Abstract
Mammary-specific genetic programs are activated during pregnancy by the common transcription factor signal transducer and activator of transcription (STAT) 5. More than one third of these genes carry nuclear factor I/B (NFIB) binding motifs that coincide with STAT5 in vivo binding, suggesting functional synergy between these two transcription factors. The role of NFIB in this governance was investigated in mice from which Nfib had been inactivated in mammary stem cells or in differentiating alveolar epithelium. Although NFIB was not required for alveolar expansion, the combined absence of NFIB and STAT5 prevented the formation of functional alveoli. NFIB controlled the expression of mammary-specific and STAT5-regulated genes and chromatin immunoprecipitation-sequencing established STAT5 and NFIB binding at composite regulatory elements containing histone H3 lysine dimethylation enhancer marks and progesterone receptor binding. By integrating previously published chromatin immunoprecipitation-sequencing data sets, the presence of NFIB-STAT5 modules in other cell types was investigated. Notably, genomic sites bound by NFIB in hair follicle stem cells were also occupied by STAT5 in mammary epithelium and coincided with enhancer marks. Many of these genes were under NFIB control in both hair follicle stem cells and mammary alveolar epithelium. We propose that NFIB-STAT5 modules, possibly in conjunction with other transcription factors, control cell-specific genetic programs.
The discovery of DNA binding activity in nuclear extracts of vertebrate cells to specific sequences in the adenovirus genome (1), the chicken lysozyme gene (2), and the immunoglobulin locus (3) led to the isolation and cloning of the first family of sequence-specific eukaryotic DNA binding proteins. The nuclear factor I (NFI) family consists of four genes, Nfia, Nfib, Nfic, and Nfix (4–6). Genetic studies in mice have established unique and cell-specific roles for each of them (7). Nfib-null mice die perinatally due to impaired lung maturation and neuronal defects (8–10). In hair follicle stem cells (HFSCs), nuclear factor I/B (NFIB) is a negative regulator of melanocyte stem cells (11).
Development and differentiation of mammary tissue during pregnancy are largely driven by progesterone and prolactin, with the latter signaling through the generic transcription factors signal transducer and activator of transcription (STAT) 5A and STAT5B (referred to as STAT5 throughout the manuscript) (12). The up to 10 000-fold induction of genes encoding milk proteins and lipid metabolism in secretory alveolar mammary epithelium during pregnancy is dependent on the presence of STAT5 and its binding to promoter and enhancer sequences (13, 14). However, it is not known to what extent additional transcription factors synergize with STAT5 in establishing cell-specific gene expression. NFIB binds to specific sequences (TGGCA) in the Wap gene promoter (15, 16) and transgenic experiments have demonstrated that these sites are required for promoter activation (17). In contrast, a disabling mutation in the juxtaposed STAT5 binding site mainly abolished increased expression (17). NFI binding sites have also been identified in other genes highly expressed in mammary epithelium, including bovine β-lactoglobulin (18), β1,4-galactosyltransferase (B4galt1) (19), carboxyl ester lipase (Cel) (20, 21), TRPM-2/clusterin (Clu) (22), p53 (Trp53) (23), and the mouse mammary tumor virus (MMTV) promoter (24–26). These findings suggest that the NFI transcription factor family may contribute to the activation of genes during pregnancy. Recent studies have also demonstrated a causative role for NFIB in estrogen receptor–negative breast cancers (27) and in small cell lung cancer (28).
This study tested the hypothesis that genes preferentially expressed in mammary epithelium depend on promoter sequences bound by STAT5 and members of the NFI family.
Materials and Methods
Mice
Mice carrying floxed Nfib (10) and Stat5 (29) genes and Wap-Cre (WC) and MMTV-Cre (MC) (30) transgenes have been described. Nfibf/f;WC females were mated and mammary tissue was harvested at specified pregnancy stages or within less than 12 hours after parturition. Nfibf/f;MC animals are not viable, and epithelial development was studied upon transplantation of embryonic anlagen as described below. Mice carrying floxed Nfib and Stat5 alleles, and the WC transgene were generated by breeding. All animals were kept in accordance with National Institutes of Health (NIH) guidelines, and the experiments were approved by the Animal Care and Use Committee of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Mammary tissue transplantation
Athymic nude mice (3 weeks old) were anesthetized with an intraperitoneal injection of avertin. Fat pads were cleared by excising the proximal part of the inguinal gland containing the mammary epithelium. Mammary buds from embryonic day 13.5 Nfibf/f;MC embryos were transplanted as described previously (31). Nfibf/+;MC anlagen were used as controls by transplantation into the contralateral fat pad. To assess complete clearing, the excised tissues were processed for whole-mount staining. Eight weeks after transplantation, fat pads were harvested either from virgin hosts, or the hosts were mated and transplanted tissue was harvested on day 6 or day 13 of pregnancy or the day of parturition within less than 12 hours after delivery.
Histology
Harvested mammary tissues were fixed in 10% formalin, dehydrated through ethanol and xylene, embedded in paraffin, and sectioned. Sections were stained with hematoxylin and eosin by standard methods. For immunostaining, antigen unmasking was performed in a decloaking chamber (Biocare Medical) using Borg Decloaker solution, pH 9.5 (Biocare Medical), at 125°C and 18 to 24 psi for 5 minutes. Sections were blocked for 30 minutes in Tris-buffered saline with Tween 20 with 3% goat serum. Primary antibodies were incubated overnight at 4°C (anti-phosphorylated STAT5, 1:200 [Cell Signaling]; anti-NKCC1, 1:1000 [a gift from Dr Jim Turner, National Institute of Dental and Craniofacial Research, NIH]; anti-perilipin 2 (PLIN2), 1:1000 [a gift from Drs Dean Landos and Alan Kimmel, NIDDK, NIH]; anti–smooth muscle actin, 1:1000 [Sigma-Aldrich]; and anti-E-cadherin, 1:200 [BD Biosciences]). Alexa Fluor 488– or 594–conjugated secondary antibodies (Invitrogen) were applied at a dilution of 1:400 for 30 minutes at room temperature.
RNA-sequencing (RNA-seq) data processing
Poly(A) RNA was purified twice from 1 μg of total RNA. cDNA was synthesized using SuperScript II (Invitrogen) and a TruSeq RNA Sample Preparation Kit (Illumina Inc) and sequenced using a HiSeq 2000 system (Illumina Inc). The single-end reads of biological replicates obtained from each sample were aligned to the mouse reference genome (mm9 assembly) using the TopHat program (32). The total number of the mapped reads on each gene was calculated with the HTSeq program (http://www-huber.embl.de/users/anders/HTSeq/). Transcript abundance was estimated by means of reads per kilobase of transcript per million mapped reads (RPKM) according to the mapped reads on exons as described previously (33). The RNA-seq data are deposited in Gene Expression Omnibus (GEO) under the accession number GSE42878. For Supplemental Figure 1, we obtained RNA-seq data from the GEO repository (GSE29278) (34) and processed them with the same procedure as outlined above.
ChIP-seq analysis
Previously published ChIP-seq (chromatin immunoprecipitation sequencing) data for STAT5 (GSE52016 and GSE40930), the progesterone receptor (PR) (GSE42887), NFIB (GSE42900), and histone H3 lysine dimethylation (H3K4me2) (GSE25105) were analyzed. NFIB antibodies were obtained from Santa Cruz Biotechnology. ChIP-seq analysis was conducted as described previously (13, 14).
Motif analysis
We used two different algorithms to pinpoint transcription factor binding motifs. To identify significantly overrepresented motifs in the promoter of NFIB- and STAT5-regulated genes (see Figure 6), we used the Pscan tool (35) with the JASPAR motif database (36). To scan all 26 237 STAT5 binding sites with 130 known position weight matrices from the JASPAR database, the MOODS algorithm was used with a P value cutoff of .001 (37). Flanking sequences of each STAT5 binding site (500 bp) were scored against the matrices, and only the maximum score within each site per matrix was used for motif score calculation. The background motif score was estimated by the same approach with a randomly generated set. The background score was subtracted before the final motif score.
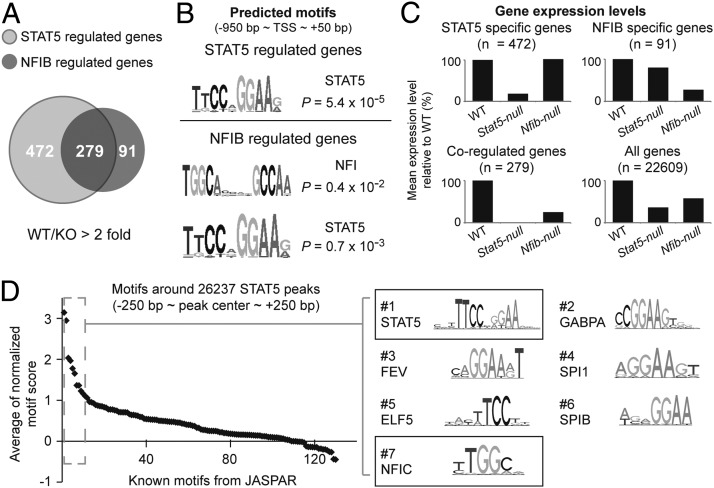
Figure 6.
NFIB binding motif significantly co-occur near in vivo STAT5 binding sites. A, Common and unique genes, whose expression is dependent on NFIB and/or STAT5. STAT5 RNA-seq data were obtained from the GEO database (GSE37646). KO, knockout. B, Significantly overrepresented motifs were identified in promoters of the NFIB- and STAT5-regulated genes. C, Gene expression levels were determined by RNA-seq and mean expression levels of gene sets are shown. D, The NFI binding motif significantly co-occurs near in vivo STAT5-binding sites. Motif analysis identified NFI and GAS motifs as highly co-occurring near in vivo STAT5 binding sites; 130 defined motifs from the JASPAR database were used. Each spot indicates a single motif and sequence logos of the top 7 motifs (dashed box) are shown (right).
Gene ontology analysis
Genes showing more than 5 RPKM and 2-fold up-regulated expression in the wild type (WT) compared with the absence of NFIB (or STAT5) were categorized into 3 groups according to coregulation (NFIB-specific, STAT5-specific, and NFIB/STAT5-coregulated gene sets). Functional annotation of the genes in each group was assessed by using the DAVID tool (38).
Data visualization
To visualize the next generation sequencing data (see Figures 7 and 8), total mapped reads of RNA-seq were scaled to 1 million and total mapped reads of ChIP-seq data were scaled to 10 million by using the HOMER program (39). Then, the integrative genomics viewer tool was used to load the scaled reads onto the mouse genome (40).
Figure 7.
NFIB and STAT5 are key regulators in mammary gland development. A, Gene ontology (GO) analysis showed that most NFIB- and STAT5-regulated genes were associated with mammary gland development. B, Expression levels of the clustered genes. C, Genome browser images of 3 representative genes are shown. The first and second tracks represent expression levels (tags were normalized to 1 million) of exons in 2 independent studies (Stat5-null and Nfib-null). Expression levels of the genes in knockout (KO; black bar graph) relative to WT (gray line graph) are shown. The rest of tracks show NFIB and STAT5 binding (day 13 of pregnancy) as well as H3K4me2 enrichment (virgin) as peaks. The STAT5 ChIP-seq and H3K4me2 data in mammary gland were obtained from a previous study (GSE40930 and GSE25105). Actual sequences of NFI and GAS motifs located in CRMs (gray bars) are represented (bottom).
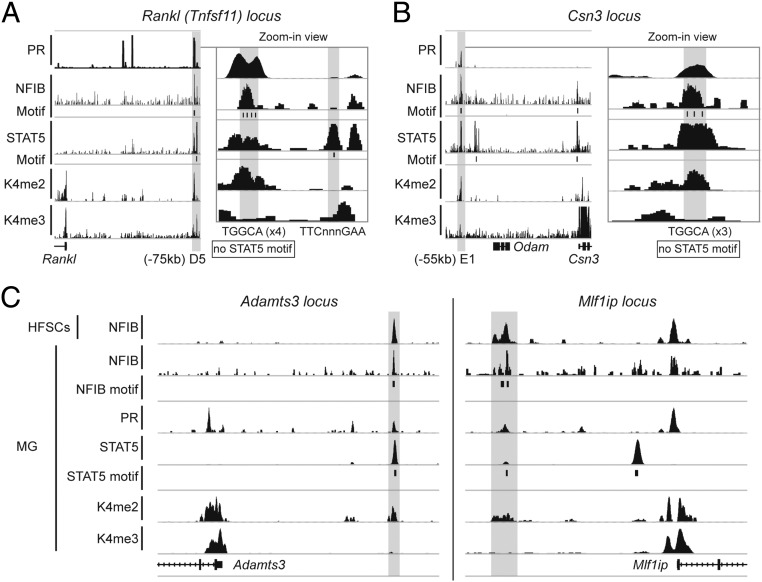
Figure 8.
Combinatorial binding of NFIB, PR, and STAT5 is observed on putative enhancers. Gray bars represent joint binding sites of the transcription factors. Genome browser images of Rankl (A), Csn3 (B), and Adamts3 and Mlf1ip (C) are shown with PR, NFIB, STAT5, H3K4me2 (K4me2), and histone H3 lysine 4 trimethylation (K4me3) ChIP-seq peaks. MG, mammary gland tissue.
Results
Deletion of Nfib from mouse mammary epithelium
Immunofluorescence (IF) demonstrated the presence of nuclear NFIB in luminal and basal mammary epithelial cells of virgin, pregnant, and lactating mice (Figure 1), and the staining intensity was similar at all stages of development. In tissue from virgin mice, some stromal cells, possibly of immunological nature, are also positive for NFIB.
Figure 1.

NFIB (red; arrowhead) is present in mammary luminal cells throughout development. A, Virgin. B, Pregnancy day 6 (p6). C, Pregnancy day 13 (p13). D, Day 1 of lactation (L1). E-cadherin (green) outlines luminal epithelial cells. Nuclei were visualized with 4′,6-diamidino-2-phenylindole (blue). The nature of the NFIB-positive stromal cells in mammary tissue from virgin mice is not clear, but they could represent immune cells.
MMTV-Cre (MC) and WAP-Cre (WC) transgenes were used to inactivate the Nfib gene in mammary stem cells and in differentiating alveolar epithelium during pregnancy, respectively. Nfibf/+;MC females appeared normal and were able to nurse their pups. Nfibf/f; MC mice were not obtained, possibly due to perinatal lethality. To bypass this obstacle, mammary anlagen isolated from Nfibf/f;MC fetuses were transplanted into cleared fat pads of recipient immunocompromised mice. Mammary tissue was analyzed at days 13 and 18 of pregnancy (Figure 2). Overall, WT and mutant alveolar epithelium had a similar appearance, including the accumulation of milk fat globules, indicating secretory activity and functional differentiation. These results demonstrate that during pregnancy NFIB is not required for alveolar development or expansion or for the initial stages of differentiation.
Figure 2.

Alveolar development during pregnancy in the absence of NFIB. Deletion of Nfib in mammary stem cells using the MMTV-Cre transgene. Alveoli (arrows) are sparser in the fat pads of Nfibf/f;MC mice (B) than in those of Nfibf/+;MC mice (A) on day 13 and day 18 (D and C) of pregnancy. Secreted material and lipid droplets are present in tissues of both genotypes.
Nfibf/f;WC females were fertile and gave birth to normal-sized litters. There was increased pup mortality and only 40% of pups born were weaned. However, the surviving pups grew at the same rate as pups reared by control dams (data not shown). Histological analysis of mammary tissue from Nfibf/f;WC females at parturition demonstrated the presence of alveoli (Figure 3). However, they were sparser and tended to be smaller in size. IF confirmed the absence of NFIB in most alveoli (Figure 4). The intensity of phosphorylated STAT5 (pSTAT5) staining at day 1 of lactation was similar in control and mutant mice (Figure 4), suggesting that prolactin signaling was intact in the absence of NFIB. WAP, a major milk protein (41), was preferentially present in the alveolar lumina of control tissue and to some extent in the cytoplasm (Figure 4). The cytoplasmic presence of WAP was more prominent in mutant tissue, suggesting impaired secretion. The distribution of PLIN2, an integral component of the lipid droplet membrane, supports the observation that secretion is impaired in mutant tissue (Figure 4). However, secretion is sufficiently intact to produce milk for pups to survive.
Figure 3.
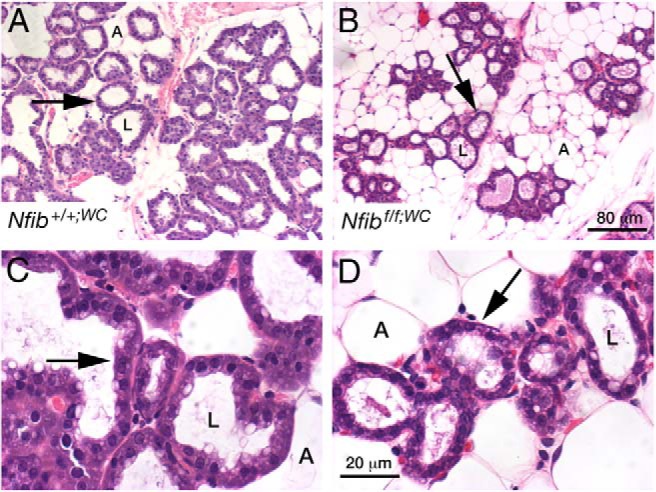
Alveolar development during pregnancy in the absence of NFIB. Deletion of Nfib in differentiating mammary alveolar cells during pregnancy using the Wap-Cre transgene. Alveoli (arrows) are sparser in the fat pads of Nfibf/f;WC mice (B and D) than in those of Nfib+/+;WC mice (A and C). A, adipocyte; L, alveolar lumen.
Figure 4.

Immunohistochemical analysis of the epithelial differentiation status of Nfibf/f;WC epithelium at day 1 of lactation. A and B, NFIB staining (in red; arrow) in control (A) and mutant (B) tissue. The broad arrow indicates incomplete deletion in cells by the WAP-Cre transgene, which is not expressed in ducts and in a mosaic fashion in luminal cells. E-cadherin (green) is used as a counterstain. C and D, pSTAT5 staining in Nfib+/+;WC (C) and Nfibf/f;WC (D) epithelium. pSTAT5 (arrow; red fluorescence) is concentrated in nuclei of luminal cells and E-cadherin is stained green. E and F, WAP (red) staining in mammary tissue from Nfib+/+;WC (E) and Nfibf/f;WC (F) mice. E-cadherin staining in green indicates epithelial cell membranes. G and H, Staining for PLIN2, a component of the milk fat globule membrane in mammary tissue from Nfib+/+;WC (G) and Nfibf/f;WC (H) mice. Green staining of smooth muscle actin (SMA) demonstrates normal development of myoepithelial cells.
The combined function of NFIB and STAT5 in lactation was investigated in mice carrying floxed Stat5 and Nfib alleles and the Wap-Cre transgene. Mammary development in these mice appeared heterogeneous, and IF analyses demonstrated large areas positive for pSTAT5 next to smaller regions that had underdeveloped alveoli devoid of pSTAT5 (Figure 5). This result provides genetic evidence that functional alveolar epithelium cannot be established in the combined absence of NFIB and STAT5 and unrecombined cells preferentially expand during pregnancy.
Figure 5.

Alveolar development at day 1 of lactation in the absence of NFIB and STAT5. A, Whole mount of mammary tissue of Nfibf/fStat5f/f;WC dams shows attenuated alveologenesis. B, Heterogeneous alveolar development and incomplete filling of the fat pad is seen. The tissue consists of a mixture of mature, expanded (arrow), and small, underdeveloped (arrowhead) alveoli. C, Presence of pSTAT5 staining (red, arrowheads) in some alveoli next to alveoli that have lost STAT5 (asterisk) indicates mosaic expression of the Wap-Cre transgene. D, Cytoplasmic localization of WAP (red, arrowhead) and lack of WAP in the alveolar lumen (asterisk) suggest impaired secretion. E-cadherin is stained green, and nuclei are stained blue with 4′,6-diamidino-2-phenylindole. E and F, Hematoxylin and eosin–stained section of mammary tissue from Nfibf/fStat5f/f;WC dams at the onset of the second lactation shows the presence of expanded alveoli (arrow) interspersed with underdeveloped alveoli (arrowhead).
Identification of NFIB target genes
RNA-seq analyses were performed from mammary tissue harvested from Nfibf/f;WC and WT mice at parturition. Expression of approximately 370 genes was reduced at least 2-fold in the absence of NFIB (Figure 6A and Supplemental Table 1). These include most of the STAT5 target genes that encode proteins linked to alveolar differentiation during pregnancy (13, 14). An unbiased de novo motif search identified GAS motifs (STAT5 binding sites) and NFIB binding motifs in promoter regions of these genes (Figure 6B). Of the NFIB-regulated genes, 75% (279 of 370) were also under the control of STAT5, and their expression was reduced between 75% and 99% in the absence of either STAT5 or NFIB (Figure 6, A and C). In contrast, expression of the 472 genes only under STAT5 control was reduced up to 80% in the absence of STAT5 but not NFIB. Similarly, reduced expression of the 91 NFIB-specific target genes was only observed in Nfib-null tissue (Figure 6C). Notably, NFIB and STAT5 had differential importance in the expression of milk protein genes (Table 1). Motif analysis was conducted on a 500-bp fragment overlapping all 26 237 in vivo STAT5 binding sites (14) using the MOODS algorithm with predefined 130 motif position weight matrices. The GAS motif was by far the most significantly enriched motif around the STAT5 binding sites (Figure 6D). The next 5 high-ranked motifs were GABPA, FEV, SPI1, ELF5, and SPIB motifs, which contain a half sequence of the GAS motif (TTC or GAA). Upon exclusion of these candidate partial GAS motifs, the known NFI binding motif was the most significantly enriched motif near the genome-wide STAT5 binding sites.
Table 1.
Expression Levels of Genes Coregulated by NFIB and STAT5
| Gene | NFIB |
STAT5 |
||||
|---|---|---|---|---|---|---|
| WTa | Nulla | Fold change | WTa | Nulla | Fold change | |
| Csn2 | 143 846 | 50 688 | 2.8 | 193 547 | 557 | 347 |
| Csn1s2a | 120 513 | 49 693 | 2.4 | 164 023 | 470 | 348 |
| Csn1s1 | 61 562 | 15 930 | 3.9 | 87 949 | 230 | 382 |
| Wap | 102 755 | 9085 | 11.3 | 109 772 | 327 | 335 |
| Csn3 | 34 488 | 8794 | 3.9 | 57 840 | 131 | 440 |
| Glycam1 | 102 755 | 8021 | 12.8 | 102 504 | 265 | 385 |
| Trf | 16 334 | 6734 | 2.4 | 21 091 | 589 | 35 |
| Expi | 7003 | 2012 | 3.5 | 11 627 | 375 | 30 |
| Lalba | 8155 | 914 | 8.9 | 8626 | 21 | 410 |
| Csn1s2b | 11 359 | 712 | 16.0 | 13 610 | 12 | 1133 |
Normalized expression level (RPKM).
Gene ontology analysis was performed with 472 STAT5-specific, 91 NFIB-specific, and 279 coregulated genes. As expected, overall functional outputs of these genes were mostly related to mammary gland development (Figure 7A). Coregulated genes appear to contain functional outputs of both NFIB- and STAT5-specific gene sets. They also contained a unique group of milk protein genes (Supplemental Table 2). Expression levels of these genes were reduced between 50% and 95% in the absence of either NFIB or STAT5 (Figure 7B). Among these 3 gene sets, the coregulated genes encompassed mammary-specific genes (Supplemental Figure 1). Manual investigation further demonstrated that a STAT5-NFIB cis-regulatory module (CRM), a DNA fragment containing both STAT5 and NFIB binding motifs, is the integral component of many genes expressed in mammary epithelium during pregnancy (Figure 7C). For instance, the Lalba, Wap, and Csn3 genes harbor1 STAT5-NFIB CRM upstream of their transcription start sites, and their expression levels were greatly reduced in the absence of either NFIB or STAT5. All of the STAT5-NFIB CRMs near the 3 representative genes consist of conventional NFIB and GAS motifs. NFIB ChIP-seq experiments confirmed NFIB binding to these CRMs (Figure 7C). NFIB binding was obtained at bona fide NFIB binding motifs (TGGCA) juxtaposed to STAT5 binding sites at the mammary-specific genes.
STAT5 binds not only to promoter proximal sequences but also to putative enhancer sequences (13, 14, 42). Because STAT5, the PR (43), and NFIB are key mammary transcription factor, it can be hypothesized that their joint binding could be a defining feature of at least some mammary enhancers. This hypothesis was explored through the integration of NFIB, STAT5 (13, 14), PR (43), and H3K4me2 (44) ChIP-seq data sets. Binding of these 3 transcription factors was located at known enhancers (Figure 8A), such as the one located 75 kbp 5′ of the Rankl (Tnfsf11) gene (45, 46). Frequently areas recognized by both NFIB and STAT5 contain either a GAS motif or an NFIB motif but not both, suggesting that either of these transcription factors can be the guide or follower. Although the mammary specificity of some milk protein gene promoters has been established in transgenic mice (47–50), genuine enhancers activating genes during pregnancy have not been identified. Based on sequence conservation and DNase I hypersensitivity, an intergenic region in the casein locus has been proposed as an enhancer (51). ChIP-seq–based data demonstrated the presence of H3K4me2 marks and binding of STAT5, NFIB, and PR to a 300-bp sequence in this region (Figure 8B). NFIB-binding motifs (TGGCA) but no GAS motifs were identified, suggesting a role for STAT5 as a cofactor.
Search for NFIB-STAT5 modules in nonmammary cells
If NFIB-STAT5 modules are operational in other cell types, skin and its appendages are qualified candidates because they are evolutionary related to the mammary lineage. NFIB ChIP-seq data from HFSCs (11) were used to search for potential NFIB-STAT5 modules. NFIB occupied 1449 sites in the genome of HFSCs, and expression of some target genes was altered in Nfib-null HFSCs (11). NFIB binding to gene regulatory regions in HFSCs coincided with STAT5 binding in mammary epithelium (Figure 8C). Moreover, these sequences were also recognized by PRs and exhibited H3K4me2 marks as shown for Adamts3 and Mlf1ip (Figure 8C). Notably, genes under NFIB control in HFSCs are also regulated by NFIB and STAT5 in mammary alveolar epithelium. Although STAT5 ChIP-seq data are not available for HFSCs, it can be predicted that, as in mammary epithelium, NFIB-STAT5 modules control the expression of specific gene sets.
Discussion
Twenty years ago, Li and Rosen (17, 52) published landmark studies demonstrating the critical importance of NFI binding sites in the rat Wap gene promoter for its expression in transgenic mice and the relevance of a juxtaposed STAT5 binding site in controlling prolactin-induced activation during pregnancy. Based on these studies, it was hypothesized that the combination of transcription factors might control expression of genes in mammary tissue. This investigation now demonstrates that more than 250 mammary-specific genes are under joint control by STAT5 and NFIB, suggesting that an NFIB-STAT5 module is key in the activation of genes in mammary alveolar epithelium during pregnancy. Moreover, genes under NFIB control in HFSCs are also regulated by NFIB and STAT5 in mammary tissue. These findings establish the concept that the NFIB-STAT5 transcription factor module is critical for the activation of genetic programs in 2 distinct, yet evolutionary related, cell types.
Deletion of Nfib from mammary stem cells and differentiating alveoli had no detrimental impact on the development of mammary alveoli, and the slightly impaired differentiation was reflected by the incomplete activation of milk protein genes. The degree by which steady-state levels of individual mRNAs were reduced possibly reflects the state of epithelial differentiation. Expression of approximately 270 genes during pregnancy is jointly controlled by STAT5 and NFIB with most them being preferentially active in mammary epithelium. In general, the impact of STAT5 on the expression of these genes greatly exceeds that of NFIB because for many of them the presence of STAT5 is mandatory for any induction during pregnancy. In contrast, pregnancy-induced expression is only attenuated in the absence of NFIB (Figure 9). The Janus tyrosine kinase 2/STAT5 pathway is probably intact in the absence of NFIB because expression of a subset of STAT5 target genes remained unaltered.
Figure 9.

Genetically proven model of mammary gland–specific gene regulation by NFIB and STAT5. The 279 genes that are cogoverned by NFIB and STAT5 are specifically expressed in mammary gland tissue (Supplemental Figure 1). Regulatory regions of most of these genes contain STAT5-NFI CRMs, which bind STAT5 in mammary tissue and also contain NFI consensus sequences (left panel). In the absence of NFIB or STAT, expression of coregulated genes is reduced up to 75% or 98%, respectively, compared with the WT (right panel).
Expression of NFIB target genes in mammary epithelium is probably also controlled by additional members of the NFI family. Support for this notion comes from studies with a mutant Wap transgene that was silenced upon disabling of NFI binding sites (17), whereas loss of NFIB does not result in the complete inactivation of the Wap gene. Although NFIB might be the most abundant member in the mammary epithelium, all four Nfi members are expressed (16). Wap transgenes containing only promoter-bound STAT5 binding sites are never fully activated during pregnancy (48, 53), and an additional putative upstream STAT5 site (14) might be required for full activation (49).
The presence of NFIB-STAT5 modules is not only based on the presence of the respective binding motifs but also mostly on the in vivo binding of these two transcription factors, and their biological significance was established in the respective gene knockout mice. NFIB-STAT5 modules are also likely to be active in HFSCs. Transcription factor modules provide an efficient means to control cell-specific genetic programs as has been shown for Smad3 and Oct4, MyoD1 and Pu.1 (54), and Nkx3.1 with Myc (7). In primary mammary cells, expression of the Tnfsf11 (RankL) gene is controlled by progesterone, and PR and STAT5 binding has been localized to at least 5 enhancers within 80 kbp (45). ChIP-seq data analyzed in this study provide evidence for the binding of STAT5, PR, and NFIB and the presence of H3K4me2 marks preferentially at the D5 region located 75 kbp upstream of the transcriptional start site, indicating that this is the key enhancer. An NFIB-STAT5-PR-H3K4me2 module also marks a putative enhancer region within the casein locus (44, 55, 56). Although it is generally believed, and correctly so, that STAT5 and NFIB bind to their respective recognition sites in regulatory regions, their interaction with enhancers is more complex. NFIB, but not STAT5, motifs are found in the Tnfsf11 and casein enhancers, suggesting that NFIB is the driver and STAT5 is the passenger. These findings narrow the knowledge gap on the role of transcription factor modules in regulating cell-specific and hormone-controlled genetic programs.
Additional material
Supplementary data supplied by authors.
Acknowledgments
We thank Dr Harold Smith from the National Institute of Diabetes and Digestive and Kidney Diseases Genomics Core Facility for expert help with sequencing.
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. Funding for B.-M.Z. came from the National Natural Science Funding of China (81273838) and the Natural Science Fund for Colleges and Universities, Department of Education, Jiangsu, China (11KJA360003). R.M.G. was supported by the National Heart, Lung, and Blood Institute, National Institutes of Health (Award HL080624) and New York State Stem Cell Science (Award C026429).
G.W.R. provided experimental design, performed mouse experiments and data analysis, and wrote the article. K.K. provided experimental design, performed ChIP-seq and RNA-seq data analyses, and wrote the article. K.H.Y. performed ChIP-seq experiments. Y.T. performed mouse and RNA-seq experiments. B.-M.Z. provided experimental advice. D.Y. provided STAT5 ChIP-seq and RNA-seq data. V.C. performed immunofluorescence experiments. S.J.J. performed genotyping and lactation studies. R.M.G. provided reagents, participated in discussion of data, and wrote the article. L.H. provided experimental design and data analysis and wrote the article.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ChIP-seq
- chromatin immunoprecipitation sequencing
- CRM
- cis-regulatory module
- GEO
- Gene Expression Omnibus
- H3K4me2
- histone H3 lysine dimethylation
- HFSC
- hair follicle stem cell
- IF
- immunofluorescence
- MMTV
- mouse mammary tumor virus
- NFI
- nuclear factor I
- NFIB
- nuclear factor I/B
- NIDDK
- National Institute of Diabetes and Digestive and Kidney Diseases
- NIH
- National Institutes of Health
- PLIN2
- perilipin 2
- PR
- progesterone receptor
- pSTAT
- phosphorylated signal transducer and activator of transcription
- RNA-seq
- RNA-sequencing
- RPKM
- reads per kilobase of transcript per million mapped reads
- STAT
- signal transducer and activator of transcription.
References
- 1. Gronostajski RM, Nagata K, Hurwitz J. Isolation of human DNA sequences that bind to nuclear factor I, a host protein involved in adenovirus DNA replication. Proc Natl Acad Sci USA. 1984;81:4013–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nowock J, Sippel AE. Specific protein-DNA interaction at four sites flanking the chicken lysozyme gene. Cell. 1982;30:607–615. [DOI] [PubMed] [Google Scholar]
- 3. Hennighausen L, Siebenlist U, Danner D, et al. High-affinity binding site for a specific nuclear protein in the human IgM gene. Nature. 1985;314:289–292. [DOI] [PubMed] [Google Scholar]
- 4. Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. [DOI] [PubMed] [Google Scholar]
- 5. Kruse U, Qian F, Sippel AE. Identification of a fourth nuclear factor I gene in chicken by cDNA cloning: NFI-X. Nucleic Acids Res. 1991;19:6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rupp RA, Kruse U, Multhaup G, Gobel U, Beyreuther K, Sippel AE. Chicken NFI/TGGCA proteins are encoded by at least three independent genes: NFI-A, NFI-B and NFI-C with homologues in mammalian genomes. Nucleic Acids Res. 1990;18:2607–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson PD, McKissic SA, Logan M, et al. Nkx3.1 and Myc crossregulate shared target genes in mouse and human prostate tumorigenesis. J Clin Invest. 2012;122:1907–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gründer A, Ebel TT, Mallo M, et al. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mech Dev. 2002;112:69–77. [DOI] [PubMed] [Google Scholar]
- 9. Steele-Perkins G, Plachez C, Butz KG, et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25:685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hsu YC, Osinski J, Campbell CE, et al. Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev Biol. 2011;354:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang CY, Pasolli HA, Giannopoulou EG, et al. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature. 2013;495:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6:715–725. [DOI] [PubMed] [Google Scholar]
- 13. Kang K, Yamaji D, Yoo KH, Robinson GW, Hennighausen L. Mammary-specific gene activation is defined by progressive recruitment of STAT5 during pregnancy and the establishment of H3K4me3 marks. Mol Cell Biol. 2014;34:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamaji D, Kang K, Robinson GW, Hennighausen L. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res. 2013;41:1622–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mukhopadhyay SS, Rosen JM. The C-terminal domain of the nuclear factor I-B2 isoform is glycosylated and transactivates the WAP gene in the JEG-3 cells. Biochem Biophys Res Commun. 2007;358:770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mukhopadhyay SS, Wyszomierski SL, Gronostajski RM, Rosen JM. Differential interactions of specific nuclear factor I isoforms with the glucocorticoid receptor and STAT5 in the cooperative regulation of WAP gene transcription. Mol Cell Biol. 2001;21:6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li S, Rosen JM. Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in transgenic mice. Mol Cell Biol. 1995;15:2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Watson CJ, Gordon KE, Robertson M, Clark AJ. Interaction of DNA-binding proteins with a milk protein gene promoter in vitro: identification of a mammary gland-specific factor. Nucleic Acids Res. 1991;19:6603–6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rajput B, Shaper NL, Shaper JH. Transcriptional regulation of murine β1,4-galactosyltransferase in somatic cells. Analysis of a gene that serves both a housekeeping and a mammary gland-specific function. J Biol Chem. 1996;271:5131–5142. [DOI] [PubMed] [Google Scholar]
- 20. Kannius-Janson M, Lidberg U, Bjursell G, Nilsson J. The tissue-specific regulation of the carboxyl ester lipase gene in exocrine pancreas differs significantly between mouse and human. Biochem J. 2000;351(Pt 2):367–376. [PMC free article] [PubMed] [Google Scholar]
- 21. Kannius-Janson M, Lidberg U, Hulten K, Gritli-Linde A, Bjursell G, Nilsson J. Studies of the regulation of the mouse carboxyl ester lipase gene in mammary gland. Biochem J. 1998;336(Pt 3):577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Furlong EE, Keon NK, Thornton FD, Rein T, Martin F. Expression of a 74-kDa nuclear factor 1 (NF1) protein is induced in mouse mammary gland involution. Involution-enhanced occupation of a twin NF1 binding element in the testosterone-repressed prostate message-2/clusterin promoter. J Biol Chem. 1996;271:29688–29697. [DOI] [PubMed] [Google Scholar]
- 23. Johansson EM, Kannius-Janson M, Bjursell G, Nilsson J. The p53 tumor suppressor gene is regulated in vivo by nuclear factor 1-C2 in the mouse mammary gland during pregnancy. Oncogene. 2003;22:6061–6070. [DOI] [PubMed] [Google Scholar]
- 24. Brüggemeier U, Rogge L, Winnacker EL, Beato M. Nuclear factor I acts as a transcription factor on the MMTV promoter but competes with steroid hormone receptors for DNA binding. EMBO J. 1990;9:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Archer TK, Cordingley MG, Wolford RG, Hager GL. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Archer TK, Lefebvre P, Wolford RG, Hager GL. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. [DOI] [PubMed] [Google Scholar]
- 27. Moon HG, Hwang KT, Kim JA, et al. NFIB is a potential target for estrogen receptor-negative breast cancers. Mol Oncol. 5:538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dooley AL, Winslow MM, Chiang DY, et al. Nuclear factor I/B is an oncogene in small cell lung cancer. Genes Dev. 2011;25:1470–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cui Y, Riedlinger G, Miyoshi K, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wagner KU, Wall RJ, St-Onge L, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson GW, Wagner KU, Hennighausen L. Functional mammary gland development and oncogene-induced tumor formation are not affected by the absence of the retinoblastoma gene. Oncogene. 2001;20:7115–7119. [DOI] [PubMed] [Google Scholar]
- 32. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. [DOI] [PubMed] [Google Scholar]
- 34. Shen Y, Yue F, McCleary DF, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bryne JC, Valen E, Tang MH, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Korhonen J, Martinmäki P, Pizzi C, Rastas P, Ukkonen E. MOODS: fast search for position weight matrix matches in DNA sequences. Bioinformatics. 2009;25:3181–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jiao X, Sherman BT, Huang da W, et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol cell. 2010;38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hennighausen LG, Sippel AE. Mouse whey acidic protein is a novel member of the family of 'four-disulfide core' proteins. Nucleic Acids Res. 1982;10:2677–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vahedi G, Takahashi H, Nakayamada S, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lain AR, Creighton CJ, Conneely OM. Research resource: progesterone receptor targetome underlying mammary gland branching morphogenesis. Mol Endocrinol. 2013;27:1743–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rijnkels M, Freeman-Zadrowski C, Hernandez J, Potluri V, Wang L, Li W, Lemay DG. Epigenetic modifications unlock the milk protein gene loci during mouse mammary gland development and differentiation. PloS One. 2013;8:e53270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Obr AE, Grimm SL, Bishop KA, Pike JW, Lydon JP, Edwards DP. Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells. Mol Endocrinol. 2013;27:1808–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martowicz ML, Meyer MB, Pike JW. The mouse RANKL gene locus is defined by a broad pattern of histone H4 acetylation and regulated through distinct distal enhancers. J Cell Biochem. 2011;112:2030–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burdon TG, Maitland KA, Clark AJ, Wallace R, Watson CJ. Regulation of the sheep β-lactoglobulin gene by lactogenic hormones is mediated by a transcription factor that binds an interferon-γ activation site-related element. Mol Endocrinol. 1994;8:1528–1536. [DOI] [PubMed] [Google Scholar]
- 48. Pittius CW, Hennighausen L, Lee E, Westphal H, Nicols E, Vitale J, Gordon K. A milk protein gene promoter directs the expression of human tissue plasminogen activator cDNA to the mammary gland in transgenic mice. Proc Natl Acad Sci USA. 1988;85:5874–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bischoff R, Degryse E, Perraud F, et al. A 17.6 kbp region located upstream of the rabbit WAP gene directs high level expression of a functional human protein variant in transgenic mouse milk. FEBS Lett. 1992;305:265–268. [DOI] [PubMed] [Google Scholar]
- 50. Lee KF, Atiee SH, Rosen JM. Differential regulation of rat beta-casein-chloramphenicol acetyltransferase fusion gene expression in transgenic mice. Mol Cell Biol. 1989;9:560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rijnkels M, Elnitski L, Miller W, Rosen JM. Multispecies comparative analysis of a mammalian-specific genomic domain encoding secretory proteins. Genomics. 2003;82:417–432. [DOI] [PubMed] [Google Scholar]
- 52. Li S, Rosen JM. Distal regulatory elements required for rat whey acidic protein gene expression in transgenic mice. J Biol Chem. 1994;269:14235–14243. [PubMed] [Google Scholar]
- 53. Pittius CW, Sankaran L, Topper YJ, Hennighausen L. Comparison of the regulation of the whey acidic protein gene with that of a hybrid gene containing the whey acidic protein gene promoter in transgenic mice. Mol Endocrinol. 1988;2:1027–1032. [DOI] [PubMed] [Google Scholar]
- 54. Mullen AC, Orlando DA, Newman JJ, et al. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147:565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rijnkels M, Kabotyanski E, Montazer-Torbati MB, et al. The epigenetic landscape of mammary gland development and functional differentiation. J Mammary Gland Biol Neoplasia. 2010;15:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rijnkels M, Kabotyanski E, Shore A, Rosen JM. The chromatin landscape of the casein gene locus. Horm Mol Biol Clin Invest. 2012;10:201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.