SUMMARY
In most bacteria cell division is mediated by a protein super-complex called the divisome that co-ordinates the constriction and scission of the cell envelope. FtsZ is the first of the divisome proteins to accumulate at the division site and is widely thought to function as a force generator that constricts the cell envelope. In this study we have used a combination of confocal fluorescence microscopy and Fluorescence Recovery After Photobleaching (FRAP) to determine if divisome proteins are present at the septum at the time of cytoplasmic compartmentalisation in Escherichia coli. Our data suggests that many are, but that FtsZ and ZapA disassemble before the cytoplasm is sealed by constriction of the inner membrane. This observation implies that FtsZ cannot be a force generator during the final stage(s) of envelope constriction in E. coli.
Keywords: Cell division, Disassembly, Escherichia coli, FRAP, FtsZ
INTRODUCTION
Cell division in the Gram-negative bacterium Escherichia coli requires constriction and eventually scission of the cell envelope. This process is mediated by a protein super-complex (the ‘divisome’), which consists of more than 24 different proteins (reviewed in (Vicente et al., 2006, de Boer, 2010, Lutkenhaus et al., 2012)). The assembly of these proteins is hierarchical and occurs in a two-step process (Aarsman et al., 2005), starting with the arrival of FtsZ at the mid-cell (reviewed in (Adams & Errington, 2009, Erickson et al., 2010)). FtsA and ZipA are simultaneously recruited, and tether FtsZ to the inner membrane (Pichoff & Lutkenhaus, 2002). Four non-essential divisome proteins (i.e. ZapA, ZapB, ZapC and ZapD) are also recruited by FtsZ-FtsA-ZipA, to form an intermediate structure called the Z-ring (Adams & Errington, 2009, Hale et al., 2011, Durand-Heredia et al., 2012, Durand-Heredia et al., 2011). In the second phase of assembly, inner membrane, periplasmic and outer membrane proteins are recruited to form the mature divisome (Aarsman et al., 2005, Vicente et al., 2006, de Boer, 2010, Lutkenhaus et al., 2012, Gerding et al., 2007, Typas et al., 2010). Once assembled, the divisome constricts the cell envelope and ultimately brings about the sequential closure of the cytoplasm and then the periplasm (Skoog et al., 2012).
FtsZ is a tubulin-like GTPase thought to play two key roles in the division process. First as an assembly platform for other cell division proteins, and then as a force generator that pulls the cell envelope inward (Mingorance et al., 2010, Erickson et al., 2010, Adams & Errington, 2009). The latter role is more controversial and is founded on two key observations. (i) FtsZ-YFP can constrict liposomes when fused to a membrane targeting sequence (Osawa et al., 2008, Osawa et al., 2009), and in the presence of FtsA* these constrictions occasionally go to completion (Osawa & Erickson, 2013). (ii) FtsZ filaments can undergo a conformational change from straight to curved in vitro (Lu et al., 2000). Curiously however, calculations derived from the in vitro analysis suggest that FtsZ proto-filaments can only curve to a diameter of ~24 nm. If the tethers provided by FtsA and ZipA are taken into account, then this calculation implies that FtsZ proto-filaments can only pull the inner membrane to a diameter of ~57 nm (not to complete closure) (Erickson et al., 2010). In this study we provide evidence that FtsZ departs from the septum before the cytoplasmic compartment has been separated. This simple observation implies that FtsZ cannot constrict the inner membrane during the final stage(s) of septal closure.
RESULTS
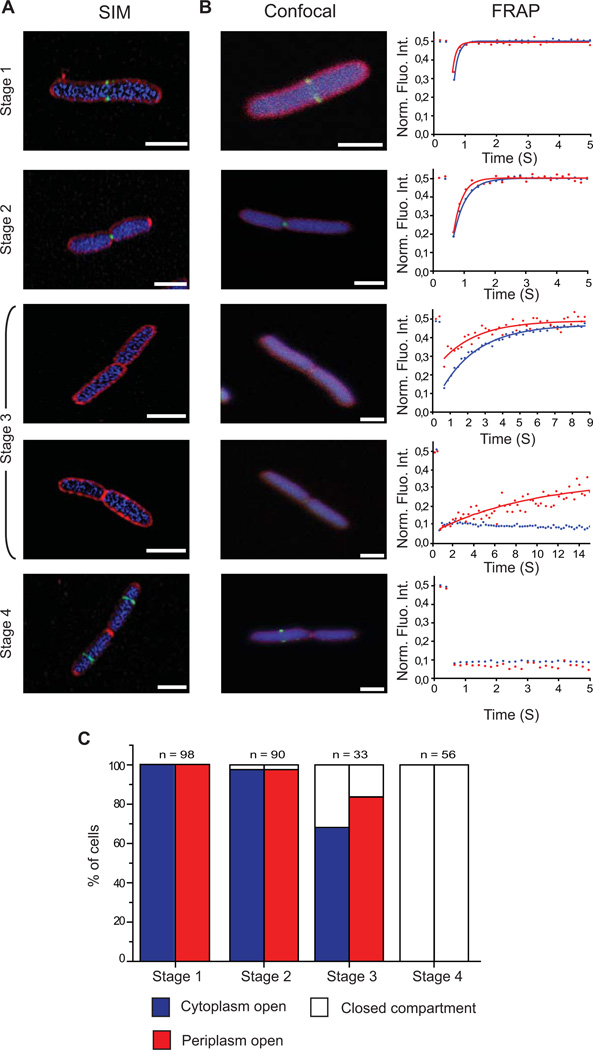
FtsZ-GFP disassembles from the divisome prior to closure of the cytoplasm
We engineered a strain of E. coli (MG1655) that simultaneously expressed three different fluorescent proteins; Cerulean in the cytoplasm (CeruleanCYTO), mCherry in the periplasm (mCherryPERI) and FtsZ-GFP. The ratio of native FtsZ : FtsZ-GFP was approximately 60:40 and the engineered strain grew similarly to the parental strain (supplementary Fig S1), indicating that cell division was not perturbed. Visual inspection of the cells by Super-Resolution Structured Illumination Microscopy (SR-SIM; (Gustafsson, 2000)) (Fig 1A) and confocal microscopy (Fig 1B) confirmed that there were different stages of FtsZ-GFP localization during the cell cycle, as reported by others (Wang et al., 2005, Den Blaauwen et al., 1999, Sun & Margolin, 1998). Briefly, FtsZ-GFP was first identified at the division septum as two dots or a bar (Stage 1). The two dots then converged to a single dot (Stage 2). In a third, more transient stage FtsZ-GFP was no longer visible at the division septum (Stage 3). Time-lapse imaging indicated that this stage represented roughly 1–2% of the cell cycle (supplementary Fig S2). Finally, in a fourth stage, FtsZ-GFP re-localized to the future division site of the daughter cells (Stage 4). To correlate the four stages of FtsZ-GFP localization with envelope constriction we combined the confocal microscopy with Fluorescence Recovery After Photobleaching (FRAP) (Stromqvist et al., 2010, Skoog et al., 2012). In the FRAP experiment we irreversibly bleached both CeruleanCYTO and mCherryPERI on one side of the division septum and monitored the diffusion of fluorescence from the non-bleached half of the cell over time (Fig 1B). Fluorescence recovery therefore indicated whether the cytoplasm and / or periplasm were still connected across the division septum (open), or whether they had been compartmentalized by the division septum (closed). In 186/188 cells where FtsZ-GFP was visualized at the division septum (Stages 1 and 2), both CeruleanCYTO and mCherryPERI were able to diffuse to the bleached half of the cell thus indicating that both compartments were open (Fig 1C). In 22/33 cells where FtsZ-GFP was absent from the division septum (Stage 3), the FRAP measurement indicated that the cytoplasm and the periplasm were still open (Fig 1C). However, in the remaining 11/33 cells the cytoplasm was closed across the division septum, and the periplasm was either open or closed (5 and 6 cells, respectively). All cells in this stage were followed from a stage where FtsZ-GFP was accumulated at the septum to a stage where it was not, to ensure sequential progression of the FtsZ-GFP localization pattern. Together, these data indicate that the cytoplasmic and periplasmic compartments are closing (or compartmentalising) at the stage when FtsZ-GFP is no longer detected at the divisome. By the time FtsZ-GFP had relocated to the future division sites in the daughter cells (Stage 4), the cytoplasm and the periplasm were both closed across the division septum (Fig 1C).
Figure 1. FtsZ-GFP dissociates from the divisome prior to cytoplasmic compartmentalization in E. coli.
Cells simultaneously co-expressing FtsZ-GFP (green), CeruleanCYTO (blue) and mCherryPERI (red) were imaged by (A) super-resolution Structural Illumination Microscopy (SIM) or (B) confocal microscopy. Following confocal imaging, cells were categorized into four different stages based on the localization pattern of FtsZ-GFP and then subjected to a FRAP measurement. In these measurements, one half of the cell was bleached and the subsequent recovery of CeruleanCYTO (blue) and mCherryPERI (red) was monitored over time (right panel). Recovery of fluorescence indicated that the compartments were still connected across the division septum (open). In contrast, no recovery indicated that the compartments were sealed by the division septum (closed). (C) Summation of FRAP data from 277 cells at different stages of division. These data indicated that FtsZ-GFP had dissociated from the septum in cells where the cytoplasm and the periplasm were still connected (stage 3). Scale bars: 2 μm.
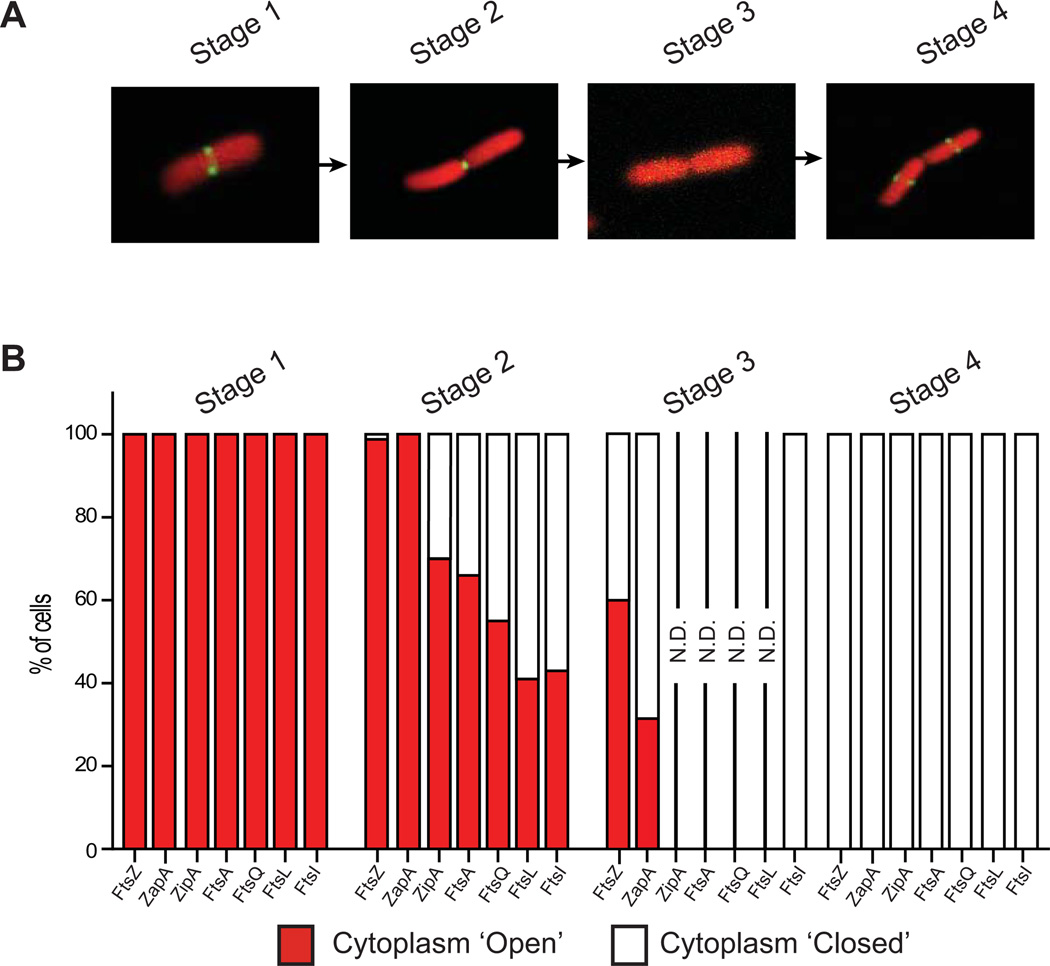
Most divisome proteins remain at the division site until after the cytoplasm is closed
To determine if other divisome proteins stayed longer at the division site we expressed GFP-ZapA, FtsA-GFP, ZipA-GFP, GFP-FtsQ, GFP-FtsL or GFP-FtsI together with mCherryCYTO (FtsZ-GFP was included as a control). The ZipA-GFP, GFP-FtsQ, and GFP-FtsI fusions were expressed at lower levels than their native counterparts (supplementary Fig S1) and all strains grew similarly to the parental strain indicating that division was not perturbed (supplementary Fig S1). The arrival and departure of all GFP-fusions at the division septum was similar to FtsZ-GFP (Fig 2A) and could be correlated with envelope constriction using the FRAP approach (Fig 2B). These data indicated that the transition from ‘cytoplasm-open’ to ‘cytoplasm-closed’ occurred whilst FtsA-GFP, ZipA-GFP, GFP-FtsQ, GFP-FtsL and GFP-FtsI were present at the division septum (Stage 2) (n > 150 for each strain) (Fig 2B). In contrast GFP-ZapA was absent from the division septum during this transition (Stage 3).
Figure 2. Most divisome proteins remain at the division septum after cytoplasmic compartmentalization.
(A) Live cells expressing GFP-ZapA, ZipA-GFP, FtsA-GFP, GFP-FtsQ, GFP-FtsL or GFP-FtsI together with mCherryCYTO were studied at different stages of division by monitoring the localization pattern of GFP. Only images for cells expressing ZipA-GFP and mCherryCYTO are shown. (B) FRAP measurements were carried out to determine if the cytoplasm was still connected across the division septum (open). Summation of FRAP data from at least 150 cells of each strain at different stages of division. GFP-ZapA behaved as FtsZ-GFP, disassembling from the divisome before the cytoplasm was compartmentalized. For all other strains the cytoplasm compartmentalised whilst the GFP fusion was present at the septum. Since compartmentalization occurred during Stage 2, we generally did not perform FRAP measurements on cells during stage 3 (N.D.= Not Determined). Cells in which the GFP localization was relocated in the daughters were always closed (Stage 4). Note that FtsZ-GFP was included as a control.
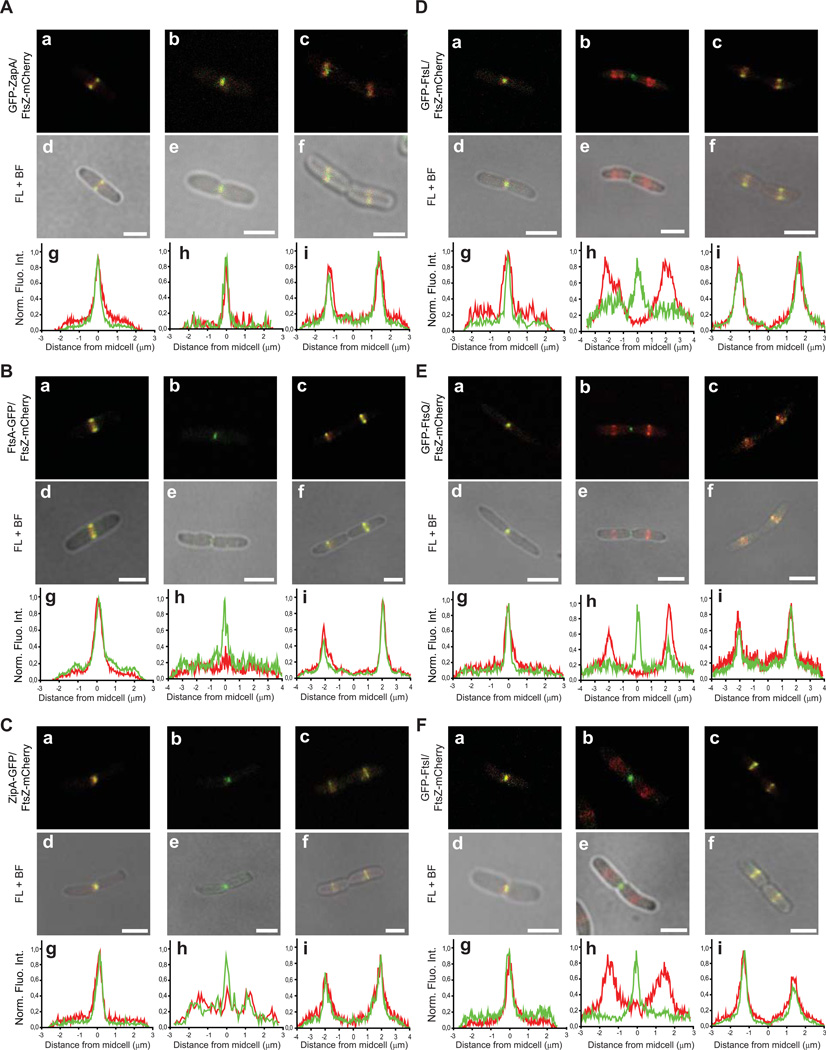
FtsZ-mCherry disassembles from the divisome before other divisome proteins
The FRAP data indicated that FtsZ-GFP and GFP-ZapA disassembled from the division site prior to compartmentalisation of the cytoplasm, whereas other divisome proteins remained until the cytoplasm compartmentalised. To substantiate this observation we monitored the localization of FtsZ-mCherry along with GFP-ZapA, ZipA-GFP, FtsA-GFP, GFP-FtsL, GFP-FtsQ or GFP-FtsI (n > 150 for all strains; Fig 3). Again, all strains grew similarly to the parental strain, indicating that division was not perturbed (supplementary Fig S3). Notably, FtsZ-mCherry and GFP-ZapA co-localized throughout the cell cycle (n = 792). This observation was expected, as ZapA is associated with the inner part of the Z-ring and should thus serve as a marker for FtsZ (Galli & Gerdes, 2010). For all other strains the mCherry and GFP signals also co-localized at the septum through most of the constriction process, however, at a late stage the mCherry signal disappeared from the division septum while the GFP signal remained. Those GFP fusion proteins that remained at the division septum were either membrane anchored (FtsA) or embedded (ZipA, FtsQ / L / I). In cells expressing GFP-FtsL, GFP-FtsQ or GFP-FtsI (but not ZipA-GFP or FtsA-GFP), the GFP signal remained at the division septum even when FtsZ-mCherry had relocated to the daughter cells. As expected all GFP fusions eventually relocated to the daughter cells to again co-localize with FtsZ-mCherry. Although we did not monitor FtsK in our experiment, Sherratt and co-workers have noted that FtsK-YFP remains at the division site longer than FtsZ-CFP (Wang et al., 2005). Taken together with the FRAP data, we conclude that FtsZ-GFP and GFP-ZapA disassemble from the division site before other division proteins. This event occurs before compartmentalization of the cytoplasm.
Figure 3. FtsZ-mCherry disassembles from the divisome before other divisome proteins.
Live cells co-expressing FtsZ-mCherry and GFP-ZapA, FtsA-GFP, ZipA-GFP, GFP-FtsL, GFP-FtsQ or GFP-FtsI were monitored throughout cell division. Representative images of the different localisation patterns are shown. The figures show fluorescence images, with and without bright field images. Beneath each image are the corresponding fluorescence profiles of FtsZ-mCherry (red) and the respective GFP fusion (green). These profiles were generated by integrating the fluorescence intensity originating from an area through the longitudinal axis of the entire cell. The width of the area varied so that it encompassed the fluorescent signal. Scale bar: 2 µm.
The localization patterns of FtsZ-GFP (and FtsZ-mCherry) reflect those of FtsZ
Although fluorescent fusions to FtsZ are not fully functional in vivo (Ma et al., 1996), they can complement an ftsZ84(Ts) mutant (Weiss et al., 1999) and their localization patterns and dynamics mimic that of the native FtsZ (see (Margolin, 2012) and references therein). To further substantiate the latter point we monitored the localization patterns of both FtsZ and GFP in a strain expressing both the native FtsZ and the FtsZ-GFP fusion by immuno-cytochemical fluorescence microscopy. In all cells analysed (n = 178) FtsZ co-localized with GFP, indicating that the native FtsZ behaved in a similar manner to FtsZ-GFP (supplementary Fig S4). One notable limitation with IFM is that it is not possible to discriminate between cells that are in Stage 3 (i.e. the protein is no longer at the septum) from cells that have not labelled with the antibodies.
DISCUSSION
FtsZ is thought to be a major force generator that pulls the inner membrane towards closure during division in E. coli (Mingorance et al., 2010, Erickson et al., 2010, Adams & Errington, 2009 2). To better understand this role, we correlated the localization of FtsZ-GFP at the division septum with envelope constriction by dual colour FRAP. Intriguingly, we noted that FtsZ-GFP disassembled from the division septum before the cytoplasmic and periplasmic compartments were sealed. To determine if other cell division proteins behaved in a similar manner to FtsZ-GFP we monitored the localization of GFP-ZapA, ZipA-GFP, FtsA-GFP, GFP-FtsL, GFP-FtsQ and GFP-FtsI during cytoplasmic compartmentalisation. GFP-ZapA behaved in a similar manner to FtsZ-GFP, departing from the divisome before cytoplasmic compartmentalisation, whilst all other proteins remained until the cytoplasm had been compartmentalised. The step-wise disassembly of the divisome was verified by carrying out dual colour fluorescence imaging. Although we have not assayed all divisome proteins in this study, other groups have noted that fluorescently labelled FtsZ, ZapA and ZapB co-localized throughout the cell cycle, but that FtsK remained longer at the division septum (Galli & Gerdes, 2010, Wang et al., 2005). Collectively, these observations are consistent with the notion that late stages of inner membrane constriction do not involve FtsZ or ZapA.
One caveat to our interpretation is that we do not know the detection limit for FtsZ-GFP. This limit will change during constriction as the amount of divisome bound FtsZ-GFP transits from approximately 33% to 0%. The possibility therefore remains that FtsZ-GFP is present in the deepest constrictions but not detected. However, this scenario seems unlikely as many other GFP-Fts fusion proteins were readily detected in deep constrictions even after FtsZ-GFP had disappeared. Some of these proteins, like GFP-FtsI and GFP-FtsQ, were more challenging to visualize than FtsZ-GFP on account of their significantly lower abundance, as documented by Western blotting and comparatively weak fluorescence during microscopy. Additionally, FtsZ-GFP can be detected at the new pole after division in Caulobacter crescentus and Agrobacterium tumefaciens (Thanbichler & Shapiro, 2006, Zupan et al., 2013), implying that it is possible to detect it in deeply constricted cells if it is still present.
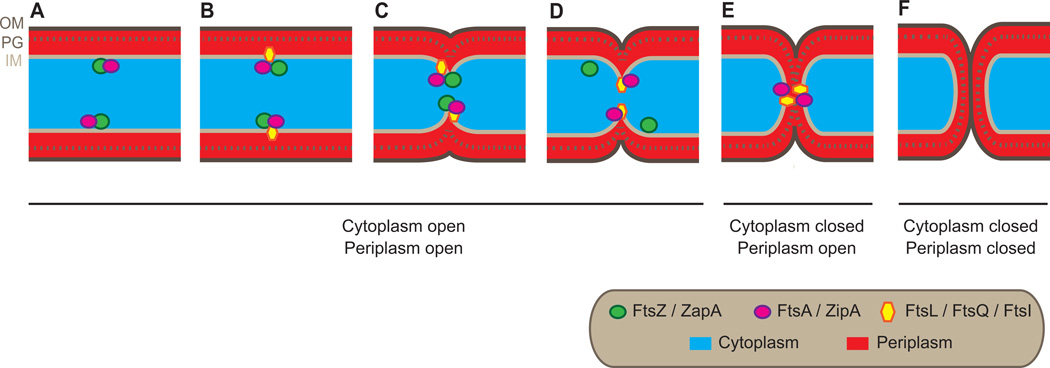
Another consideration in the interpretation of our data is whether a GFP fusion protein reflects the behaviour of the native protein in every detail. GFP-ZapA, GFP-FtsQ, GFP-FtsL and GFP-FtsI fusions can complement their respective null mutants indicating that they are fully functional (Weiss et al., 1999, Ghigo et al., 1999, Chen et al., 1999, Galli & Gerdes, 2010). It therefore seems reasonable to assume that the localization patterns of these fusions are similar to the native protein. FtsZ-GFP, FtsA-GFP, ZipA-GFP cannot fully complement their respective null mutants but their fluorescence localizations patterns and dynamics are thought to follow those of the native versions ((Margolin, 2012, Hale & de Boer, 1997, Ma et al., 1996) and references therein)). For FtsZ-GFP we re-evaluated this point by immuno-cytochemical fluorescence microscopy. Considering all the data, we suggest that the native FtsZ and ZapA also dissociate from the division site before cytoplasmic compartmentalization, whilst native FtsA, ZipA, FtsQ, FtsL and FtsI do not (Fig 4). The disassembly of FtsZ-GFP before cytoplasmic compartmentalisation was unexpected, as Osawa and Erickson have noted that FtsZ-YFP and FtsA* can (in rare cases) divide liposomes (Osawa & Erickson, 2013). However it is worth noting that there are no negative-regulatory systems (like Noc and MinCDE) acting on FtsZ-YFP in the liposomes, so it is possible that FtsZ proto-filaments persist longer than they would do in vivo.
Figure 4. A ‘first-in, first-out’ model for FtsZ at the division septum in E. coli.
(A and B) Prior to constriction of the cell envelope FtsZ and other cell division proteins sequentially assemble at the division septum. (C) Constriction of the envelope is thought to initiate when all divisome proteins are present. (D) At a late stage of inner membrane constriction, FtsZ and ZapA disassemble from the septum. (E) Closure of the cytoplasm by constriction of the inner membrane occurs in the absence of FtsZ and ZapA. (F) Following cytoplasmic compartmentalization, ZipA, FtsA, FtsQ, FtsL and FtsI disassemble from the septum.
Our data do not enable us to accurately determine when FtsZ-GFP and GFP-ZapA disassembled, but we estimate that it occurs at a diameter >12 nm. This estimation is based on calculations that GFP can freely diffuse through a ring-diameter of >12 nm (Stromqvist et al., 2010). Our estimate is consistent with in vitro experiments and calculations done by Erickson and colleagues (Erickson et al., 2010, Erickson et al., 1996, Lu et al., 2000, Chen & Erickson, 2009). They estimate that highly curved FtsZ proto-filaments could bend to a minimum diameter of ~24 nm, which would correspond to a diameter of ~57 nm for the inner membrane (Erickson et al., 2010). In this scenario FtsZ would be unable to complete the closure of the inner membrane. We speculate that disassembly of FtsZ (and ZapA) from the leading edge of the constricting inner membrane might occur so that fusion of the lipid bilayer can proceed. In principle, fusion could be driven by lipid synthesis, as shown in the L-form of Bacillus subtilus (Mercier et al., 2013), or by inward growth of the peptidoglycan. Further work is required to gain insight into this fundamental aspect of division.
EXPERIMENTAL PROCEDURES
Strain and Plasmid construction
The locus encoding the GFP fusions from strains EC448 (MC4100 ∆[λattL-lom]::bla lacIq P208-ftsZ-gfp), EC450 (MC4100 ∆[λattL-lom]::bla lacIq P208-zipA-gfp), EC447 (MC4100 ∆[λattL-lom]::bla lacIq P210-ftsA-gfp), EC442 (MC4100 ∆[λattL-lom]::bla lacIq P207-gfp-ftsQ), EC438 (MC4100 ∆[λattL-lom]::bla lacIq P207-gfp-ftsL), and EC436 (MC4100 ∆[λattL-lom]::bla lacIq P207-gfp-ftsI) (Ghigo et al., 1999, Chen et al., 1999, Weiss et al., 1999) were transferred to MG1655 by P1 phage transduction, selecting for AmpR, to create strains BS001, BS002, BS003, BS004, BS005 and BS007, respectively (supplementary Table S1).
The gfp-zapA fusion was constructed by PCR amplifying zapA from E. coli MG1655 chromosomal DNA using primers zapA_up (CACGAATTCAACAACAACATGTCTGCACAACCCGTCGATATC) and zapA_down (CACAAGCTTCATTCAAAGTTTTGGTTAGTTTTTTCG). The 356 bp product was cut with EcoRI and HindIII (sites underlined in primers) and ligated into the GFP fusion vector pDSW207 (Weiss et al., 1999). The resulting plasmid was verified by sequencing and designated pDSW1646. Next, pDSW1646 was digested with SphI and ScaI to obtain a ~ 3.4 kb restriction fragment carrying lacIq and P204::gfp-zapA. This fragment was ligated into SphI/ScaI-digested pJC69 (Chen & Beckwith, 2001) to produce pDSW1655. Finally, plasmid pDSW1655 was integrated into the phi 80 phage attachment site of MG1655 as described (Haldimann & Wanner, 2001). The final strain was designated EC3408 and has the following genotype: MG1655 Φ80::pDSW1655(lacIq P204::gfp-zapA SpcR).
The pRha-BAD plasmid was constructed by PCR amplifying the multiple cloning site of pBAD24, and cloning it into a non-coding region of the pRha67 plasmid (between the ColE1 origin of replication and the multiple cloning site). The pRha67 (Giacalone et al., 2006) plasmid was obtained from Jan Willem de Gier (Stockholm University). The pKS1 plasmid was constructed by cloning the gene for the Cerulean fluorescent protein (Rizzo et al., 2004) downstream of the rhamnose inducible promoter in pRha-BAD. The gene encoding mCherry (Shaner et al., 2004) was fused to the region encoding the signal sequence of TorA as described previously (Skoog et al., 2012), and cloned downstream of the arabinose inducible promoter in pRha-BAD. The pKS2 plasmid was constructed by cloning the gene for mCherry downstream of the rhamnose inducible promoter in the pRha67 plasmid. All cloning was carried out using the uracil-excision method (Norholm, 2010). Both constructs were verified by DNA sequencing (MWG, Germany). A complete list of the strains and plasmids used in this study is provided in supplementary Table S1.
Bacterial growth and fluorescent protein expression
To create the strain expressing FtsZ-GFP, CeruleanCYTO and mCherryPERI, we transformed BS001 with pKS1. Cells were grown as described previously (Skoog et al., 2011, Thomas et al., 2001), with the addition of 2.5 µM Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Weiss et al., 1999) to induce expression of FtsZ-GFP. To create the strains expressing the GFP fusions and mCherryCYTO, we transformed BS002, BS003, BS004, BS005, BS007 and EC3408 with pKS2. Overnight cultures were back-diluted 1:400 in fresh LB media supplemented with appropriate antibiotics [BS002, BS003, BS004, BS005, BS007: 25 µg mL−1 Ampicillin; EC3408: 15 µg mL−1 Spectinomycin; 60 µg mL−1 Kanamycin], 12 mM L-rhamnose and IPTG (BS007 and BS005, 2.5 µM; BS004 and EC3408, 5 µM; BS002, 50 µM; BS003, 100 µM) (Ghigo et al., 1999, Chen et al., 1999, Weiss et al., 1999). The cultures were incubated at 37 °C until an OD600 of ~ 0.4 was reached.
Strains BS002, BS003, BS004, BS005, BS007 and EC3408 were transformed with pEG4 (7) to produce cells co-expressing ZipA-GFP, FtsA-GFP, GFP-FtsL, GFP-FtsQ, GFP-FtsI or GFP-ZapA and FtsZ-mCherry. Cells were grown and induced essentially as described before (Galli & Gerdes, 2010). Cells were grown in M9 minimal media at 30 °C supplemented with 0.2% glucose,1 µg ml−1 thiamine and 0.1% casamino acids, with the addition of appropriate antibiotics and IPTG (BS007 and BS005, 2.5 µM; BS004 and EC3408, 5 µM; BS002, 50 µM; BS003, 100 µM) (Ghigo et al., 1999, Chen et al., 1999, Weiss et al., 1999). The cultures were incubated until an OD600 of ~ 0.2 was reached.
Western blotting
A volume of cells corresponding to 0.2 OD600 units was collected from cell cultures. The samples were suspended in loading buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred to nitrocellulose membranes using a semi-dry Transfer-Blot apparatus (Bio-Rad). The membranes were blocked in 5% (w/v) milk and probed with anti-sera to FtsZ (Agrisera, Sweden), FtsI (Weiss et al., 1999), ZipA (Hale & de Boer, 1997) or FtsQ (Dopazo et al., 1992).
Imaging
Live cell imaging and FRAP measurements
~ 6 µl of cell culture were placed on a microscope glass coated with thin pre-made agarose pads (1% (w/v) agarose). A cover slip was added and the cells were left for ~ 5 minutes so that they were immobilised. Confocal imaging and FRAP measurements were performed on a Zeiss LSM700 System (Carl Zeiss, Jena, Germany). Image acquisition time in confocal mode was < 1 sec (512 × 512). In FRAP mode the resolution was lowered (256 × 256) in order to gain scanning speed. All FRAP data were analysed in Origin 8.5 (OriginLab Corporation, Northampton). SIM images were acquired on a Zeiss LSM780 ELYRA PS.1 system. For each fluorophore the illumination grids were phase shifted 5 times over the field of view (1024×1024) and then rotated 180/5 degrees (i.e. five rotations per image) to generate a data set containing 25 raw images per fluorophore. In general, acquisition time for a single data set was 3–6 seconds. The SIM images were reconstructed and subsequently analysed in the Zeiss software ZEN2011 black. The calibrated system gave a lateral (xy−) resolution of ~100 nm and a height (z−) resolution of ~275 nm. All other images were analysed using ImageJ (N.I.H. U.S.A.). In all imaging and FRAP measurements Cerulean was excited at a wavelength of 405 nm, GFP at 488 nm and mCherry at 555 nm (for SIM imaging mCherry was excited with a 561 nm laser). Appropriate filter settings for the respective fluorophores were chosen in the software (ZEN2011 Black). To reduce bleaching the laser power output was kept below 1% for the 405 nm and 488 nm lasers, and below 5 % for the 555/561 nm lasers (total laser power was 5 mW, 10 mW and 10 mW, respectively). Objectives used were either 100X or 63X NA 1.4 oil immersion plan-Apochromat for SIM imaging and 63X NA 1.4 oil immersion plan-Apochromat for confocal imaging and FRAP measurements.
Due to limitations in the ZEN2011 software it was not possible to carry out FRAP and time-lapse imaging simultaneously. Therefore we manually followed cells by acquiring confocal images as they progressed from Stage 2 (one dot) into Stage 3 (no dot), then immediately carried out the FRAP measurement (the timing between the last acquired confocal image and the FRAP measurement was < 10 seconds). For bleaching during the FRAP measurements the laser power was set to 100 % and applied for ~195 ms. Fluorescence recovery was monitored by acquiring an image every ~190 ms for a total of 6 to 18 seconds. During the FRAP measurements the green channel was disconnected to gain scanning speed. In the FRAP analysis we followed Skoog et. al. (Skoog et al., 2012), in short; the recovery of fluorescence intensity was recorded within two equally sized circular regions (r ≈ 0.2 µm), one in the middle of the bleached compartment and the other in the middle of the unbleached compartment. To address the fact that the fluorophores were subject to constant photobleaching, the fluorescence recovery data were normalized to the total remaining fluorescence at any given time point t. The normalized fluorescence intensity in the bleached compartment was thus
where F1(t) denotes the fluorescence intensity in the bleached compartment at time t and F2(t) is the fluorescence intensity in the unbleached compartment at that time.
For the cell length measurements, cells were harvested at an OD600 of 0.2 or 0.4 and fixed in the same way as for the immunofluorescence measurements. All strains were imaged under bright field illumination and cell length statistics were acquired using the built in ImageJ plug-in. Cell lengths were determined for more than 100 cells (collected form at least 3 different liquid cultures).
Immunofluorescence microscopy
The protocol of Addinall (Addinall et al., 1996) was followed with minor adjustments. After fixing [2.6% (vol/vol) paraformaldehyde, 0.04% (vol/vol) glutaraldehyde, 32.25 mM Na3PO4, pH 7.4] in room temperature for 15 minutes samples were incubated for 20 minutes on ice. Cells were washed 3 times in PBS and resuspended in GTE buffer (50 mM glucose, 10 mM EDTA, 20 mM Tris-HCl pH 7.5). The samples were incubated for 1–2 minutes with lysozyme (10 mg mL−1) and washed twice in PBS. Cells were resuspended in PBS containing 2 % BSA and blocking was carried out for 15 minutes. The cells were washed with PBS and incubated with primary antibodies against FtsZ (Agrisera, Sweden) and GFP (Sigma Aldrich, USA) diluted 1:100 in the same buffer, and then incubated overnight at 4 °C. Alexa Fluor 647-conjugated anti-rabbit sera (Molecular Probes) and Oregon Green 488-conjugated anti-mouse sera (Molecular Probes), both at a 1:500 dilution (in PBS containing 2 % BSA) were used as secondary antibody against FtsZ and GFP, respectively. After incubation the cells were washed at least 5 times in PBS and mounted on glass slides pre-coated with 1 % agarose pads. Imaging was performed on a Zeiss LSM700 using appropriate laser and filter settings.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Kenn Gerdes (Newcastle University) for the pEG4 plasmid, Piet de Boer (Case Western Reserve University) for the ZipA anti-sera and Miguel Vicente (Centro National de Biotecnologia) for the FtsQ anti-sera. BS acknowledges support from Kungliga Vetenskapsakademien, Stiftelsen Längmanska Kulturfonden and the Swedish Association for Cytoskeletal Research. The work was supported by grants from the Swedish Foundation for Strategic Research (GvH), the European Research Council (GvH; ERC-2008-AdG 232648), the Swedish Research Council (GvH, DOD) and the US National Institutes of Health (DSW, GM083975).
Footnotes
The authors declare that they have no conflict of interest.
REFERENCES
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Molecular Microbiology. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Addinall SG, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. Journal of Bacteriology. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Molecular Microbiology. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- Chen JC, Weiss DS, Ghigo JM, Beckwith J. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. Journal of Bacteriology. 1999;181:521–530. doi: 10.1128/jb.181.2.521-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Erickson HP. FtsZ filament dynamics at steady state: subunit exchange with and without nucleotide hydrolysis. Biochemistry. 2009;48:6664–6673. doi: 10.1021/bi8022653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PAJ. Advances in understanding E. coli cell fission. Curr Opin Microbiol. 2010;13:730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Blaauwen T, Buddelmeijer N, Aarsman ME, Hameete CM, Nanninga N. Timing of FtsZ assembly in Escherichia coli. Journal of Bacteriology. 1999;181:5167–5175. doi: 10.1128/jb.181.17.5167-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopazo A, Palacios P, Sanchez M, Pla J, Vicente M. An amino-proximal domain required for the localization of FtsQ in the cytoplasmic membrane, and for its biological function in Escherichia coli. Molecular Microbiology. 1992;6:715–722. doi: 10.1111/j.1365-2958.1992.tb01520.x. [DOI] [PubMed] [Google Scholar]
- Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. Journal of Bacteriology. 2012;194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. Journal of Bacteriology. 2011;193:1405–1413. doi: 10.1128/JB.01258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci U S A. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli E, Gerdes K. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Molecular Microbiology. 2010;76:1514–1526. doi: 10.1111/j.1365-2958.2010.07183.x. [DOI] [PubMed] [Google Scholar]
- Gerding MA, Ogata Y, Pecora ND, Niki H, De Boer PAJ. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Molecular Microbiology. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo JM, Weiss DS, Chen JC, Yarrow JC, Beckwith J. Localization of FtsL to the Escherichia coli septal ring. Molecular Microbiology. 1999;31:725–737. doi: 10.1046/j.1365-2958.1999.01213.x. [DOI] [PubMed] [Google Scholar]
- Giacalone MJ, Gentile AM, Lovitt BT, Berkley NL, Gunderson CW, Surber MW. Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. Biotechniques. 2006;40:355–364. doi: 10.2144/000112112. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. Journal of Bacteriology. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Hale CA, Shiomi D, Liu B, Bernhardt TG, Margolin W, Niki H, de Boer PAJ. Identification of Escherichia coli ZapC (YcbW) as a Component of the Division Apparatus That Binds and Bundles FtsZ Polymers. Journal of Bacteriology. 2011;193:1393–1404. doi: 10.1128/JB.01245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton (Hoboken) 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci U S A. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. The price of tags in protein localization studies. Journal of Bacteriology. 2012;194:6369–6371. doi: 10.1128/JB.01640-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Kawai Y, Errington J. Excess membrane synthesis drives a primitive mode of cell proliferation. Cell. 2013;152:997–1007. doi: 10.1016/j.cell.2013.01.043. [DOI] [PubMed] [Google Scholar]
- Mingorance J, Rivas G, Velez M, Gomez-Puertas P, Vicente M. Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol. 2010;18:348–356. doi: 10.1016/j.tim.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Norholm MH. A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 2010;10:21. doi: 10.1186/1472-6750-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28:3476–3484. doi: 10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci U S A. 2013;110:11000–11004. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Skoog K, Söderström B, Widengren J, von Heijne G, Daley DO. Sequential closure of the cytoplasm and then the periplasm during cell division in Escherichia coli. Journal of Bacteriology. 2012;194:584–586. doi: 10.1128/JB.06091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromqvist J, Skoog K, Daley DO, Widengren J, von Heijne G. Estimating Z-ring radius and contraction in dividing Escherichia coli. Mol Microbiol. 2010;76:151–158. doi: 10.1111/j.1365-2958.2010.07087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Margolin W. FtsZ dynamics during the division cycle of live Escherichia coli cells. Journal of Bacteriology. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Daniel RA, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Molecular Microbiology. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Possoz C, Sherratt DJ. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 2005;19:2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. Journal of Bacteriology. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente M, Rico AI, Martinez-Arteaga R, Mingorance J. Septum enlightenment: assembly of bacterial division proteins. Journal of Bacteriology. 2006;188:19–27. doi: 10.1128/JB.188.1.19-27.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan JR, Cameron TA, Anderson-Furgeson J, Zambryski PC. Dynamic FtsA and FtsZ localization and outer membrane alterations during polar growth and cell division in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A. 2013;110:9060–9065. doi: 10.1073/pnas.1307241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.