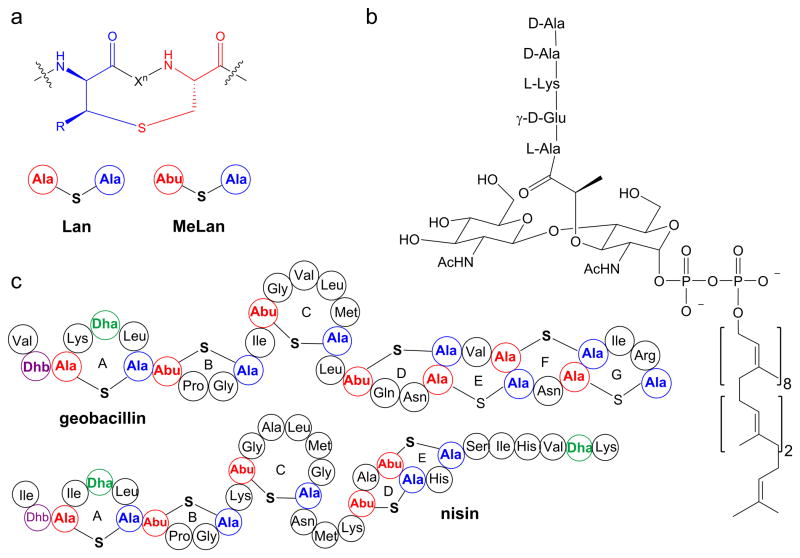
Lanthipeptides are lanthionine- and methyllanthionine- containing peptides that are ribosomally-synthesized and post-translationally modified.1 Lanthipeptides that possess antimicrobial activity are called lantibiotics.2 Lanthionines consist of two alanine residues that are linked through a thioether that connects their β-carbons, and methyllanthionines contain an additional methyl group (Figure 1a). Nisin is the best studied and longest known lantibiotic and has been used as a food preservative for over 50 years.3,4 Nisin displays antibacterial activity against clinically important pathogens such as methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococci, Streptococcus pneumoniae, and food-borne pathogens such as Clostridium botulinum, and Listeria monocytogenes.5–7 Despite its use for over 50 years, reports of resistance against nisin have been scarce.8–12 The slow development of resistance may stem from the dual mode-of-action of nisin. Nisin exhibits antimicrobial activity by binding to the pyrophosphate moiety of lipid II (Figure 1b),13,14 a membrane-bound advanced intermediate involved in the biosynthesis of the cell wall. By doing so, nisin inhibits the transglycosylation step in cell wall biogenesis and sequesters lipid II from its functional location.15,16 Furthermore, the nisin-lipid II complex leads to formation of pores in the membrane causing cell death.17
Figure 1.
Structures of a) lanthionine and methyllanthionine, b) lipid II, and c) geobacillin I and nisin. The shorthand notation for lanthionine (Lan) and methyllanthionine (MeLan) depicted in panel a is used in panel c.
Recently, we characterized two lanthipeptides, geobacillin I and geobacillin II, from the thermophilic bacterium Geobacillus thermodenitrificans NG80-2.18 Geobacillin I contains seven thioether bridges, one dehydroalanine (Dha), and one dehydrobutyrine (Dhb) (Figure 1c). The N-terminal A and B rings of geobacillin I are very similar to the corresponding rings of nisin but the C-terminal structures are very different (Figure 1c). The nisin A and B rings are involved in lipid II binding,13 and hence we anticipated that geobacillin I might also bind lipid II. The three amino acid linker peptide between the C and D rings of nisin has been shown to be indispensable for pore formation activity.17,19–21 For instance, the ΔN20ΔM21 and N20P/M21P mutants of nisin lost pore formation ability against Gram-positive bacteria.17,19 Geobacillin I has only a single amino acid between the C and D rings, similar to the ΔN20ΔM21 mutant of nisin. Thus, based on the available data on nisin, we anticipated that geobacillin I would bind to lipid II, but not form pores in the membrane of Gram-positive bacteria. In this work we tested these expectations experimentally.
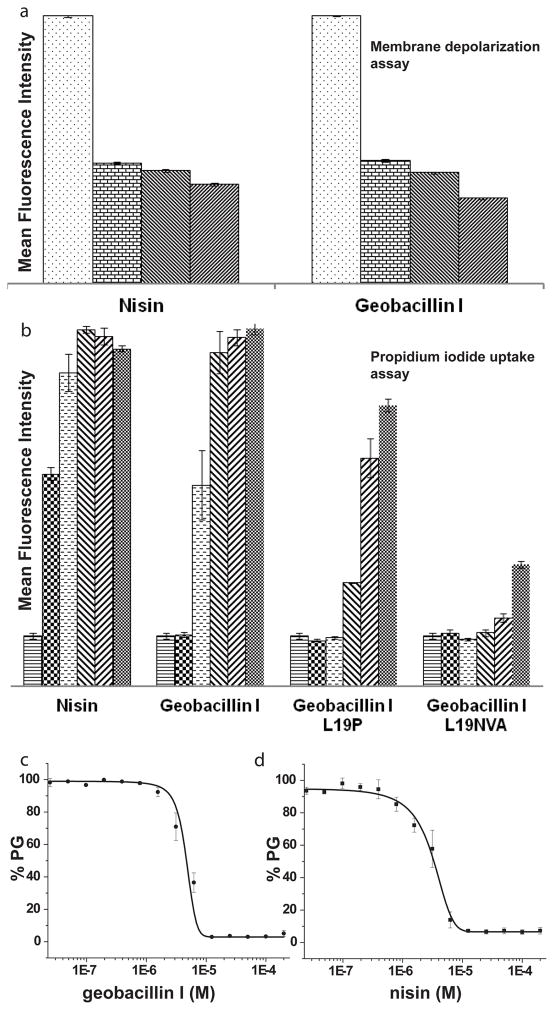
We first conducted antimicrobial activity assays in liquid medium. In these assays, geobacillin I exhibited a four-fold higher minimal inhibitory concentration (MIC) against Bacillus subtilis ATCC 6633 compared to nisin (Table 1). Flow cytometry was then used to examine changes in the polarization of the bacterial membrane of B. subtilis ATCC 6633 upon incubation with geobacillin I using the membrane potential sensitive dye 3,3′-diethyloxacarbocyanine iodide (DiOC2).22 Incubation with geobacillin I resulted in a significant decrease in mean fluorescence intensity (MFI), similar to the observations when the same experiments were carried out with nisin (Figure 2a and Supplementary Figures S1 and S2). The unexpected ability of geobacillin I to form pores despite the single amino acid linker between rings C and D may be a consequence of the overall differences between the C-terminal region of geobacillin I and nisin (Figure 1c).
Table 1.
Specific activity of nisin, geobacillin I and the geobacillin I analogues generated in this study against B. subtilis ATCC6633.
| Peptide | IC50 (μM) | MIC (μM) |
|---|---|---|
| Nisin | 0.12 ± 0.01 | 0.25 |
| Geobacillin I | 0.56 ± 0.01 | 1.0 |
| Geobacillin-L19P | 1.00 ± 0.03 | 2.0 |
| Geobacillin-L19NVA | 5.70 ± 0.08 | 8.0 |
| Geobacillin-Dha5F/L6I | 0.45 ± 0.09 | 1.0 |
Figure 2.
The effect of geobacillin I and nisin on the membrane integrity of B. subtilis ATCC 6633. (a) Average mean fluorescence intensity (MFI) of triplicate flow cytometry measurements with different concentrations of nisin and geobacillin I using DiOC2 as indicator of membrane potential. The different bars represent concentrations of 0, 0.2, 2, and 20 μM (left to right). For a representation using the ratio of fluorescence at 610 and 530 nm, see Supplementary Figure S2. (b) Increase in MFI resulting from propidium iodide uptake by B. subtilis ATCC 6633 in response to treatment with nisin and geobacillin I (average of three measurements). The different bars represent concentrations of 0, 0.31, 1.25, 2.5, 5 and 10 μM (left to right). (c) Inhibition of PBP1b-catalyzed peptidoglycan (PG) formation by geobacillin I and nisin, at a lipid II concentration of 4 μM and a PBP1b concentration of 100 nM. Error bars represent the standard deviation from triplicate experiments.
We also investigated the efficiency of pore formation by geobacillin I using propidium iodide (PI), a membrane impermeable fluorescent dye. Upon pore formation or membrane disruption, PI can enter the cell, resulting in an increase in fluorescence intensity because of the interaction of PI with nucleic acids. PI uptake was monitored at nine different concentrations with each experiment conducted in triplicate (Figure 2b and Supplementary Figure S3). The data showed only two-fold lower efficiency in PI uptake for geobacillin I, with IC50 values for nisin at 0.3 μM compared to 0.6 μM for geobacillin I (Figure 2b and Supplementary Figure S3).
Previously, site-saturation mutagenesis was performed on the amino acids in the linker between the C- and D-rings of nisin. The antibiotic activity of the nisin mutants N20P, M21V, K22T, and K21A was increased against several pathogenic bacteria.23 Conversely, introduction of a double Pro in this region (N20P/M21P) strongly decreased pore formation;17 this double mutant as well as the single M21P mutant also had strongly reduced antimicrobial activities. 17,23 To evaluate how such mutations would affect the activity of geobacillin I, site-directed mutagenesis was used in this study to replace the naturally occuring Leu19 in geobacillin I with Pro, and to introduce the tripeptide AsnValAla as linker between the C- and D-rings, thus generating a linker sequence that combines two of the mutations in the nisin variants with improved activity. These analogs were generated by co-expression of mutants of the precursor peptide GeoAI with the modification enzymes GeoB and GeoC in Escherichia coli as previously reported for the production of wild type geobacillin I (Supplementary Figure S4; Tables S1 and S2).18 Compared to wild type geobacillin, the analogues with NVA and P as the linker between the C and D rings had eight-fold and two-fold increased MIC values, respectively (Table 1). The ability to induce pore formation by these analogues was also investigated. Although the efficiency of pore formation was strongly reduced, replacement of Leu19 with Pro did not abolish this activity (Figure 2b). Introduction of the amino acid residues NVA in this region region also greatly reduced formation of pores in the bacterial cell membrane by geobacillin I. Thus, mutations in the linker peptide between the C- and D-rings affect the activities of nisin and geobacillin quite differently, suggesting that the detailed mechanism of pore formation by geobacillin I differs from that of nisin. These findings also suggest that the structure of the C-terminus of class I lantibiotics may vary significantly while retaining pore formation activity.
The ability of geobacillin I to bind lipid II was investigated next using in vitro inhibition of the transglycosylation reaction catalyzed by penicillin-binding protein 1b (PBP1b) from E. coli. PBP1b uses lipid II as a substrate for glycan polymerization.24 Geobacillin I inhibited PBP1b-catalyzed peptidoglycan formation using 4 μM heptaprenyl lipid II with a half-maximal inhibitory concentration (IC50) of 4.6 ± 0.8 μM (Figure 2c). For comparison, inhibition by nisin under the same conditions displayed an IC50 of 2.9 ± 0.6 μM (Figure 2c). Thus, the inhibitory activity of the two peptides is very similar.
Geobacillin I has higher stability at physiological pH compared to nisin.18 Although higher stability for a compound from a thermophile is not unexpected, the higher stability was somewhat surprising because nisin degradation at neutral pH is believed to be caused by non-enzymatic hydrolysis at Dha5,25 a residue that is also present in geobacillin I. We wondered whether the stability and hence antimicrobial activity of geobacillin could be further improved by mutation of Dha5 in light of a previous report that the nisin analog I4K/Dha5F/L6I had higher antimicrobial activity against various bacteria.26 Geobacillin I already has a Lys at position 4, and hence the mutant Dha5F/L6I was generated, but it proved to be only slightly more stable than the wild type geobacillin I (Supplementary Figure S4) while displaying similar MIC values (Table 1).
Compared to nisin, ring C of geobacillin I is reduced in size by one amino acid, the region between rings C and D is reduced in length by two amino acids, and ring E is a lanthionine ring as opposed to a methyllanthionine ring (Figure 1c). Furthermore, geobacillin I contains two additional thioether bridges at its C-terminus. However, the antimicrobial activity and the overall mode of action of the two lantibiotics appear to be quite similar: binding to lipid II and pore formation.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01 GM58822 to W.A.V. and R01 GM076710 to S.W.). C.G.G was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award 5 T32 GM070421 from the National Institute of General Medical Sciences
Footnotes
This study is dedicated to Prof. Christopher T. Walsh for his inspiring leadership in natural product biosynthesis and mechanistic enzymology.
Supplementary Information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja).
References
- 1.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 2.Schnell N, et al. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature. 1988;333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 3.Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek. 1996;69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 4.Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci. 2008;65:455–476. doi: 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brumfitt W, Salton MR, Hamilton-Miller JM. Nisin, alone and combined with peptidoglycan-modulating antibiotics: activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci. J Antimicrob Chemother. 2002;50:731–734. doi: 10.1093/jac/dkf190. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein BP, Wei J, Greenberg K, Novick R. Activity of nisin against Streptococcus pneumoniae, in vitro, and in a mouse infection model. J Antimicrob Chemother. 1998;42:277–278. [PubMed] [Google Scholar]
- 7.Mota-Meira M, LaPointe G, Lacroix C, Lavoie MC. MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob Agents Chemother. 2000;44:24–29. doi: 10.1128/aac.44.1.24-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breukink E, de Kruijff B. Lipid II as a target for antibiotics. Nat Rev Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 9.Schneider T, Sahl HG. Lipid II and other bactoprenol-bound cell wall precursors as drug targets. Curr Opin Investig Drugs. 2010;11:157–164. [PubMed] [Google Scholar]
- 10.Kramer NE, et al. Increased D-alanylation of lipoteichoic acid and a thickened septum are main determinants in the nisin resistance mechanism of Lactococcus lactis. Microbiology. 2008;154:1755–1762. doi: 10.1099/mic.0.2007/015412-0. [DOI] [PubMed] [Google Scholar]
- 11.Gravesen A, Sørensen K, Aarestrup FM, Knøchel S. Spontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibiotics. Microb Drug Resist. 2001;7:127–135. doi: 10.1089/10766290152045002. [DOI] [PubMed] [Google Scholar]
- 12.Khosa S, Alkhatib Z, Smits SH. NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol Chem. 2013 doi: 10.1515/hsz-2013-0167. [DOI] [PubMed] [Google Scholar]
- 13.Hsu ST, et al. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol. 2004;11:963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 14.Brötz H, et al. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol. 1998;30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 15.Breukink E, et al. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 16.Hasper HE, et al. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science. 2006;313:1636–1637. doi: 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- 17.Wiedemann I, et al. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem. 2001;276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 18.Garg N, Tang W, Goto Y, Nair SK, van der Donk WA. Lantibiotics from Geobacillus thermodenitrificans. Proc Natl Acad Sci USA. 2012;109:5241–5246. doi: 10.1073/pnas.1116815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasper HE, de Kruijff B, Breukink E. Assembly and stability of nisin-lipid II pores. Biochemistry. 2004;43:11567–11575. doi: 10.1021/bi049476b. [DOI] [PubMed] [Google Scholar]
- 20.Brötz H, Sahl HG. New insights into the mechanism of action of lantibiotics--diverse biological effects by binding to the same molecular target. J Antimicrob Chemother. 2000;46:1–6. doi: 10.1093/jac/46.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Yuan J, Zhang ZZ, Chen XZ, Yang W, Huan LD. Site-directed mutagenesis of the hinge region of nisinZ and properties of nisinZ mutants. Appl Microbiol Biotechnol. 2004;64:806–815. doi: 10.1007/s00253-004-1599-1. [DOI] [PubMed] [Google Scholar]
- 22.Oman TJ, van der Donk WA. Insights into the Mode of Action of the Two-Peptide Lantibiotic Haloduracin. ACS Chem Biol. 2009;4:865–874. doi: 10.1021/cb900194x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Field D, Connor PM, Cotter PD, Hill C, Ross RP. The generation of nisin variants with enhanced activity against specific gram-positive pathogens. Mol Microbiol. 2008;69:218–230. doi: 10.1111/j.1365-2958.2008.06279.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, et al. Vancomycin analogues active against vanA-resistant strains inhibit bacterial transglycosylase without binding substrate. Proc Natl Acad Sci USA. 2003;100:5658–5663. doi: 10.1073/pnas.0931492100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lian LY, et al. Solution structures of nisin A and its two major degradation products determined by n.m.r. Biochem J. 1992;283:413–420. doi: 10.1042/bj2830413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rink R, et al. Dissection and modulation of the four distinct activities of nisin by mutagenesis of rings A and B and by C-terminal truncation. Appl Environ Microbiol. 2007;73:5809–5816. doi: 10.1128/AEM.01104-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.