SUMMARY
LSD1 is a critical chromatin modulator controlling cellular pluripotency and differentiation through the demethylation of H3K4me1/2. Overexpression of LSD1 has been observed in many types of tumors and is correlated with its oncogenic effects in tumorigenesis. However, the mechanism leading to LSD1 upregulation in tumors remains unclear. Using an unbiased siRNA screening against all the human deubiquitinases, we identified USP28 as a bona fide deubiquitinase of LSD1. USP28 interacted with and stabilized LSD1 via deubiquitination. USP28 overexpression correlated with LSD1 upregulation in multiple cancer cell lines and breast tumor samples. Knockdown of USP28 resulted in LSD1 destabilization, leading to the suppression of cancer stem cell (CSC)-like characteristics in vitro and inhibition of tumorigenicity in vivo, which can be rescued by ectopic LSD1 expression. Our study reveals a critical mechanism underlying the epigenetic regulation by USP28 and provides a new treatment approach against breast cancer.
Posttranslational modifications, such as phosphorylation, acetylation and methylation of histone tails are crucial epigenetic marks that regulate diverse cellular processes. Methylation, in particular, has a global effect on cell cycle control, tumor progression and embryonic stem cell (ESC) self-renewal and differentiation (Klose and Zhang, 2007; Shi, 2007). The best characterized modifications are the methylation of the Lys9 and Lys27 residues of histone H3 (H3K9me2/3 and H3K27me3), which associates with heterochromatin and generally represses transcription, and H3K4me2/3, which often associates with actively transcribed gene promoters. These epigenetic modifications create unique promoter architectures that control gene expression.
Lysine methylation is a dynamic and reversible process. Lysine-specific demethylase 1 (LSD1) was the first discovered histone demethylase, which specifically removes H3K4me1/2 through flavin adenine dinucleotide (FAD)-dependent oxidative reaction (Shi et al., 2004). LSD1 is also involved in the demethylation of H3K9 when associated with androgen receptor (Metzger et al., 2005). LSD1 exhibits its functions in gene repression by operating as a key component of several co-repressor complexes, such as Co-REST, NuRD, CtBP, HDAC or Sirt1 (Mulligan et al., 2011; Nicholson and Chen, 2009). Genetic ablation of LSD1 in mice causes embryonic lethality and LSD1-deficient ESCs have cell defects and global DNA hypomethylation (Wang et al., 2009; Wang et al., 2007), suggesting that LSD1 is required for the maintenance and differentiation of stem cells. In addition, LSD1 and several master transcriptional factors of ESC, such as Oct4, Sox2 and Nanog, co-occupy the high-confidence promoter and enhancer regions of a subset of key developmental genes during early embryonic development to control the balance between self-renewal and differentiation (Foster et al., 2010; Whyte et al., 2012). Interestingly, aberrant elevation of LSD1 has been observed in poorly differentiated neuroblastoma, sarcoma and neuroendocrine carcinomas (Bennani-Baiti et al., 2012; Magerl et al., 2010; Schulte et al., 2009). LSD1 is also overexpressed in breast, bladder, lung and colon cancers (Hayami et al., 2011; Lim et al., 2010). Despite the fact that LSD1 is an important determinant of stem cell pluripotency and differentiation and its critical role in tumorigenesis, the mechanism leading to the aberrant upregulation of LSD1 in tumor remains unclear.
Protein abundance is tightly regulated by transcriptional and post-transcriptional mechanisms. Previous studies have suggested that LSD1 is an unstable protein and subjected to proteasome degradation (Lin et al., 2010; Shi et al., 2005). Although ubiquitination plays a central role in controlling the rapid turnover of many key molecules to regulate a diverse array of cellular processes (Cardozo and Pagano, 2004), de-ubiquitination, the reverse process that mediates by deubiquitinases (DUBs), has gained increasing appreciation as an important mechanism in controlling protein turnover (Reyes-Turcu et al., 2009; Sowa et al., 2009). DUBs are emerging as important modulators of carcinogenesis by controlling several key signaling pathways. For example, CYLD is a key DUB for removing the ubiquitination of several key components of NF-κB signaling pathway, including TRAF2/6 and NEMO. Loss of CYLD activity allows prolonged NF-κB activation and neoplasm transformation (Brummelkamp et al., 2003).
To better understand the mechanism underlying the upregulation of LSD1 in cancer, we undertook the unbiased approach of screening siRNA library of DUBs to identify the specific DUBs of LSD1. We found that USP28 is a bona fide DUB of LSD1 and plays a critical role in breast cancer.
RESULTS
USP28 stabilizes LSD1 at protein level
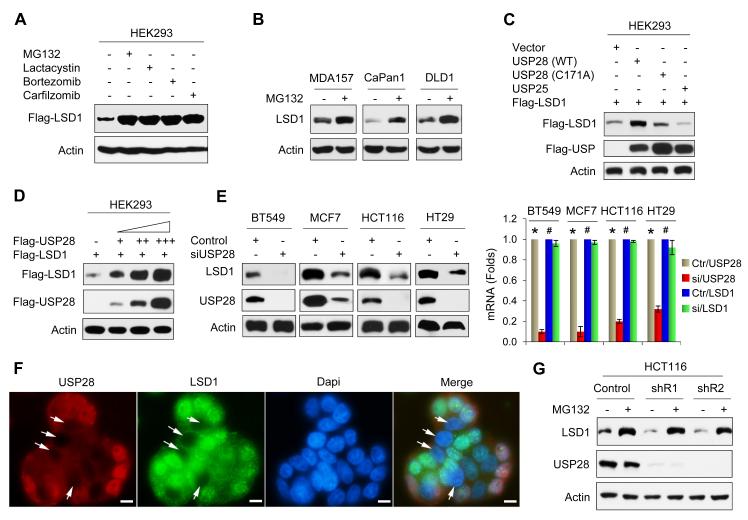
Previous studies suggested that LSD1 was subjected to proteasome degradation (Lin et al., 2010; Shi et al., 2005). To extend this observation, we expressed Flag-LSD1 in HEK293 cells followed by treatment with several proteasome inhibitors, including MG132, Lactacystin, Bortezomib or Carfilzomib, for 6 h. LSD1 became significantly stabilized by about 5 fold after the treatment with these pharmacological inhibitors that have different modes of action in suppressing proteasome-mediated protein degradation (Figure 1A). We treated breast cancer (MDA-MB157), pancreatic cancer (Capan-1), and colon cancer (DLD1) cells with MG132 and noticed that the level of endogenous LSD1 was also stabilized by MG132 (Figure 1B). Protein ubiquitination is a dynamic process, involving enzymes that add ubiquitin (ubiquitin ligases) and enzymes that remove ubiquitin (DUBs). To identify the potential DUB of LSD1, we screened cells with a siRNA library that consists of four non-overlapping siRNAs targeting 99 known or putative DUBs (data not shown). This initial screen identified 24 genes that may directly or indirectly control LSD1 stability. When these DUBs were co-expressed with LSD1 in HEK293 cells, we noticed that USP28 significantly increased LSD1 level, similar to that treated with MG132 (Figure S1A). However, expression of a mutant USP28, in which the catalytic cysteine has been replaced by alanine (C171A), failed to stabilize LSD1 (lane 3, Figure 1C). In addition, USP25, a DUB that most closely related to USP28, could not stabilize LSD1 (lane 4, Figure 1C). The steady-state level of LSD1 was enhanced by ectopic USP28 expression in a dose-dependent manner (Figure 1D). Conversely, knockdown of endogenous USP28 resulted in a drastic decrease of endogenous LSD1 protein but not mRNA in breast (BT549 and MCF7) and colon (HCT116 and HT29) cancer cells (Figure 1E). This observation was further confirmed with immunofluorescent analysis showing that the loss of LSD1 staining co-localized with USP28-knockdown in nuclei (Figure 1F). Interestingly, the downregulation of LSD1 by two independent USP28 shRNAs in HCT116 cells could be restored by MG132 treatment (Figure 1G), indicating that USP28 enhances LSD1 stabilization through a deubiquitination event. Because LSD1 is a major demethylase for H3K4me2, we examined the global levels of H3K4me2 in HCT116 cells with USP28-knockdown. H3K4me2 was elevated in two clones with USP28-knockdown (Figure S1B). The elevation of H3K4me2 after USP28-knockdown is likely mediated by the downregulation of LSD1, as knockdown of LSD1 independently also increased H3K4me2. However, knockdown of USP28 or LSD1 did not affect the levels of H3K9me2 and H3K27me3. Together, these data indicated that USP28 is a specific DUB that controls the level and activity of LSD1.
Figure 1. USP28 stabilizes LSD1.
(A) Flag-LSD1 was expressed in HEK293 cells. After treating cells with 10 μM of MG132, Lactacystin, Bortezomib or Carfilzomib for 6 h, LSD1 expression was analyzed by Western blotting.
(B) Tumor cell lines were treated with 10 μM MG132 for 6 h. Endogenous LSD1 was examined by Western blotting.
(C) Flag-LSD1 was co-expressed with either Flag-tagged USP28 (wild-type or catalytic inactive C171A mutant) or USP25 in HEK293 cells. Expression of LSD1 and USP were analyzed by Western blotting.
(D) Flag-LSD1 was co-expressed with increasing amounts of Flag-USP28 in HEK293 cells. Lysates were subjected to analysis by Western blotting.
(E) Tumor cell lines were transfected with control or USP28 siRNA, expression of endogenous LSD1 and USP28 was examined by Western blotting. The mRNA level of endogenous LSD1 and USP28 was also determined by real time RT-PCR (right panel) (mean ± SD from two independent experiments with triplicate). *p value < 0.01 when USP28 mRNA from control siRNA group was compared with that from USP28 siRNA group. #p value > 0.05 when LSD1 mRNA from control siRNA group was compared with that from USP28 siRNA group.
(F) USP28 was knocked down by USP28 siRNA in HT29 cells. After fixation, expression of endogenous USP28 (red) and LSD1 (green) was analyzed by immunofluorescent staining (nuclei were stained with Dapi; blue). Arrowhead points to USP28-knockdown cells. Scale bar = 10 μm.
(G) HCT116 cells stably transfected with control or two individual USP28 shRNAs were treated with 10 μM MG132 for 6 h; expression of LSD1 and USP28 was analyzed by Western blotting.
The protein but not the mRNA levels of USP28 and LSD1 are coordinately overexpressed in tumors
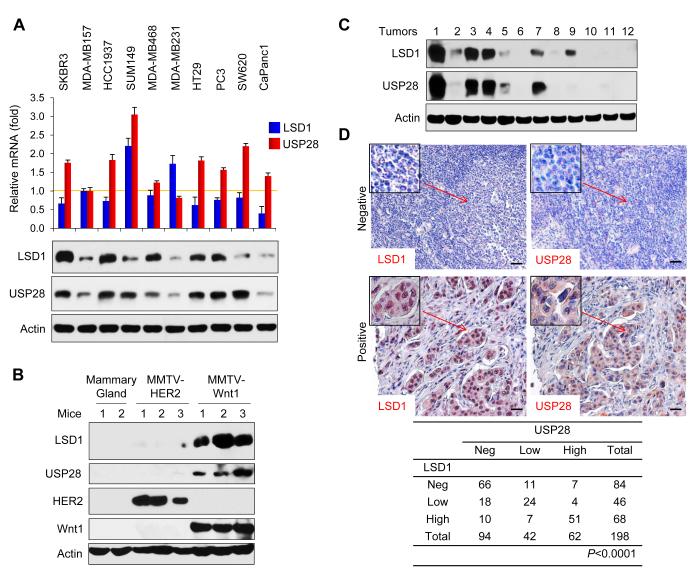
To further examine the USP28-LSD1 relationship, we analyzed the expression of USP28 and LSD1 from two gene expression datasets, which contain 118 and 149 breast tumor samples, respectively (Figure S2). We did not notice any correlation in the expression of USP28 and LSD1 at the mRNA level in these two gene expression datasets. We also examined the mRNA levels of USP28 and LSD1 in ten tumor cell lines and did not found any correlation in the expression of these two molecules at the mRNA level (top panel, Figure 2A). However, we found that the protein levels of USP28 and LSD1 were positively correlated in the majority of these tumor cell lines (except SW620) (bottom panel, Figure 2A). In addition, we analyzed the protein level of USP28 and LSD1 from mouse mammary gland and from breast tumors derived from MMTV-Wnt1 and MMTV-HER2 transgenic mice. Although we did not observe upregulation of USP28 and LSD1 from mouse mammary gland and tumors of MMTV-HER2 mice, we found an increased USP28 positively correlated with LSD1 upregulation in tumors collected from three individual MMTV-Wnt1 transgenic mice (Figure 2B). Furthermore, we examined the protein levels of USP28 and LSD1 in twelve fresh-frozen breast tumor samples. Although one sample did not match well (sample 9, Figure 2C), the majority of these tumor samples showed a positive correlation in the expression of USP28 and LSD1 at the protein level. The correlation of USP28 and LSD1 was further validated by examining the expression of these two molecules in 198 cases of primary breast tumor samples using immunohistochemical staining (Figure 2D). It has been reported that USP28 expression was low and existed in the cytoplasm in normal epithelial cells, however, strong nuclei staining of USP28 was found in majority of tumor cells in invasive ductal carcinomas (IDC) (Popov et al., 2007). Our results showed that the intensity of USP28 was positively correlated with a strong LSD1 staining in the nucleus in these IDC samples. Together, these results on the positive correlated expression between USP28 and LSD1 on protein but not mRNA level from various tumor cell lines tumor samples strengthen our observation that USP28 is critical in controlling the protein level of LSD1.
Figure 2. Expression of USP28 and LSD1 are positively correlated at the protein level in cancer cell lines and breast tumor samples.
(A) Levels of USP28 and LSD1 mRNA in various tumor cell lines were analyzed by real-time RT-PCR. Values are normalized to Actin (upper panel; mean ± SD from two experiments in triplicate). Expression of endogenous USP28 and LSD1 in these tumor cell lines was analyzed by Western blotting (bottom panel).
(B) Cell extracts prepared from normal mammary glands of two individual mice and from breast tumors of three individual MMTV-HER2 and MMTV-Wnt1 transgenic mice were analyzed for the expression of USP28 and LSD1 by Western blotting.
(C) Expression of USP28 and LSD1 from 12 cases of fresh frozen human breast tumors was examined by Western blotting (for tumor information, see Supplemental Table S2).
(D) The 198 surgical specimens of breast cancer were immunostained using antibodies against USP28, LSD1, and the control serum (data not shown). Representative images of IHC staining from the same tumor samples are shown in the top panel (Scale bar = 50 μm) and statistical analysis is shown in the bottom panel.
USP28 interacts with LSD1
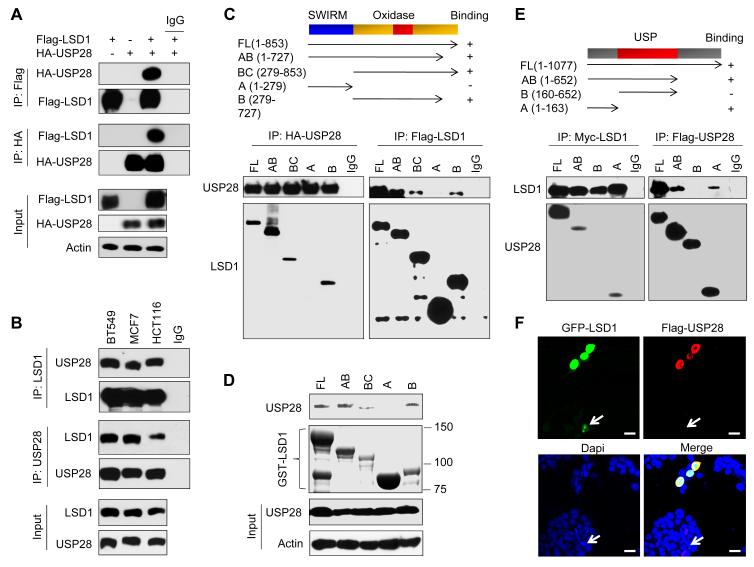
To investigate whether USP28 interacted with LSD1, we co-expressed Flag-LSD1 and HA-USP28 in HEK293 cells. After immunoprecipitating LSD1, we detected the associated USP28, and vice versa (Figure 3A). The interaction between LSD1 and USP28 is specific, as LSD1 could not interact with USP7 and USP25 under similar conditions (Figure S3A). We also immunoprecipitated endogenous LSD1 and USP28 from BT549, MCF7 and HCT116 cells and detected the presence of endogenous USP28 and LSD1, respectively (Figure 3B).
Figure 3. USP28 interacts with LSD1.
(A) Flag-LSD1 was co-expressed with HA-USP28 in HEK293 cells. LSD1 and USP28 were immunoprecipitated with Flag or HA antibody, respectively, and the associated USP28 and LSD1 were analyzed by Western blotting using either HA or Flag antibody.
(B) Endogenous LSD1 and USP28 were immunoprecipitated from BT549, MCF7 and HCT116 cells, and bound endogenous USP28 and LSD1 were examined by Western blotting.
(C) Schematic diagram showing the structure of LSD1 and deletion constructs used (top panel). Flag-tagged full-length or deletion mutants of LSD1 were co-expressed with HA-USP28 in HEK293 cells. Extracts were immunoprecipitated with Flag or HA antibody, and bound USP28 or LSD1 was examined by Western blotting using HA or Flag antibody (for input control, see Figure S3B).
(D) Lysate from HEK293 cells expressing HA-USP28 was mixed with WT or different deletion mutants of GSTLSD1. After pull-down by glutathione-agarose, the associated USP28 was examined by Western blotting using HA antibody.
(E) Schematic diagram showing the structure of USP28 and deletion constructs used (top panel). Flag-tagged full-length or deletion mutants of USP28 were co-expressed Myc-LSD1 in HEK293 cells. Extracts were immunoprecipitated with Flag or Myc antibody, and bound LSD1 or USP28 was examined by Western blotting using either Myc or Flag antibody (for input control, see Figure S3C).
(F) GFP-LSD1 was co-expressed with Flag-USP28 in HEK293 cells. After fixation, the cellular location of LSD1 (green) and USP28 (red) was examined by immunofluorescent staining (nuclei were stained with Dapi; blue). Arrowhead points to cell expressed GFP-LSD1 only. Scale bar = 25 μm.
The N-terminal one-third of LSD1 contains a SWIRM domain and the C-terminal two-thirds of LSD1 comprise an amine oxidase (AO) domain that shares extensive sequence homology with FAD-dependent amine oxidase (Figure 3C). To identify the region that is responsible for the LSD1 interaction with USP28, we generated LSD1 domain-deletion mutants and co-expressed them with USP28 in HEK293 cells. Immunoprecipitation of USP28 revealed the association with full length LSD1. A small C-terminal deletion mutant of LSD1 and the AO domain retained the ability to interact with USP28 (Figure 3C and Figure S3B). The N-terminal region of the SWIRM domain, however, was incapable of interacting with USP28. In the reciprocal immunoprecipitation experiment, immunoprecipitation of full-length and small C-terminal–deleted LSD1, and the AO domain, but not the SWIRM domain, revealed the association of USP28. These results indicate that the AO domain is required for the interaction of LSD1 with USP28. Consistent with this observation, when wild-type and deletion mutants of GST-LSD1 were pulled-down from cell lysates, the SWIRM domain of LSD1 failed to interact with USP28 (Figure 3D). To determine which region of USP28 is responsible for the interaction with LSD1, we generated several USP28 deletion mutants and co-expressed them with LSD1 in HEK293 cells. We found that the N-terminal region of USP28 retained the ability to interact with LSD1 (Figure 3E and Figure S3C). The C-terminal region and the middle USP domain, however, were not required for the interaction with LSD1. Consistent with the interaction of these two molecules in vitro and in vivo, when USP28 was co-expressed with GFP-LSD1 in HEK293 cells, we found that USP28 co-localized with and stabilized GFP-LSD1 in the nucleus (Figure 3F). Together, these data indicate that USP28 interacts with LSD1 and this interaction is mediated through the AO domain of LSD1 and N-terminal region of USP28.
USP28 stabilizes LSD1 through deubiquitination
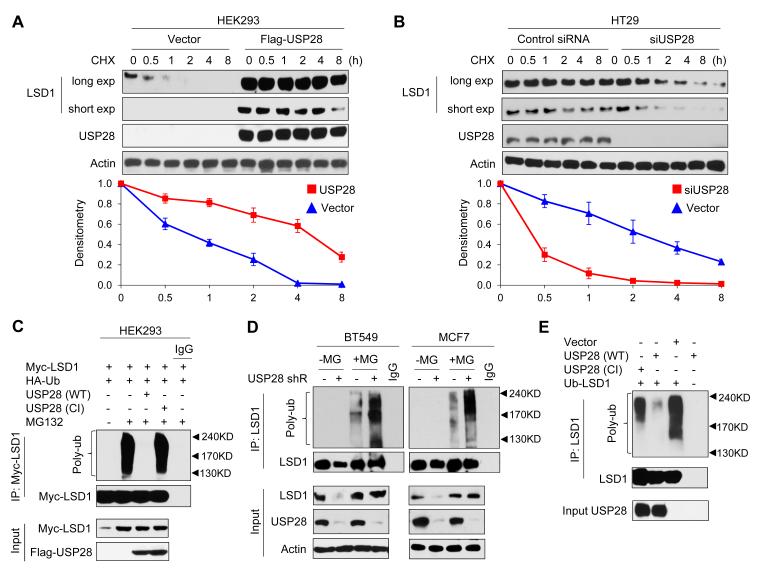
To test whether USP28 directly regulates protein stability of LSD1, we co-expressed LSD1 with USP28 or vector control in HEK293 cells and examined the degradation of LSD1. After treatment with translational inhibitor cycloheximide to block newly protein synthesis, LSD1 degraded rapidly from the cells transfected with a control vector (Figure 4A). However, LSD1 became stabilized in the presence of USP28 and this effect can last up to 4 h in the presence of cycloheximide. To test whether endogenous LSD1 is also subjected to similar regulation by USP28, we knocked down endogenous USP28 in HT29 cells, and found that endogenous LSD1 became unstable and degraded rapidly (Figure 4B). To further investigate whether this USP28 effect is mediated through the de-ubiquitination of LSD1, we co-expressed myc-LSD1 and HA-ubiquitin with either wild-type (WT) or catalytic inactive (CI, C171A) mutant of USP28 in HEK293 cells. After immunoprecipitating LSD1 from cells treated with MG132, we found that LSD1 was heavily ubiquitinated (lane 2, Figure 4C). However, co-expression of WT-USP28, but not CI-USP28, almost completely abolished LSD1 ubiquitination (lane 3 vs. lane 4, Figure 4C). Conversely, LSD1 ubiquitination significantly increased in USP28-knockdown BT549 and MCF7 cells after MG132 treatment (Figure 4D). These results indicate that USP28 stabilizes LSD1 by removing its ubiquitination through the deubiquitinase activity of USP28. To further extend this contention, we purified WT-USP28 and CI-USP28 and incubated them separately with poly-ubiquitinated LSD1 in an in vitro deubiquitination assay as described by Dupont et al (Dupont et al., 2009). We found that WT-USP28, but not CI-USP28, specifically removed LSD1 ubiquitination (Figure 4E), indicating that USP28 directly deubiquitinated LSD1.
Figure 4. USP28 deubiquitinates LSD1.
(A) Myc-LSD1 was co-expressed with vector or Flag-USP28 in HEK293 cells. After treating cells with cycloheximide (CHX) for indicated time intervals, expression of LSD1 and USP28 was analyzed by Western blotting (top panel) using Myc and Flag antibodies, respectively. The intensity of LSD1 expression for each time point was quantified by densitometry and plotted (bottom panel). Experiment was repeated three times and a representative experiment is presented. Exp stands for exposure.
(B) HT29 cells were transfected with control or USP28 siRNA. After cells were treated with CHX, expression of endogenous LSD1 and USP28 was analyzed by Western blotting (top panel); the intensity of LSD1 expression for each time point was quantified by densitometry and plotted (bottom panel). Experiment was repeated three times and a representative experiment is presented.
(C) Myc-LSD1 and HA-ubiquitin were co-expressed with wild-type or catalytic inactive (CI, C171A) mutant of USP28 in HEK293 cells. After cells were treated with or without 10 μM MG132 for 6 h, LSD1 was immunoprecipitated and the poly-ubiquitination of LSD1 was detected by Western blotting using HA antibody. Immunoprecipitated LSD1 was blotted using Myc antibody.
(D) BT549 and MCF7 cells stably transfected with control, USP28 shRNA were treated with or without MG132 for 6 h. Extracts were immunoprecipitated with LSD1 antibody and the poly-ubiquitination of LSD1 was examined by Western blotting using ubiquitin antibody.
(E) Ubiquitinated LSD1 was purified from MG132 treated HEK293 cells expressing Myc-LSD1, and then incubated with purified Flag-tagged wild-type USP28 or CI-USP28 in a deubiquitination assay as described in Experimental Procedures. The poly-ubiquitinated state of LSD1 was examined by Western blotting using HA antibody. Immuno-purified LSD1 and USP28 used in this assay were analyzed using Myc and Flag antibodies, respectively.
Knockdown of USP28 directly increases the expression of differentiation genes but indirectly suppresses the expression of pluripotent molecules
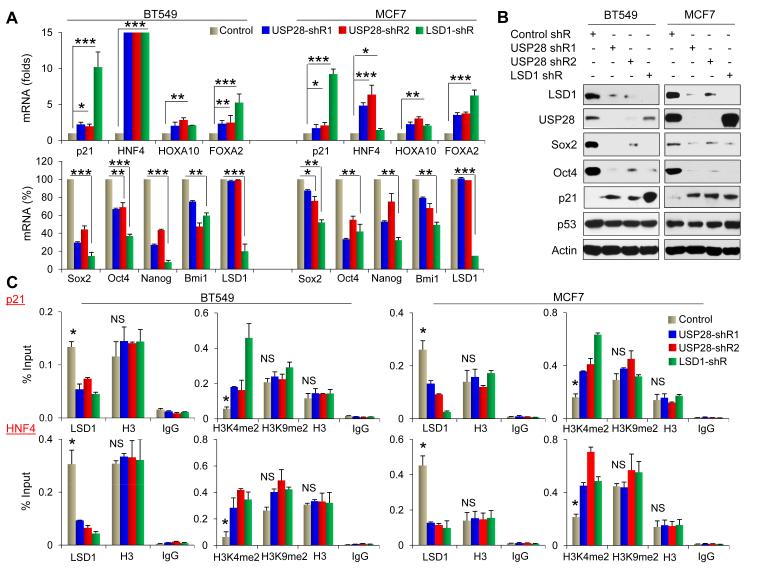
LSD1 is critical in controlling cellular differentiation and pluripotency by regulating the expression of several differentiation genes (such as p21Cif1/Waf1, HNF4, HoxA10 and FoxA2) and pluripotent molecules (such as Sox2, Oct4, Nanog and Bmi-1) (Adamo et al., 2011). To investigate whether knockdown of USP28 affects the expression of these LSD1 downstream targets, we established stable clones with knockdown of either USP28 or LSD1 expression in breast cancer BT549 and MCF7 cells and achieved about 90% knockdown efficiency in these cells. When measuring the mRNA level of these genes by real-time PCR, we found the increase of several key lineage-specific differentiation genes with USP28-knockdown (top panel, Figure 5A). In particular, USP28-knockdown led to an increased expression of p21Cif1/Waf1 in both mRNA and protein level (Figure 5A & 5B). The upregulation of p21Cif1/Waf1 was not due to the change of p53 as no difference in p53 expression was found in cells with knockdown of USP28 or LSD1 (Figure 5B). To strengthen the concept that USP28 affects the expression of these differentiation genes through LSD1, we performed chromatin immunoprecipitation (ChIP) to measure H3K4me2 at the promoters of several of these target genes (p21Cif1/Waf1, HNF4, HoxA10 and FoxA2). We found a great decrease of LSD1-occupancy at the promoters of these targets in cells with USP28-knockdown (Figure 5C and Figure S5A). Consistent with this observation, H3K4me2 was significantly increased at the promoters of these genes. The reduced occupancy of LSD1 as well as increased H3K4me2 at these target gene promoters was specific due to the loss of LSD1, as cells with LSD1-knockdown had similar effects.
Figure 5. Knockdown of USP28 alters the expression of LSD1-targeted genes.
(A) Levels of mRNA of several differentiation and pluripotent genes from BT549 and MCF7 cells stably expressed control, USP28, or LSD1 shRNAs were quantified by real-time RT-PCR. Values are normalized to Actin (mean ± SD from three experiments in triplicate). *p value < 0.05; **p value < 0.01; and ***p value < 0.001 when the control group is compared with two individual clones expressing USP28 shRNAs and/or one clone expressing LSD1 shRNA.
(B) Cells above were analyzed for the expression USP28 and LSD1 as well as several pluripotent and differentiation molecules by Western blotting.
(C) The association of LSD1 and the chromatin methylation marks (H3K4me2 and H3K9me2) at the LSD1-targeted gene promoters were analyzed by ChIP in cells stably transfected control, USP28, or LSD1 shRNAs. Levels were determined by qPCR and expressed as percentage of input (mean ± SD from three experiments in duplicate). Histone H3 and IgG were served as a control in ChIP. *p value < 0.01 when the control group is compared with two individual clones expressing USP28 shRNAs and one clone expressing LSD1 shRNA. NS means not significant.
We also observed the decrease of mRNA of several pluripotent molecules (Sox2, Oct4, Nanog and Bmi1) in two independent clones with USP28-knockdown (bottom panel, Figure 5A). Similar results were also found in cells with LSD1-knockdown. These results indicate that the effect of USP28 is specifically mediated through the downregulation of the protein level of LSD1 in these cells, because the mRNA level of LSD1 remained intact in two individual clones with USP28-knockdown. The mRNA downregulation of Sox2 and Oct4 correlated with the decreased occupancy of RNA polymerase II at their promoters in BT549 and MCF7 cells with knockdown expression of either USP28 or LSD1 (Figure S5B). Downregulation of Sox2 and Oct4 were further validated by Western blot analysis (Figure 5B). However, we could not detect an association of LSD1 at the Sox2 and Oct4 promoters by ChIP (Figure S5B), suggesting that LSD1 indirectly regulates the expression of Sox2 and Oct4 in these cells. In addition, knockdown of USP28 or LSD1 did not significantly alter the level of H3K9me2 on most of these promoters in both cell lines (Figure 5C and Figure S5A); this is consistent with the observation that USP28-knockdown did not impair the global levels of H3K9me2 (Figure S1B). Together, these data indicate that USP28 regulates the expression of differentiation genes directly and pluripotency activators indirectly through LSD1 stabilization.
Knockdown of USP28 induces differentiation and suppresses self-renewal in CSCs derived from MMTV-Wnt1 mice
Phenotypic and functional heterogeneity is a defining feature of leukemia and seen in many solid tumors. It is believed that only a subset of CSCs is responsible for the heterogeneity and efficient propagation of bulky tumors, since CSCs exhibit the ability for unlimited self-renewal and the capacity for differentiation (Nguyen et al., 2012). Because LSD1 is critical in controlling the expression of differentiation genes directly and pluripotency activators indirectly, we investigated whether the USP28-LSD1 axis was involved in the regulation of CSCs self-renewal and differentiation. To this end, we selected cells isolated from MMTV-Wnt1 tumor model, in which Wnt1 preferentially targets mammary stem cells and leads to a profound expansion of mammary stem cells in tumors (Herschkowitz et al., 2007; Liu et al., 2004; Shackleton et al., 2006). Consistent with the our contention that the USP28-LSD1 axis is critical in regulating CSC properties, expression of USP28 and LSD1 was significantly higher in cells isolated from MMTV-Wnt1 tumors than those from normal mammary glands or tumors from MMTV-HER2 mice (Figure 2 B). Cells isolated from MMTV-Wnt1 tumor grew as sphere-cluster structures in monolayer; USP28-knockdown conferred morphological changes as the cells became disassociated and scattered, a phenomenon associated with cellular differentiation (Figure 6A) (Guo et al., 2012; Jeselsohn et al., 2010). In addition, USP28-knockdown resulted in fewer and smaller tumorspheres than parental cells under non-adherent condition, suggesting that USP28-knockdown impairs the self-renewal, and enhances differentiation of CSCs from MMTV-Wnt1 tumors (Figure 6B & 6C). Consistent with this finding, the population of non-CSCs, those with a CD49flow/CD24high expression profile, greatly increased in USP28-knockdown MMTV-Wnt1 CSCs (Figure 6D and Figure S6).
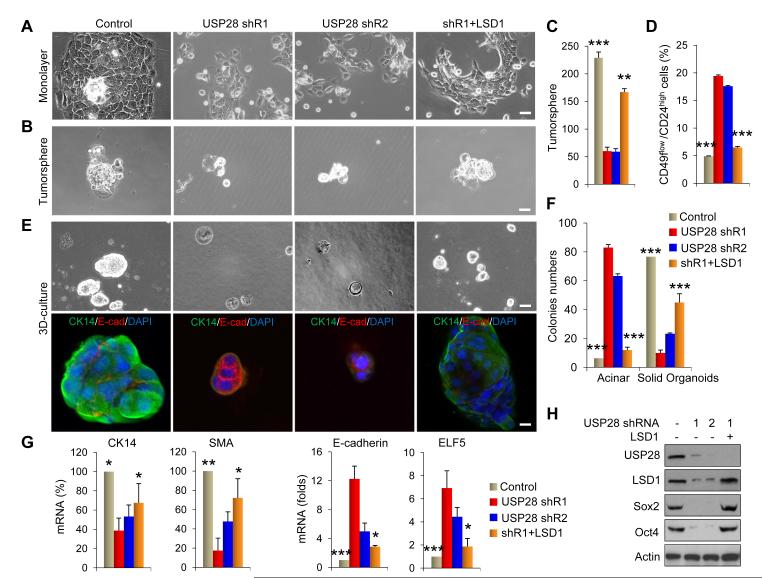
Figure 6. USP28 is required to maintain CSC-like features in breast cancer cells via LSD1.
(A) Phase-contrast micrograph of MMTV-Wnt1 cells stably transfected with control vector, USP28 shRNA or in a USP28-knockdown clone with LSD1-rescued expression. Scale bar = 100 μm.
(B) Tumorsphere-formation is examined in cells above. Representative images of tumorsphere were shown. Scale bar = 100 μm.
(C) Quantification of tumorspheres from cells in (B) was plotted and shown.
(D) CD49flow/CD24high population in cells above was analyzed by FACS.
(E) Representative images of the 3D structures formed by cells above in Matrigel (top panel) were presented. Cellular structures were stained for CK14 and E-cadherin (bottom panel; nuclei were stained with Dapi). Scale bar = 20 μm.
(F) Data presented are a quantification of the numbers of solid organoid and acini-forming structures from (E).
(G) mRNA levels of several basal and luminal genes from cells above were quantified by real-time RT-PCR. Values were normalized to 18S.
(H) Lysates from cells above were analyzed by Western blotting.
In panel C, D, F, and G, data presented are representative of three experiments performed in duplicates as the mean ± SD. *p value < 0.05; **p value < 0.01; and ***p value < 0.001 when the control group or rescued group was compared with two individual clones expressing USP28 shRNAs.
We further examined colony formation in Matrigel to assess the differentiation event associated with USP28-knockdown. Colonies from parental MMTV-Wnt1 CSCs grew as a large solid organoid with apparent expression of CK14 (basal marker) and lack of expression of E-cadherin (luminal marker). However, USP28-knockdown not only reduced the size of colonies but also shifted these colonies from solid organoid to a more acinar-like structure, accompanied by a reduced CK14 expression and increased E-cadherin expression (Figure 6E & 6F). Real-time PCR further validated the reduced expression of basal markers (CK14 and smooth muscle actin SMA) and increased expression of luminal molecules (E-cadherin and ELF5) in USP28-knockdown colonies (Figure 6G). Notably, LSD1-rescued expression reversed morphological changes, restored tumorsphere-formation, reduced the non-CSC population, and returned expression of basal markers in USP28-knockdown cells (Figure 6A-6G). Consistent with these findings, USP28-knockdown reduced the expression of Sox2 and Oct4 whereas LSD1-rescued expression restored their expressions (Figure 6H). Together, our data indicate that the USP28-LSD1 axis is required for regulating the self-renewal and differentiation properties of CSCs derived from MMTVWnt1 tumors.
Knockdown of USP28 inhibits CSC-like traits in vitro and suppresses tumorigenicity in vivo
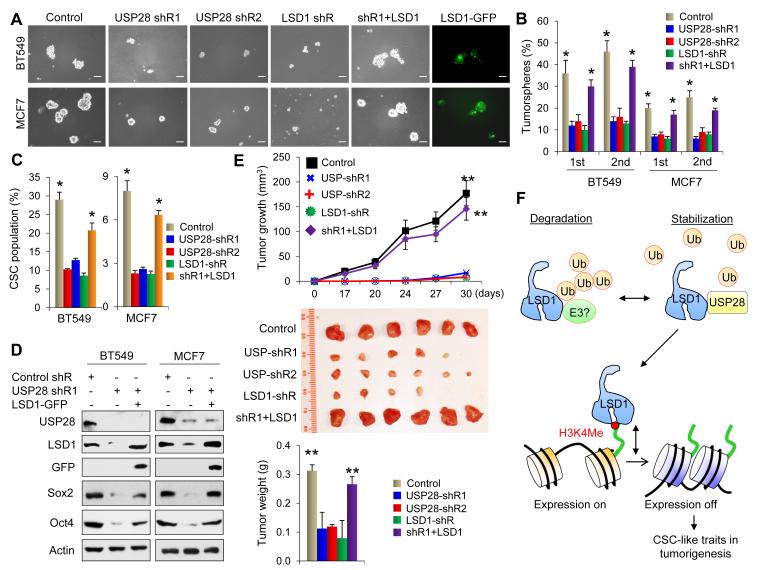
To further examine the USP28-LSD1 axis in regulating CSC-like properties in human breast cancer, we measured tumorsphere formation in BT549 and MCF7 cells with knockdown of USP28. We found that USP28-knockdown reduced the number and size of primary tumorspheres in BT549 and MCF7 cells (Figure 7A & 7B). Knockdown of USP28 also reduced the number and size of secondary tumorspheres (Figure 7B). This function of USP28 is likely mediated through the regulation of LSD1, as knockdown of LSD1 also inhibited the number and size of tumorspheres to a similar extent in these two cell lines (Figure 7A & 7B). Consistent with this idea, USP28-mediated inhibition on tumorsphere-formation was largely restored by LSD1-rescued expression (vector expressing LSD1-IRES-GFP) in these cells (Figure 7A & 7B). As human breast CSCs are enriched in CD44high/CD24low population (Fillmore and Kuperwasser, 2007), we measured the CD44high/CD24low population in BT549 and MCF7 cells with knockdown of USP28 or LSD1. We found that USP28-knockdown reduced CD44high/CD24low population in both BT549 and MCF7 cells (Figure 7C and Figure S7A); similar observations were noted with LSD1-knockdown in these two cell lines. Again, the reduction of CSC population in USP28-knockdown clone could be restored by LSD1-rescued expression. To further confirm this, we also used EpCAM+/CD49fhigh/CD24low as cell-surface markers in sorting cells that are known to be enriched with CSCs in breast cancer (Plaks et al., 2013; Stingl et al., 2006; Van Keymeulen et al., 2011). As expected, similar results were obtained (Figure S7B). The difference of CSC populations among vector-control cells, USP28-knockdown cells and LSD1-rescued cells is unlikely to be due to the variable gating in FACS analysis as same gating was used in each assay (Figure S7C). In line with the critical role of the USP28-LSD1 axis in regulating CSC-like characteristics, USP28-knockdown reduced the expression of Sox2 and Oct4 and LSD1-rescued expression restored their downregulations (Figure 7D).
Figure 7. Knockdown of USP28 inhibits CSC-like traits in vitro and suppresses tumorigenicity in vivo.
(A) Tumorsphere-formation was examined in BT549 and MCF7 cells stably expressing control, USP28, and LSD1 shRNAs and in a USP28-knockdown clone with LSD1-rescued expression (LSD1-IRES-GFP). Representative images of primary tumorspheres from these cells were shown. Scale bar = 100 μm.
(B) Quantification of primary and secondary tumorsphere from above experiments were plotted and shown (mean ± SD from three independent experiments). *p value < 0.01 when control or rescued group was compared with two groups with USP28 shRNAs and one group with LSD1 shRNA.
(C) CD44high/CD24low population in cells above was examined by FACS analysis. Quantification of CD44high/CD24low population was presented (mean ± SD from three independent experiments). *p value < 0.01 when control or rescued group was compared with two groups with USP28 shRNAs and one group with LSD1 shRNA. (D) Lysate from cells above were analyzed by Western blotting.
(E) MCF7 cells stably transfected with control, USP28 or LSD1 shRNAs or a USP28-knockdown cells with LSD1-rescued expression were injected into mammary fat pad of female SCID mice. Xenograft tumor size was measured twice a week. Data points represent the mean ± SD of 6 mice per group. The mean of tumor growth was presented in the top panel and tumors from these mice were shown in the middle panel. The mean of tumor weight was presented in the bottom panel. Statistical analysis was performed using Student’s t-test. **P value < 0.001; comparisons include: group from vector control cells or USP28-knockdown cells with LSD1 expression compared with two groups with USP28 shRNAs and one group with LSD1 shRNA.
(F) A proposed model to illustrate USP28 induces LSD1 stabilization through a deubiquitination event, and thus results in the maintenance of pluripotency by blocking cellular differentiation via suppressing H3K4me2 on LSD1-targeted differentiation gene promoters.
To further extend our findings in vivo, we implanted the following cells in the mammary fat pad of female SCID mice: (1) vector control MCF7 cells; (2) USP28- or LSD1-knockdown MCF7 cells; and (3) LSD1-rescued expression in USP28-knockdown MCF7 cells. Strikingly, knockdown of USP28 or LSD1 significantly inhibited tumor growth in vivo (Figure 7E). The sizes of tumors from these mice were significantly smaller than that from vector control mice. We also found that the level of Sox2 and Oct4 were greatly reduced in tumors with knockdown of USP28 or LSD1 (Figures S7D & S7E). Consistent with results obtained in cell culture, LSD1-rescued expression largely restored tumor growth and tumor formation (Figure 7E). In addition, expression of Sox2 and Oct4 was also restored in these tumors (Figure S7D & S7E). Furthermore, the proliferation index Ki67, which was significantly reduced in tumors from USP28- and LSD1-knockdown cells, increased in tumors derived from LSD1-rescued cells (Figure S7E). Together, these results indicate that USP28 is critical in controlling CSC-like properties and tumorigenicity through LSD1.
DISCUSSION
In this study, we found that USP28 interacted with and stabilized LSD1 from ubiquitination. We showed that USP28 was overexpressed in breast cancer cells and conferred them with CSC-like features that are mediated through LSD1 stabilization. Our study has revealed several new insights into the ubiquitination and chromatin regulation in breast cancer. First, our data indicates that USP28 is a specific DUB for LSD1, as knockdown of USP28 reduced LSD1 stability, whereas overexpression of USP28 stabilized LSD1. This event required the catalytic activity of USP28 because a catalytically inactive mutant could not stabilize LSD1. In addition, USP25, the most related member of this subtype, could not stabilize LSD1, indicating the specificity of USP28 for LSD1. We further mapped and identified the AO domain of LSD1 and the N-terminal region of USP28 as required for their mutual interactions. Although we did not notice any correlation between the mRNA of USP28 and LSD1 in several microarray datasets and various tumor cell lines, we did find a significant correlation between USP28 and LSD1 in protein level on cancer cell lines and tumors from mouse models and human breast cancer specimens, confirming that the regulation of LSD1 by USP28 is a post-translational event. In agreement with this notion, knockdown of USP28 reduced the protein level and functional activity of LSD1, but these could be rescued by exogenous LSD1 expression (Figure 7F).
The limited numbers of DUBs in the human genome suggests that each DUB may have several substrates. Indeed, USP28 has been shown to de-ubiquitinate c-Myc and p53BP1 (Popov et al., 2007; Zhang et al., 2006). Different from the interaction of USP28 with LSD1, USP28-mediated p53BP1 stabilization only occurs after DNA damage and no effect is found in the absence of DNA damage, suggesting that the interaction between USP28 with p53BP1 is regulated by DNA damaging. On the other hand, USP28 does not directly interact with Myc, but rather associates with an E3 ligase of Myc, Fbw7, to counteract Fbw7-mediated Myc ubiquitination (Popov et al., 2007). Knockdown of USP28 shifts the balance toward Fbw7-mediated ubiquitination. In our study, we found that knockdown of Myc did not affect the binding between USP28 and LSD1 (Figure S7F). In addition, LSD1-rescued expression restored, to large extent, the functions mediated by USP28-knockdown. Although our results indicate that LSD1 is a major target of USP28 in breast cancer, it does not rule out the possibility that USP28 promotes CSC-like traits through both LSD1 stabilization and inhibition of Myc degradation.
Second, our study indicates that USP28 provides an additional layer of control on CSC-like properties through LSD1 stabilization. In ESCs, the promoters of developmental regulators are simultaneously poised with bivalent marks, H3K4me3 and H3K27me3, representing both active and repressive modifications that are controlled by trithorax and polycomb (PcG) complexes, respectively. During differentiation, pluripotent genes lose active (H3K4me3) modification while maintaining the repressor mark (H3K27me3); whereas the lineage-specific developmental genes, silent in pluripotent ESCs, retain the activation modification and lose the repressive mark for expression upon differentiation. Recent studies indicate that LSD1 occupies the promoter of a subset of developmental genes containing bivalent domains (Adamo et al., 2011). High levels of LSD1 are required to tightly control the level of H3K4 methylation at the regulatory regions of developmental genes for the maintenance of pluripotency and suppression of differentiation. Consistent with this notion, knockdown of USP28 greatly enhanced H3K4me2 at the promoter region of p21Cif1/Waf1, HNF4, HoxA10 and FoxA2 and upregulated the expression of these differentiation genes directly. Although we noticed reduced occupancy of RNA polymerase II at the promoters of Sox2 and Oct4 and a corresponding downregulation of mRNA and protein of these molecules in USP28-knockdown cells, we could not detect an association of LSD1 at the Sox2 and Oct4 promoters in ChIP experiments. Our results suggest that LSD1 does not directly regulate the expression of Sox2 and Oct4. Recent studies also show that LSD1-knockdown significantly reduces the expression of Sox2 and Oct4 and that LSD1 does not directly control the expression of these genes during cellular differentiation (Adamo et al., 2011; Wang et al., 2011). Although the detailed mechanism remains elusive, it has been proposed that the differentiation genes that are directly controlled by LSD1 can indirectly suppress the expression of Sox2 and Oct4. Future thorough investigations using various cellular differentiation systems are required to provide a comprehensive understanding regarding this intricate regulation by LSD1.
In agreement with these molecular changes, the USP28-LSD1 axis is critical in controlling the differentiation and self-renewal of breast CSCs from mouse model and human cancer. Intriguingly, increased USP28 expression positively correlated with LSD1 upregulation in tumors from MMTV-Wnt1 mice but not from normal mouse mammary gland or tumors from MMTV-HER2 mice. This observation is compatible with the notion that Wnt signaling is critical in self-renewal and the maintenance of cellular pluripotency. USP28-knockdown suppressed the self-renewal abilities and enhanced differentiation capacities in CSCs from MMTV-Wnt1 tumors. USP28-knockdown also suppressed the CSC-like traits in human breast cancer cell lines in vitro and inhibited tumorigenicity in vivo. LSD1-rescued expression largely restored these defects. These data provide substantial support to our contention that USP28 is critical for controlling CSC-like properties via LSD1 stabilization in breast cancer (Figure 7F). Similar to our finding, Williams et al has shown that USP1 preserved a mesenchymal stem cell program through deubiquitination of ID protein (Williams et al., 2011). In addition, USP1 is overexpressed in gastric cancer and correlates with transformation and aggressiveness of this disease (Luise et al., 2011). Thus, our study, and those of others, point to the critical function of USP in controlling CSC-like characteristics through the regulation of several key transcriptional molecules.
Third, our study indicates that USP28 is a druggable target for inhibiting breast CSC-like characteristics (Figure 7F). CSCs are rare subpopulation that is intrinsically resistant to therapy and allows tumor re-growth and metastasis. Targeting this subpopulation of breast CSCs, as well as the other tumor cells, presents a winning approach to prevent tumor relapse. Consistent with the critical role of LSD1 in regulating CSC-like properties, inhibition of LSD1 using Parnate induced differentiation and expression retinoic acid receptor in acute myeloid leukemia (AML), and thus sensitized AML cells to growth suppression by all-trans-retinoic acid (Schenk et al., 2012). In addition, inhibition of LSD1 using Parnate suppresses colon cancer and neuroblastoma growth in vitro and in xenograft mouse models (Huang et al., 2007; Huang et al., 2009; Schulte et al., 2009). These pre-clinical studies are in line with the observation that LSD1 is overexpressed in multiple types of cancer and has tumor-promoting effects. However, commonly used LSD1 inhibitors, such as Parnate, were originally developed to inhibit Monoamine oxidase A and B for the treatment of Parkinson’s disease and other neurodegenerative disorders. They are non-selective amine oxidase inhibitors and induce substantial toxicity in vitro and in vivo. In this study, we found that USP28 was the major factor responsible for LSD1 stabilization in cancer cell lines and tumor samples, and that knockdown of USP28 decreased the level of LSD1 and suppressed CSC-like properties in vitro and tumorigenicity in vivo. Notably, copy number gained of USP28 was identified as one of the six genes that presented in both primary tumor and metastatic lesions from patients with invasive lobular breast cancer on a genome-wide sequencing study (Shah et al., 2009). In addition, expression of USP28 was strongly elevated in human breast and colon carcinoma (Popov et al., 2007). Together, our study and those of others indicate that USP28 represents a key modulator in facilitating tumor development and progression. Therapeutics targeting USP28 provides an excellent alternative approach for overcoming the nonspecific side-effects of LSD1 inhibitors in treating patients with breast cancer.
EXPERIMENTAL PROCEDURES
Antibodies and plasmids
Detailed information is provided in the Supplemental Experimental Procedures.
Small Interfering RNA (siRNA) library Screening
The human deubiquitinating enzyme siGENOME RTF Library was purchased from Dharmacon (Chicago, IL). The screen was performed according to manufacturer’s instructions. Briefly, HEK293 cells were added to the rehydrated Dharmacon RTF siRNA library plates (96-well plates). Two days later, cell lysates were extracted and the expression of endogenous LSD1 was examined by Western blotting.
Matrigel Organoid Culture
MMTV-Wnt1 tumor cells were cultured with DMEM/F12 medium supplemented with 5% Matrigel, 5 ng/ml EGF, 5% horse serum, 0.5 μg/ml hydrocortisone, 10 μg/ml insulin, 100 ng/ml Cholera toxin. Cells were seeded at 25,000/well in 24-well plate. The number of organoid and acinar srtuctures was counted 5-7 days after seeding by visual inspection.
In vivo Ubiquitination Assay
HEK293 cells were transfected with HA-ubiquitin, Myc-LSD1 and Flag-USP plasmids as indicated. The cells were treated for 6 h with 10 μM MG132 at 42 h post-transfection, and then lysed. The samples were immunoprecipitated using anti-Myc agarose (Sigma).
In vitro Deubiquitination Assay
The in vitro deubiquitination was performed as described (Dupont et al., 2009). Briefly, HA-ubiquitin and Myc-LSD1 were co-expressed in HEK293 cells. At 42 hours post-transfection, cells were treated with 10 μM MG132 for 6 hours to enrich the ubiquitinated form of LSD1. Myc-tagged ubiquitinated LSD1 was purified by immunoprecipitation with anti-Myc-agarose bead (Sigma) followed by elution with Myc peptide. In a parallel experiment, Flag-USP28 (wild-type and catalytic-inactive mutant) was expressed in HEK293 cells for 48 hours. Flag-USP28 was purified by immunoprecipitation with anti-Flag-agarose bead (Sigma) followed by elution with Flag peptide. For in vitro deubiquitination assay, purified ubiquitinated Myc-LSD1 was incubated with purified Flag-USP28 in a deubiquitination reaction buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 5% glycerol, 5mM MgCL2, 1 mM ATP and 1 mM DTT) at 300C as described by Dupont (Dupont et al., 2009). The reaction mixture was dissolved in SDS-PAGE buffer and analyzed by Western blotting.
Other methods are included in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Cathy Anthony for critical reading and editing of this manuscript. We also thank Dr. Q. Wang and Dr. C. Liu for providing DUBs plasmids. This work was supported by grants from NIH (RO1CA125454), Susan G Komen Foundation (KG081310), Mary Kay Ash Foundation (to B.P. Zhou), and American Cancer Society Research Scholar Award (RSG13187-01 to Y Wu).
REFERENCES
- Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Bennani-Baiti IM, Machado I, Llombart-Bosch A, Kovar H. Lysine-specific demethylase 1 (LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug target in chondrosarcoma, Ewing’s sarcoma, osteosarcoma, and rhabdomyosarcoma. Hum Pathol. 2012 doi: 10.1016/j.humpath.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- Fillmore C, Kuperwasser C. Human breast cancer stem cell markers CD44 and CD24: enriching for cells with functional properties in mice or in man? Breast Cancer Res. 2007;9:303. doi: 10.1186/bcr1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CT, Dovey OM, Lezina L, Luo JL, Gant TW, Barlev N, Bradley A, Cowley SM. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol Cell Biol. 2010;30:4851–4863. doi: 10.1128/MCB.00521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA., Jr. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci U S A. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA., Jr. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin Cancer Res. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeselsohn R, Brown NE, Arendt L, Klebba I, Hu MG, Kuperwasser C, Hinds PW. Cyclin D1 kinase activity is required for the self-renewal of mammary stem and progenitor cells that are targets of MMTV-ErbB2 tumorigenesis. Cancer Cell. 2010;17:65–76. doi: 10.1016/j.ccr.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci U S A. 2004;101:4158–4163. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luise C, Capra M, Donzelli M, Mazzarol G, Jodice MG, Nuciforo P, Viale G, Di Fiore PP, Confalonieri S. An atlas of altered expression of deubiquitinating enzymes in human cancer. PLoS One. 2011;6:e15891. doi: 10.1371/journal.pone.0015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magerl C, Ellinger J, Braunschweig T, Kremmer E, Koch LK, Holler T, Buttner R, Luscher B, Gutgemann I. H3K4 dimethylation in hepatocellular carcinoma is rare compared with other hepatobiliary and gastrointestinal carcinomas and correlates with expression of the methylase Ash2 and the demethylase LSD1. Hum Pathol. 2010;41:181–189. doi: 10.1016/j.humpath.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell. 2011;42:689–699. doi: 10.1016/j.molcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- Nicholson TB, Chen T. LSD1 demethylates histone and non-histone proteins. Epigenetics. 2009;4:129–132. doi: 10.4161/epi.4.3.8443. [DOI] [PubMed] [Google Scholar]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, Werb Z. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 2013;3:70–78. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T, et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71:7238–7249. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, Cao TC, Carano RA, Dixit VM. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 2011;146:918–930. doi: 10.1016/j.cell.2011.07.040. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.