Abstract
MicroRNAs (miRNAs) are epigenetic regulators of eukaryotic gene expression and play key roles in many cellular processes. However, the role of miRNAs for replicative senescence of normal human keratinocytes (NHKs) remains unknown. Thus, we examined the expression profiles of 847 miRNAs in exponentially replicating and senescent NHKs and identified 126 senescence-associated miRNAs (SA-miRs). Among SA-miRs, 117 miRNAs (93%) were upregulated and 9 miRNAs (7%) were downregulated in senescent NHKs compared to those of exponentially replicating cells. Among the above miRNAs, we selected two miRNAs, miR-137 and miR-668, for further investigation because they were consistently upregulated with replicative senescence of three independent NHK cultures. Ectopic overexpression of miR-137 or miR-668 induced senescence in rapidly proliferating NHKs; a notable increase in senescence-associated β-galactosidase activity, p16INK4A and p53 was observed, indicating that they are novel senescence-inducing miRNAs. In addition, these senescence-inducing miRNAs were gradually increased during organismal aging of normal human oral epithelia. We also detected downregulation of miR-137 and miR-668 in many tested human head and neck squamous cell carcinoma cell lines. Since senescence would be viewed as a potent tumor suppressive pathway, the newly identified senescence-inducing miRNAs deserve to be further investigated for their therapeutic application in cancer treatment.
Keywords: senescence, microRNA, normal human keratinocytes
Introduction
MicroRNAs (miRNAs) have recently come into focus as a novel class of epigenetic regulators of eukaryotic gene expression (1). They are short 18–24nt RNA molecules that post-transcriptionally regulate protein expression through direct binding to specific mRNA molecules. The specific binding of miRNAs to the cognate mRNA can either promote mRNA degradation or prevent translation of the mRNA. miRNA regulation of gene expression plays a role in a variety of cellular processes, and dysregulation of miRNAs have been reported to be involved in the pathogenesis of age-related human diseases, such as cancer, heart disease and Alzheimer’s disease (2).
Replicative senescence entails an irreversible arrest of cell division; mitogenic stimulation of senescent cells does not evoke entry into the S phase (3). Senescent cells exhibit senescence-associated β-galactosidase (SA β-gal) activity (4). Previously, we characterized the phenotypic alterations that occur in normal human keratinocytes (NHK) during in vitro replication (5). Primary NHK underwent 22±3 population doublings (PDs) when serially subcultured. At the end of their replicative life span, these cells spontaneously ceased replicating and exhibited morphological and cytochemical features of replicative senescence. We also reported that the onset of senescence in NHK is associated with altered expression of the senescence-associated genes, including cell cycle regulators, G protein-coupled receptors, apolipoproteins, matrix metalloproteinases, and mitochondrial proteins (6). Although our previous studies provide evidence of the complex changes in gene expression profiles during senescence of NHK, the role of miRNAs in senescence of NHK has not been investigated.
Recently, several lines of evidence have supported the role of miRNAs in senescence. First, overexpression of miR-20a induced senescence in mouse embryonic fibroblasts (7). Second, miRNA profiling assay identified specific subsets of miRNAs as significantly altered during senescence of mesenchymal stem cells (8), diploid fibroblasts (9), trabecular meshwork cells (9), and endothelial cells (10). Third, overexpression of miR-34a and miR-449a induced senescence-like phenotypes in human cancer cells (11,12). Although recent studies suggested potential roles of miRNAs in senescence, the role of miRNA in senescence of normal human keratinocytes has been poorly documented (13).
Here, we report the identification of novel senescence-inducing miRNAs in NHK. miR-137 and miR-668 were upregulated during both replicative and organismal aging of NHK. Overexpression of miR-137 and miR-668 markedly increased SA β-gal activity and senescence markers, p16INK4A and p53 in exponentially replicating NHK. Moreover, both senescence-inducing miRNAs were frequently downregulated in human head and neck squamous cell carcinoma (HNSCC) cells compared to NHK. Since senescence is considered as a tumor suppressor mechanism (14), the newly identified senescence-inducing miRNAs need to be tested for novel senescence markers and therapeutic application for cancer.
Materials and methods
Cell culture
Primary NHK were prepared from separated epithelial tissue and serially subcultured in Keratinocyte Growth Medium (Loza) as described previously (5). Six HNSCC cell lines (SCC-4, SCC-9, SCC-15, 1483, Tu-139 and Tu-177) were used in this study. Detailed culture condition for the cell lines can be found in our previous publication (15).
miRNA expression profiling
miRNA expression profiling of 847 human miRNAs was performed in NHK at the exponential replicating (PDs 9; young) and senescent (PDs 20; old) phases using the Genome-wide qPCR microRNA Array Panels by a service provider (System Biosciences).
Quantitative real-time RT-PCR analysis of miRNA
Total RNA was extracted from cells using Tri zol Reagent (Invitrogen). A cDNA pool of miRNAs was synthesized by QuantiMir cDNA Kit (System Biosciences) according to the manufacturer’s protocol. Human miRNAs were amplified as described in the SYBR Green I Master PCR Mix (Roche) with the LightCycler 480 II real-time PCR system (Roche). For miRNA amplification, one primer is miRNA specific and the other is a universal primer. U6 small nuclear RNA (snRNA) level was used as internal control for the starting amount of cDNA. Primer sequences are available upon request.
Overexpression of miRNA
Lentiviral vectors expressing miR-137 or miR-668 were obtained from SBI’s microRNA Precursor Construct Collection (System Biosciences). Detailed methods of lentivirus production can be found in our previous publications (16). Exponentially replicating NHK were transduced with the lentiviruses that express miR-137 (LV-miR137), miR-668 (LV-miR668) or green fluorescent protein alone (LV-GFP) as a control in the presence of polybrene (6 μg/ml) for 3 h. Efficient transduction and overexpression of the miRNAs were determined by GFP epifluorescence and quantitative real-time RT-PCR, respectively.
SA β-gal activity
Cells were fixed in 2% formaldehyde/0.2% glutaraldehyde for 3–5 min at room temperature. The cells were then stained for SA β-gal activity in freshly prepared staining solution. The presence of SA β-gal activity was evidenced by a dark-green color in the perinuclear cytoplasmic region. SA β-gal positive cells were scored by counting at least 1,000 cells from random microscope fields to allow for statistical validation. The detailed experimental procedure has been previously described (5).
Cell proliferation
To investigate the effect of miRNA on the kinetics of replication, the growth rates of NHK transduced with LV-GFP or LV-miR were compared. Two days post-transduction, cells were plated at 2×104 cells per plate into 60-mm Petri dishes and incubated at 37°C. The number of cells was counted at 2 and 4 days post-subculture.
Western blotting
Whole-cell extracts were isolated, separated by SDS-PAGE and transferred to Immobilon protein membranes (Millipore). Antibodies for p16INK4A (C-20), p53 (DO-1), CDK5 (DC-17), CDK6 (C-21), hTERT (H-231), p21 (C-19), TGF-β (V), actin (I-19) were purchased from Santa Cruz Biotechnology. The detailed procedures have been previously described (16).
Laser-capture microdissection of normal human oral epithelium
Normal human oral epithelia were microdissected using a Pixcell II Laser Capture Microscope (Arcturus Engineering) in the UCLA Human Tissue Research Center Core Facility. From multiple sections of each case, ~4,000–5,000 cells were microdissected from the 5-μm-thick histologic sections and subjected to RNA isolation with High Pure RNA Isolation Kit (Roche).
Results
Identification of senescence-associated miRNAs (SA-miRs) in NHK
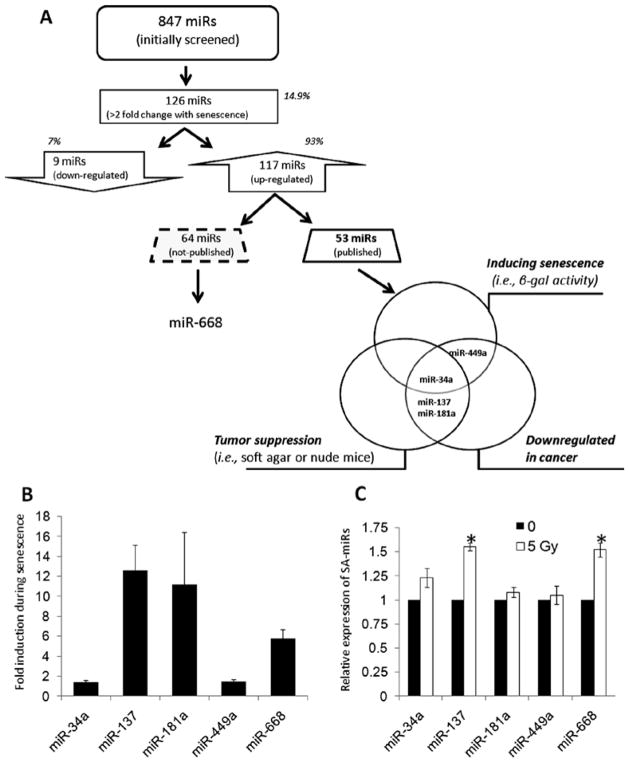
In order to understand the miRNA signature that is specific for the senescence of NHK, we compared the expression profiling of human miRNAs in NHK at the exponentially replicating (young; 9 PDs) and senescent (old; 20 PDs) phases using the Genome-wide qPCR microRNA Array Panels (System Biosciences). The assay revealed global up-regulation of miRNA expression during senescence in NHK. We identified 126 SA-miRs altered >2-fold with senescence. Among them, 117 SA-miRs (93%) were up-regulated and 9 SA-miRs (7%) were downregulated in senescent NHK compared to exponentially replicating NHK (Fig. 1A).
Figure 1.
Identification of senescence-associated miRNAs (SA-miRs) in NHK. (A) Schematic strategy for the identification of SA-miRs in NHK. (B) Confirmation of the up-regulation of five SA-miRs (miR-34a, 137, 181a, 449a, and 668) during senescence of NHK. Three independent cultures of NHK derived from different donors were serially propagated and used in this assay. The levels of mature SA-miR expression were measured by quantitative real-time RT-PCR. For each sample, the expression of miRNA was normalized to U6 snRNA. The value for miRNA from exponential replicating NHK was set at 1, and the relative amounts of miRNA from senescent NHK were plotted as fold induction. The graph shows the mean of three experiments. Error bar represents the standard deviation. (C) Regulation of SA-miRs in IR-induced premature senescence. Exponential replicating NHK were exposed to 0 or 5 Gy and harvested for determining the levels of the SA-miRs. The value for miRNA from control NHK (0 Gy) was set at 1, and the relative amounts of miRNA from NHK exposed IR were calculated. *Significantly different (p<0.0, Student’s t-test) from the control expression.
Of these 117 up-regulated SA-miRs, we found that to date 53 (45.3%) were previously reported and 64 (54.7%) were not reported. Cellular senescence is viewed as a potent tumor suppressor mechanism (14). Thus, we further classified the reported 54 SA-miRs into three different groups: i) miRNAs reported to induce senescence phenotype in human cells; ii) miRNAs with tumor suppressive activity against human cancer; and iii) miRNAs reported to be up-regulated in human cancer. Fig. 1A summarizes the result and reveals that miR-34a, miR-137, miR-181a, and miR-449a belong to at least 2 different groups.
Confirmation of expression profiles of SA-miRs
In order to confirm the up-regulation of miR-34a, miR-137, miR-181a, and miR-449a found in the microRNA array experiment and rule out the donor-to-donor variation, three independent cultures of NHK derived from different donors were obtained and serially propagated. Three sets of exponentially replicating and senescent NHK cultures were used to determine the levels of miR-34a, miR-137, miR-181a, and miR-449a using quantitative real-time RT-PCR. We also included miR-668 which is most upregulated miRNA among the not-reported miRNAs. The expression of all tested miRNAs was consistently enhanced in three different senescent NHK cultures compared to their corresponding actively replicating NHK cultures, consistent with the profiling results (Fig. 1B). These data indicated that the SA-miR expression indeed increases during replicative senescence of NHK and is independent of the donor variation.
To further understand the expressional status of the SA-miRs in stress-induced premature senescence, we irradiated actively replicating NHK with 5 Gy and determined the levels of the five SA-miRs. Our unpublished data showed that 5 Gy of ionizing radiation (IR) induced premature senescence in NHK (unpublished data). Among the 5 SA-miRs, miR-137 and miR-668 were significantly up-regulated by IR (Fig. 1C). However, the magnitudes of induction were much less than those observed in replicative senescence. Our results indicate that up-regulation of miR-137 and miR-668 are common regulatory event shared between replicative senescence and IR-induced premature senescence.
Overexpression of miR-137 or miR-668 induces senescence in NHK
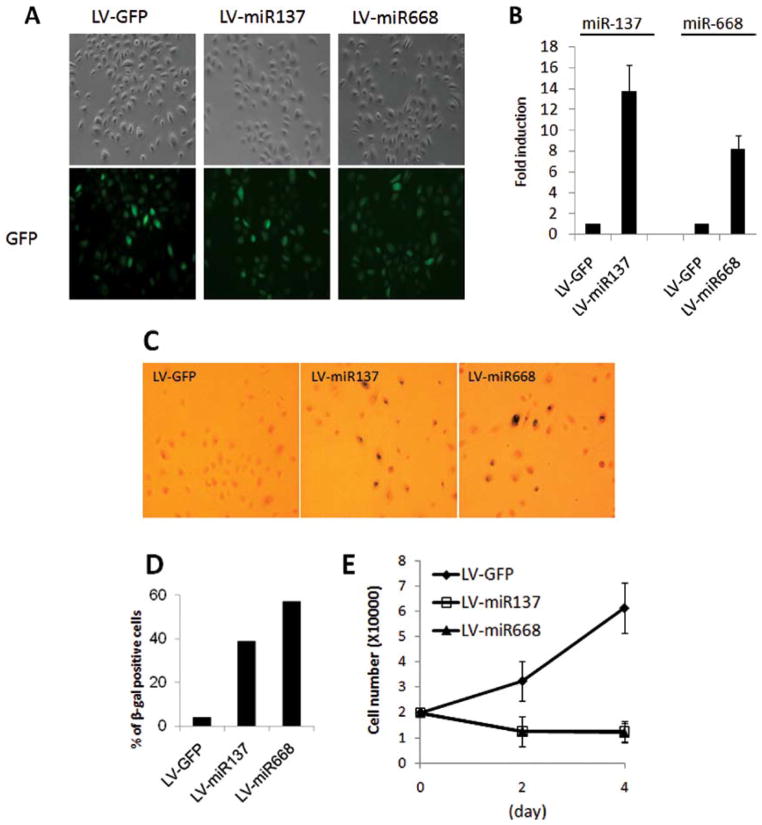
To investigate the role of miR-137 and miR-668 on senescence of NHK, we constructed lentiviral vectors capable of expressing miR-137 (LV-miR137) or miR-668 (LV-miR668) using the pMIRNA1 plasmid (System Biosciences), which also contains GFP expression cassette under a heterologous promoter. Actively replicating NHK (7 PDs) were transduced with LV-miR137, LV-miR668, or control LV-GFP, expressing GFP alone. At 3 days post-transduction, >95% of the cultures demonstrated green fluorescence (Fig. 2A), indicating efficient transduction of NHK. Also, the expression levels of mature miR-137 and miR-668 were increased by 14- and 8-fold, respectively, in NHK transduced with LV-miRs compared to the LV-GFP-transduced cells (Fig. 2B).
Figure 2.
Overexpression of miR-137 or miR-668 causes senescence in NHK. (A) Rapidly replicating NHK were transduced by lentiviruses that express miR-137 (LV-miR137), miR-668 (LV-miR668), or GFP alone (LV-GFP) for 3 h. Three days post-transduction, phase contrast photographs and GFP epifluorescence images were obtained. GFP fluorescence revealed those cells that were transduced by the lentiviruses. Original magnification x100. (B) Three days post-transduction of NHK, the levels of miR-137 and miR-668 expression were measured by quantitative real-time RT-PCR. The value for miRNA from NHK transduced with LV-GFP was set at 1, and the relative amounts of miRNA from NHK transduced with LV-miR137 or LV-miR668 were plotted as fold induction. (C) SA β-gal staining of each culture was obtained 3 days post-transduction. Original magnification x100. (D) SA β-gal positive cells were scored by counting 1,000 cells with normal light microscopy. (E) Proliferation kinetics of NHK transduced with LV-GFP, LV-miR137, or LV-miR668 was determined. One day post-transduction, 2×104 cells were plated, and the number of cells was counted after 2 and 4 days of incubation. Results were expressed as the average number of triplicate cultures.
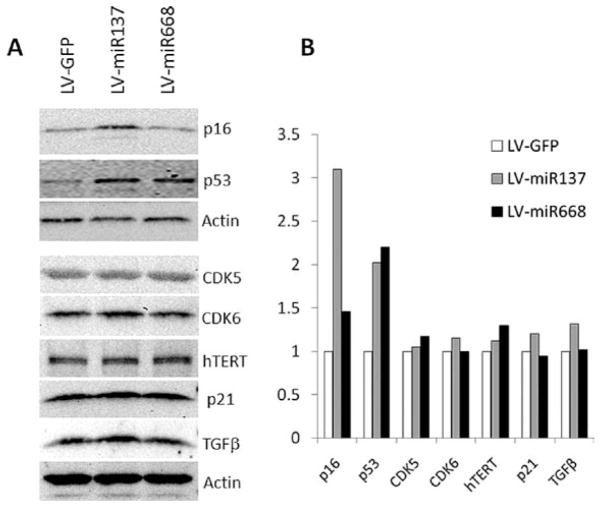
To evaluate the effect of miR-137 and miR-668 on senescence in NHK, SA β-gal activity, a marker for senescence, was determined at 3 days post-transduction. Overexpression of miR-137 or miR-668 increased the percentage of SA β-gal positive cells by 8- to 11-fold over control and markedly inhibited cell proliferation in actively replicating NHK (Fig. 2C–E). We also tested whether key senescence markers, p16INK4A and p53, are induced during the senescence mediated by miR-137 and miR-668. We found that miR-137 increased both p16INK4A and p53, and that miR-668 induced p53 in NHK (Fig. 3). Similar observations were obtained using an independent NHK culture from a separate donor (data not shown). We also investigated the effect of miR-137 and miR-668 on other senescence-associated proteins, such as CDK5, CDK6, hTERT, p21, and TGF-β (Fig. 3). None of the tested proteins was significantly altered by either miRNA. Our findings indicated that miR-137 and miR-668 induced premature senescence in NHK, suggesting that they are novel senescence-inducing miRNAs.
Figure 3.
Effect of miR-137 and miR-668 on the expression of senescence-associated proteins. (A) The expression levels were determined by Western blotting at 3 days post-transduction. (B) The band intensities (shown in A) were quantitated and plotted by Scion Image software against those of actin.
Levels of miR-137 and miR-668 expression in organismal aging and human HNSCC
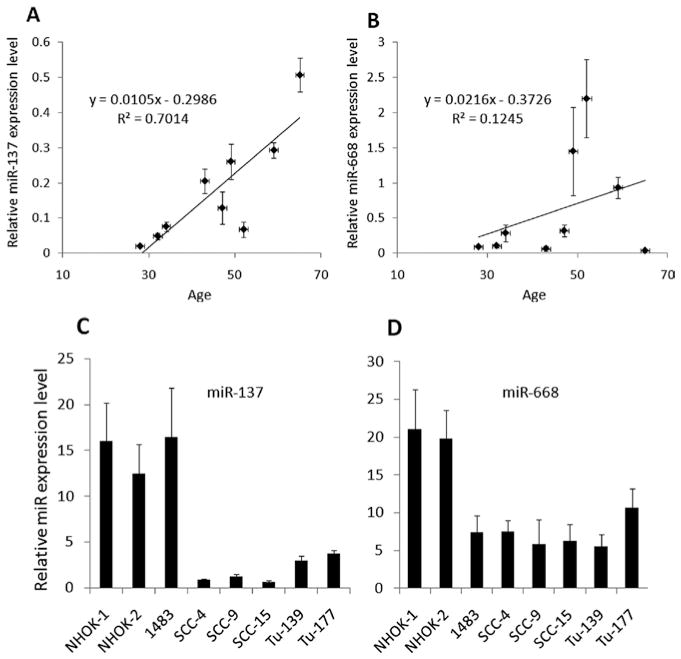
To further investigate whether the up-regulation of miR-137 and miR-668 also occurs in organismal aging, we measured the levels of the miRNAs in normal human oral epithelia obtained from individuals of various ages. Although the sample size was rather small in the study, our data indicate that an increase in the level of miR-137 is associated with an increase in age [correlation coefficients (R2) = 0.7014; Fig. 4A]. This indicates that the up-regulation of miR-137 is a common event during replicative senescence and organismal aging of human oral epithelial cells. However, such association was not observed in the levels of miR-668 in the tissues (R2 = 0.1245; Fig. 4B).
Figure 4.
Status of miR-137 and miR-668 in organismal aging and HNSCC cell lines. (A and B) Age-dependent expression of miR-137 and miR-668 in vivo. Normal epithelial cells were microdissected from normal human oral epithelium. The levels of mature miR-137 (A) and miR-668 (B) expression were measured by quantitative real-time RT-PCR. R2, coefficient of determination. y, the equation of a line. (C and D) Downregulation of miR-137 and miR-668 expression in HNSCC. The levels of miR-137 (C) and miR-668 (D) were measured by quantitative real-time RT-PCR in normal and cancer cell lines. Two independent cultures of NHK (NHK-1 and -2) and six HNSCC cell lines (1483, SCC-4, SCC-9, SCC-15, Tu-139, and Tu-177) were used.
The ability of miR-137 and miR-668 to induce premature senescence and growth inhibition in NHK indicates possible tumor suppressive effects, as many of known tumor suppressor genes, e.g., p53, pRb, and p16INK4A, are also involved in senescence pathway (14). Therefore, to examine whether miR-137 and miR-668 are implicated in human HNSCC, we measured the levels of the miRNAs in six HNSCC cell lines (1483, SCC-4, SCC-9, SCC-15, Tu-139, and Tu-177) and compared them to that in NHK (Fig. 4C and D). To rule out donor variation in the levels of miRNAs, two independent NHK cultures (NHK-1 and -2) derived from different donors were used for this experiment. Our results showed that miR-137 and miR-668 were significantly downregulated in the majority of the tested cancer cell lines compared with the two NHK cultures (Fig. 4C and D). This suggests that downregulation of miR-137 and miR-668 could be involved in head and neck carcinogenesis.
Discussion
The major findings of the present study are: i) global up-regulation of miRNA expression during senescence of NHK; ii) identification of senescence-associated miRNAs in NHK; iii) up-regulation of miR-137 and miR-668 during replicative and organismal aging; iv) their ability to induce senescence in young NHK with a concomitant induction of senescence markers, p53 and p16INK4A; and v) frequent downregulation of miR-137 and miR-668 in HNSCC.
In this study, we identified senescence-associated miRNAs, using the well-characterized senescence model of NHK (5). In our model, we defined in vitro replication of NHK into three distinct phases: exponential, senescing, and senescent phases (5). Unlike most human somatic cells, NHK demonstrated telomerase activity and replicated in vitro with delayed or undetectable level of telomere shortening (17). Thus, the senescence mechanism in NHK appears to be specific and different from that of telomerase-negative primary human cells, e.g., human diploid fibroblasts (17). Initial miRNA profiling assay revealed that a subset of miRNA contributes to senescence of NHK. Approximately 90% of the tested miRNAs were upregulated in senescent NHK, indicating global up-regulation of miRNA during replicative senescence of NHK. This finding also suggests that miRNAs may be regulated by epigenetic mechanism, i.e., DNA methylation. Indeed, many studies have reported that global DNA demethylation during cellular and organismal senescence (18–20) and miRNA regulation by DNA methylation (21,22).
Antiproliferative effects of senescence pathways have been viewed as a potent tumor suppressor mechanism (14). Cellular senescence can be mediated by the two main tumor suppressor pathways of the cell, the ARF/p53 and the p16INK4A/RB pathways. Loss of these tumor suppressor genes is critical for the establishment of in vitro human cell cultures, and is frequently observed in human cancer cells, underlining the linkage between senescence and tumor suppressor pathways. Recent studies suggest that miRNAs contribute to the development of human cancers by functioning as tumor suppressors or oncogenes (23). In this regard, tumor suppressive miRNAs are often downregulated in tumors, and the reduction in the expression may cause increased expression of the oncogenic target gene(s) (24,25). Since senescence is mediated by the major tumor suppressive pathways, senescence-inducing miRNAs may have tumor suppressive effects. However, the presence of senescence-inducing miRNAs and the functional linkage between the miRNAs and tumor suppressor pathways have yet to be established.
The ability of miR-137 and miR-668 to induce premature senescence and growth inhibition in NHKs suggests possible tumor suppressive effects, as many of the known tumor suppressor genes, such as p53, pRb, and p16, are also involved in the senescence pathway (14). In agreement with a previous report by Kozaki et al, our study also revealed frequent reduction of miR-137 in human head and neck cancer cell lines. miR-137 is known as a tumor suppressive miRNA and frequently downregulated in various human cancer, including oral cancers (22). Furthermore, the overexpression of miR-137 inhibited cancer cell growth in vitro (22). The possible role of miR-137 in the inhibition of cancer cell growth is also supported by its ability to induce cell cycle G1 arrest (22,26). The cell cycle ability of miR-137 was explained by identification of its targets, CDK6 and Cdc42 (22,26). However, we failed to observe the same effect of miR-137 on CDK6 in NHK, indicating the cell type specific effect of miRNAs. Indeed, we observed that miR-137 downregulated CDK6 expression in the SCC-4 cell line (data not shown).
miR-668 is located in chromosomal region 14q32.31 and significantly up-regulated in the two senescence states, suggesting that the up-regulation of miR-668 may progressively lead NHK to the senescent state. This supports the notion that the transient expression of miR-668 induces growth arrest and premature senescence in NHK. In order to understand a possible role of miR-668 in senescence, we performed database search of the term miR-668, using pubmed and google scholar (as of June 22, 2010). The literature search revealed 2 studies with plausible evidence for regulation of senescence by miR-668. miR-668 was upregulated by benzene, known oxidative DNA damaging agent and carcinogen (27). Since DNA damage is an important senescence-inducing endogenous stimulus, increased miR-668 expression during senescence is possibly through the generation of oxidative DNA damage during replicative senescence (28,29). Although there is no experimental evidence, integrating of miRNA cluster information with protein-protein interaction network data speculated that circadian rhythm is modulated by miR-668 (30). Aging is associated with a variety of alterations of circadian rhythms (31). Therefore, aberrant miR-668 expression could impair circadian rhythmicity and lead to cellular senescence (32).
In conclusion, our study demonstrated that miRNAs are critical modulators of cellular senescence in normal human epithelial cells. We further identified miR-137 and miR-668 as novel senescence-inducing miRNAs. Our study also suggests the possible use of miR-137 and miR-668 as novel biomarkers of cellular aging and therapeutic application for HNSCC.
Acknowledgments
This study was supported by UCLA School of Dentistry faculty seed grant (to K.-H.S,), Department of Pathology and Laboratory Medicine, Research Service Research Fund (to K.-H.S.), R01DE18295 (to M.K.K.), K02DE18959 (to M.K.K.) and K08DE17121 (to R.H.K.) from the National Institute of Dental and Craniofacial Research (NIDCR).
References
- 1.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garofalo M, Condorelli G, Croce CM. MicroRNAs in diseases and drug response. Curr Opin Pharmacol. 2008;8:661–667. doi: 10.1016/j.coph.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 4.Dimri GP, Lee X, Basile G, Ascosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, Peacocke M, Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang MK, Bibb C, Baluda MA, Rey O, Park N-H. In vitro replication and differentiation of normal human oral keratinocytes. Exp Cell Res. 2000;258:288–297. doi: 10.1006/excr.2000.4943. [DOI] [PubMed] [Google Scholar]
- 6.Kang MK, Kameta A, Shin K-H, Baluda MA, Park N-H. Senescence associated genes in normal human oral keratinocytes. Exp Cell Res. 2003;287:272–281. doi: 10.1016/s0014-4827(03)00061-2. [DOI] [PubMed] [Google Scholar]
- 7.Poliseno L, Pitto L, Simili M, Mariani L, Riccardi L, Ciucci A, Rizzo MM, Evangelista M, Mercatanti A, Pandolfi PP, Rainaldi G. The proto-oncogene LRF is under post-transcriptional control of MiR-20a: implications for senescence. PLoS One. 2008;3:e2542. doi: 10.1371/journal.pone.0002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V, Ho AD. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008;3:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G, Luna C, Qiu JD, Epstein L, Gonzalez P. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing. 2009;130:731–741. doi: 10.1016/j.mad.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Mück C, Laschober GT, Lepperdinger G, Sampson N, Berger P, Herndler-Brandstetter D, Wieser H, Kühnel M, Strasser A, Rinnerthaler M, Breitenbach M, Mildner M, Eckhart L, Tschachler E, Trost A, Bauer JW, Papak C, Trajanoski Z, Scheideler M, Grillari-Voglauer R, Grubeck-Loebenstein B, Jansen-Dürr P, Grillari J. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 2010;9:291–296. doi: 10.1111/j.1474-9726.2010.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR 449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 13.Lafferty-Whyte K, Cairney CJ, Jamieson NB, Oien KA, Keith WN. Pathway analysis of senescence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim Biophys Acta. 2009;1792:341–352. doi: 10.1016/j.bbadis.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:27–31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- 15.Min BM, Baek JH, Shin K-H, Gujuluva CN, Cherrick HM, Park N-H. Inactivation of the p53 gene by either mutation or HPV infection is extremely frequent in human oral squamous cell carcinoma cell lines. Eur J Cancer (Oral Oncol) 1994;30B:338–345. doi: 10.1016/0964-1955(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 16.Shin K-H, Kim RH, Kang MK, Kim R, Kim S, Lim PK, Yochim JM, Baluda MA, Park N-H. p53 promotes the fidelity of DNA end-joining activity by, in part, enhancing the expression of heterogeneous nuclear ribonucleoprotein G. DNA Repair. 2007;6:830–840. doi: 10.1016/j.dnarep.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang K, Guo W, Park N-H. Replicative senescence of normal human oral keratinocytes is associated with the loss of telomerase activity without shortening of telomeres. Cell Growth Differ. 1998;9:85–95. [PubMed] [Google Scholar]
- 18.Zhang W, Ji W, Yang J, Yang L, Chen W, Zhuang Z. Comparison of global DNA methylation profiles in replicative versus premature senescence. Life Sci. 2008;8:475–480. doi: 10.1016/j.lfs.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Mays-Hoopes LL. DNA methylation in aging and cancer. J Gerontol. 1989;44:35–36. doi: 10.1093/geronj/44.6.35. [DOI] [PubMed] [Google Scholar]
- 20.Wilson L, Smith RA, Mag S, Richard CG. Genomic 5-methyl-deoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- 21.Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455–1459. [PubMed] [Google Scholar]
- 22.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. exploration of tumor suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 24.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 25.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q, Chen J, Chen X, Zhang S, Liu Z, Zhou J, Zhu Y, Deng Y, Zheng Y, Bi F. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest, and inhibits invasion in colorectal cancer cells. Int J Cancer. 2011;128:1057–1068. doi: 10.1002/ijc.25452. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, McHale CM, Rothman N, Li G, Ji Z, Vermeulen R, Hubbard AE, Ren X, Shen M, Rappaport SM, North M, Skibola CF, Yin S, Vulpe C, Chanock SJ, Smith MT, Lan Q. Systems biology of human benzene exposure. Chem Biol Interact. 2010;184:86–93. doi: 10.1016/j.cbi.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen Y, Shen HM, Shi CY, Ong CN. Benzene metabolites enhance reactive oxygen species generation in HL60 human leukemia cells. Hum Exp Toxicol. 1996;15:422–427. doi: 10.1177/096032719601500510. [DOI] [PubMed] [Google Scholar]
- 29.Singh R, Sram R, Binkova B, Kalina I, Popov TA, Georgieva T, Garte S, Taioli E, Farmer PB. The relationship between biomarkers of oxidative DNA damage, polycyclic aromatic hydrocarbon DNA adducts, antioxidant status and genetic susceptibility following exposure to environmental air pollution in humans. Mutat Res. 2007;620:83–92. doi: 10.1016/j.mrfmmm.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 30.Yuan X, Liu C, Yang P, He S, Liao Q, Kang S, Zhao Y. Clustered microRNAs’ coordination in regulating protein-protein interaction network. BMC Syst Biol. 2009;3:65. doi: 10.1186/1752-0509-3-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinert D. Age-dependent changes of the circadian system. Chronobiol Int. 2000;17:261–283. doi: 10.1081/cbi-100101048. [DOI] [PubMed] [Google Scholar]
- 32.Kunieda T, Minamino T, Katsuno T, Tateno K, Nishi J, Miyauchi H, Orimo M, Okada S, Komuro I. Cellular senescence impairs circadian expression of clock genes in vitro and in vivo. Circ Res. 2006;98:532–539. doi: 10.1161/01.RES.0000204504.25798.a8. [DOI] [PubMed] [Google Scholar]