Abstract
Histone H1 and the high-mobility group (HMG) proteins are chromatin binding proteins that regulate gene expression by modulating the compactness of the chromatin fiber and affecting the ability of regulatory factors to access their nucleosomal targets. Histone H1 stabilizes the higher-order chromatin structure and decreases nucleosomal access, while the HMG proteins decrease the compactness of the chromatin fiber and enhance the accessibility of chromatin targets to regulatory factors. Here we show that in living cells, each of the three families of HMG proteins weakens the binding of H1 to nucleosomes by dynamically competing for chromatin binding sites. The HMG families weaken H1 binding synergistically and do not compete among each other, suggesting that they affect distinct H1 binding sites. We suggest that a network of dynamic and competitive interactions involving HMG proteins and H1, and perhaps other structural proteins, constantly modulates nucleosome accessibility and the local structure of the chromatin fiber.
The chromatin fiber is a dynamic, malleable structure that is targeted by numerous regulatory factors that modify the histones and the DNA and remodel the structure of the nucleosome. The dynamics of the chromatin fiber reflect the combined action of numerous chromatin modifiers, including architectural proteins such as histone H1 and members of the high-mobility group (HMG) protein superfamily. The interaction of histone H1 with nucleosomes stabilizes the higher-order, compact chromatin structure (57, 61), thereby restricting the ability of regulatory factors, nucleosome remodeling complexes, and histone modifiers to access their chromatin binding sites (17, 27, 28, 30, 32, 34). Loss of H1 results in both up regulation and down regulation of specific gene expression (2, 26, 53, 56), suggesting that the protein affects the action of both positive and negative transcriptional regulators. Experiments with H1 knockout mice demonstrate the existence of cellular mechanisms that strive to maintain a constant level of H1 and that reduction of H1 beyond a critical point is not compatible with normal embryonic development (21). Thus, factors that modulate the interaction of H1 with nucleosomes may affect a variety of chromatin-related processes and participate in genetic regulatory mechanisms.
The HMG superfamily is composed of three families: HMGB, HMGA, and HMGN, each family being characterized by a distinct DNA or chromatin binding motif (12). These non-histones decompact the higher-order chromatin structure and promote the binding of nuclear regulatory factors (1, 12, 49, 58). Footprinting analysis and in vitro binding assays (29, 64) showed that the chromatin binding sites of the HMG proteins are similar to those of H1 and suggested that HMG proteins and H1 compete for the same binding sites, and recent photobleaching experiments demonstrated that HMGN proteins affect the binding of H1 to chromatin in living cells (16). Photobleaching techniques like fluorescence recovery after photobleaching (FRAP), which is used to monitor the intracellular mobility of molecules in living cells, revealed that the interaction of chromatin binding proteins with their nuclear targets is temporary and that these proteins constantly move throughout the entire nucleus (45). The mobility of H1-green fluorescent protein (GFP) and other chromatin-interacting proteins such as heterochromatin binding protein 1 or HMGN is a direct indicator of their chromatin binding, since modifications that weaken this binding increase their intranuclear mobility (18, 20, 36, 42, 48). At any given time, most of the H1 is bound to chromatin; however, the association of a specific H1 molecule with a specific nucleosome is transient (20, 36, 42). The transient dissociation of H1 from nucleosomes provides a window of opportunity for regulatory factors to access chromatin. HMG proteins move very rapidly throughout the nucleus (45, 52), raising the possibility that they can access the nucleosomal sites temporarily vacated by H1 and modulate the interaction of H1 with chromatin.
Here we used FRAP to show that through a dynamic competition process, members of each of the three HMG families weaken the binding of H1 to chromatin. Competition with H1 is dose dependent and is observed both in euchromatin and in heterochromatin. We detect competition for chromatin binding sites among members of the same HMG family, but not among members of different HMG families. The results indicate that each HMG family competes with H1 through distinct chromatin binding sites, without affecting the binding of members of a different HMG family. Our data indicate that H1 binding to chromatin is modulated at least in part by a competition process involving the HMG proteins and suggest that a dynamic and competitive network of chromatin interactions involving H1, HMG proteins, and perhaps other structural proteins plays a key role in regulating the local structure and activity of the chromatin fiber.
MATERIALS AND METHODS
Cell lines, vectors, and purified proteins.
Experiments were performed either with a BALB/c 3T3 cell line expressing H1°-GFP or with mouse embryonic fibroblasts as previously described (16, 42). pEGFP-HMGB1 was kindly provided by P. Scaffidi (52). Mouse embryonic fibroblasts were transfected with pEGFP-HMGB1 24 h before a FRAP experiment, with Fugene 6 reagent (Roche) in accordance with the supplier's recommendations. HMGN and fluorescently labeled HMGN proteins were prepared as described previously (16, 42). The HMGA1a, HMGB1, and HMGB-box mutant proteins were obtained as described previously (37, 50). Protein concentrations were determined by the Bradford assay (Bio-Rad) and by Coomassie blue staining of SDS-PAGE gels.
Microinjection into cultured cells.
Cells were plated 2 days prior to injection. Before injection, the medium was replaced with Dulbecco modified Eagle medium completed with a 5 mM final concentration of HEPES-Na, pH 7.4. Cells were injected at 37°C with a Micromanipulator 5171 and a Microinjector 5246 (Eppendorf) as described previously (16) and incubated at 37°C in 5% CO2 for at least 30 min before a FRAP experiment. Fluorescent Texas red-labeled high-molecular-weight dextran (Molecular Probes) was added to the injection mixture to identify injected cells. On average, cells were injected with 10 to 20 fl of solutions ranging from 0.05 to 0.4 mM in HMG proteins. Injection of 15 fl of a 0.4 mM solution introduces 3.6 × 106 molecules of HMG into the nucleus.
Microscopy and FRAP.
Immunostainings were performed with rabbit anti-HMGB1 (11) or rabbit anti-HMGA1 (50) and anti-rabbit secondary antibody conjugated to either Alexa-Fluor 488 or Alexa-Fluor 594 (Molecular Probes) as described previously (47). DNA was stained with Hoechst 33342 (Molecular Probes). Imaging and FRAP experiments were performed with a Zeiss LSM 510 confocal microscope as described previously (16, 42). Typically, five prebleach images were collected, followed by a 135-ms bleach pulse with a spot 3 μm in diameter. Bleaching was performed with the 488-nm line of a 25-mW argon laser set to 100% and the 543- and the 633-nm lines of two HeNe lasers. Single images were collected with the 488-nm line of the argon laser (set to 1%) every 296 ms for 20 s in short-time experiments and every 3 s for 200 s in long-time experiments. To increase the sensitivity of FRAP experiments performed on HMGB1-GFP, half of the nucleus was used as a bleached area. Pictures were then collected every 296 ms for 45 s. FRAP recovery curves were generated from images with the background subtracted. Total fluorescence was determined for each image and compared to the total prebleach fluorescence to determine the amount of fluorescence lost during the imaging. The fluorescence intensity in the bleached area was normalized to the initial fluorescence in the bleached area. In a typical experiment, 8 to 15 cells were used for FRAP. Each experiment was repeated at least three times. The Student t test was used to determine the significance of the results.
RESULTS
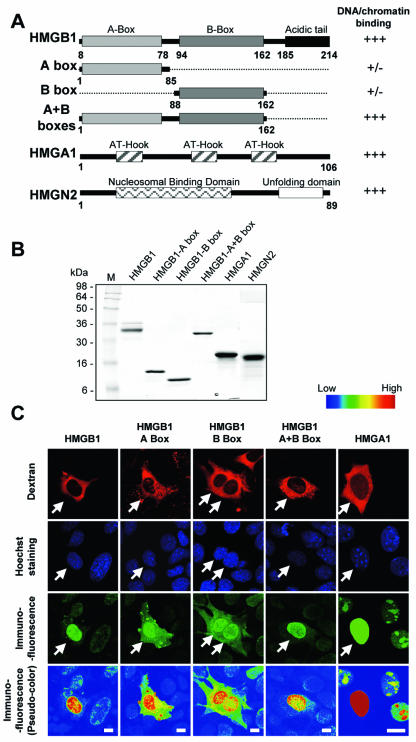
To test for competition among nuclear proteins for chromatin binding sites, we microinjected a potential competitor protein into the cytoplasm of cells expressing a GFP fusion protein and used FRAP to monitor the mobility of the GFP fusion protein in the injected cells and uninjected control cells. By microinjecting H1 or HMGN1 into the cytoplasm of cells expressing H1°-GFP or fluorescent HMGN1, we already demonstrated that this approach is suitable to monitor competition between a chromatin binding protein and its fluorescently labeled counterpart, i.e., proteins that bind to identical binding sites (16). To test whether the HMG proteins and protein fragments that we planned to use here (Fig. 1A and B) do indeed enter the nucleus, we microinjected proteins into the cytoplasm of cells and used immunofluorescence to visualize the location of the protein in the injected cells. The injected cells were easily identifiable by the Texas red-labeled high-molecular-weight dextran that was coinjected and remained in the cytoplasm (Fig. 1C). We found that within 1 h of injection, most if not all of the injected protein accumulated in the nucleus (Fig. 1C) (47).
FIG. 1.
(A) Competitors used in this study. A representation of the proteins and protein fragments injected is shown. The relative DNA or chromatin binding activity of each protein is indicated on the right (see text for reference). (B) Coomassie blue staining of an SDS-PAGE gel containing the proteins used in this study. M, molecular size markers (sizes are in kilodaltons). (C) All of the injected HMG proteins accumulate within the nucleus (for HMGN, see reference 47). HMG proteins or fragments, each at a 0.4 mM final concentration, were mixed with Texas red-labeled dextran and injected into the cytoplasm of mouse fibroblasts. The HMG injected into the cell is indicated at the top of each column. The cells were fixed 1 h after injection, immunostained with appropriate antibodies, and examined by confocal microscopy. The left side of each row indicates the image visualized. The pseudocolored images of anti-HMG immunostaining emphasize the significant increase in the protein concentration in the nuclei of the injected cells. Note that these are confocal images and do not accurately reflect the quantitative increase in protein. The scale indicates the color code used, from blue (low intensity) to red (high intensity). Bars, 5 μm.
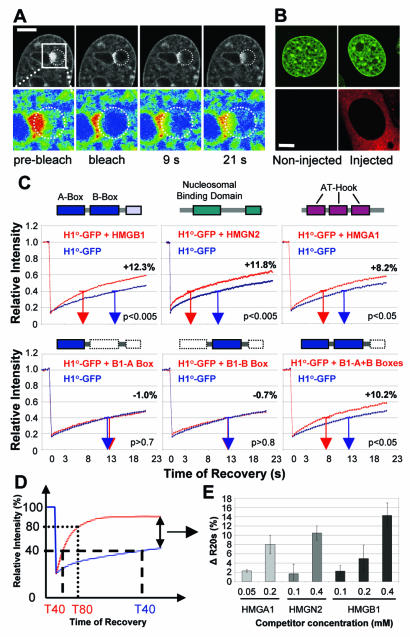
To test whether HMG proteins could affect the binding of H1 to chromatin, we microinjected purified proteins from each of the three HMG families, HMGB, HMGN, and HMGA (Fig. 1A and B) into the cytoplasm of H1°-GFP-expressing cells and used FRAP (Fig. 2A) to compare the mobility of H1°-GFP in injected cells to that in uninjected control cells (Fig. 2B). We assessed the effect of the injected proteins by measuring the time needed to recover either 40% (t40) or 80% (t80) of the H1°-GFP prebleach fluorescence intensity and by determining the amount of fluorescent intensity recovered within 20 s (R20s) of photobleaching (Fig. 2D and Table 1). In control uninjected cells, the average t40 was 12.5 s (range, 11 to 14 s) and the R20s was less than 50% (Fig. 2C, blue curves). In cells injected with HMGB1 protein, the R20s increased by 12.3% (P < 0.005) and the t40 decreased by 6.8 s (P < 0.005) (Fig. 2C). Similarly, injection of HMGN2 or HMGA1 protein significantly increased the R20s values for H1°-GFP recovery by 11.8 and 8.2% (P < 0.005) and decreased the t40 to 4.5 and 7 s (P < 0.005), respectively. The effect was not due to the microinjection technique, since injection of neither Texas red-labeled dextran nor histone H2B affected H1°-GFP recovery or global localization (data not shown, Fig. 2B, and reference 16). The decrease in H1°-GFP binding is due to competition for chromatin binding sites, since injection of the HMGB1 deletion mutants HMG AB1-box and HMG BB1-box (Fig. 1A), which bind DNA and chromatin weakly (24, 44, 62), did not affect H1 mobility (P > 0.7) (Fig. 2C) while the HMGB1-A+B mutant, which binds DNA and chromatin as strongly as does wild-type HMGB1 (24, 44, 62), increased the mobility of H1°-GFP (Fig. 2C). These results are consistent with previous findings that the injection of an HMGN point mutant form that does not bind to chromatin did not affect H1°-GFP mobility (16). Thus, the increase in H1 mobility is specific to the injected HMG protein and involves HMG interactions with chromatin.
FIG. 2.
HMG proteins decrease H1°-GFP binding to chromatin. (A) FRAP assay of H1°-GFP-expressing cells showing the fluorescence signal (top) and enlarged pseudocolored images of the boxed area (bottom). Bleached areas are circled. Bar, 5 μm. (B) Confocal imaging of live H1°-GFP-expressing cells injected with a solution containing Texas red-labeled injection marker. (C) Quantitative FRAP analysis of H1°-GFP-expressing cells either left uninjected (blue curves) or injected (red curves) with a 0.4 mM solution of HMGB1, HMGN2, HMGB1-A box, HMGB1-B box, or HMGB1-A+B box or a 0.2 mM solution of HMGA1. The time required to reach 40% recovery (t40) is indicated by blue and red arrows, and the statistical significance (t test) is indicated. The difference in recovery between the two curves (ΔR20s) is indicated next to the curves. The boxes above each graph of the upper panel outline the main functional domains characteristic of each injected HMG family. The boxes above each graph of the lower panel outline the HMGB1 segments injected. (D) Graph outlining the kinetic parameters measured (see text). (E) Dose-dependent increase in ΔR20s of H1°-GFP upon injection of the indicated concentration of an HMG protein.
TABLE 1.
HMGB and HMGA increase the intranuclear mobility of H1°-GFPa
| Expt | Chromatin domain | t40
|
R20s
|
t80
|
R200s
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Avg time (s) ± SD | P value | Avg % ± SD | P value | Avg time (s) ± SD | P value | Avg % ± SD | P value | ||
| H1°-GFP | Mixed | 14.7 ± 4.27 | <0.005 | 46.4 ± 6.2 | <0.005 | ||||
| H1°-GFP + HMGB1 | 7.9 ± 2.3 | 58.7 ± 6.5 | |||||||
| H1°-GFP | Mixed | 11.4 ± 4.9 | <0.005 | 46.7 ± 5.6 | <0.005 | ||||
| H1°-GFP + HMGB1-AB boxes | 7.0 ± 1.8 | 57.8 ± 5.2 | |||||||
| H1°-GFP | Mixed | 11.4 ± 4.9 | 0.78 | 46.7 ± 5.6 | 0.70 | ||||
| H1°-GFP + HMGB1-A box | 13.1 ± 6.0 | 46.6 ± 7.2 | |||||||
| H1°-GFP | Mixed | 11.4 ± 4.9 | 0.89 | 46.7 ± 5.6 | 0.69 | ||||
| H1°-GFP + HMGB1-B box | 12.7 ± 3.8 | 47.0 ± 4.3 | |||||||
| H1°-GFP | Mixed | 12.4 ± 5.0 | <0.03 | 49.2 ± 7.1 | <0.04 | ||||
| H1°-GFP + HMGA1 | 7.6 ± 2.7 | 57.4 ± 6.9 | |||||||
| H1°-GFP | Euchromatin | 102 ± 40 | <0.005 | 87.9 ± 7.4 | <0.05 | ||||
| H1°-GFP + HMGB1 | 45.0 ± 17 | 92.7 ± 5.4 | |||||||
| H1°-GFP | Heterochromatin | 125 ± 44 | <0.05 | 84.6 ± 5.9 | <0.05 | ||||
| H1°-GFP + HMGB1 | 81.3 ± 34 | 88.4 ± 4.9 | |||||||
| H1°-GFP | Euchromatin | 102 ± 40 | <0.005 | 87.9 ± 7.4 | <0.05 | ||||
| H1°-GFP + HMGB1-AB boxes | 55.4 ± 27 | 94 ± 8.9 | |||||||
| H1°-GFP | Heterochromatin | 128 ± 47 | 0.1 | 84.6 ± 5.9 | 0.16 | ||||
| H1°-GFP + HMGB1-AB boxes | 104 ± 42 | 87.2 ± 6.2 | |||||||
| H1°-GFP | Euchromatin | 102 ± 35 | <0.05 | 87.9 ± 7.4 | 0.3 | ||||
| H1°-GFP + HMGA1 | 49.7 ± 42 | 90.4 ± 7.1 | |||||||
| H1°-GFP | Heterochromatin | 128 ± 47 | <0.005 | 84.5 ± 5.9 | <0.05 | ||||
| H1°-GFP + HMGA1 | 82.8 ± 26 | 88.6 ± 6.7 | |||||||
The time required to reach 40% (t40) or 80% (t80) of the prebleach fluorescence was determined for each curve. The recoveries 20 s (R20s) or 200 s (R200s) after bleaching are means of the last five images of each curve. Results are from 8 to 15 cells. The statistical significance of the t40, t80, R20s, and R200s values relative to those of controls was determined by the Student t test.
The levels of HMG proteins vary among cells and change during development and differentiation. Ultimately, these variations may affect the interaction of H1 with chromatin. We therefore tested if the HMG proteins would increase the mobility of H1 in a dose-dependent manner. Injection of a 0.05 or 0.1 mM solution of HMGA1 increased the R20s of H1°-GFP by 2.3% ± 0.26% or 8.0% ± 1.98%, respectively. Likewise, injection of increasing concentrations of HMGN2 or HMGB1 resulted in increased R20s values for H1°-GFP (Fig. 2E). The results show a dose-dependent effect of HMG proteins on H1 mobility and are consistent with the view of a competition mechanism.
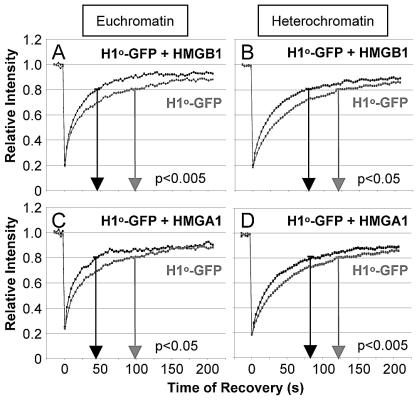
We next tested whether the state of chromatin compaction affects the HMG-H1 competition. We identified euchromatin and heterochromatin domains as regions stained weakly or strongly by H1°-GFP and Hoechst (42) and by their pattern of staining with antibodies to either acetylated or methylated lysine 9 of histone H3 (data not shown). In condensed heterochromatin, the H1°-GFP mobility is lower than in euchromatin, suggesting stronger H1 binding in the former. The injected HMGB and HMGA increased the mobility of H1°-GFP in euchromatin (t80 changed from 100 s to less than 50 s, P < 0.005) and also in heterochromatin (t80 changed from about 125 s to about 80 s, P < 0.05) with little effect on the immobile H1 fraction (t > 200 s) (Fig. 3 and Table 1). Together with our previous findings on HMGN (16), these results demonstrate that all of the three HMG families decrease the chromatin residence time of H1 in both euchromatin and heterochromatin. Thus, the HMG proteins can compete for H1 binding sites regardless of the degree of chromatin compaction.
FIG. 3.
HMG proteins decrease the H1°-GFP binding in euchromatin and heterochromatin. Quantitative FRAP analysis of H1°-GFP-expressing cells either left uninjected (gray curves) or injected (black curves) with a 0.4 mM solution of HMGB1 (A and B) or with 0.2 mM HMGA1 (C and D). FRAP analysis was performed on either euchromatin (A and C) or heterochromatin (B and D). Gray and black arrows indicate the t80 values, and the statistical significance (t test) is indicated.
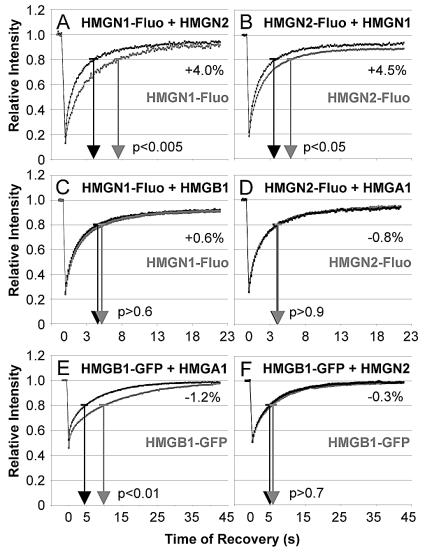
Our finding that each of the HMG proteins increased the mobility of H1 raises the possibility that the various HMG families interact with the same H1 binding sites on the nucleosomes and thus cross compete for chromatin binding. We therefore performed experiments in which we targeted HMGN or HMGB1 proteins, rather than H1. Microinjection experiments targeting fluorescently labeled HMG proteins revealed no competition among HMG proteins from different families. Whereas members of one family such as HMGN1 and HMGN2 cross compete for chromatin binding sites (Fig. 4A and B), neither HMGB1 nor HMGA1 affected the chromatin residence of HMGN2 (Fig. 4C and D and Table 2). Likewise, HMGN2 did not affect the chromatin residence time of HMGB1 (Fig. 4F and Table 2). HMGA1 reproducibly increased the chromatin residence time of HMGB1 (Fig. 4E), suggesting that the DNA-unbending activity of HMGA1 (49) affects the chromatin binding of HMGB1. Our finding that HMG proteins from different families do not compete for chromatin binding sites suggests that each family binds to different types of binding sites. Taken together, these findings indicate that each of the HMG families weakens H1 binding through competition for distinct H1 chromatin binding sites.
FIG. 4.
HMG proteins from different families do not compete with each other. The fluorescent target and the fluorescence recovery curves of the target determined in the absence of an injected competitor are gray. The fluorescence recovery curves determined in the presence of an indicated competitor are black. Gray and black arrows indicate the t80, and the statistical significance (t test) is indicated. The ΔR20s is indicated next to the curves.
TABLE 2.
Absence of competition for chromatin binding between HMG familiesa
| Exptc | t80
|
R20sb
|
Effect on mobility | ||
|---|---|---|---|---|---|
| Avg time (s) ± SD | P value | Avg % ± SD | P value | ||
| HMGN1 | 8.81 ± 4.2 | <0.005 | 90.2 ± 7.0 | 0.12 | Increase |
| HMGN1 + HMGN2 | 4.17 ± 1.4 | 94.2 ± 3.76 | |||
| HMGN2 | 5.87 ± 1.9 | <0.02 | 88.6 ± 5.1 | 0.10 | Increase |
| HMGN2 + HMGN1 | 3.77 ± 0.9 | 93.1 ± 3.8 | |||
| HMGN1 | 5.40 ± 1.6 | >0.6 | 91.0 ± 2.0 | >0.5 | None |
| HMGN1 + HMGB1 | 4.68 ± 0.95 | 91.6 ± 4.0 | |||
| HMGN2 | 5.04 ± 0.8 | >0.9 | 94.5 ± 2.9 | >0.5 | None |
| HMGN2 + HMGA1 | 5.08 ± 1.0 | 93.7 ± 3.5 | |||
| HMGB1b | 5.20 ± 2.8 | <0.01 | 98.5 ± 2.0b | >0.4 | Decrease |
| HMGB1 + HMGA1 | 10.20 ± 2.3 | 97.3 ± 2.4 | |||
| HMGB1b | 5.78 ± 2.0 | >0.7 | 98.7 ± 3.0b | >0.8 | None |
| HMGB1 + HMGN2 | 5.39 ± 2.6 | 98.4 ± 0.9 | |||
Analysis was done as for Table 1.
For HMGB1, the R was measured at 45 s rather than at 20 s (i.e., R45s rather than R20s).
Mixed chromatin domains were tested in each experiment.
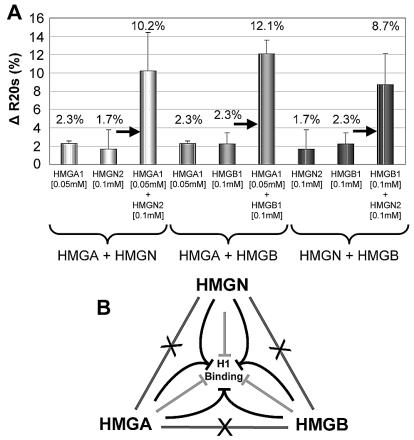
Lack of competition among the HMG families and the HMGA1-mediated increase in the chromatin binding of HMGB1 raise the possibility that the effect of the HMG proteins on the binding of H1 to chromatin is additive, or even synergistic. We therefore measured the intranuclear mobility of H1°-GFP in cells injected with HMGA1, HMGB1, or HMGN2 protein either alone or in pairs. We used a protein concentration of 0.05 or 0.1 mM since within this concentration range the increase in the mobility of H1 is approximately proportional to the amount of injected protein (Fig. 2E). At these low concentrations, injection of only one HMG protein resulted in a small but reproducible increase in H1°-GFP mobility (1.7 to 2.3% increase in the R20s) (Fig. 5A). Upon injection of an HMGB1-HMGN2 mixture, the R20s of H1°-GFP increased by 8.7% ± 3.4% rather than 4%, as would have been expected had the effect been additive. Likewise, upon injection of an HMGN2-HMGA1 or HMGA1-HMGB1 mixture, the H1°-GFP R20s increased by 10.2% ± 4.2% and 12.1% ± 1.5%, respectively (Fig. 5A). Thus, coinjection of proteins from different HMG families produced a synergistic rather than an additive effect on the mobility of H1°-GFP, suggesting that the HMG proteins function cooperatively to weaken the binding of H1 to chromatin (Fig. 5B).
FIG. 5.
HMG proteins act synergistically to compete with H1 for chromatin binding. (A) Shown are the ΔR20s values between uninjected and injected cells expressing H1°-GFP (see Fig. 2D). The concentration of the injected proteins is indicated below each bar. (B) Scheme of the H1-HMG chromatin binding network. The lines connecting the HMG families, crossed by an X, indicate lack of competition between members of different HMG families. The light gray lines indicate that, by itself, each of the HMG families reduces the binding of H1 to chromatin. The converging black lines indicate that members of distinct HMG families weaken the binding of H1 to chromatin synergistically. The scheme points emphasize that H1 is a target common to all of the HMG proteins.
DISCUSSION
Our main finding is that members of all of the canonical HMG protein families compete with the linker histone H1 for chromatin binding sites. Our studies are relevant to the understanding of molecular mechanisms regulating chromatin dynamics and the local compaction of the chromatin fiber. The results raise the possibility that the network of dynamic competition between histone H1 and HMG proteins modulates the local compactness of the chromatin fiber and the ability of regulatory factors to access their nucleosomal targets. Given that many nuclear proteins are in a constant state of flux, and their binding to chromatin is transient (T. Misteli, unpublished observation; 39), it is likely that competition for chromatin binding plays a major role in chromatin dynamics and genetic regulatory mechanisms.
Several in vitro studies have suggested that competition with nuclear proteins, mainly HMG proteins, might affect the binding of H1 to chromatin (12, 29, 43, 64). Our studies indicate that such a competition occurs in living cells. The average nucleus contains 107 nucleosomes, an equal number of H1 molecules (59), and approximately 106 HMGB, 105 HMGN, and 104 HMGA molecules (12, 31). While the amount of H1 varies little among cells, the cellular amounts of the various HMG proteins fluctuate significantly, especially during development (14, 19, 25, 33, 35). Certain malignancies are associated with significant increases in the levels of HMGA2 (22, 23, 49). Furthermore, immunofluorescence analysis indicates that the HMG proteins are clustered into foci, suggesting that at selected nuclear loci there is a significantly higher concentration of these proteins (40, 46). By injecting 0.05 to 0.4 mM protein solutions, we introduce 4 × 105 to 3.6 × 106 HMG molecules into the nucleus, a quantity that corresponds to 4 to 35% of the potential nucleosomal and H1 binding targets. Thus, even though histone H1 is in excess over HMG and the chromatin interactions of H1 are stronger than those of HMG proteins, the amounts of HMG protein introduced into the cell are sufficient to cause detectable alteration in the chromatin binding of H1.
Thus, our results demonstrate that, indeed, in living cells all members of the HMG superfamily can compete with H1 and weaken its binding to chromatin. The competition involves chromatin binding since point or deletion mutant forms of HMG that do not bind to chromatin or to DNA do not affect the intranuclear mobility of H1. We cannot exclude the possibility that, in addition, HMG-induced changes in chromatin modify the binding of H1. This possibility is still compatible with the main conclusion that HMG proteins may modulate the binding of H1 to chromatin. The effects are seen in both euchromatin and heterochromatin, an indication that HMG proteins can affect H1 binding throughout the entire nucleus, regardless of the state of chromatin compaction and histone modification. Thus, the effect of HMG seems to be genome wide rather than gene or locus specific, which is consistent with the observation that HMG proteins bind chromatin without DNA sequence specificity (12). The ability of HMG proteins to affect nucleosome accessibility at many chromatin loci may explain their involvement in a wide range of nuclear activities, including transcription, replication, repair, and recombination (1, 8, 12, 14, 29, 43, 49, 64). Previous FRAP experiments indicated that the in vivo chromatin interactions of the various H1 variants are indistinguishable (36, 42), suggesting that HMG proteins could compete with all of the cellular H1 variants.
Convincing experiments in numerous laboratories have demonstrated that the linker H1 promotes and stabilizes chromatin compaction. Although Tetrahymena sp. survives complete deletion of its particular H1 form (54), overexpression of the linker histone in yeast (41) or drastic reduction of H1 levels in mice (21) is lethal. Invariably, loss of H1 is associated with noticeable changes in gene expression. Although H1 was initially considered strictly a transcriptional repressor, it is now clear that loss of H1 leads to both up and down regulation of gene expression (2, 26, 53). Thus, H1 impedes the action of both transcriptional activators and repressors, most probably by promoting chromatin compaction (10, 57), stabilizing nucleosomal structures (60), and obstructing nucleosomal access (27). It follows that factors that modulate the interaction of H1 with chromatin may regulate nucleosomal access.
Although in some cases loss of HMG is not compatible with normal development (3, 15), it is important to note that their action may not always be necessary for cell survival (4, 6, 9, 15, 51, 63). By modulating the interaction of H1, and perhaps other proteins, with chromatin, HMG proteins optimize the cellular response to changing external or internal conditions. For example, loss of HMGN1 reduced, rather than abolished, the rate of DNA repair and therefore decreased, rather than abolished, the ability of a cell to survive UV damage (9). Significantly, it was shown that a change in the balance between H1 and HMG proteins caused by injection of either H1 or HMGA into mouse embryos (5), and more recently of HMGN into Xenopus laevis embryos (33), induces gene expression modification and chromatin structure changes that can be accompanied by developmental defects.
Although each of the HMG proteins affects the chromatin binding of H1 and members of the same HMG family compete with each other for chromatin sites, different types of HMG proteins do not compete with each other. Several reasons could account for the lack of competition among different types of HMG proteins. One possibility is that the major chromatin binding sites of each family localize to distinct nuclear compartments or to unique chromatin domains. Another possibility is that each type of HMG protein recruits specific protein partners that target the HMG protein to unique chromatin regions (38, 49). Finally, since histone H1 has a tripartite structure (7, 13) and binds to nucleosomes through multiple sites (57, 61), it is conceivable that each HMG family affects a distinct set of H1 nucleosome binding sites. The last possibility is compatible with our observation of the synergistic effect of distinct HMG families on H1 binding. The functional interplay between HMG proteins and H1 is also fully compatible with the available data showing that loss of H1, or addition of HMG proteins, reduces the compaction of chromatin, changes the transcriptional profile, and enhances the accessibility of nucleosomal components to various regulatory factors.
Our study emphasizes that binding of H1 to chromatin is modulated not only by posttranslational modifications (20, 30, 36) but also by a dynamic, stochastic process in which HMG proteins continuously weaken the binding of H1 to chromatin. The probability of one competition event (i.e., the binding of one HMG molecule instead of one H1 molecule) will depend on the relative quantity of each HMG protein present in the vicinity of a given nucleosome. Control of the local HMG concentration (for example, by up or down regulation of HMG gene expression) will thus increase or decrease the probability of the “stochastic” binding event. The dynamic properties of both H1 and HMG proteins are the driving force facilitating the competition between these chromatin binding proteins. We suggest that during the transient dissociation of the entire, or even part of, the H1 molecule from its nucleosomal binding site, the rapidly moving HMG proteins bind to the vacated site, thereby hindering H1 rebinding or providing a temporary wedge that further weakens the binding of H1. By reducing the binding of H1 to chromatin, the HMG proteins increase the ability of positive or negative regulatory factors to access their nucleosomal targets.
The example we provide with histone H1 and the HMG proteins is likely to reflect a general mechanism. Indeed, with the exception of core histone proteins, all of the chromatin-associated proteins (Misteli, unpublished observation) are highly mobile within the nucleus and bind to chromatin transiently. Chromosomal proteins are thus able to roam an entire nucleus within seconds and scan the nucleus for potential binding sites. Since the binding of proteins to chromatin is transient, binding sites become frequently and repeatedly unoccupied and thus constantly available for binding by proteins roaming in the vicinity. The pool of competitors locally available will stochastically modulate the occupancy of the binding site. The pool of competitors may vary depending on their level of expression and their concentration or sequestration within specific nuclear compartments (55). The network of continuous, dynamic interaction of chromatin binding proteins with their nucleosomal targets provides flexibility to the structure and activity of the chromatin fiber.
Acknowledgments
We thank D. Brown for generating the H1°-GFP-expressing cells, Paola Scaffidi and Marco Bianchi for the HMGB1-GFP constructs, and Susan H. Garfield and Stephen M. Wincovitch (Laboratory of Experimental Carcinogenesis, Center for Cancer Research, National Cancer Institute) for help with the confocal microscopy.
REFERENCES
- 1.Agresti, A., and M. E. Bianchi. 2003. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13:170-178. [DOI] [PubMed] [Google Scholar]
- 2.Alami, R., Y. Fan, S. Pack, T. M. Sonbuchner, A. Besse, Q. Lin, J. M. Greally, A. I. Skoultchi, and E. E. Bouhassira. 2003. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc. Natl. Acad. Sci. USA 100:5920-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashar, H. R., M. Schoenberg Fejzo, A. Tkachenko, X. Zhou, J. A. Fletcher, S. Weremowicz, C. C. Morton, and K. Chada. 1995. Disruption of the architectural factor HMGI-C: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domain. Cell 82:57-65. [DOI] [PubMed] [Google Scholar]
- 4.Battista, S., F. Pentimalli, G. Baldassarre, M. Fedele, V. Fidanza, C. M. Croce, and A. Fusco. 2003. Loss of Hmga1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. FASEB J. 17:1496-1498. [DOI] [PubMed] [Google Scholar]
- 5.Beaujean, N., C. Bouniol-Baly, C. Monod, K. Kissa, D. Jullien, N. Aulner, C. Amirand, P. Debey, and E. Kas. 2000. Induction of early transcription in one-cell mouse embryos by microinjection of the non-histone chromosomal protein HMG-I. Dev. Biol. 221:337-354. [DOI] [PubMed] [Google Scholar]
- 6.Beitzel, B., and F. Bushman. 2003. Construction and analysis of cells lacking the HMGA gene family. Nucleic Acids Res. 31:5025-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bharath, M. M., S. Ramesh, N. R. Chandra, and M. R. Rao. 2002. Identification of a 34 amino acid stretch within the C-terminus of histone H1 as the DNA-condensing domain by site-directed mutagenesis. Biochemistry 41:7617-7627. [DOI] [PubMed] [Google Scholar]
- 8.Bianchi, M. E., and M. Beltrame. 2000. Upwardly mobile proteins. Workshop: the role of HMG proteins in chromatin structure, gene expression and neoplasia. EMBO Rep. 1:109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birger, Y., K. L. West, Y. V. Postnikov, J. H. Lim, T. Furusawa, J. P. Wagner, C. S. Laufer, K. H. Kraemer, and M. Bustin. 2003. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 22:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, D. T. 2003. Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol. 81:221-227. [DOI] [PubMed] [Google Scholar]
- 11.Bustin, M. 1989. Preparation and application of immunological probes for nucleosomes. Methods Enzymol. 170:214-251. [DOI] [PubMed] [Google Scholar]
- 12.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bustin, M., S. C. Rall, R. H. Stellwagen, and R. D. Cole. 1969. Histone structure: Asymmetric distribution of lysine residues in lysine-rich histone. Science 163:391-393. [DOI] [PubMed] [Google Scholar]
- 14.Bustin, M., and R. Reeves. 1996. High mobility group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 54:35-100. [DOI] [PubMed] [Google Scholar]
- 15.Calogero, S., F. Grassi, A. Aguzzi, T. Voigtlander, P. Ferrier, S. Ferrari, and M. E. Bianchi. 1999. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 22:276-280. [DOI] [PubMed] [Google Scholar]
- 16.Catez, F., D. T. Brown, T. Misteli, and M. Bustin. 2002. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 3:760-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung, E., A. S. Zarifyan, and W. L. Kraus. 2002. Histone H1 represses estrogen receptor alpha transcriptional activity by selectively inhibiting receptor-mediated transcription initiation. Mol. Cell. Biol. 22:2463-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheutin, T., A. J. McNairn, T. Jenuwein, D. M. Gilbert, P. B. Singh, and T. Misteli. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721-725. [DOI] [PubMed] [Google Scholar]
- 19.Chiappetta, G., V. Avantaggiato, R. Visconti, M. Fedele, S. Battista, F. Trapasso, B. M. Merciai, V. Fidanza, V. Giancotti, M. Santoro, A. Simeone, and A. Fusco. 1996. High level expression of the HMGI (Y) gene during embryonic development. Oncogene 13:2439-2446. [PubMed] [Google Scholar]
- 20.Dou, Y., J. Bowen, Y. Liu, and M. A. Gorovsky. 2002. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J. Cell Biol. 158:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan, Y., T. Nikitina, E. M. Morin-Kensicki, J. Zhao, T. R. Magnuson, C. L. Woodcock, and A. I. Skoultchi. 2003. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 23:4559-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fedele, M., S. Battista, G. Manfioletti, C. M. Croce, V. Giancotti, and A. Fusco. 2001. Role of the high mobility group A proteins in human lipomas. Carcinogenesis 22:1583-1591. [DOI] [PubMed] [Google Scholar]
- 23.Giancotti, V., B. Pani, P. D'Andrea, M. T. Berlingieri, F. P. Di, A. Fusco, G. Vecchio, R. Philp, R. C. Crane, R. H. Nicolas, et al. 1987. Elevated levels of a specific class of nuclear phosphoproteins in cells transformed with v-ras and v-mos oncogenes and by cotransfection with c-myc and polyoma middle T genes. EMBO J. 6:1981-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grasser, K. D., S. H. Teo, K. B. Lee, R. W. Broadhurst, C. Rees, C. H. Hardman, and J. O. Thomas. 1998. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1). Eur. J. Biochem. 253:787-795. [DOI] [PubMed] [Google Scholar]
- 25.Guazzi, S., A. Strangio, A. T. Franzi, and M. E. Bianchi. 2003. HMGB1, an architectural chromatin protein and extracellular signalling factor, has a spatially and temporally restricted expression pattern in mouse brain. Gene Expr. Patterns 3:29-33. [DOI] [PubMed] [Google Scholar]
- 26.Hellauer, K., E. Sirard, and B. Turcotte. 2001. Decreased expression of specific genes in yeast cells lacking histone H1. J. Biol. Chem. 276:13587-13592. [DOI] [PubMed] [Google Scholar]
- 27.Herrera, J. E., K. L. West, R. L. Schiltz, Y. Nakatani, and M. Bustin. 2000. Histone H1 is a specific repressor of core histone acetylation in chromatin. Mol. Cell. Biol. 20:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill, D. A., and A. N. Imbalzano. 2000. Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry 39:11649-11656. [DOI] [PubMed] [Google Scholar]
- 29.Hill, D. A., and R. Reeves. 1997. Competition between HMG-I(Y), HMG-1 and histone H1 on four way junction DNA. Nucleic Acids Res. 25:3523-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horn, P. J., L. M. Carruthers, C. Logie, D. A. Hill, M. J. Solomon, P. A. Wade, A. N. Imbalzano, J. C. Hansen, and C. L. Peterson. 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol. 11:11. [DOI] [PubMed] [Google Scholar]
- 31.Johns, E. W. 1982. The HMG chromosomal proteins. Academic Press, London, England.
- 32.Koop, R., L. Di Croce, and M. Beato. 2003. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J. 22:588-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korner, U., M. Bustin, U. Scheer, and R. Hock. 2003. Developmental role of HMGN proteins in Xenopus laevis. Mech. Dev. 120:1177-1192. [DOI] [PubMed] [Google Scholar]
- 34.Laybourn, P. J., and J. T. Kadonaga. 1991. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 254:238-245. [DOI] [PubMed] [Google Scholar]
- 35.Lehtonen, S., V. M. Olkkonen, M. Stapleton, M. Zerial, and E. Lehtonen. 1998. HMG-17, a chromosomal non-histone protein, shows developmental regulation during organogenesis. Int. J. Dev. Biol. 42:775-782. [PubMed] [Google Scholar]
- 36.Lever, M. A., J. P. Th'ng, X. Sun, and M. J. Hendzel. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408:873-876. [DOI] [PubMed] [Google Scholar]
- 37.Li, J., R. Kokkola, S. Tabibzadeh, R. Yang, M. Ochani, X. Qiang, H. E. Harris, C. J. Czura, H. Wang, L. Ulloa, H. S. Warren, L. L. Moldawer, M. P. Fink, U. Andersson, K. J. Tracey, and H. Yang. 2003. Structural basis for the proinflammatory cytokine activity of high mobility group box 1. Mol. Med. 9:37-45. [PMC free article] [PubMed] [Google Scholar]
- 38.Lim, J. H., M. Bustin, V. V. Ogryzko, and Y. V. Postnikov. 2002. Metastable macromolecular complexes containing high mobility group nucleosome-binding chromosomal proteins in HeLa nuclei. J. Biol. Chem. 277:20774-20782. [DOI] [PubMed] [Google Scholar]
- 39.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell. Biol. 2:444-456. [DOI] [PubMed] [Google Scholar]
- 40.Martelli, A. M., M. Riccio, R. Bareggi, G. Manfioletti, G. Tabellini, G. Baldini, P. Narducci, and V. Giancotti. 1998. Intranuclear distribution of HMGI/Y proteins: an immunocytochemical study. J. Histochem. Cytochem. 46:863-864. [DOI] [PubMed] [Google Scholar]
- 41.Miloshev, G., P. Venkov, K. van Holde, and J. Zlatanova. 1994. Low levels of exogenous histone H1 in yeast cause cell death. Proc. Natl. Acad. Sci. USA 91:11567-11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 43.Nightingale, K., S. Dimitrov, R. Reeves, and A. P. Wolffe. 1996. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 15:548-561. [PMC free article] [PubMed] [Google Scholar]
- 44.Pallier, C., P. Scaffidi, S. Chopineau-Proust, A. Agresti, P. Nordmann, M. E. Bianchi, and V. Marechal. 2003. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol. Biol. Cell 14:3414-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 46.Postnikov, Y. V., J. E. Herrera, R. Hock, U. Scheer, and M. Bustin. 1997. Clusters of nucleosomes containing chromosomal protein HMG-17 in chromatin. J. Mol. Biol. 274:454-465. [DOI] [PubMed] [Google Scholar]
- 47.Prymakowska-Bosak, M., R. Hock, F. Catez, J. H. Lim, Y. Birger, H. Shirakawa, K. Lee, and M. Bustin. 2002. Mitotic phosphorylation of chromosomal protein HMGN1 inhibits nuclear import and promotes interaction with 14.3.3 proteins. Mol. Cell. Biol. 22:6809-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prymakowska-Bosak, M., T. Misteli, J. E. Herrera, H. Shirakawa, Y. Birger, S. Garfield, and M. Bustin. 2001. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 21:5169-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeves, R. 2001. Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277:63-81. [DOI] [PubMed] [Google Scholar]
- 50.Reeves, R., and M. S. Nissen. 1999. Purification and assays for high mobility group HMG-I(Y) protein function. Methods Enzymol. 304:155-188. [DOI] [PubMed] [Google Scholar]
- 51.Ronfani, L., M. Ferraguti, L. Croci, C. E. Ovitt, H. R. Scholer, G. G. Consalez, and M. E. Bianchi. 2001. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development 128:1265-1273. [DOI] [PubMed] [Google Scholar]
- 52.Scaffidi, P., T. Misteli, and M. E. Bianchi. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418:191-195. [DOI] [PubMed] [Google Scholar]
- 53.Shen, X., and M. A. Gorovsky. 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86:475-483. [DOI] [PubMed] [Google Scholar]
- 54.Shen, X., L. Yu, J. W. Weir, and M. A. Gorovsky. 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell 82:47-56. [DOI] [PubMed] [Google Scholar]
- 55.Spector, D. L. 2003. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72:573-608. [DOI] [PubMed] [Google Scholar]
- 56.Takami, Y., R. Nishi, and T. Nakayama. 2000. Histone H1 variants play individual roles in transcription regulation in the DT40 chicken B cell line. Biochem. Biophys. Res. Commun. 268:501-508. [DOI] [PubMed] [Google Scholar]
- 57.Thomas, J. O. 1999. Histone H1: location and role. Curr. Opin. Cell Biol. 11:312-317. [DOI] [PubMed] [Google Scholar]
- 58.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 59.van Holde, K. E. 1988. Chromatin. Springer Verlag, New York, N.Y.
- 60.Vicent, G. P., M. J. Melia, and M. Beato. 2002. Asymmetric binding of histone H1 stabilizes MMTV nucleosomes and the interaction of progesterone receptor with the exposed HRE. J. Mol. Biol. 324:501-517. [DOI] [PubMed] [Google Scholar]
- 61.Wolffe, A. P., S. Khochbin, and S. Dimitrov. 1997. What do linker histones do in chromatin? Bioessays 19:249-255. [DOI] [PubMed] [Google Scholar]
- 62.Yoshioka, K., K. Saito, T. Tanabe, A. Yamamoto, Y. Ando, Y. Nakamura, H. Shirakawa, and M. Yoshida. 1999. Differences in DNA recognition and conformational change activity between boxes A and B in HMG2 protein. Biochemistry 38:589-595. [DOI] [PubMed] [Google Scholar]
- 63.Zhou, X., K. F. Benson, H. R. Ashar, and K. Chada. 1995. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376:771-774. [DOI] [PubMed] [Google Scholar]
- 64.Zlatanova, J., and K. van Holde. 1998. Linker histones versus HMG1/2: a struggle for dominance? Bioessays 20:584-588. [DOI] [PubMed] [Google Scholar]