centromeres. Therefore, we examined the functions of the fission yeast Hir proteins Slm9 and the previously uncharacterised protein Hip1. Deletion of hip1+ resulted in phenotypes that were similar to those described previously for slm9Δ cells: a cell cycle delay, synthetic lethality with cdc25-22, and poor recovery from nitrogen starvation. However, while it has previously been shown that Slm9 is not required for the periodic expression of histone H2A, we found that loss of Hip1 led to derepression of core histone genes expression outside of S phase. Importantly, we found that deletion of either hip1+ or slm9+ resulted in increased rates of chromosome loss, increased sensitivity to spindle damage, and reduced transcriptional silencing in the outer centromeric repeats. Thus, S. pombe Hir proteins contribute to pericentromeric heterochromatin, and our data thus suggest that Hir proteins may be required for the function of metazoan centromeres.
Homologues of the human gene HIRA have been identified in a range of eukaryotic organisms, including yeasts, worms, flies, and mammals (29, 31, 58). In higher cells, Hir proteins have been linked to embryogenesis. Both mouse and chicken HIRA proteins exhibit highly regulated patterns of expression during embryogenesis (53, 68), and targeted disruption of mouse HIRA has demonstrated that it is essential for embryonic development (54). The human HIRA gene was originally identified as a candidate for the developmental disorder DiGeorge syndrome (19), and although recent evidence indicates that hemizygosity of HIRA is not required for most of the phenotypes associated with DiGeorge syndrome, it is thought to contribute to the human disease (32, 33). Studies have also linked Hir proteins to control of cell cycle progression because HIRA is a cdk2-cyclin E target and overexpression of HIRA causes a delay in S phase (20).
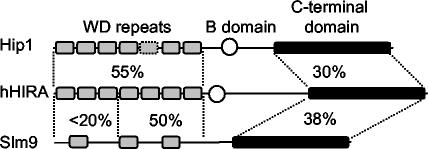
In higher organisms, these proteins are encoded by a single gene and have a highly conserved structure. The N-terminal domain consists of WD repeat motifs that have homology to the Saccharomyces cerevisiae Tup1 repressor and the p60 subunit of CAF-I (28, 59). The C-terminal region contains no obvious structural motifs but is known to interact with histones, the nonhistone chromatin protein HIRIP3, and the developmental transcription factor Pax3 (34, 35). In contrast to higher eukaryotes, S. cerevisiae possesses two Hir proteins, called Hir1 and Hir2, which are most similar to the N-terminal and C-terminal regions of HIRA, respectively (58). The fission yeast Schizosaccharomyces pombe also contains two related Hir proteins: Slm9, which has greatest similarity to S. cerevisiae Hir2 (26), and a hypothetical protein, here designated Hip1, which is unlike other yeast Hir proteins because both its N-terminal and C-terminal domains exhibit extensive homology to those of higher eukaryotic Hir proteins.
Understanding of Hir protein function at the molecular level has come predominantly from analysis of S. cerevisiae, which has revealed that they organize repressive chromatin structures and are unusual because they both contribute to transcriptional silencing at heterochromatic loci and repress the transcription of specific euchromatic genes. Indeed, S. cerevisiae Hir1 and Hir2 were first identified as repressors that restrict the expression of six of eight core histone genes to S phase (58, 60). Relief of Hir1- and Hir2-mediated repression is thought to involve the recruitment of the Swi-Snf complex because Hir2 interacts with Snf2 and expression of HTA1 is reduced in snf2 and snf5 mutants (10).
It has recently been demonstrated that Xenopus laevis HIRA is a critical component of a replication-independent nucleosome assembly pathway (52). Moreover, S. cerevisiae Hir1 and Hir2 are part of a nucleosome assembly pathway that functionally overlaps chromatin assembly factor I (CAF-I), encoded by the CAC genes (56, 57). These pathways are required for the assembly and/or maintenance of heterochromatin, because combined mutations in the CAC and HIR genes result in a synergistic decrease in silencing at both mating type and telomeric loci (27, 51). Furthermore, cac hir double mutants have defective centromeric chromatin that impairs kinetochore assembly and results in an anaphase delay (57). Thus, Hir1 and Hir2 contribute to the function of the point centromeres of S. cerevisiae, but whether they are required for the function of the complex centromeres of other eukaryotes remains to be determined.
S. pombe has complex centromeres that occupy between approximately 35 and 110 kb; they are arranged with a central core region flanked by arrays of variable elements that are assembled into chromatin, reminiscent of centromeric heterochromatin in metazoans (48). Furthermore, the domain structure of S. pombe centromeres is conserved with that of human centromeres (30). Therefore, we analyzed the functions of the S. pombe Hir proteins Slm9 and Hip1. slm9+ was originally cloned in a genetic screen for mutations that are synthetically lethal with cdc25-22, a temperature-sensitive allele of cdc25 (26). The Cdc25 phosphatase activates the cyclin-dependent kinase Cdc2 and thereby promotes the G2/M transition (38). Here we found that deletion of hip1+ led to a number of defects that were similar to those associated with loss of slm9+, namely, a cell cycle delay, synthetic lethality with cdc25-22, and poor recovery from nitrogen starvation. However, unlike Slm9, loss of Hip1 resulted in the derepression of core histone genes outside of S phase. We also found that Hip1 and Slm9 contributed to the function of centromeric chromatin, because loss of either protein resulted in sensitivity to spindle damage, increased rates of chromosome loss, and reduced transcriptional silencing in the outer repeat region. These results suggest that Hir proteins may also contribute to the function of pericentromeric heterochromatin in higher cells.
MATERIALS AND METHODS
Strains and plasmids.
Routine culture of S. pombe and general genetic methods were performed as described previously (39). The strains used in this study are described in Table 1. The hip1+ gene was disrupted with a PCR-based approach as described previously (2). Oligonucleotides 5′ KO (5′-GCCCATTCCATTTAAGAGCTTATAAAAGTTTAGACTTCATTTTACGACCACACTTCACAAGACTCCTTCGCACAAATCCCGCTTAGCTACAAATCCCACT-3′) and KO 3′ (5′-CTATTCAGATTTTTCCAATGAGTATTGTATTAGACTTAAACAGAATGCAAGAAAGAAAAGTCAAATTTAAAATAGCCTCATCTGACATAAAACGCCTAGG-3′) were used to amplify a 1.6-kb ura4+-containing fragment from pRep42 (3). The amplified fragment was used to transform a diploid SW4/SW5 strain to Ura+, and integration at the correct locus was confirmed by PCR analysis. In order to tag the genomic locus of hip1+ with three copies of the protein kinase (Pk) epitope at the carboxy terminus, the ars1 sequence from pRep42 PkC (9) was removed by EcoRI digestion, filling in with Klenow, and religation to give pRip42-PkC. The carboxy-terminal region of the hip1+ open reading frame was amplified by PCR with primers PKhip1fwd (5′-TACTCTGCAGCAACAATGTTACCATTGAAAAC-3′) and PKhip1rvs (5′-TGGATCCTCGAGGTTGATGCGTATTTTTCTATT-3′), cleaved with PstI and XhoI, and cloned into the PstI and XhoI sites of the pRip42-PkC vector. The resulting plasmid was linearized with Bst98I and used to transform strains SW48 and NT4. Stable integration was confirmed by PCR and Western blotting.
TABLE 1.
Strains Used
| Strain | Relevant genotype | Source |
|---|---|---|
| 972 | h− | Lab stock |
| SW5 | h− ade6-M216 leu1-32 ura4-D18 | Lab stock |
| SW4 | h+ ade6-M210 leu1-32 ura4-D18 | Lab stock |
| SW61 | h− leu1-32 ura4-D18 | Lab stock |
| SW137 | h− ade6-210 leu1-32 ura4-D18 hip1::ura4+ | This study |
| JK2246 | h− leu1-32 ura4-D18 slm9::ura4+ | 26 |
| SW152 | h− leu1-32 ura4-D18 slm9::ura4+ hip1::ura4+ | This study |
| SW159 | h+ ade6-M210 leu1-32 ura4-D18 hip1-Pk(ura4+) cdc25-22 | This study |
| SW187 | h+ ade6-M210 leu1-32 ura4-D18 hip1-Pk(ura4+) | This study |
| SW48 | h+ ade6-M210 leu1-32 ura4-D18 cdc25-22 | Lab stock |
| SW155 | h− ade6-M210 leu1-32 ura4-D18 hip1-Pk(ura4+)slm9HA6H(ura4+) cdc25-22 | This study |
| SW47 | h+ leu1-32 cdc10-129 | Lab stock |
| SW168 | h+ leu1-32 cdc10-129 ura4-D18 hip1::ura4 | This study |
| SW165 | h− ade6-M210 leu1-32 ura4-D18 hip1::ura4+ cdc25-22 (pRep41FLAG Hip1) | This study |
| SW115 | h− ade6-M210 leu1-32 ura4-D18 (Ch16-216-LEU2) | This study |
| SW114 | h+ ade6-M210 leu1-32 ura4-D18 hip1::ura4+ (Ch16-216-LEU2) | This study |
| SW113 | h− ade6-M210 leu1-32 ura4-D18 slm9::ura4+ (Ch16-216-LEU2) | This study |
| FY1182 | h+ ade6-M210 leu1-32 ura4-D18 otr1R(SphI)::ade6+ | 12 |
| SW118 | h+ ade6-M210 leu1-32 ura4-D18 slm9::ura4+ otr1R(SphI)::ade6+ | This study |
| SW151 | h+ ade6-M210 leu1-32 ura4-D18 hip1::ura4+ otr1R(SphI)::ade6+ | This study |
| PG1672 | mat1-PΔ17::LEU2 mat3-M(EcoRV)::ade6 ade6-M210 leu1-32 ura4-D18 | 64 |
| SW149 | mat1-PΔ17::LEU2 mat3-M(EcoRV)::ade6 ade6-M210 leu1-32 ura4-D18 slm9::ura4+ | This study |
| SW150 | mat1-PΔ17::LEU2 mat3-M(EcoRV)::ade6 ade6-M210 leu1-32 ura4-D18 hip1::ura4+ | This study |
| mad2Δ | h− ade6-M210 leu1-32 ura4-D18 mad2::ura4+ | 23 |
| SW178 | h− ade6-M210 leu1-32 ura4-D18 mad2::ura4+ slm9::ura4+ | This study |
| SW179 | h+ ade6 M210 leu1-32 ura4-D18 mad2::ura4+ hip1::ura4+ | This study |
| 393 | h− ade6-M216 ura4DS/E leu1-32 his1-102 bub1::ura4+ | 6 |
| SW182 | h− ade6-M216 leu1-32 ura4− bub1::ura4+ slm9::ura4+ | This study |
| SW181 | h+ ade6-M216 leu1-32 ura4− bub1::ura4+ hip1::ura4+ | This study |
| FY86 | h+ his3-D1 | 43 |
| FY1862 | h90 ade6-M210 leu1-32 ura4-D18 his3-D1 otr1R(SphI)::ade6+ tel1L::his3+ tel2L::ura4+ | 43 |
| SW188 | h− leu1-32 his3-D1 tel1L::his3+ slm9::ura4+ | This study |
| SW189 | h90 leu1-32 his3-D1 tel1L::his3+ hip1::ura4+ | This study |
| 1645 | h+ ade6-210 ura4-D18 leu1-32 arg3-D4 his3-D1 | 49 |
| 2221 | h− cnt1::arg3 ade6-210 ura4-D18 leu1-32 arg3-D4 his3-D1 | 49 |
| 6243 | h− cnt1::arg3 hip1::ura4+ ade6-210 ura4-D18 leu1-32 arg3-D4his3-D1 | This study |
| 6246 | h− cnt1::arg3 slm9::ura4+ ade6-210 ura4-D18 leu1-32 arg3-D4 his3-D1 | This study |
| 4462 | h− cnt1::arg3 sim2-76 cnt3 ade6 otr2 ura4 his3::tel1L ade6-210 ura4-D18 leu1-32 arg3-D4 his3-D1 | 49 |
| FY2214 | h− ade6-210 leu1-32 ura4-D18 ars1(MluI)::pREP81Xgfpswi6-LEU2 | 25 |
| SW193 | h− ade6-210 leu1-32 ura4-D18 ars1(MluI)::pREP81Xgfpswi6-LEU2 hip1::ura4+ | This study |
| SW194 | h− ade6-210 leu1-32 ura4-D18 ars1(MluI)::pREP81Xgfpswi6-LEU2 slm9::ura4+ | This study |
Plasmids expressing the hip1+ open reading frame were constructed. The amino-terminal region of hip1+ (corresponding to amino acids 1 to 391) was amplified from an S. pombe cDNA library with primers Hip1 5′ opt (5′-GTAGCAGCGGCCGCAATGAAAATTAAAAAAATTCCATGGTTAGGACAC-3′) and Hip1 Xho site (5′-GGCTACGTCGACCTCAAGCTCGAGTTGCTTTGCAGATTC-3′). The carboxy-terminal region of hip1+ (corresponding to amino acids 367 to 932) was amplified from genomic DNA with primers Hip1 3′ fwd (5′-CGATCAGCGGCCGCGTTGGCTAAATATGGTCATGGTC-3′) and Hip1 3′ rvs (5′-GGACTGGTCGACCTCAATTTTTTTCAGGTTGATGC-3′). The resulting PCR products were cleaved with NotI and SalI and cloned into the NotI and SalI sites of pRep41FLAG to give plasmids pRep41F-Hip1-N and pRep41F-Hip1-C, respectively. A plasmid expressing full-length hip1+ was constructed by subcloning the NotI/XhoI fragment from pRep41F-Hip1-N into the NotI and XhoI sites of pREP41F-Hip1-C to give plasmid pRep41F-Hip1. This plasmid was sequenced to check for in-frame reassembly of the Hip1 coding sequence.
FACS.
DNA content analysis was performed by fluorescence-activated cell sorting (FACS) with a Becton Dickinson FACScan as described previously (62).
RNA analysis.
RNA samples were prepared from 0.25 × 109 to 0.5 × 109 cells. Cell pellets were washed in H2O and resuspended in 200 μl of RNA buffer (50 mM Tris HCl [pH 8.0], 100 mM NaCl, 50 mM EDTA [pH 8.0], 0.25% sodium dodecyl sulfate) with 200 μl of phenol-chloroform. Cells were disrupted with approximately 0.5 ml of glass beads (0.5 mm; Biospec) in a Hybaid Ribolyser. A further 0.6 ml of RNA buffer was added, followed by centrifugation at 13,000 rpm for 10 min. The aqueous layer was subjected to two further phenol-chloroform extractions before the RNA was precipitated with 0.1 volume of sodium acetate (pH 5.2) and 0.6 volume of isopropanol. RNA pellets were washed in 70% ethanol and resuspended in H2O. RNA analysis was performed as described previously (66). Briefly, a 10- to 15-μg sample of total RNA was denatured with glyoxal, separated on a 1.2% agarose gel prepared in 15 mM sodium phosphate (pH 6.5), and transferred to a GeneScreen hybridization membrane (Dupont NEN Research Products). Gene-specific probes were produced by PCR amplification from genomic DNA with the appropriate primers. All probes were labeled with [α-32P]dCTP with a Prime-a-Gene labeling kit (Promega).
Protein extraction and coprecipitations.
Whole-cell extracts were prepared as described previously (67) with some modification. Cultures were grown to mid log phase (optical density at 595 nm, 0.25 to 0.5) in Edinburgh minimal medium (EMM) (39). Cells were harvested, washed once, and snap frozen. Cell pellets were washed in 1 ml of lysis buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 0.5% NP-40, 10 mM imidazole, 2 μg of pepstatin per ml, 2 μg of leupeptin per ml, 2 μg of aprotinin per ml, 100 μg of phenylmethylsulfonyl fluoride per ml, 50 mM NaF, 0.1 mM NaVO3), resuspended in 200 μl of lysis buffer, and disrupted with 2 ml of glass beads by vortexing twice for 45 s. Protein extracts were recovered and clarified by centrifugation at 13,000 rpm for 10 min at 4°C. Protein precipitations were performed by adding 25 μl of nickel-agarose (50% slurry in lysis buffer) to 1 mg of whole protein extract and incubating at 4°C for 1 h with gentle agitation. Precipitates were recovered by centrifugation and washed four times with lysis buffer containing 200 mM NaCl and 20 mM imidazole. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to Western blotting with polyclonal antihemagglutinin (YY1) antibody (Santa Cruz) or monoclonal Pk antibody (Serotec).
Chromatin immunoprecipitations.
Chromatin immunoprecipitations were performed as previously described (49). Multiplex PCR analysis was performed as described previously (25).
Fluorescence microscopy.
Immunolocalization of Pk-tagged Hip1 was performed essentially as described previously (18). Ten-milliliter aliquots of exponentially growing cells were collected and fixed in 3.7% formaldehyde freshly prepared in PEM [100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 1 mM EGTA, 1 mM MgSO4 (pH 6.9)] for 10 min. Cells were washed in PEM and resuspended in PEMS (PEM plus 1.2 M sorbitol) containing 0.25 mg of zymolase per ml and incubated at 37°C for 70 min. Cells were washed in PEM, resuspended in PEMBAL (PEM plus 1% globulin-free bovine serum albumin [Sigma], 0.1% NaN3, 100 mM lysine hydrochloride) for 30 min. Cells were pelleted and incubated with a 1:1,000 dilution of Pk antibody (Serotec) at 4°C overnight. Cells were washed in PEM and resuspended in a 1:50 dilution of goat anti-mouse immunoglobulin fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Sigma) for 1 h at room temperature. Cells were washed sequentially in PEMBAL and phosphate-buffered saline and resuspended in phosphate-buffered saline before being spread onto poly-l-lysine-coated coverslips and mounted onto slides with Vectashield mounting medium (Vector Laboratories) containing 1.5 mg of 4′,6′-diamidino-2-phenylindole (DAPI). For green fluorescent protein experiments, mid-log-phase cells were incubated in Hoechst stain (0.5 μg/ml) for 10 min at room temperature before being processed for microscopy. Fluorescence was captured by exciting cells at the appropriate wavelength with a Zeiss Axioskop microscope with a 63× oil immersion objective and Axiovision imaging software.
RESULTS
Disruption of hip1+ results in growth defects.
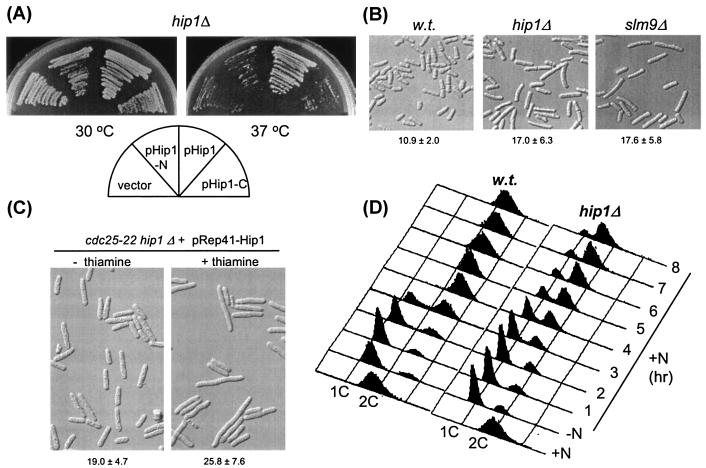
Sequencing of the S. pombe genome revealed that fission yeast has two members of the HIRA (Hir) protein family: Slm9, which shows greatest similarity to S. cerevisiae Hir2, and a hypothetical protein (SPBC31F10.13c), which we have designated Hip1 (HIRA in S. pombe 1). Hip1 is unlike other yeast Hir proteins (S. cerevisiae Hir1 and Hir2 and S. pombe Slm9) because both its N-terminal and C-terminal regions show similarity to higher eukaryotic Hir proteins (Fig. 1). As a first step to understanding the function of this protein, we deleted hip1+ by one-step gene replacement. hip1Δ cells were viable but exhibited a number of defects. They were slow growing at 30°C and were temperature sensitive, having a limited ability to grow at elevated temperatures (>35°C) (Fig. 2A). These growth phenotypes were rescued by ectopically expressing full-length hip1+ but not either the N-terminal or C-terminal half of hip1+. Microscopic examination of hip1Δ cells revealed that they were elongated, a phenotype that was exacerbated by incubation at 36°C and is indicative of a cell cycle delay (Fig. 2B). Importantly, these phenotypes are highly similar to those that result from the loss of Slm9 function (26). However, ectopic expression of slm9+ did not rescue the defects of hip1Δ cells or vice versa (data not shown). Also, high-level overexpression of slm9+ or hip1+ did not result in any detectable phenotypes (data not shown).
FIG. 1.
Schematic comparison of the S. pombe Hir proteins Slm9 and Hip1 with human HIRA. The percentage of similarity between regions of Hip1 and Slm9 with the equivalent regions of human HIRA is shown. The conserved C-terminal domain is shaded black, and the B-domain (29) is represented with an open circle. WD repeat motifs are represented as grey boxes. Note that the motif in Hip1 that is represented with a dashed line exhibits significant homology with the equivalent WD repeat in human HIRA but falls below the threshold to be designated a WD repeat as defined by Pfam (4).
FIG. 2.
Deletion of hip1+ results in growth defects. (A) A hip1Δ strain (SW137) was transformed with the indicated plasmid and cultured on EMM agar at 30 or 37°C for 3 to 4 days. (B) Wild-type (SW5), hip1Δ (SW137), and slm9Δ (JK2246) cells were grown overnight at 36°C and then processed for microscopy. Mean cell length (micrometers) ± standard deviation is indicated. (C) hip1 interacts genetically with cdc25. A cdc25-22 hip1Δ strain containing pRep41F-Hip1 was grown to early log phase at 25°C. Incubation was continued in either the presence or absence of thiamine (2 μM). Mean cell length (micrometers) ± standard deviation is indicated. (D) hip1Δ cells are defective in recovery from nitrogen starvation. DNA content analysis of wild-type (SW5) and hip1Δ (SW137) cells. Cells were grown to log phase in EMM medium (+N) and shifted to nitrogen-free EMM medium for 16 h (−N). Cells were then shifted to EMM medium and harvested at the indicated intervals.
slm9+ was identified in a screen for mutations that were synthetically lethal with cdc25-22, a temperature-sensitive allele of cdc25 (26). The similarity in the morphology of hip1Δ and slm9Δ cells suggested that mutation of hip1+ might also be lethal in a cdc25-22 background. To test this possibility, we created a cdc25-22 hip1Δ double mutant strain covered with a plasmid ectopically expressing hip1+ from the thiamine-repressible nmt41 promoter (3). When this strain was cultured at the permissive temperature (25°C) in the absence of thiamine, the cells had an elongated shape that is characteristic of cdc25-22 mutants. However, addition of thiamine to the medium caused a marked increase in cell length, indicating that the loss of Hip1 function exacerbates the cell cycle delay in this background (Fig. 2C). That the cells continued to grow in the presence of thiamine is likely to be due to residual expression of hip1+ from the nmt41 promoter. However, despite repeated attempts with a number of independent isolates of the cdc25-22 hip1Δ strain, we were unable to identify cells that had lost the covering plasmid. The simplest explanation of these findings is that, like deletion of slm9+, deletion of hip1+ is synthetically lethal in combination with cdc25-22.
slm9Δ cells are defective in recovery from G1 arrest following nitrogen starvation (26). This prompted us to examine whether Hip1 was also required for efficient reentry into the cell cycle. Following nitrogen starvation, both wild-type and hip1Δ cells arrested with a 1C DNA peak, although we noted some delay in the time taken to arrest in the hip1Δ strain (data not shown). Upon addition of nitrogen, wild-type cells reentered the cell cycle, and FACS analysis demonstrated that the first round of DNA replication occurred within 4 h (Fig. 2D). In contrast, a significant proportion of hip1Δ cells remained with a 1C peak up to 8 h after the readdition of nitrogen, indicating that Hip1 is also required for efficient recovery from nitrogen starvation.
Subcellular localization and levels of Hip1.
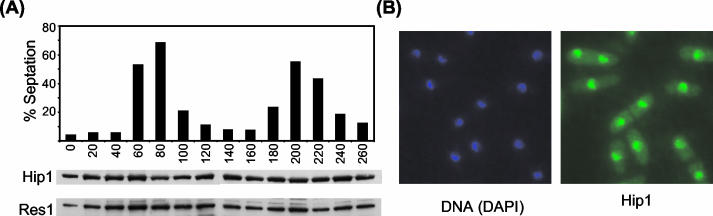
We next examined whether Hip1 protein levels and/or subcellular localization were regulated in response to cell cycle progression. In order to facilitate this analysis, the chromosomal copy of hip1+ was epitope tagged with three copies of the Pk epitope (9). A synchronous culture expressing Hip1-Pk was then produced with a cdc25-22 block-and-release protocol (Fig. 3A). Western analysis of extracts prepared from this culture demonstrated that the level and mobility of Hip1-Pk were essentially unaffected by cell cycle progression. We used immunofluorescence microscopy to determine the subcellular localization of Hip1-Pk. In cells expressing Hip1-Pk, we observed strong nuclear fluorescence that was unaffected by the stage of the cell cycle (Fig. 3B). Thus, the level and the subcellular localization of Hip1 remain relatively constant throughout the cell cycle.
FIG. 3.
Level and localization of Hip1 are not affected by cell cycle progression. (A) cdc25-22 hip1-Pk cells were grown to early log phase in EMM medium at 25°C before being shifted to 34°C for 4 h to arrest cells at the G2/M transition. Cells were rapidly cooled to 25°C and harvested at 20-min intervals. Cell cycle synchronicity was determined by counting the percentage of cells with a septum (top panel). Whole-cell extracts were prepared from each sample and subjected to Western blotting with anti-Pk and anti-Res1 antibodies. (Res1 levels are known to remain constant throughout the cell cycle.) (B) Hip1-Pk localizes to the nucleus. hip1-Pk (SW187) cells were grown to mid-log phase in YE5S medium at 30°C. Cells were stained with DAPI and anti-Pk antibodies and processed for indirect immunofluorescence.
Hip1 and Slm9 interact.
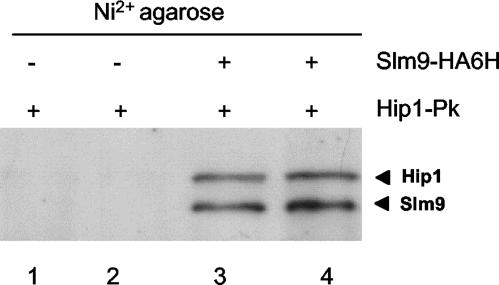
The similarity in the cell cycle defects resulting from the loss of Hip1 and Slm9 suggested that these proteins may function in the same pathway. Consistent with this, we found that a hip1Δ slm9Δ double mutant strain had cell length and temperature sensitivity phenotypes that were no more severe than those of the parental strains (data not shown). Therefore, we next investigated the ability of Hip1 and Slm9 to interact with a strain that expressed both Pk-tagged Hip1 and six-His--HA-tagged Slm9 at native levels. Slm9 was precipitated from extracts derived from this strain with Ni2+-agarose, and Western analysis indicated that Hip1 coprecipitated with Slm9 (Fig. 4). This interaction was specific, as Hip1-Pk was absent in precipitates derived from cells that did not contain 6His-HA-tagged Slm9. These results suggest that not only do Hip1 and Slm9 function on the same pathway, they also have the potential to form a complex.
FIG. 4.
Hip1 and Slm9 interact. hip1-Pk (SW159) (lanes 1 and 2) and hip1-Pk slm9-HA6H (SW155) (lanes 3and 4) cells were grown to mid-log phase. Whole-cell extracts were prepared and partially purified with Ni2+-agarose, and the precipitates were analyzed by Western blotting with anti-Pk and anti-HA antibodies.
Slm9 and Hip1 are not functionally identical.
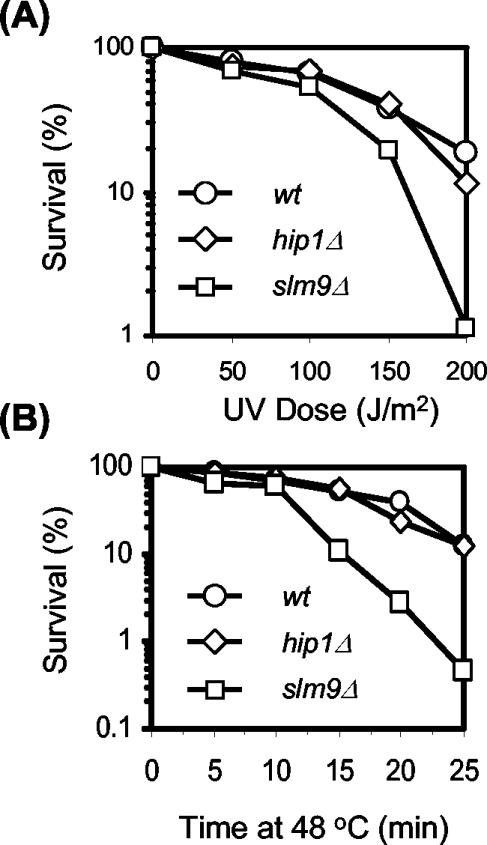
Loss of Hip1 or Slm9 results in a number of similar phenotypes; however, we found that they are not identical. We found that hip1Δ cells are severely comprised in their ability to undergo mating but that slm9Δ cells mate efficiently (data not shown). Furthermore, while slm9Δ cells have reduced tolerance to UV irradiation and to a 48°C heat shock (26), hip1Δ cells exhibit wild-type levels of tolerance to both of these stresses (Fig. 5A and B). Thus, Hip1 and Slm9 are functionally distinct.
FIG. 5.
hip1Δ and slm9Δ cells have different sensitivities to heat shock and UV. (A) Wild-type (SW5), hip1Δ (SW137), and slm9Δ (JK2246) strains were grown to log phase, plated onto YE5S agar, and exposed to UV irradiation (254 nm) at the indicated dose with a Stratalinker. (B) Wild-type (SW5), hip1Δ (SW137), and slm9Δ (JK2246) strains were grown to log phase at 30°C and then shifted to 48°C. Aliquots were taken at the indicated intervals, diluted in cold YE5S medium, and then plated onto YE5S agar. Colony numbers were determined after 4 days of incubation at 30°C.
Hip1 represses histone gene expression in G1-arrested cells.
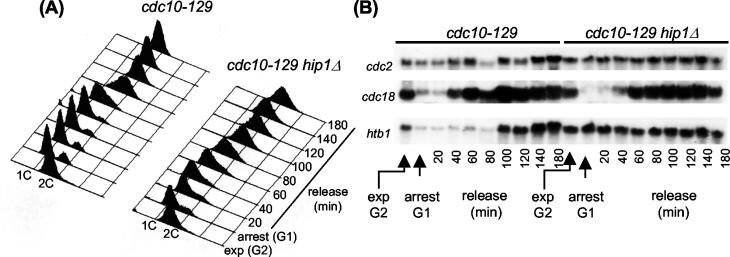
Hir proteins were originally identified as regulators of histone gene transcription in S. cerevisiae (58). However, it is not clear whether they regulate this class of gene in all eukaryotes, and indeed, loss of Slm9 was found not to influence the pattern of H2A expression in S. pombe (26). Nonetheless, our data indicates that Hip1 and Slm9 are not functionally identical, and so we investigated whether Hip1 regulates histone gene transcription. We monitored histone mRNA levels during a cell cycle block-and-release experiment with the cdc10-129 allele. Mid-log-phase cells growing at the permissive temperature (25°C) were arrested in G1 (prior to start) by being shifted to the restrictive temperature (34°C) for 4 h. FACS analysis confirmed that the majority of both the hip1+ and the hip1Δ cells arrested with a 1C DNA content (Fig. 6A). We did note that in the case of the hip1Δ strain, a small fraction of cells were found to be in S phase, suggesting that a small number of cells leaked through the block. Cells were released from the G1 block by reducing the incubation temperature, and FACS analysis confirmed that both wild-type and mutant cells had completed S phase within 120 min following release. We monitored mRNA levels throughout the course of the experiment by Northern blotting and first measured cdc18+ transcript levels, which are Cdc10 dependent and peak during G1 and S phases (5). Similar patterns of cdc18+ expression were observed in both the hip1+ and hip1Δ strains; shifting cells to the restrictive temperature resulted in a large drop in cdc18+ mRNA levels, but, following release from the block, transcript levels increased as the cells progressed through G1 and S phases (Fig. 6B).
FIG. 6.
Hip1 is required for repression of histone mRNA levels in G1-arrested cells. Cells were grown to mid-log phase at 25°C (exp G2) and then shifted to 34°C for 4 h (arrest G1). The cells were then shifted back to 25°C, and samples were taken at the indicated time points. DNA content was analyzed by flow cytometry (A), and gene expression was monitored by Northern blotting with the indicated probes (B). The strains used were SW47 (cdc10-129) and SW168 (cdc10-129 hip1Δ).
Having confirmed the efficacy of the block-and-release protocol and demonstrated that Hip1 is not required for the periodic expression of cdc18+, we next monitored the expression of core histone genes. In hip1+ cells, histone H2B (htb1+) levels were low in G1-arrested cells but increased dramatically 100 min following release as the cells progressed through S phase. In contrast, htb1+ transcript levels were not reduced in G1-arrested hip1Δ cells, and constitutively high levels of htb1+ transcripts were observed throughout the course of the experiment (Fig. 6B). The core histone genes in S. pombe are thought to be coordinately regulated (37), and indeed, we found that H2A mRNA expression patterns were similar to those of H2B (data not shown). These results suggest that Hip1 is required to restrict the expression of histone genes to S phase.
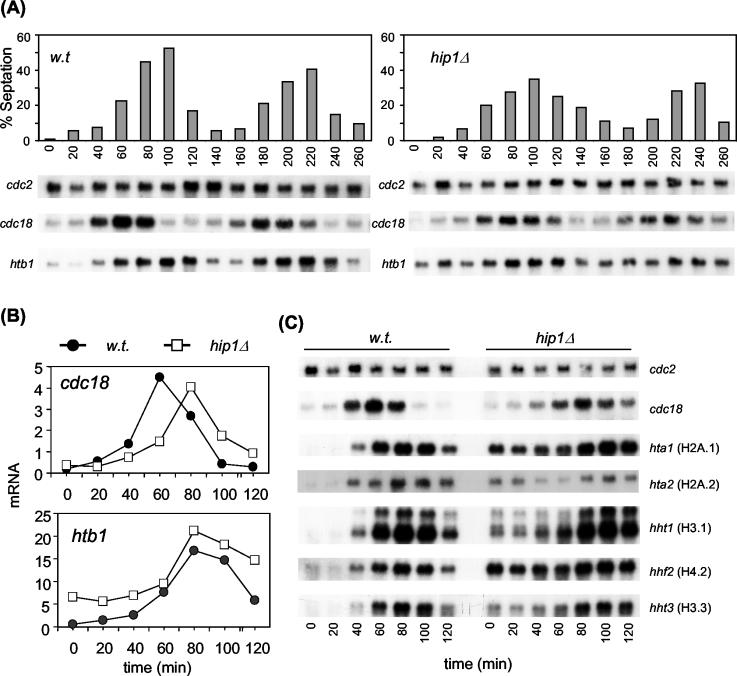
In order to confirm the findings of the block-and-release experiment, we synchronized wild-type and hip1Δ cells in early G2 by centrifugal elutriation and monitored gene expression through two synchronous rounds of cell division (Fig. 7A). The level of synchronicity was determined by counting the percentage of cells with a septum (septation index), which peaks at the G1/S transition. In wild-type cells, the expression of cdc18+ was clearly periodic and, as expected, peaked just prior to the peak of septation. The accumulation of cdc18+ transcripts was also periodic in a hip1Δ background but peaked approximately 20 min later than in wild-type cells, which is consistent with the extended cell cycle in this background. However, we again observed a striking difference in the patterns of histone gene expression. In wild-type cells, htb1+ mRNA levels were low during G2 and peaked during S phase, but in hip1Δ cells, high levels of htb1+ transcripts were observed throughout the cell cycle (Fig. 7A).
FIG. 7.
Loss of Hip1 leads to constitutive expression of histone genes. (A) Wild-type (SW5) and hip1Δ (SW137) cells were grown to mid-log phase in YE5S medium at 30°C. Cells were synchronized in early G2 by centrifugal elutriation, inoculated into fresh YE5S medium, and sampled at 20-min intervals as indicated. The synchronicity of the cultures was determined by counting the percentage of cells with a septum (top panels). RNA was prepared from each sample and analyzed by Northern blotting with the indicated probes. cdc2+ mRNA levels served as a loading control. (B) Samples from the first cell cycle (0 to 120 min) were analyzed side by side as described above, and mRNA levels relative to the loading control were determined with a Phosphorimager. (C) Samples from the first cell cycle (0 to 120 min) were analyzed by Northern blotting with the indicated probes.
In order to directly compare expression levels in wild-type and hip1Δ strains, samples from the first cell cycle were loaded on the same gel and htb1+ mRNA levels were quantified. This revealed that deletion of hip1+ led to a 10-fold increase in the level of transcripts during early G2, confirming that Hip1 is required for the repression of htb1+ gene expression outside of S phase (Fig. 7B). Furthermore, loss of Hip1 had a similar influence on the expression patterns of other core histone genes (Fig. 7C). This analysis also revealed that even in a hip1Δ strain, histone mRNA expression was induced as cells entered S phase (Fig. 7B and C), indicating that S. pombe histone transcript levels are also subject to positive control.
Hip1 and Slm9 are required for accurate chromosome segregation.
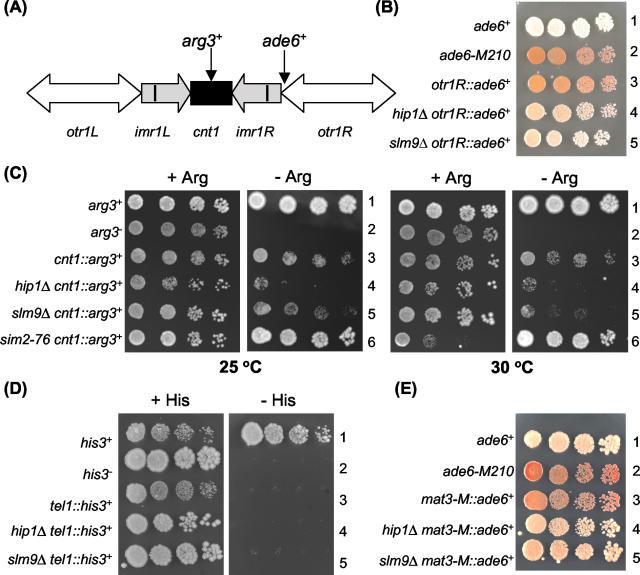
S. cerevisiae Hir1 and Hir2 have been shown to contribute to the structure of centromeric chromatin. However, it is unclear whether this is a conserved function of Hir proteins because budding yeast has point centromeres that are contained within 125 bp of DNA and are structurally different from those of metazoans. In contrast, S. pombe has complex centromeres that occupy between approximately 35 and 110 kb and are arranged with a central core region that is flanked by arrays of variable elements (48). Thus, fission yeast provides an excellent system to examine the role of Hir proteins in the function of complex centromeres. Therefore, we first investigated whether or not S. pombe Hir proteins are required for accurate chromosome segregation by measuring the rate of loss of a nonessential minichromosome (Ch16) (44).
The Ch16 minichromosome contains the ade6-216 allele, which complements the ade6-210 allele. Therefore, in an ade6-210 background, cells that contain Ch16 are Ade+ and form white colonies on adenine-limited medium, while loss of Ch16 results an Ade− phenotype and red colonies. Ch16 loss rates were measured at 30°C and also at 33°C, which exacerbates the elongated phenotype of hip1Δ and slm9Δ cells (Table 2). As previously reported, the Ch16 minichromosome was faithfully segregated in a wild-type background with loss rates of 0.025% per division at 33°C and less than 0.029% per division at 30°C. In an slm9Δ background, loss rates were significantly increased, being 0.21% at 30°C and 0.32% when cells were incubated at 33°C. Loss of Hip1 also resulted in elevated rates of chromosome loss (0.38% at 30°C and 0.25% at 33°C). These results indicate that S. pombe Hir proteins are required for accurate chromosome transmission and are consistent with a role of these proteins in the function of centromeres.
TABLE 2.
Ch16 minichromosome loss ratesa
| Genotype | Ch16 loss (%) at:
|
|
|---|---|---|
| 30°C | 33°C | |
| Wild type | <0.029 (3,488) | 0.025 (3,943) |
| slm9Δ | 0.21 (6,072) | 0.32 (4,429) |
| hip1Δ | 0.38 (5,792) | 0.25 (8,011) |
Ch16-containing colonies were plated on adenine-limited plates at the indicated temperature, and the number of half-sectored colonies was determined. The number of colonies counted is indicated in parentheses.
slm9Δ and hip1Δ mutants are sensitive to spindle damage.
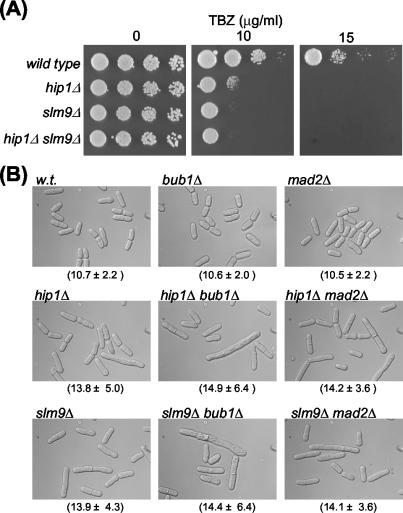
Mutations that disrupt centromere function often result in increased sensitivity to drugs such as thiabendazole (TBZ) that destabilize microtubules and so cause damage to mitotic spindles. Indeed, when we compared the ability of wild-type, slm9Δ, and hip1Δ cells to grow on rich agar plates containing TBZ, we found that loss of either Hir protein resulted in hypersensitivity to this spindle poison (Fig. 8A).
FIG. 8.
hip1Δ and slm9Δ cells are sensitive to spindle damage. (A) Strains SW5 (wild type) SW137 (hip1Δ), JK2246 (slm9Δ), and SW152(slm9Δ hip1Δ) were grown to log phase in YE5S medium. Cultures were diluted to approximately 0.4 × 107 cells/ml, subjected to fivefold serial dilutions, and spotted on YE5S agar or YE5S agar supplemented with TBZ at the indicated concentration. Plates without TBZ were incubated at 30°C for 3 days, while plates supplemented with TBZ were incubated at 30°C for 6 days. (B) The elongated cell morphology of slm9Δ and hip1Δ cells is independent of Mad2 and Bub1. Comparison of the morphology of wild-type (w.t.), slm9Δ, hip1Δ, mad2Δ slm9Δ, mad2Δ hip1Δ, bub1Δ slm9Δ, and bub1Δ hip1Δ cells is shown. Cultures were grown at 30°C, and the mean cell length (micrometers) ± standard deviation is indicated.
S. cerevisiae cac hir double mutants have an anaphase delay that is partially dependent upon the spindle assembly checkpoint (57). Therefore, we reasoned that the cell cycle delay associated with the loss of S. pombe Hir proteins may also be dependent upon the integrity of the spindle assembly checkpoint. However, deletion of spindle checkpoint genes such mad2+ (23) and bub1+ (6) did not suppress the elongated cell phenotype of hip1Δ or slm9Δ cells (Fig. 8B). We also found that the cell cycle delay was independent of Mph1 kinase (22) (data not shown). Thus, the cell cycle delay that is associated with loss of Slm9 or Hip1 is independent of the spindle assembly checkpoint.
Slm9 and Hip1 contribute to transcriptional silencing.
S. pombe centromeres are organized into two distinct transcriptionally silent domains: a central region that comprises the central core (cnt) and the inner parts of the imr sequences, and an outer domain that includes the outer imr sequences and the otr repeats (45) (Fig. 9A). The central core is associated with Mal2, Mis6, and Cnp1 (the centromere-specific H3 histone variant) (25, 55, 61), and mutation of their genes alleviates silencing in the core but not the outer repeats (45, 49). Conversely, mutations in swi6+, rik1+, clr4+, chp1+, and 12 csp genes disrupt silencing in the outer repeats but do not influence silencing in the central core (1, 12, 13, 15, 45). We measured silencing in slm9 and hip1 null backgrounds to determine whether or not Hir proteins contribute to either region of centromeric chromatin. Silencing in the outer domain was assayed with a strain that contains the ade6+ marker gene in otr1R of cen1 (12). This ade6+ marker gene is subjected to strong transcriptional silencing, and as a result this strain forms red colonies on adenine-limited medium (12). However, deletion of either slm9+ or hip1+ in this background resulted in the formation of light pink colonies, and so loss of Hir proteins results in a reduction in transcriptional silencing in the otr centromeric repeats (Fig. 9B). This suggests that loss of Hir proteins disturbs pericentromeric heterochromatin.
FIG. 9.
Loss of Slm9 or Hip1 impairs transcriptional silencing. (A) Schematic of cen1 showing the relative insertion sites of the arg3+ and ade6+ marker genes. (B) Silencing in the outer repeats. Cells containing the otrR1::ade6+ allele in combination with the appropriate deletions were grown to log phase, subjected to 10-fold serial dilutions, and spotted onto YE5S agar lacking adenine. Plates were incubated for 3 to 4 days at 30°C. Row 1, SW26; row 2, SW4; row 3, FY1181; row 4, SW151; and row 5, SW118. (C) Silencing in the central core (cnt1). Cells containing the cnt1::arg3+ allele were spotted onto EMM plates lacking arginine (−Arg) or supplemented with arginine (+Arg) and incubated at either 25 or 30°C as indicated. Strains: row 1, 972; row 2, 1645; row 3, 2221; row 4, 6243; row 5, 6246; row 6, 4462. (D) Telomeric silencing. Cells containing the tel1L::his3+ insertion were spotted onto EMM plates lacking histidine (−His) or supplemented with histidine (+His) and incubated at 30°C. The strains used were: row 1, SW4; row2, FY86; row 3, FY1862; row 4, SW189; and row 5, SW188. (E) Silencing at the mating type loci. Cells containing the mat3-M::ade6+ allele in combination with the appropriate deletions were spotted onto YE5S agar lacking adenine. The strains were: row 1, SW26; row 2, SW4; row 3, PG1672; row 4, SW150; and row 6, SW149.
Silencing in the central domain was assayed with a strain carrying the arg3+ marker in the central core of cen1 (cnt1::arg3+) that grows very slowly on arginine-limited medium (47) (Fig. 9C). As previously reported, mutation of Cnp1, the centromere-specific histone H3 variant (sim2-72), alleviated central core silencing at both 25 and 30°C. In contrast, the slow-growth phenotype was maintained in both slm9Δ and hip1Δ mutants, indicating that Hir proteins are not required for silencing in this region (Fig. 9C).
In addition to centromeres, telomeres and the mating type region are also subject to transcriptional silencing (11, 42, 63), although there is a degree of specialization in the proteins required for heterochromatin at these loci (24). Therefore, we examined telomeric silencing with a strain that has the his3+ marker inserted adjacent to a telomere (his3+::tel1L) and is unable to grow on minimal agar plates lacking histidine (43). We crossed hip1Δ and slm9Δ mutations into this background, but the resulting strains retained a His− phenotype, indicating that silencing was maintained (Fig. 9D). To investigate the influence of Hip1 and Slm9 on the heterochromatin in the mating type region, we employed a strain in which the ade6+ marker gene was inserted next to the mat3-M locus (64). As previously reported, this strain formed red colonies on adenine-limited medium, indicating that the marker gene is silenced, but marker strains lacking Hip1 or Slm9 showed severely reduced silencing and formed light pink colonies (Fig. 9E). Thus, Hip1 and Slm9 contribute to the function of heterochromatin at mating type loci as well as in the outer repeats of centromeres.
Hip1 and Slm9 are not required for the localization of Cnp1 and Swi6.
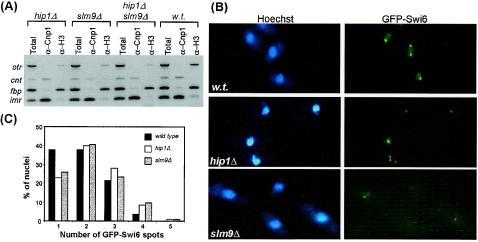
The nucleosomes associated with the central core (cnt) and inner imr sequences contain the histone H3 variant Cnp1, which is the fission yeast homologue of CENP-A (61). Deposition of Cnp1 during G1/S plays a key role in the function of the central core, as its mutation leads to changes in chromatin structure and defects in chromosome segregation (49, 61). Mutation of hip1+ results in the expression of core histone genes outside of S phase, and so we tested whether this deregulation led to the abnormal replacement of Cnp1 with histone H3.
We used chromatin immunoprecipitation assays and multiplex PCR analysis to probe the association of histone H3 and Cnp1 with centromeric sequences in hir mutant backgrounds (Fig. 10A). In the wild-type background, cnt and imr sequences but not otr sequences were enriched relative to the amounts in the euchromatic control (fbp) in Cnp1 chromatin immunoprecipitation. Importantly, this was also the case in the hip1Δ, slm9Δ, and hip1Δ slm9Δ backgrounds, demonstrating that Cnp1 deposition does not require Hip1 or Slm9. As expected, histone H3 chromatin immunoprecipitation with wild-type cells did not enrich imr and cnt sequences, and this was also the case for hip1Δ, slm9Δ, and the double-deleted cells. Thus, the deregulation of histone expression that is associated with loss of Hip1 does not result in the invasion of the central domain by histone H3. Indeed, coupled with the silencing experiments, the data indicate that the structure of the central region is not affected in hir mutants.
FIG. 10.
Hip1 and Slm9 are not required for the localization of Cnp1 or Swi6. (A) Chromatin immunoprecipitation analysis. The association of histone H3 and Cnp1 with centromeric sequences was compared in wild-type (SW61), hip1Δ (SW147), slm9Δ (JK2246), and hip1Δ slm9Δ (SW153) backgrounds. The positions of the specific PCR products are indicated, and fbp served as a euchromatic control. (B) Cells expressing green fluorescent protein-Swi6 were grown to mid-log phase at 30°C and analyzed by fluorescence microscopy. The wild-type (FY2214), hip1Δ (SW193), and slm9Δ (SW194) strains were used. (C) The frequency of wild-type, hip1Δ, and slm9Δ cells with 1, 2, 3, 4, or 5 Swi6 signals is shown. More than 100 cells were examined for each background.
Swi6 is associated with heterochromatin and is visible as up to five discrete spots per haploid nucleus: up to three major spots correspond to centromeres, while other minor spots correspond to the mat loci and telomeres (13, 14). Since failure to correctly localize Swi6 at heterochromatic loci is accompanied by diffuse nuclear staining (14), we compared green fluorescent protein-Swi6 localization patterns in wild-type and hir mutant cells. Consistent with previous reports, Swi6 was visible as several discrete spots in wild-type cells, and furthermore a similar pattern of punctate staining was observed in both the hip1Δ and slm9Δ backgrounds (Fig. 10B and C). This indicates that Hip1 and Slm9 are not required for the localization of Swi6 to heterochromatic regions, and so in hir mutants, heterochromatin is defective despite being associated with Swi6.
DISCUSSION
Here we have identified overlapping and distinct functions for the S. pombe Hir proteins Hip1 and Slm9. Like Slm9, Hip1 is required for normal cell cycle progression and efficient recovery from a G1 arrest. In addition, both proteins are required for the faithful segregation of chromosomes and contribute to the function of pericentromeric heterochromatin. However, Hip1 is distinct from Slm9 in that it is required for the cell cycle-dependent repression of core histone genes.
Hir protein complexes.
Our data indicate that Hip1 and Slm9 form a complex that is required for normal mitotic progression. In support of this, Hip1 and Slm9 copurify in size exclusion gel filtration chromatography with an apparent molecular mass of ≈2 MDa and are therefore associated with a high-molecular-weight protein complex (our unpublished results). However, the phenotypes associated with hip1Δ cells and slm9Δ cells are not identical, and Hip1 has functions that are independent of Slm9. This raises the possibility that Hip1 (and Slm9) exists in a number of distinct protein complexes. There is clear precedence for such a phenomenon, for instance, the histone acetylase Gcn5 exists in at least two distinct complexes, SAGA and ADA (17), and compositional heterogeneity of the RSC chromatin remodeling complex has also been reported (7).
Regulation of histone gene expression.
Although Hir proteins were originally identified as repressors of histone gene expression in budding yeast, it has not been clear if this is a conserved function of these proteins. Indeed, the involvement of mammalian HIRA in embryonic development (54) and its interaction with the developmental regulator Pax3 (35) have led to the proposal that they regulate the expression of developmental genes. Furthermore, the finding that Slm9 is not required for the proper expression of histone H2A in fission yeast suggested that the role of Hir proteins in regulating this class of gene expression was limited to budding yeast. However, recent evidence indicates that HIRA functions as a repressor at some histone genes in human cells (20, 41), and here we demonstrate that Hip1 is necessary for the periodic expression of core histone genes in S. pombe.
Hip1-mediated repression is not the only mechanism by which core histone mRNA levels are controlled, because transcript levels increase during S phase in the absence of Hip1. This induction may be mediated by a transcriptional activator, as is the case in mammalian cells, where Oct1, NPAT, and YY1 have all been implicated in the control of specific histone gene sets (8, 36). Histone mRNA levels are also controlled posttranscriptionally in both yeasts and mammals, and thus the increase in S. pombe histone transcripts during S phase may reflect an increase in mRNA stability.
Hir proteins and heterochromatin.
Our demonstration that loss of Slm9 or Hip1 impairs centromere function in fission yeast is important because fission yeast has complex centromeres that provide a highly useful model for those of metazoans. Our data indicate that the Slm9 and Hip1 proteins contribute to the function of the heterochromatin that is associated with the otr repeat regions that flank the central core. This domain, which is subject to strong transcriptional silencing dependent upon Swi6, a homologue of mammalian heterochromatin protein 1 (HP1), has no direct equivalent in budding yeast but appears to be related to pericentromeric heterochromatin in metazoans. Importantly, Kanoh and Russell have demonstrated that loss of Slm9 does not influence the expression of core histone genes (26), and so the increased rates of chromosome missegregation associated with slm9Δ cells are unlikely to be due to altered histone levels. Our data indicate that S. pombe Hir proteins are not necessary for silencing in the central core or at telomeres, suggesting that they are not required for the integrity of the chromatin at these loci. Nonetheless, it is worth noting that, in S. cerevisiae, a role for Hir proteins in silencing is only revealed in the absence of functional CAF-I. It is possible that CAF-I and Hir proteins also have overlapping functions in S. pombe, and so a role for Hip1 and Slm9 at telomeres and/or the central core, albeit redundant, cannot at present be ruled out.
The establishment of silent chromatin in the otr repeats involves the methylation of lysine 9 on histone H3 by Clr4, which allows the association of Swi6 (14, 40). Recent evidence has also revealed a requirement for components of the RNA interference machinery in the establishment of heterochromatin in this region (50, 65). Hir proteins are not required for Swi6 localization, so what are their roles in this process? Mutations in a large number of genes (>20) affect otr silencing, and it is possible that Slm9 or Hip1 regulates the expression of one or more of these genes. However, other evidence is suggestive of a more direct mode of action. Xenopus HIRA is an essential component of a replication-independent nucleosome assembly pathway (52), and S. cerevisiae Hir1 localizes at centromeric regions (57). Thus, Slm9 and Hip1 may be required to assemble nucleosomes in the otr repeat regions of fission yeast centromeres. Furthermore, there are data that suggest that centromeric chromatin undergoes reversible deformation that is caused by spindles (21, 46), and it has been proposed that this may cause loss of nucleosomes, imposing a need for continual Hir-dependent nucleosome assembly (57). Other evidence supports this dynamic view of heterochromatin because recent experiments have shown that mammalian HP1 is highly mobile within heterochromatic domains (16).
HIRA is essential for murine embryogenesis (54), and it has been thought that this reflects a vital role for HIRA in the regulation of developmental gene expression. Here we find that that in fission yeast, mutation of hir genes increases chromosome loss rates 10-fold. While this level of missegregation is tolerable for a unicellular eukaryote, it is unlikely to be tolerable in a multicellular organism. Therefore, it will be important to determine whether the essential requirement for murine HIRA is the result of its contribution to the faithful segregation of chromosomes.
Acknowledgments
We are very grateful to Iain Hagan at the Cancer Research UK-supported Paterson Institute for fission yeast elutriation. We thank Jan Quinn and Elizabeth Veal for comments on the manuscript and Paul Russell and Genevieve Thon for providing strains.
This work was supported by the BBSRC, by Cancer Research UK, and by the Wellcome Trust.
REFERENCES
- 1.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 2.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, 3rd, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 3.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum, B., J. Wuarin, and P. Nurse. 1997. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 16:4676-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard, P., K. Hardwick, and J. P. Javerzat. 1998. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143:1775-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, S. G., M. Li Del Olmo, P. Beglan, and U. Bond. 2002. A sequence element downstream of the yeast HTB1 gene contributes to mRNA 3′ processing and cell cycle regulation. Mol. Cell. Biol. 22:8415-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craven, R. A., D. J. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan, and A. M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221:59-68. [DOI] [PubMed] [Google Scholar]
- 10.Dimova, D., Z. Nackerdien, S. Furgeson, S. Eguchi, and M. A. Osley. 1999. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol. Cell 4:75-83. [DOI] [PubMed] [Google Scholar]
- 11.Egel, R. 1981. Mating-type switching and mitotic crossing-over at the mating-type locus in fission yeast. Cold Spring Harb. Symp. Quant. Biol. 45:1003-1007. [DOI] [PubMed] [Google Scholar]
- 12.Ekwall, K., G. Cranston, and R. C. Allshire. 1999. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics 153:1153-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekwall, K., J. P. Javerzat, A. Lorentz, H. Schmidt, G. Cranston, and R. Allshire. 1995. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269:1429-1431. [DOI] [PubMed] [Google Scholar]
- 14.Ekwall, K., E. R. Nimmo, J. P. Javerzat, B. Borgstrom, R. Egel, G. Cranston, and R. Allshire. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localization of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109:2637-2648. [DOI] [PubMed] [Google Scholar]
- 15.Ekwall, K., T. Olsson, B. M. Turner, G. Cranston, and R. C. Allshire. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91:1021-1032. [DOI] [PubMed] [Google Scholar]
- 16.Festenstein, R., S. N. Pagakis, K. Hiragami, D. Lyon, A. Verreault, B. Sekkali, and D. Kioussis. 2003. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299:719-721. [DOI] [PubMed] [Google Scholar]
- 17.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 18.Hagan, I. M., and K. R. Ayscough. 2000. Fluorescence microscopy in yeast, p. 179-206. In V. J. Allan (ed.), Protein localization by fluorescence microscopy. Oxford University Press, New York, N.Y.
- 19.Halford, S., R. Wadey, C. Roberts, S. C. Daw, J. A. Whiting, H. O'Donnell, I. Dunham, D. Bentley, E. Lindsay, A. Baldini, et al. 1993. Isolation of a putative transcriptional regulator from the region of 22q11 deleted in DiGeorge syndrome, Shprintzen syndrome and familial congenital heart disease. Hum. Mol. Genet. 2:2099-2107. [DOI] [PubMed] [Google Scholar]
- 20.Hall, C., D. M. Nelson, X. Ye, K. Baker, J. A. DeCaprio, S. Seeholzer, M. Lipinski, and P. D. Adams. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21:1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, X., S. Asthana, and P. K. Sorger. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101:763-775. [DOI] [PubMed] [Google Scholar]
- 22.He, X., M. H. Jones, M. Winey, and S. Sazer. 1998. Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J. Cell Sci. 111:1635-1647. [DOI] [PubMed] [Google Scholar]
- 23.He, X., T. E. Patterson, and S. Sazer. 1997. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 94:7965-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y. 2002. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 30:1465-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin, Q. W., A. L. Pidoux, C. Decker, R. C. Allshire, and U. Fleig. 2002. The Mal2p protein is an essential component of the fission yeast centromere. Mol. Cell. Biol. 22:7168-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanoh, J., and P. Russell. 2000. Slm9, a novel nuclear protein involved in mitotic control in fission yeast. Genetics 155:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufman, P. D., J. L. Cohen, and M. A. Osley. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18:4793-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufman, P. D., R. Kobayashi, N. Kessler, and B. Stillman. 1995. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell 81:1105-1114. [DOI] [PubMed] [Google Scholar]
- 29.Kirov, N., A. Shtilbans, and C. Rushlow. 1998. Isolation and characterization of a new gene encoding a member of the HIRA family of proteins from Drosophila melanogaster. Gene 212:323-332. [DOI] [PubMed] [Google Scholar]
- 30.Kniola, B., E. O'Toole, J. R. McIntosh, B. Mellone, R. Allshire, S. Mengarelli, K. Hultenby, and K. Ekwall. 2001. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell 12:2767-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamour, V., Y. Lecluse, C. Desmaze, M. Spector, M. Bodescot, A. Aurias, M. A. Osley, and M. Lipinski. 1995. A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum. Mol. Genet. 4:791-799. [DOI] [PubMed] [Google Scholar]
- 32.Lindsay, E. A., A. Botta, V. Jurecic, S. Carattini-Rivera, Y. C. Cheah, H. M. Rosenblatt, A. Bradley, and A. Baldini. 1999. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature 401:379-383. [DOI] [PubMed] [Google Scholar]
- 33.Lindsay, E. A., F. Vitelli, H. Su, M. Morishima, T. Huynh, T. Pramparo, V. Jurecic, G. Ogunrinu, H. F. Sutherland, P. J. Scambler, A. Bradley, and A. Baldini. 2001. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410:97-101. [DOI] [PubMed] [Google Scholar]
- 34.Lorain, S., J. P. Quivy, F. Monier-Gavelle, C. Scamps, Y. Lecluse, G. Almouzni, and M. Lipinski. 1998. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol. Cell. Biol. 18:5546-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnaghi, P., C. Roberts, S. Lorain, M. Lipinski, and P. J. Scambler. 1998. HIRA, a mammalian homologue of Saccharomyces cerevisiae transcriptional co-repressors, interacts with Pax3. Nat. Genet. 20:74-77. [DOI] [PubMed] [Google Scholar]
- 36.Marzluff, W. F., and R. J. Duronio. 2002. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14:692-699. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto, S., and M. Yanagida. 1985. Histone gene organization of fission yeast: a common upstream sequence. EMBO J. 4:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millar, J. B., C. H. McGowan, G. Lenaers, R. Jones, and P. Russell. 1991. p80cdc25 mitotic inducer is the tyrosine phosphatase that activates p34cdc2 kinase in fission yeast. EMBO J. 10:4301-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 40.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 41.Nelson, D. M., X. Ye, C. Hall, H. Santos, T. Ma, G. D. Kao, T. J. Yen, J. W. Harper, and P. D. Adams. 2002. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 22:7459-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimmo, E. R., G. Cranston, and R. C. Allshire. 1994. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 13:3801-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nimmo, E. R., A. L. Pidoux, P. E. Perry, and R. C. Allshire. 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392:825-828. [DOI] [PubMed] [Google Scholar]
- 44.Niwa, O., T. Matsumoto, Y. Chikashige, and M. Yanagida. 1989. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 8:3045-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Partridge, J. F., B. Borgstrom, and R. C. Allshire. 2000. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 14:783-791. [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson, C. G., P. S. Maddox, E. D. Salmon, and K. Bloom. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pidoux, A., and R. Allshire. 2003. Chromosome Segregation: clamping down on deviant orientations. Curr. Biol. 13:R385-R387. [DOI] [PubMed] [Google Scholar]
- 48.Pidoux, A. L., and R. C. Allshire. 2000. The structure of yeast centromeres and telomeres and the role of silent heterochromatin, p. 212-245. In P. Fantes and J. Beggs (ed.), The yeast nucleus. Oxford University Press, Oxford, England.
- 49.Pidoux, A. L., W. Richardson, and R. C. Allshire. 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161:295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Provost, P., R. A. Silverstein, D. Dishart, J. Walfridsson, I. Djupedal, B. Kniola, A. Wright, B. Samuelsson, O. Radmark, and K. Ekwall. 2002. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl. Acad. Sci. USA 99:16648-16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian, Z., H. Huang, J. Y. Hong, C. L. Burck, S. D. Johnston, J. Berman, A. Carol, and S. W. Liebman. 1998. Yeast Ty1 retrotransposition is stimulated by a synergistic interaction between mutations in chromatin assembly factor I and histone regulatory proteins. Mol. Cell. Biol. 18:4783-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ray-Gallet, D., J. P. Quivy, C. Scamps, E. M. Martini, M. Lipinski, and G. Almouzni. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9:1091-1100. [DOI] [PubMed] [Google Scholar]
- 53.Roberts, C., S. C. Daw, S. Halford, and P. J. Scambler. 1997. Cloning and developmental expression analysis of chick Hira (Chira), a candidate gene for DiGeorge syndrome. Hum. Mol. Genet. 6:237-245. [DOI] [PubMed] [Google Scholar]
- 54.Roberts, C., H. F. Sutherland, H. Farmer, W. Kimber, S. Halford, A. Carey, J. M. Brickman, A. Wynshaw-Boris, and P. J. Scambler. 2002. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell. Biol. 22:2318-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saitoh, S., K. Takahashi, and M. Yanagida. 1997. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell 90:131-143. [DOI] [PubMed] [Google Scholar]
- 56.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 57.Sharp, J. A., A. A. Franco, M. A. Osley, and P. D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherwood, P. W., S. V. Tsang, and M. A. Osley. 1993. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:28-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith, R. L., and A. D. Johnson. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25:325-330. [DOI] [PubMed] [Google Scholar]
- 60.Spector, M. S., A. Raff, H. DeSilva, K. Lee, and M. A. Osley. 1997. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi, K., E. S. Chen, and M. Yanagida. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288:2215-2219. [DOI] [PubMed] [Google Scholar]
- 62.Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida, and N. Jones. 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14:6193-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thon, G., A. Cohen, and A. J. Klar. 1994. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thon, G., and J. Verhein-Hansen. 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155:551-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 66.White, J. H., D. G. Barker, P. Nurse, and L. H. Johnston. 1986. Periodic transcription as a means of regulating gene expression during the cell cycle: contrasting modes of expression of DNA ligase genes in budding and fission yeast. EMBO J. 5:1705-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitehall, S., P. Stacey, K. Dawson, and N. Jones. 1999. Cell cycle-regulated transcription in fission yeast: Cdc10-Res protein interactions during the cell cycle and domains required for regulated transcription. Mol. Biol. Cell 10:3705-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilming, L. G., C. A. Snoeren, A. van Rijswijk, F. Grosveld, and C. Meijers. 1997. The murine homologue of HIRA, a DiGeorge syndrome candidate gene, is expressed in embryonic structures affected in human CATCH22 patients. Hum. Mol. Genet. 6:247-258. [DOI] [PubMed] [Google Scholar]