Abstract
Herpesvirus saimiri (HVS) encodes seven Sm-class small nuclear RNAs, called HSURs (for Herpesvirus saimiri U RNAs), that are abundantly expressed in HVS-transformed, latently infected marmoset T cells but are of unknown function. HSURs 1, 2, and 5 have highly conserved 5′-end sequences containing the AUUUA pentamer characteristic of AU-rich elements (AREs) that regulate the stability of many host mRNAs, including those encoding most proto-oncogenes and cytokines. To test whether the ARE-containing HSURs act to sequester host proteins that regulate the decay of these mRNAs, we demonstrate their in vivo interaction with the ARE-binding proteins hnRNP D and HuR in HVS-transformed T cells using a new cross-linking assay. Comprehensive Northern and microarray analyses revealed, however, that the levels of endogenous ARE-containing mRNAs are not altered in T cells latently infected with HVS mutants lacking HSURs 1 and 2. HSUR 1 binds the destabilizing ARE-binding protein tristetraprolin induced following activation of HVS-transformed T cells, but even in such stimulated cells, the levels of host ARE-containing mRNAs are not altered by deletion of HSURs 1 and 2. Instead, HSUR 1 itself is degraded by an ARE-dependent pathway in HVS-transformed T cells, suggesting that HVS may take advantage of the host ARE-mediated mRNA decay pathway to regulate HSUR expression. This is the first example of posttranscriptional regulation of the expression of an Sm small nuclear RNA.
Herpesvirus saimiri strain A11 (HVS) is a gammaherpesvirus which causes aggressive T-cell leukemias and lymphomas in some nonhuman New World primates (reviewed in reference 29) and can efficiently transform cultured T cells of the common marmoset, Callithrix jacchus, to continuous interleukin-2 (IL-2)-independent growth (22). HVS-transformed marmoset T cells have the same phenotype as those derived from tumors of infected animals, predominantly CD8+ T cells with natural killer (NK) cell activity (reviewed in references 6, 35, and 74), providing an ideal system in which to study HVS gene function. HVS encodes seven small nuclear RNAs (snRNAs) of the Sm class, called HSURs for Herpesvirus saimiri U RNAs (2, 44, 56, 77). The HSURs are classified as Sm snRNAs since, similar to cellular snRNAs that function in RNA processing, they are transcribed by RNA polymerase II, acquire a trimethylguanosine cap, and assemble with Sm core proteins (44, 45). The HSURs show no extensive sequence similarity to any cellular snRNA and are the only Sm snRNAs known to be encoded by a virus.
The HSURs are the most abundant of the very few viral gene products expressed in latently infected, transformed T cells (56). HSURs 1 and 4 are present at ∼20,000 copies per cell, whereas the remaining five HSURs exist at ∼2,000 copies per cell (44). Deletion studies have demonstrated that the HSURs are not essential for viral replication or for transformation of cultured marmoset T cells by HVS (24, 56, 57); the tumorigenicity of the deletion mutants has not been reported for primates. Their abundance and conservation in related viruses suggest that the HSURs play a critical function in latently infected, transformed T cells.
HSURs 1 and 2 share an AU-rich sequence at their 5′ ends that is almost perfectly conserved among different HVS strains and in a closely related virus, Herpesvirus ateles (HVA) (1); HSUR 5 contains a similar but less conserved sequence (Fig. 1). By contrast, the remainder of the HSUR 1, 2, and 5 sequences is only about 50% identical between virus strains. The AU-rich sequences in HSURs 1, 2, and 5 show striking similarity to destabilizing AU-rich elements (AREs) found in the 3′ untranslated region (UTR) of mRNAs of a number of highly regulated cellular genes, primarily those for cytokines and growth factors and proto-oncogenes (9, 12, 16, 44, 67).
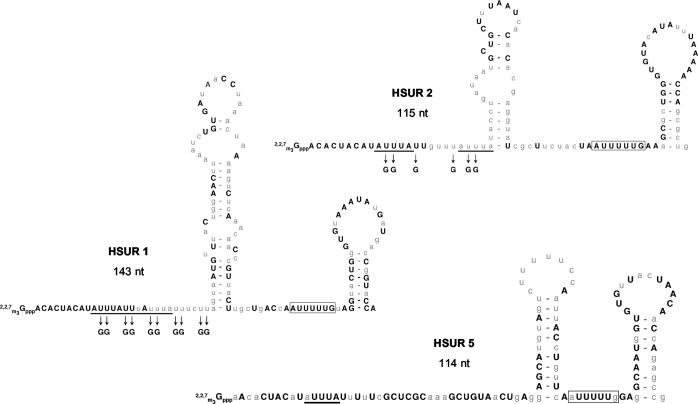
FIG. 1.
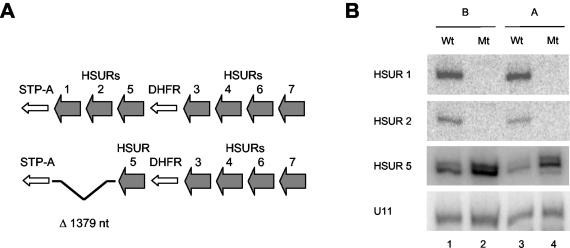
Conservation of the 5′ AU-rich sequences in HSURs 1, 2, and 5. The sequences of the ARE-containing HSURs 1, 2, and 5 were compared in HVS strains A (A11), B (B-SMHI), and C (C488-77) and the closely related strain HVA (A810). Depicted are the sequences and predicted secondary structures of the HVS A11 HSURs. For HSURs 1 and 2, nucleotides perfectly conserved in all four strains are shown in bold capital letters. For HSUR 5, the nucleotides in capital letters are conserved in the three HVS strains; however, HSUR 5 is not present in the HVA genome. The AUUUA pentamers in the AREs are underlined; the Sm-binding sites are boxed. Mutations made to disrupt the AREs in HSUR 1 (mutant HSUR 1) and HSUR 2 (mutant HSUR 2) for assays in Fig. 2, 7, and 8 are indicated. See the work of Lee and Steitz (45) for the sequences of HSURs 3 and 4 and that of Albrecht and Fleckenstein (2) for the sequences of HSURs 6 and 7. nt, nucleotides.
The stability and steady-state levels of ARE-containing mRNAs in a cell are tightly regulated and can be selectively altered in response to specific intra- and extracellular stimuli (9, 68). For example, activation of T-cell receptor signaling by using antibodies to cross-link CD3 and CD28 membrane receptors leads to stabilization and an increase in the steady-state levels of the ARE-containing mRNAs for IL-2, gamma interferon, tumor necrosis factor alpha (TNF-α), and granulocyte-macrophage colony-stimulating factor (GM-CSF) but not of those encoding c-myc and c-fos (47). Stabilization of ARE-containing mRNAs can be similarly achieved in T cells (7, 67, 79) and other cell types (32, 38, 70) by stimulation with phorbol esters or calcium ionophores, which simulate activation through the T-cell receptor by activating protein kinase C and increasing intracellular Ca2+ levels, respectively (73). Conversely, inhibition of these pathways with immunosuppressive agents can destabilize ARE-containing mRNAs (5, 59). Regulation of mRNA stability by these intracellular and extracellular signals is currently poorly understood, but several different kinase pathways are known to be involved (10, 30, 33, 51-53, 60, 71, 76, 83).
The effects of signaling pathways on ARE-containing mRNAs are mediated by specific ARE-binding proteins (8, 13, 50, 63, 82), which exhibit differential activation of the cellular mRNA decay machinery (15, 34, 43, 55). A regulatory role in the ARE-mediated mRNA decay pathway has been established in vivo for four proteins: HuR, hnRNP D (also called AUF1), and the related proteins TTP (tristetraprolin) and BRF1 (butyrate response factor 1). HuR is a predominantly nuclear, nucleocytoplasmic shuttling protein that stabilizes ARE-containing mRNAs when overexpressed (9, 27, 62). The zinc finger proteins TTP (11, 14, 41), which is inducible (14, 63), and the more recently described BRF1 (42, 69) can be found in both the nucleus and cytoplasm and promote the degradation of ARE-containing mRNAs. hnRNP D was identified as a destabilizing factor for ARE-containing mRNAs (43, 49, 64), although it appears to have a stabilizing effect in some cells (84). There are four alternatively spliced isoforms of hnRNP D, with the p37 and p42 isoforms having the greatest activity, both stabilizing and destabilizing (49, 64, 84). The expression and activity of these and other ARE-binding proteins coordinately regulate the stability of ARE-containing mRNAs in the cell.
Almost 1,500 genes that specify mRNAs with an ARE in the 3′ UTR have been identified in the human genome (4). Based on similarities in sequence and decay kinetics, these ARE-containing mRNAs have been divided into three classes (16, 61) that may be regulated independently in vivo (17, 47, 66, 71). Class I AREs are characterized by scattered AUUUA motifs within a U-rich region (e.g., c-fos and c-myc mRNAs) while class II AREs have a minimum of two tandem, overlapping copies of the AUUUA pentamer (e.g., mRNAs for many cytokines such as TNF-α and IL-3). Class III AREs contain a U-rich region but lack the AUUUA motif (e.g., c-jun mRNA) (61). By these criteria, HSUR 1, which contains three tandem AUUUA pentamers, contains a class II ARE, and HSURs 2 and 5, which have two and one AUUUA pentamer(s) in a U-rich region, respectively, contain class I AREs.
Cultured T cells transformed by HVS show modified expression of cytokines and other ARE-containing genes, including IL-2, IL-3, gamma interferon, TNF-α, TNF-β, and a homolog of IL-10 (19, 39, 74). Thus, it has been proposed that the ARE-containing HSURs compete with less abundant host mRNAs for host cell ARE-binding proteins in latently infected, transformed T cells (26, 58). This sequestration could lead to altered expression of cytokines or proto-oncogenes, perhaps enhancing cell proliferation or evasion of the host immune response. Accordingly, Lee et al. (44) found that the AREs of HSURs 1 and 2 are accessible for interactions within snRNPs by targeted RNase H cleavage, and Myer et al. (58) demonstrated that the ARE-containing HSURs can bind the ARE-binding protein HuR in vitro.
Here, we have directly tested the model that the ARE-containing HSURs 1 and 2 act to alter the stability and steady-state levels of cellular ARE-containing mRNAs in HVS-transformed marmoset T cells. We have extended the analysis of proteins bound to HSUR 1 both to the in vivo situation and after stimulation of T cells. Marmoset T cells transformed by the wild type versus HVS with HSURs 1 and 2 deleted were then compared by Northern and microarray analyses but showed no difference in the steady-state levels of endogenous ARE-containing mRNAs. Instead, we observe that the ARE in HSUR 1 leads to reduced steady-state levels of HSUR 1 in latently infected, HVS-transformed T cells, arguing that HSURs, as snRNAs, may make novel use of the ARE-dependent degradation pathway to posttranscriptionally regulate their own expression.
MATERIALS AND METHODS
HVS-transformed T-cell lines.
The HVS-transformed T-cell tumor cell line 1670 was cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. T cells of the common marmoset (C. jacchus) were isolated and transformed by wild-type HVS A11 and HSUR 1/2 deletion mutant HVS A11 (Δ2a in reference 57) as previously described (22) and were the generous gift of R. C. Desrosiers and D. P. Silva. They were grown in RPMI 1640 medium supplemented with 20% fetal bovine serum and 2 μM β-mercaptoethanol. The HSUR 1/2 deletion mutant virus in cell set B was found to contain a second, spontaneous deletion encompassing open reading frames 50 to 65.
Plasmids.
Plasmids used in transient transfections to test HSUR stability contain wild-type or ARE mutant HSUR 1 or HSUR 2 between the U1 promoter and U1 3′-end box in the pUC-U1 plasmid between the BglII and SalI sites. The pUC-U1 HSUR 1, HSUR 1 Mut, and HSUR 2 plasmids are described in the work of Fan et al. (26). The pUC-U1 Mut HSUR 2 plasmid was created by introducing mutations in the 5′ ARE of HSUR 2 (Fig. 1) into the pUC-U1 HSUR 2 plasmid by oligonucleotide-directed PCR mutagenesis. Sense and antisense HSUR transcripts were prepared using plasmids containing HSURs 1 to 5 (58) or ARE mutant HSUR 1 cloned between the EcoRI and HindIII sites of pSP64 (Promega) or ARE mutant HSUR 2 cloned between BglII and SalI in pSP73 (Promega). HSUR transcripts were created for in vitro cross-linking by linearization with HindIII and transcription with T7 RNA polymerase. Antisense transcripts for Northern blotting were created by linearization with EcoRI and transcription with SP6 RNA polymerase. Antisense transcripts for HSURs 1 and 2 lacking the 5′-end sequences were created by linearization with DdeI (removing the first 26 nucleotides of HSUR 1) and MnlI (removing the first 55 nucleotides of HSUR 2) instead of EcoRI. Antisense U11 transcripts were prepared from pSPU11 (78). Plasmids obtained from the American Type Culture Collection containing c-myc (ATCC 292217), TNF-α (ATCC 251049), and β-actin (ATCC 3708136) and a PCR fragment containing exon 3 of human IL-10 amplified from genomic DNA were used to create DNA probes for Northern blotting by random priming. Northern blotting for Pim-1 was performed using a kinase-treated 70-mer oligonucleotide complementary to nucleotides 2518 to 2577 of the 3′ UTR of the human sequence.
hnRNP D antibody.
Polyclonal rabbit anti-hnRNP D antibodies were raised against a C-terminal peptide of hnRNP D as described in the work of Dempsey et al. (20) except that they were not passed over poly(G)-agarose.
Western analyses.
Western blotting was performed using the following concentrations of primary and secondary antibody: a 1:2,000 concentration of anti-hnRNP D, a 1:20,000 concentration of anti-HuR monoclonal antibody 3A2 (31), a 1:10,000 concentration of anti-TTP (11), and a 1:5,000 concentration of donkey anti-rabbit antibody or a 1:5,000 concentration of goat anti-mouse secondary antibody (Pierce).
In vitro cross-linking and immunoprecipitation.
In the investigation of hnRNP D binding, 3 × 105 cpm of in vitro-transcribed HSUR RNAs was incubated with 33% 1670 cell nuclear extract, prepared by the method of Dignam et al. (23), together with 3.2 mM MgCl2, 2 mM ATP, 20 mM creatine phosphate, and 80 μg of cRNA/ml at 30°C for 30 min. The mixture was cross-linked on ice with a 254-nm UV Stratalinker 2400 (Stratagene) at 2 × 0.86 J and digested with 0.5 μg of RNase A/ml at 37°C for 30 min prior to electrophoresis. Assays investigating TTP interactions were performed in the same manner with the following exceptions. The RNAs were incubated with 90 μg of total cell extract prepared from unstimulated HVS in vitro-transformed marmoset T cells or cells stimulated with 25 ng of phorbol 12-myristate 13-acetate (PMA)/ml and 625 ng of ionomycin/ml for 6 h by cell lysis in 20 mM Tris-Cl (pH 8.0)-138 mM NaCl-10% glycerol-1% NP-40-1× protease inhibitor cocktail (Calbiochem)-1 mM phenylmethylsulfonyl fluoride-20 mM NaF-2 mM NaOVa4 on ice for 10 min, followed by centrifugation at 4°C. The cRNA in the cross-linking reaction mixtures was reduced to 40 μg/ml, and the reaction mixtures were incubated at 30°C for 15 min. Immunoprecipitation of cross-linked proteins was performed following RNase A digestion with use of the anti-hnRNP D antibody described above or corresponding rabbit preimmune serum, rabbit polyclonal antibody to HuR described in the work of Fan and Steitz (27), or affinity-purified rabbit antibody to TTP (11).
In vivo cross-linking.
For each interaction tested, 107 1670 cells were collected by centrifugation, washed twice with cold phosphate-buffered saline, and resuspended in 2 ml of phosphate-buffered saline in a 10-cm-diameter plate (depth, 1 to 3 mm). Cells were irradiated (or not irradiated as a negative control) on ice with 254-nm UV light at a distance of 4 cm for 10 to 15 min and collected by scraping and centrifugation in Eppendorf tubes. The cell pellets were resuspended with vortexing in 200 μl of lysis buffer (0.5% sodium dodecyl sulfate, 50 mM Tris-Cl [pH 8], 1 mM EDTA, 1 mM dithiothreitol) and boiled at 95°C for 5 min. After dilution with 4 volumes of correction buffer (1.25% NP-40, 0.625% sodium deoxycholate, 62.5 mM Tris-Cl [pH 8], 1.75 mM EDTA, 187.5 mM NaCl), each sample was gently sonicated two or three times and centrifuged for 90 min with 1× protease inhibitor cocktail (Calbiochem) and 10 mM vanadyl ribonucleoside complex (Gibco-BRL). The supernatants were directly subjected to immunoprecipitation with anti-hnRNP D (described above), anti-HuR (described above), or mouse monoclonal anti-Sm (Y12) (46) antibodies for 1 to 1.5 h at 4°C. Pellets were washed four to six times with NET-2 and digested with proteinase K with tumbling at 37°C for 1 h. Following perchloric acid extraction and ethyl alcohol precipitation, RNA was analyzed by Northern blotting with probes for HSURs 1 and 4 described above.
Microarray analyses.
Total RNA was isolated from wild-type and mutant HVS-transformed T-cell lines (stimulated and unstimulated as indicated elsewhere in text) with the use of Trizol (Invitrogen), and poly(A)+ RNA was selected using either the PolyATract mRNA isolation system III (Promega) or oligo(dT) cellulose (Amersham Pharmacia) according to the manufacturers' protocols. For each microarray, independent preparations of 5 μg of poly(A)+ RNA from each cell line were reverse transcribed with Superscript II reverse transcriptase (Invitrogen) with 4 μg of oligo(dT25V) (where V = A, G, or C) and 5 μg of random hexamers (Amersham Pharmacia) in the presence of deoxynucleoside triphosphates and either aminoallyl dUTP or Cy5- or Cy3-dUTP. cDNAs containing aminoallyl dUTP were subsequently labeled with either Cy5 or Cy3 monofunctional dyes. The labeled cDNAs were purified on Microcon YM-30 filter columns (Millipore), combined, denatured, and hybridized overnight at 42°C to human 16K 70-mer oligonucleotide arrays (OHU16K) prepared by the Keck DNA Microarray Resource at Yale University. The arrays were scanned, and the intensities were analyzed using Genepix 3.0. Lowess normalization and biostatistical analysis of the arrays were performed using GeneSpring 5.0. In Fig. 6, genes were considered to be expressed if the feature intensity on the microarray was ≥800 or ≥8-fold over background in at least three of the four microarrays for each cell set. The expression criteria were also lowered to ≥500 or ≥5-fold over background to examine weakly expressed ARE-containing mRNAs.
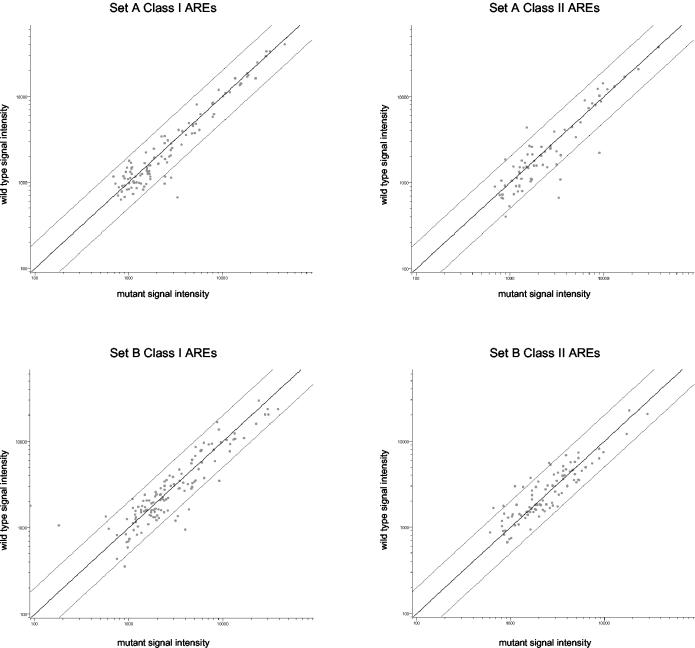
FIG. 6.
HSURs 1 and 2 do not regulate host ARE-containing mRNA levels. Polyadenylated RNA from wild-type and mutant cell lines of cell set A (top plots) and cell set B (bottom plots) was independently reverse transcribed, labeled with Cy3 or Cy5, and hybridized to microarrays. Four microarrays were performed for each cell set. The levels of more than 180 class I (left plots) and class II (right plots) ARE-containing mRNAs in each cell line were averaged across the four microarrays, and the average signal intensity of each ARE-containing mRNA (each dot) in the wild-type cell line (vertical axes) was plotted against that in the mutant cell lines (horizontal axes). The center diagonal line represents a ratio of 1 for expression in the wild-type cells compared with that in mutant transformed cells; the lines above and below the diagonal represent a twofold increase or decrease, respectively, in the levels in the wild-type cells relative to those in the mutant cells.
HSUR stability experiments.
Deletion mutant HVS-transformed marmoset T cells (107) were cotransfected with 20 μg of pUC-U1 HSUR 1 or mutant HSUR 1 plasmid and 20 μg of pUC-U1 HSUR 2 or mutant HSUR 2 by electroporation at 1,050 μF and 260 V in the presence of 1.25% dimethyl sulfoxide. The medium (without dimethyl sulfoxide) was replaced after 15 to 20 h, and total RNA was harvested 72 h after transfection, resolved in an 8% polyacrylamide gel, and analyzed by Northern analyses, with antisense RNA probes lacking the 5′ end of the HSURs where the sequences differ between wild-type and ARE mutant RNAs as described above.
RESULTS
HSURs bind hnRNP D and HuR in vivo as well as in vitro.
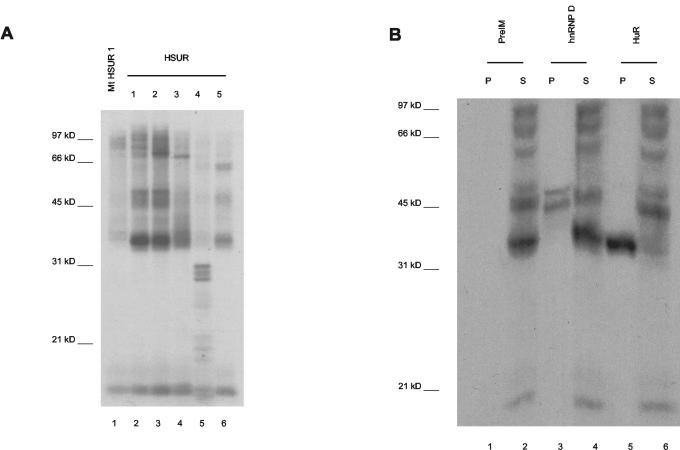
We first performed in vitro cross-linking experiments to ask if hnRNP D, like HuR (58), associates with ARE-containing HSURs. In vitro-transcribed, internally 32P-labeled HSUR 1 to 5 transcripts were incubated with cell extract from the HVS-transformed tumor cell line 1670. Exposure to 254-nm UV light then induced covalent cross-links, and after digestion with RNase A, the proteins were resolved by electrophoresis and visualized by autoradiography (Fig. 2A). In addition to the prominent HuR band at 35 kDa, proteins of the sizes expected for hnRNP D isoforms (37 to 45 kDa) (75) become cross-linked to HSURs 1 and 2, which contain multiple AUUUA repeats, and to a lesser extent to HSUR 5, which has a single AUUUA motif (lanes 2, 3, and 6). By contrast, HSURs 3 and 4, which do not contain AUUUA, show little cross-linking in the 45-kDa range (lanes 4 and 5). An HSUR 1 mutant in which the AUUUA motifs are disrupted by guanosine residues serves as a negative control (Fig. 1) and shows very weak cross-linking in the 45-kDa range, demonstrating that the cross-links are AUUUA specific (lane 1).
FIG. 2.
The ARE-containing HSURs cross-link to hnRNP D isoforms in vitro. In vitro-transcribed HSURs internally labeled with [α-32P]UTP were incubated with nuclear extract from 1670 cells, and RNA-protein cross-links were induced by irradiation of the mixture with 254-nm UV light. Following RNase A digestion, the labeled proteins were resolved in a 12% polyacrylamide gel. (A) Proteins cross-linked to the ARE-containing HSURs 1, 2, and 5 (lanes 2, 3, and 6, respectively) were compared to those cross-linked to HSURs 3 and 4 (lanes 4 and 5, respectively) or to a mutant HSUR 1 (Fig. 1) in which the ARE was disrupted by guanosine residues (lane 1). (B) Following RNase A digestion, reaction mixtures containing cross-linked wild-type HSUR 1 were subjected to immunoprecipitation with antibodies to hnRNP D (lanes 3 and 4) or anti-HuR (lanes 5 and 6) and preimmune serum (lanes 1 and 2) as positive and negative controls, respectively. Equal amounts of the pellet (P) and supernatant (S) were loaded.
Proteins cross-linked to HSUR 1 were then subjected to immunoprecipitation with rabbit polyclonal antibodies to hnRNP D. As seen in Fig. 2B, two labeled proteins were specifically immunoprecipitated (lane 3) whereas no precipitation was observed with rabbit preimmune serum (lane 1). Anti-HuR selectively precipitated a cross-linked protein of about 35 kDa, providing a positive control (lane 5). Because this anti-hnRNP D antibody quantitatively immunoprecipitates all four isoforms of hnRNP D under these conditions (data not shown), the immunoprecipitation of two cross-linked hnRNP D isoforms suggests that the interaction with the ARE-containing HSUR 1 is specific for two hnRNP D isoforms. We cannot rule out, however, that more isoforms bind but cannot be resolved following cross-linking.
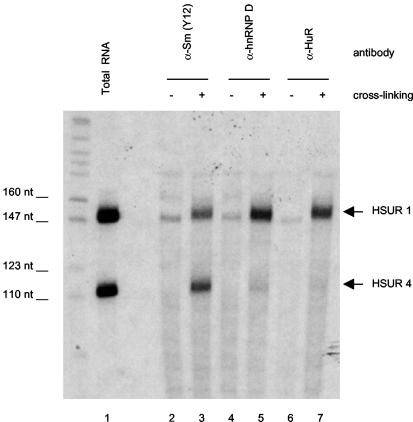
To extend these in vitro data to HSUR interactions in virally transformed T cells, we developed a new cross-linking assay that allows detection of specific RNA-protein interactions in vivo. Intact, untreated cells of the HVS-transformed tumor T-cell line 1670 were exposed to 254-nm UV light to induce RNA-protein cross-links and then lysed under denaturing conditions. Immunoprecipitations with antibodies to HuR, hnRNP D, or Sm proteins were followed by proteinase K treatment and Northern analysis to detect RNAs that were covalently cross-linked to immunoprecipitated proteins. We compared HSUR 1, which has the greatest number of AUUUA repeats, and HSUR 4, which contains no AUUUA motif, since they are both abundantly expressed at similar levels in HVS-transformed T cells. As seen in Fig. 3, both hnRNP D and HuR cross-link to 2 to 3% of HSUR 1 but not to HSUR 4 (compare lanes 5 and 7 with total RNA in lane 1), while the Sm core proteins cross-link to both HSUR 1 and HSUR 4 (likewise at 2 to 3%, lane 3). Importantly, no HSUR 1 or HSUR 4 coprecipitated with antibodies to HuR, hnRNP D, or Sm proteins in the absence of cross-linking (lanes 2, 4, and 6). We were unable to detect in vivo cross-links of HSURs 2 and 5 to HuR or hnRNP D (data not shown), presumably because of their lower abundance. We conclude that endogenous HSUR 1 directly binds HuR and several hnRNP D isoforms in HVS-transformed T cells.
FIG. 3.
HSUR 1 interacts with hnRNP D and HuR in vivo. Intact HVS-transformed 1670 cells were irradiated with 254-nm UV light to induce cross-links between RNA and protein interacting in vivo (lanes 3, 5, and 7). Cells treated identically, but not cross-linked, served as negative controls (lanes 2, 4, and 6). Cell extracts were prepared under strongly denaturing conditions and subjected to immunoprecipitation with antibody to hnRNP D (lanes 4 and 5), HuR (lanes 6 and 7), or Sm proteins (lanes 2 and 3). Following digestion with proteinase K, immunoprecipitated RNA was isolated from the pellets and resolved along with 5% of total RNA (lane 1) in a 8% polyacrylamide gel. Northern blotting detected the ARE-containing HSUR 1 or HSUR 4 as a control for ARE-binding specificity. Small amounts of nonspecific RNAs of 150, 160, and 200 nucleotides (nt) were observed in all immunoprecipitation reactions.
T cells transformed by an HVS deletion mutant virus provide a system to examine the effects of HSURs 1 and 2.
Since HSURs are expressed at significantly higher levels than are individual ARE-containing mRNAs in HVS-transformed T cells, it has been proposed that they act to sequester host ARE-binding proteins and thereby disrupt normal regulation of the stability of host ARE-containing mRNAs (26, 58). Although previous studies using transient transfection of mouse L929 cells showed no change in the stability of a β-globin reporter mRNA that contained the ARE from GM-CSF in its 3′ UTR when HSUR 1 was coexpressed (26), it remained possible that the function of the HSURs depends on pathways or mRNAs specific to T cells or requires other HSURs or viral proteins expressed during latency.
To test this hypothesis, we used T cells transformed in vitro with either wild-type HVS strain A11 (wild-type cell line) or a mutant A11 virus (mutant cell line) containing a 1,379-nucleotide deletion covering the HSUR 1 and 2 loci (Fig. 4A) (57). HVS lacking HSURs 1 and 2 was chosen for deletion studies because HSURs 1 and 2 are the most conserved and have the greatest number of AUUUA repeats and because HSUR 1 is the most abundant ARE-containing HSUR. Two sets of wild-type and mutant cell lines were the kind gift of R. C. Desrosiers and D. P. Silva. One set (cell set A) was described earlier and represents T cells from different monkeys (57). A second set (cell set B) was transformed in parallel with T cells from the same marmoset to avoid artifacts due to differences in monkeys, transformation conditions, or cell passage. Cell set B, however, contains a second, undesired deletion which removes viral genes required for reactivation to the lytic cycle (see Materials and Methods), a deletion previously observed in cells transformed by HVS both in vivo and in vitro (21, 80). All analyses were performed with both sets of cell lines.
FIG. 4.
Wild-type and HSUR 1/2 deletion mutant HVS-transformed marmoset T cells. (A) Schematic of the left ends of the genomes of wild-type HVS A11 (top) and mutant HVS A11 (bottom), which lacks a 1,379-nucleotide (nt) region precisely deleting HSURs 1 and 2, used to transform marmoset T cells in vitro. HSURs 5, 3, 4, 6, and 7 remain. STP-A, saimiri transformation-associated protein; DHFR, dihydrofolate reductase. (B) Northern blots of total RNA isolated from wild-type (Wt) and mutant (Mt) cells of both cell set A (lanes 3 and 4) and cell set B (lanes 1 and 2) for HSUR 1, HSUR 2, HSUR 5, and cellular U11. Three different forms of HSUR 5 ranging between 111 and 114 nucleotides have been described elsewhere (77).
As demonstrated in the Northern blots in Fig. 4B, HSURs 1 and 2 are not expressed in T cells transformed by the HVS deletion mutants (lanes 2 and 4). Northern and Western analyses on both cell sets further confirmed that the expression of the genes immediately upstream and downstream of the deletion, HSUR 5 (Fig. 4B) and STP (H. L. Cook et al., unpublished data), respectively, is not inhibited by the deletion. Deletion of HSURs 1 and 2, in fact, slightly (two- to threefold) upregulates HSURs 3, 4, 5, 6, and 7 (Cook et al., unpublished).
HSURs 1 and 2 have no effect on the steady-state levels of more than 180 class I or class II ARE-containing mRNAs in host T cells.
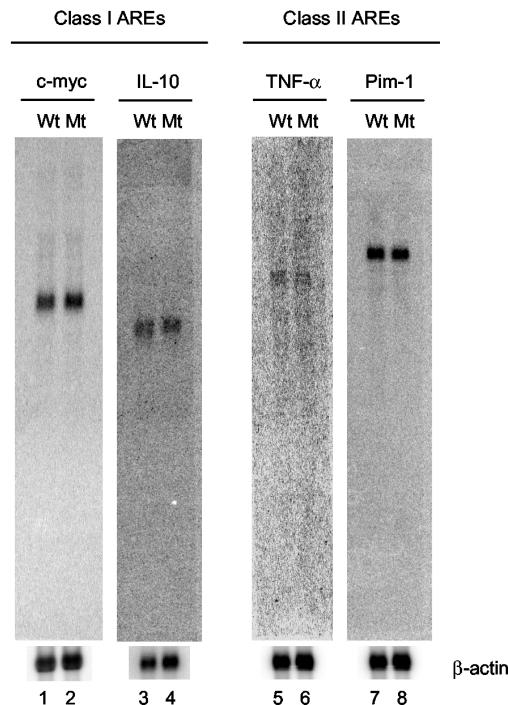
Since HSUR 1 contains a class II ARE and HSUR 2 contains a class I ARE, we began by comparing the levels of endogenous ARE-containing mRNAs of each class in the wild-type and mutant HVS-transformed T-cell lines by Northern analyses. The class I ARE-containing c-myc and IL-10 mRNAs and the class II TNF-α and Pim-1 mRNAs were chosen for analysis. These mRNAs contain varying numbers of the AUUUA motif in their 3′ UTRs, are expressed by CD8+ T cells, and are logical targets for regulation by HVS, having established roles in the proliferation of CD8+ T cells or antiviral defense in the host (18, 37, 39, 48, 54, 65, 85). Northern blotting performed on polyadenylated RNAs isolated from the wild-type and mutant cell lines (Fig. 5) revealed that the steady-state levels of mRNAs for c-myc and IL-10, as well as those for TNF-α and Pim-1, are unaffected by the absence of HSURs 1 and 2.
FIG. 5.
Northern analyses of host class I and class II ARE-containing mRNAs. Polyadenylated RNAs isolated from wild-type (Wt) and mutant (Mt) HVS-transformed T-cell lines were resolved in a 1% formaldehyde agarose gel, and Northern blotting was performed for the class I ARE-containing mRNAs c-myc (lanes 1 and 2) and IL-10 (lanes 3 and 4) and for the class II ARE-containing mRNAs TNF-α (lanes 5 and 6) and Pim-1 (lanes 7 and 8). The same membranes were probed for β-actin as a loading control. The membrane used for TNF-α was stripped and reprobed for Pim-1.
To extend our analysis to a greater number of ARE-containing mRNAs, we examined the levels of endogenous mRNAs in the presence and absence of HSURs 1 and 2 using microarray analyses. DNA microarrays comprised of 70-mer DNA oligonucleotides complementary to 16,659 human genes were hybridized with cDNA prepared from polyadenylated RNA isolated from wild-type and mutant HVS-transformed marmoset T-cell lines. Four microarrays were analyzed for each of the two sets of wild-type and mutant cell lines (four replicates for each of cell sets A and B). We required changes to be present in both sets of wild-type and mutant cells to ensure that observed differences were due only to the expression of HSURs 1 and 2. Wild-type and mutant cells were grown under the same conditions and were at the same density prior to RNA isolation. Independent poly(A) RNA preparations were used for each array. The number of cell passages was kept to a minimum but did not affect observed results. For each cell set, two arrays utilized wild-type cDNAs labeled with Cy5 and the mutant labeled with Cy3, while the fluorophores were reversed for the remaining two arrays.
Careful analysis of the microarray data revealed no significant differences in the steady-state levels of ARE-containing mRNAs. Of the 1,470 mRNAs identified by Bakheet et al. (4) in the ARE database (∼7% of all human genes), 1,419 (96.5%) are present on the microarrays used (∼8.5% of genes on the microarray). Of these, 181 ARE-containing mRNAs (109 class I AREs and 72 class II AREs) were expressed in cell set A and 222 ARE-containing mRNAs (126 class I AREs and 96 class II AREs) were expressed in cell set B on at least three of the four arrays for each cell set (∼7% of all genes expressed). Those mRNAs previously assayed by Northern blotting are included (Fig. 5). The intensity data for each expressed class I and class II ARE-containing gene were averaged across the four microarrays for each cell set and are presented in the scatter plots seen in Fig. 6. Clearly, HSURs 1 and 2 have no general effect on the steady-state levels of mRNAs containing either class I or class II AREs. Although a few statistically significant changes were seen for specific ARE-containing mRNAs in each cell set (Fig. 6, see dots above and below lines representing a twofold change), no changes of twofold or greater were observed in both cell sets for any ARE-containing mRNA. Because these mRNAs are of low abundance, we relaxed our criteria (see Materials and Methods) to include weakly expressed ARE-containing mRNAs but found no significant differences. We conclude that HSURs 1 and 2 have no effect on the steady-state levels of more than 180 tested ARE-containing mRNAs in latently infected, transformed T cells.
HSUR 1 can bind TTP following stimulation of HVS-transformed T cells.
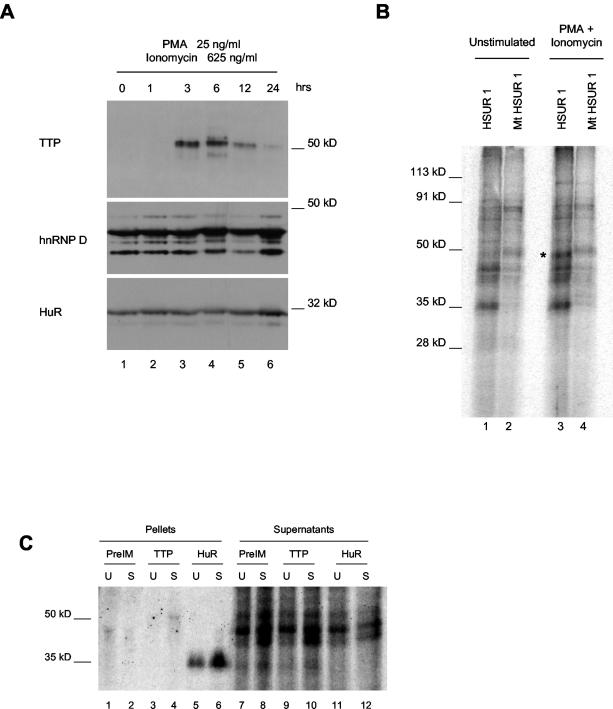
To simulate stimuli likely to be received in an organism but not under normal tissue culture conditions, we treated our latently infected T cells with PMA and ionomycin. This procedure is commonly used to activate T cells (73, 79) and has been shown to upregulate pathways that alter both the stability of ARE-containing mRNAs and the expression and/or binding activity of the ARE-binding proteins HuR and TTP (3, 63). Prior to stimulation, immunoblots revealed no detectable expression of TTP (Fig. 7A, lane 1), despite the fact that our HVS-transformed T cells appear to have an activated phenotype in many other respects (74). After stimulation of both wild-type and mutant HVS-transformed T cells of both cell sets, TTP expression was seen, with maximal induction occurring with 25 ng of PMA/ml and 625 ng of ionomycin/ml for 3 to 6 h (Fig. 7A, lanes 3 and 4). By contrast, treatment with PMA and ionomycin did not change the expression levels of HuR or hnRNP D over a 24-h period (Fig. 7A).
FIG. 7.
TTP is induced following cell stimulation and binds HSUR 1. (A) Wild-type HVS-transformed T cells were stimulated with 25 ng of PMA/ml and 625 ng of ionomycin/ml for 0, 1, 3, 6, 12, or 24 h and analyzed by Western blotting for changes in ARE-binding protein expression. The same membrane was probed with antibodies to TTP, HuR, and hnRNP D. (B) In vitro-transcribed HSUR 1 (lanes 1 and 3) and ARE mutant (Fig. 1) HSUR 1 (lanes 2 and 4) internally labeled with [α-32P]UTP were incubated with total cell extract from unstimulated HVS-transformed marmoset T cells (lanes 1 and 2) or cells stimulated with 25 ng of PMA/ml and 625 ng of ionomycin/ml for 6 h. RNA-protein cross-links were induced by irradiation with 254-nm UV light. Following RNase A digestion, the labeled proteins were resolved in a 10% polyacrylamide gel. The asterisk indicates a 50-kDa ARE-specific cross-link seen with stimulated cell extract. (C) Following RNase A digestion, proteins cross-linked to in vitro-transcribed HSUR 1 in extracts from stimulated (S) and unstimulated (U) cells prepared as in panel B above were immunoprecipitated with antibody to TTP (lanes 3, 4, 9, and 10) or anti-HuR (lanes 5, 6, 11, and 12) and preimmune serum (lanes 1, 2, 7, and 8) as controls. Pellets (lanes 1 to 6) and 25% of corresponding supernatants (lanes 7 to 12) are shown. The immunoprecipitation of low levels of nonspecific proteins by the preimmune serum is probably due to the amount of immunoglobulin G being significantly higher than that in the immunoprecipitations with affinity-purified anti-TTP antibody.
To confirm that TTP binds the ARE-containing HSURs, as it does ARE-containing mRNAs, we performed in vitro cross-linking assays using extracts from stimulated and unstimulated cells as described above. Figure 7B shows the reproducible cross-linking of an ARE-binding protein of the size expected for TTP (50 kDa) to HSUR 1 but not to the HSUR 1 ARE mutant (compare lane 3 to lane 4) in extract from cells treated with PMA and ionomycin. Consistent with TTP expression in Fig. 7A, no such cross-link was seen in extract from unstimulated cells (Fig. 7B, lanes 1 and 2). We then subjected the proteins cross-linked to wild-type HSUR 1 to immunoprecipitation with affinity-purified, polyclonal rabbit antibodies to TTP (a generous gift from W. Rigby) (11). As seen in Fig. 7C, a cross-linked band of approximately 50 kDa was immunoprecipitated specifically from extract from stimulated cells (lane 4), but not unstimulated cells (lane 3), nor from rabbit preimmune serum (lanes 1 and 2). HuR was immunoprecipitated as a positive control (lanes 5 and 6). Because of the limited availability of anti-TTP antibodies, we were unable to confirm an interaction between endogenous TTP and HSUR 1 under stimulation conditions by in vivo cross-linking. We conclude that HSUR 1 can interact with TTP induced following stimulation of latently infected T cells.
Since TTP is a known destabilizer of ARE-containing mRNAs, we repeated the microarray analyses using polyadenylated RNA isolated from wild-type and mutant cells that had been stimulated with 25 ng of PMA/ml and 625 ng of ionomycin/ml for 6 h. We performed two microarray analyses, one for each set of treated wild-type and mutant cells, with the fluorophores reversed in the second cell set to control for differences in label incorporation. We were able to confirm expression of TTP mRNA by microarray analysis; however, no significant differences were observed in the steady-state levels of ARE-containing messages in the presence and absence of HSURs 1 and 2 (data not shown). We conclude that, even in the presence of TTP, the ARE-containing HSURs do not detectably regulate the stability of host ARE-containing mRNAs.
HSUR 1 is degraded by an ARE-specific pathway in HVS-transformed T cells.
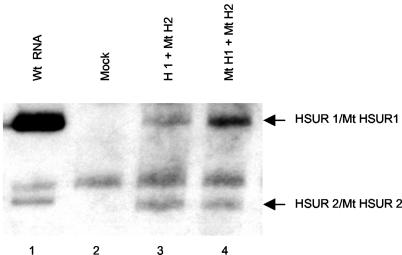
HSUR 1 transfected into mouse L929 cells was previously shown to be degraded by an ARE-specific pathway (26). However, it was important to examine latently infected, HVS-transformed T cells as well, since the stability of the ARE-containing HSURs might be affected by other viral gene products or by different activation states of the host cell. We therefore compared the levels of wild-type HSURs 1 and 2 and HSUR mutants in which the AUUUA motifs were disrupted by G residues (Fig. 1), similar to mutations known to stabilize ARE-containing RNAs (26, 67). T cells transformed by the HSUR 1/2 deletion mutant HVS were transiently transfected with plasmids containing either wild-type or mutant HSUR 1. In both cases, cotransfections with plasmids containing mutant HSUR 2 provided a stable control for transfection efficiency against which the levels of wild-type and mutant HSUR 1 were compared. All HSURs were under the transcriptional control of the cellular U1 promoter and 3′-end box and were previously shown to have comparable transcription rates by run-on transcription assays (26). After normalization to mutant HSUR 2, the observed steady-state levels of wild-type HSUR 1 were 3.4- ± 0.7-fold lower than levels of mutant HSUR 1 (Fig. 8, compare lanes 3 and 4). Similar experiments examining HSUR 2 stability by cotransfection with either wild-type or mutant HSUR 2 and mutant HSUR 1 for normalization showed no difference in the steady-state levels of wild-type HSUR 2 and the stable mutant HSUR 2 (1 ± 0.1) (data not shown). These results demonstrate that HSUR 1 is degraded by an ARE-dependent pathway during latency in HVS-transformed T cells, while HSUR 2 is stable in HVS-transformed T cells in cultured cells.
FIG. 8.
HSUR 1 is degraded in HVS-transformed T cells. HSUR 1/2 deletion mutant HVS-transformed T cells (total RNA, lane 2) were transiently transfected by electroporation with either wild-type HSUR 1 (lane 3) or ARE mutant HSUR 1 (lane 4) together with ARE mutant HSUR 2 to control for transfection efficiency and analyzed by Northern blotting. Figure 1 shows ARE mutant HSUR 1 and HSUR 2 sequences. Lane 1 shows HSUR 1 levels in wild-type transformed cells.
DISCUSSION
We show that the ARE-containing HSURs expressed in HVS-transformed marmoset T cells interact in vivo with HuR and hnRNP D, two proteins that regulate the stability of ARE-containing mRNAs (9, 27, 49, 62, 64). Yet, Northern blot and microarray experiments reveal that these interactions do not alter the steady-state levels of host ARE-containing mRNAs. Activation of HVS-transformed T cells with PMA and ionomycin induced expression of the ARE-binding protein TTP, and we demonstrate by in vitro cross-linking that the ARE-containing HSUR 1 can bind TTP. However, even in these stimulated cells, the presence of HSURs 1 and 2 does not alter ARE-containing mRNA stability. Instead, we observe that the level of HSUR 1 itself is reduced by an ARE-dependent pathway in HVS-transformed T cells. These findings indicate that the ARE sequences in HSUR 1, and perhaps also in HSURs 2 and 5, regulate the expression of the HSURs themselves. This is the first example of posttranscriptional regulation of the expression of an Sm snRNA.
The ARE-containing HSURs 1 and 2 recruit host ARE-binding proteins.
We demonstrate here, using a powerful in vivo cross-linking assay that allows detection of direct interactions between a specific RNA and proteins, that HSUR 1 is bound by hnRNP D and HuR, host proteins known to regulate the stability of ARE-containing mRNAs (Fig. 3). Interactions captured with this technique identify genuine RNP complexes present in HVS latently infected transformed T cells rather than artifactual binding produced by incubation of in vitro-transcribed RNAs in extracts from lysed cells. We dissected these interactions further by in vitro cross-linking to show that HSUR 1 binds two isoforms of hnRNP D (Fig. 2), as well as TTP induced upon T-cell stimulation with PMA and ionomycin (Fig. 7). Since we do not possess isoform-specific antibodies, we can only speculate that the hnRNP D isoforms are p37 and p42, reported by Loflin et al. (49) to rescue degradation of ARE-containing mRNAs in K562 cells.
The ARE-containing HSURs could associate with these ARE-binding proteins either during their biogenesis or as fully assembled, Sm-containing small nuclear ribonucleoprotein (Sm snRNP) particles. We assume that the HSURs share a common biogenesis pathway with cellular Sm snRNAs: following transcription by RNA polymerase II, Sm snRNAs are exported to the cytoplasm, where they assemble with Sm core proteins and acquire hypermethylated 5′ caps before being imported back into the nucleus, where they function in RNA processing (81). Interaction of ARE-binding proteins with newly transcribed, immature HSURs would be consistent with a role for the ARE sequences in regulating the levels of HSUR RNPs formed, since assembly with Sm proteins is generally believed to stabilize snRNAs (28, 36, 72). Indeed, actinomycin D decay assays performed on HSUR 1-transfected cells indicate that it is susceptible to degradation only during the first 2 h and is stable up to 20 h thereafter (25). Conversely, association of ARE-binding proteins with fully formed Sm snRNPs might suggest a role for the ARE sequences in HSUR function. Identification of other proteins present in cellular complexes containing the HSURs and ARE-binding proteins and determination of the subcellular localization of these interactions should provide additional insights into their function.
HSURs 1 and 2 do not regulate the steady-state levels of host ARE-containing mRNAs.
Our analysis of representative class I and class II ARE-containing mRNAs by Northern blotting did not reveal a change in their steady-state levels in cells transformed by virus lacking the most abundant ARE-containing HSURs, HSURs 1 and 2 (Fig. 5). These observations were extended by comprehensive microarray analyses that uncovered no effect on class I or II ARE-containing mRNAs (Fig. 6). Even after stimulation of HVS latently infected T cells with PMA and ionomycin, a procedure known to stabilize ARE-containing mRNAs (7, 67), induce TTP (Fig. 7A), and later limit the duration of T-cell activation by destabilizing ARE-containing messages (63), no effects on host ARE-containing mRNAs attributable to HSURs 1 and 2 were observed. Immunofluorescence studies likewise revealed no significant nuclear relocalization of hnRNP D (HuR is already predominantly nuclear) in HVS-transformed cells or of TTP induced following cell stimulation in the presence versus the absence of HSURs 1 and 2 (data not shown).
Microarray analyses enabled the simultaneous comparison of the steady-state levels of more than 180 ARE-containing mRNAs, representing ∼7% of expressed genes, and revealed no differences dependent on HSURs 1 and 2 (Fig. 6). We cannot rule out an HSUR effect on the stability of one or more host ARE-containing mRNAs that were not detected, due either to low abundance or to differences in the marmoset and human sequences. Since the ARE database compiled by Bakheet et al. (4) estimates that approximately 7% of all genes contain 3′-UTR AREs, it is reasonable to conclude that we detected the vast majority of this mRNA class. Additionally, all C. jacchus mRNA sequences available in GenBank show 85 to 95% nucleotide identity to their human orthologs, arguing that most marmoset cDNAs should hybridize to the human array.
We find it unlikely that the continued expression of HSUR 5 in transformed cells accounts for our negative findings, even though HSUR 5 expression is somewhat upregulated (approximately threefold [Fig. 4]) with the HSUR 1/2 deletion for several reasons. HSUR 5 has only one AUUUA pentamer (Fig. 1) and binds ARE-binding proteins much less avidly than HSURs 1 and 2 do (approximately 5- to 10-fold less [Fig. 2]). Moreover, even when upregulated in the HSUR 1/2 deletion, HSUR 5 is threefold less abundant (10-fold less abundant in wild-type cells) than HSUR 1 would have been (44). Finally, HSUR 5 is not encoded by HVA, a close relative of HVS, which encodes homologs of only HSURs 1 and 2. Together these data support our conclusion that the interaction of the HSURs with ARE-binding proteins does not affect the stability of cellular ARE-containing mRNAs.
HSUR 1 levels are posttranscriptionally regulated by the ARE-mediated decay pathway.
We show that, upon transient expression in HVS-transformed marmoset T cells, HSUR 1 accumulates to lower levels than does HSUR 1 mutated in its 5′ ARE (Fig. 8). This finding extends previous observations with the same HSUR constructs in mouse L cells (25, 26): nuclear run-on and actinomycin D transcription inhibition experiments indicated comparable transcription and decay rates, except during the first 2 h, when the ARE-containing HSUR 1 decayed significantly more rapidly than the mutant did.
It is interesting that the ARE-containing HSURs bind proteins implicated in both the stabilization and destabilization of ARE-containing mRNAs (Fig. 2, 3, and 7), yet only HSUR 1 is degraded by an ARE-dependent pathway (Fig. 8). Furthermore, although we demonstrate by in vitro cross-linking that HSUR 1 can bind the induced TTP in stimulated cell extracts (Fig. 7), treatment of HVS-transformed T cells with PMA and ionomycin does not detectably alter the endogenous levels of ARE-containing HSURs (data not shown). This is in apparent contrast to the response of ARE-containing mRNAs when levels of ARE-binding proteins are manipulated. For instance, overexpression of HuR stabilizes reporter mRNAs containing either class I or class II AREs (27, 62), and increased levels of either hnRNP D or TTP have been shown to significantly enhance the decay of ARE-containing mRNAs (14, 49). In fact, TTP can even mediate degradation of an ARE-containing histone mRNA and other mRNAs designed with a terminal 3′ stem-loop in place of a poly(A) tail (40). A likely explanation is that TTP affects only the small fraction of HSURs that are newly transcribed and not yet assembled into stable Sm snRNPs.
The ARE-containing HSURs are the first snRNAs identified as containing cis-acting sequences implicated in the posttranscriptional regulation of expression. Although ARE-dependent decay of only HSUR 1 has been observed in vitro (in both L929 cells [26] and HVS-transformed T cells [Fig. 8]), the ARE sequences in HSURs 2 and 5 might similarly regulate the stability of these snRNAs in vivo. Unfortunately, a direct comparison of the decay rates of newly transcribed HSURs is not possible, since no regulatable RNA polymerase II promoter exists for expression of snRNAs. Future development of such an inducible system should reveal whether the Sm snRNA HSUR 1 is regulated posttranscriptionally by the same extracellular stimuli that alter the stability of ARE-containing mRNAs.
Acknowledgments
We are most grateful to Ronald Desrosiers and Daniel Silva for the creation of the wild-type and mutant HVS-transformed marmoset T-cell lines examined in this study. We thank Irena Tikhonova and Aiping Lin at the Keck microarray facility for expert assistance with the microarrays and biostatistical analyses. We thank William Rigby for generously providing anti-TTP antibody. We thank Jens Lykke-Andersen and members of the Steitz lab, especially Nick Conrad, for essential materials and valuable advice and discussion throughout this work.
The Keck DNA Microarray Resource at Yale University is supported by the Anna and Argall Hull Fund, the NIH/NIDDK Microarray Biotechnology Center Grant (NIH 5 U24 DK58776; Principal Investigator, Kenneth Williams), the C. G. Swebilius Trust U.W., and the Yale University Department of Pathology. H.L.C. was a Howard Hughes Medical Institute Predoctoral Fellow, and J.A.S. is an investigator of the Howard Hughes Medical Institute. This work was supported by grant CA 16038 from the NIH.
REFERENCES
- 1.Albrecht, J. C. 2000. Primary structure of the Herpesvirus ateles genome. J. Virol. 74:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J. C., and B. Fleckenstein. 1992. Nucleotide sequence of HSUR 6 and HSUR 7, two small RNAs of herpesvirus saimiri. Nucleic Acids Res. 20:1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atasoy, U., J. Watson, D. Patel, and J. D. Keene. 1998. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 111:3145-3156. [DOI] [PubMed] [Google Scholar]
- 4.Bakheet, T., B. R. Williams, and K. S. Khabar. 2003. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 31:421-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banholzer, R., A. P. Nair, H. H. Hirsch, X. F. Ming, and C. Moroni. 1997. Rapamycin destabilizes interleukin-3 mRNA in autocrine tumor cells by a mechanism requiring an intact 3′ untranslated region. Mol. Cell. Biol. 17:3254-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertram, K. A., and G. R. Pearson. 1983. Natural killer cells in relation to Herpesvirus saimiri-induced disease course and manifestations in owl monkeys. J. Natl. Cancer Inst. 70:147-155. [PubMed] [Google Scholar]
- 7.Bickel, M., R. B. Cohen, and D. H. Pluznik. 1990. Post-transcriptional regulation of granulocyte-macrophage colony-stimulating factor synthesis in murine T cells. J. Immunol. 145:840-845. [PubMed] [Google Scholar]
- 8.Bohjanen, P. R., B. Petryniak, C. H. June, C. B. Thompson, and T. Lindsten. 1991. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3′ untranslated region of lymphokine mRNA. Mol. Cell. Biol. 11:3288-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brook, M., G. Sully, A. R. Clark, and J. Saklatvala. 2000. Regulation of tumour necrosis factor alpha mRNA stability by the mitogen-activated protein kinase p38 signalling cascade. FEBS Lett. 483:57-61. [DOI] [PubMed] [Google Scholar]
- 11.Brooks, S. A., J. E. Connolly, R. J. Diegel, R. A. Fava, and W. F. Rigby. 2002. Analysis of the function, expression, and subcellular distribution of human tristetraprolin. Arthritis Rheum. 46:1362-1370. [DOI] [PubMed] [Google Scholar]
- 12.Caput, D., B. Beutler, K. Hartog, R. Thayer, S. Brown-Shimer, and A. Cerami. 1986. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. USA 83:1670-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carballo, E., H. Cao, W. S. Lai, E. A. Kennington, D. Campbell, and P. J. Blackshear. 2001. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 276:42580-42587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 15.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 16.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 17.Chen, C. Y., N. Xu, and A. B. Shyu. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15:5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, W. F., and A. Zlotnik. 1991. IL-10: a novel cytotoxic T cell differentiation factor. J. Immunol. 147:528-534. [PubMed] [Google Scholar]
- 19.De Carli, M., S. Berthold, H. Fickenscher, I. M. Fleckenstein, M. M. D'Elios, Q. Gao, R. Biagiotti, M. G. Giudizi, J. R. Kalden, and B. Fleckenstein. 1993. Immortalization with Herpesvirus saimiri modulates the cytokine secretion profile of established Th1 and Th2 human T cell clones. J. Immunol. 151:5022-5030. [PubMed] [Google Scholar]
- 20.Dempsey, L. A., L. A. Hanakahi, and N. Maizels. 1998. A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J. Biol. Chem. 273:29224-29229. [DOI] [PubMed] [Google Scholar]
- 21.Desrosiers, R. C. 1981. Herpesvirus saimiri DNA in tumor cells—deleted sequences and sequence rearrangements. J. Virol. 39:497-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desrosiers, R. C., D. P. Silva, L. M. Waldron, and N. L. Letvin. 1986. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J. Virol. 57:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ensser, A., A. Pfinder, I. Muller-Fleckenstein, and B. Fleckenstein. 1999. The URNA genes of herpesvirus saimiri (strain C488) are dispensable for transformation of human T cells in vitro. J. Virol. 73:10551-10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan, X. C. 1998. Ph.D. thesis. Yale University, New Haven, Conn.
- 26.Fan, X. C., V. E. Myer, and J. A. Steitz. 1997. AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev. 11:2557-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer, U., J. Heinrich, K. van Zee, E. Fanning, and R. Luhrmann. 1994. Nuclear transport of U1 snRNP in somatic cells: differences in signal requirement compared with Xenopus laevis oocytes. J. Cell Biol. 125:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleckenstein, B., and R. Desrosiers. 1983. Herpesvirus saimiri and herpesvirus atales, p. 253-332. In B. Roizman (ed.), The herpesviruses, vol. 1. Plenum Press, New York, N.Y.
- 30.Frevel, M. A., T. Bakheet, A. M. Silva, J. G. Hissong, K. S. Khabar, and B. R. Williams. 2003. p38 mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 23:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 97:3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorospe, M., S. Kumar, and C. Baglioni. 1993. Tumor necrosis factor increases stability of interleukin-1 mRNA by activating protein kinase C. J. Biol. Chem. 268:6214-6220. [PubMed] [Google Scholar]
- 33.Holtmann, H., R. Winzen, P. Holland, S. Eickemeier, E. Hoffmann, D. Wallach, N. L. Malinin, J. A. Cooper, K. Resch, and M. Kracht. 1999. Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways. Mol. Cell. Biol. 19:6742-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarrousse, A. S., F. Petit, C. Kreutzer-Schmid, R. Gaedigk, and H. P. Schmid. 1999. Possible involvement of proteasomes (prosomes) in AUUUA-mediated mRNA decay. J. Biol. Chem. 274:5925-5930. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, D. R., and M. Jondal. 1981. Herpesvirus-transformed cytotoxic T-cell lines. Nature 291:81-83. [DOI] [PubMed] [Google Scholar]
- 36.Jones, M. H., and C. Guthrie. 1990. Unexpected flexibility in an evolutionarily conserved protein-RNA interaction: genetic analysis of the Sm binding site. EMBO J. 9:2555-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitagawa, N., M. Goto, K. Kurozumi, S. Maruo, M. Fukayama, T. Naoe, M. Yasukawa, K. Hino, T. Suzuki, S. Todo, and K. Takada. 2000. Epstein-Barr virus-encoded poly(A)(−) RNA supports Burkitt's lymphoma growth through interleukin-10 induction. EMBO J. 19:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein, N., A. M. Curatola, and R. J. Schneider. 1999. Calcium-induced stabilization of AU-rich short-lived mRNAs is a common default response. Gene Expr. 7:357-365. [PMC free article] [PubMed] [Google Scholar]
- 39.Knappe, A., S. Hor, S. Wittmann, and H. Fickenscher. 2000. Induction of a novel cellular homolog of interleukin-10, AK155, by transformation of T lymphocytes with herpesvirus saimiri. J. Virol. 74:3881-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai, W. S., and P. J. Blackshear. 2001. Interactions of CCCH zinc finger proteins with mRNA: tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. J. Biol. Chem. 276:23144-23154. [DOI] [PubMed] [Google Scholar]
- 41.Lai, W. S., E. Carballo, J. R. Strum, E. A. Kennington, R. S. Phillips, and P. J. Blackshear. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19:4311-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 43.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 44.Lee, S. I., S. C. Murthy, J. J. Trimble, R. C. Desrosiers, and J. A. Steitz. 1988. Four novel U RNAs are encoded by a herpesvirus. Cell 54:599-607. [DOI] [PubMed] [Google Scholar]
- 45.Lee, S. I., and J. A. Steitz. 1990. Herpesvirus saimiri U RNAs are expressed and assembled into ribonucleoprotein particles in the absence of other viral genes. J. Virol. 64:3905-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lerner, E. A., M. R. Lerner, C. A. Janeway, Jr., and J. A. Steitz. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA 78:2737-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindsten, T., C. H. June, J. A. Ledbetter, G. Stella, and C. B. Thompson. 1989. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244:339-343. [DOI] [PubMed] [Google Scholar]
- 48.Lindsten, T., C. H. June, and C. B. Thompson. 1988. Multiple mechanisms regulate c-myc gene expression during normal T cell activation. EMBO J. 7:2787-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loflin, P., C. Y. Chen, and A. B. Shyu. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13:1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahtani, K. R., M. Brook, J. L. Dean, G. Sully, J. Saklatvala, and A. R. Clark. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ming, X. F., M. Kaiser, and C. Moroni. 1998. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 17:6039-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ming, X. F., G. Stoecklin, M. Lu, R. Looser, and C. Moroni. 2001. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell. Biol. 21:5778-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montero, L., and Y. Nagamine. 1999. Regulation by p38 mitogen-activated protein kinase of adenylate- and uridylate-rich element-mediated urokinase-type plasminogen activator (uPA) messenger RNA stability and uPA-dependent in vitro cell invasion. Cancer Res. 59:5286-5293. [PubMed] [Google Scholar]
- 54.Moroy, T., A. Grzeschiczek, S. Petzold, and K. U. Hartmann. 1993. Expression of a Pim-1 transgene accelerates lymphoproliferation and inhibits apoptosis in lpr/lpr mice. Proc. Natl. Acad. Sci. USA 90:10734-10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee, D., M. Gao, J. P. O'Connor, R. Raijmakers, G. Pruijn, C. S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murthy, S., J. Kamine, and R. C. Desrosiers. 1986. Viral-encoded small RNAs in Herpesvirus saimiri induced tumors. EMBO J. 5:1625-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murthy, S. C., J. J. Trimble, and R. C. Desrosiers. 1989. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J. Virol. 63:3307-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Myer, V. E., S. I. Lee, and J. A. Steitz. 1992. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc. Natl. Acad. Sci. USA 89:1296-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nair, A. P., S. Hahn, R. Banholzer, H. H. Hirsch, and C. Moroni. 1994. Cyclosporin A inhibits growth of autocrine tumour cell lines by destabilizing interleukin-3 mRNA. Nature 369:239-242. [DOI] [PubMed] [Google Scholar]
- 60.Pages, G., E. Berra, J. Milanini, A. P. Levy, and J. Pouyssegur. 2000. Stress-activated protein kinases (JNK and p38/HOG) are essential for vascular endothelial growth factor mRNA stability. J. Biol. Chem. 275:26484-26491. [DOI] [PubMed] [Google Scholar]
- 61.Peng, S. S., C. Y. Chen, and A. B. Shyu. 1996. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol. Cell. Biol. 16:1490-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng, S. S., C. Y. Chen, N. Xu, and A. B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raghavan, A., R. L. Robison, J. McNabb, C. R. Miller, D. A. Williams, and P. R. Bohjanen. 2001. HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. J. Biol. Chem. 276:47958-47965. [DOI] [PubMed] [Google Scholar]
- 64.Sarkar, B., Q. Xi, C. He, and R. J. Schneider. 2003. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 23:6685-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schiavi, S. C., J. G. Belasco, and M. E. Greenberg. 1992. Regulation of proto-oncogene mRNA stability. Biochim. Biophys. Acta 1114:95-106. [DOI] [PubMed] [Google Scholar]
- 66.Schuler, G. D., and M. D. Cole. 1988. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell 55:1115-1122. [DOI] [PubMed] [Google Scholar]
- 67.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 68.Shim, J., and M. Karin. 2002. The control of mRNA stability in response to extracellular stimuli. Mol. Cells 14:323-331. [PubMed] [Google Scholar]
- 69.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoecklin, G., S. Hahn, and C. Moroni. 1994. Functional hierarchy of AUUUA motifs in mediating rapid interleukin-3 mRNA decay. J. Biol. Chem. 269:28591-28597. [PubMed] [Google Scholar]
- 71.Stoecklin, G., P. Stoeckle, M. Lu, O. Muehlemann, and C. Moroni. 2001. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA 7:1578-1588. [PMC free article] [PubMed] [Google Scholar]
- 72.Terns, M. P., E. Lund, and J. E. Dahlberg. 1993. A pre-export U1 snRNP in Xenopus laevis oocyte nuclei. Nucleic Acids Res. 21:4569-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Truneh, A., F. Albert, P. Golstein, and A. M. Schmitt-Verhulst. 1985. Early steps of lymphocyte activation bypassed by synergy between calcium ionophores and phorbol ester. Nature 313:318-320. [DOI] [PubMed] [Google Scholar]
- 74.Tsygankov, A. Y., and G. Romano. 1999. Mechanisms of cell transformation by Herpesvirus saimiri. Anticancer Res. 19:973-983. [PubMed] [Google Scholar]
- 75.Wagner, B. J., C. T. DeMaria, Y. Sun, G. M. Wilson, and G. Brewer. 1998. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48:195-202. [DOI] [PubMed] [Google Scholar]
- 76.Wang, S. W., J. Pawlowski, S. T. Wathen, S. D. Kinney, H. S. Lichenstein, and C. L. Manthey. 1999. Cytokine mRNA decay is accelerated by an inhibitor of p38-mitogen-activated protein kinase. Inflamm. Res. 48:533-538. [DOI] [PubMed] [Google Scholar]
- 77.Wassarman, D. A., S. I. Lee, and J. A. Steitz. 1989. Nucleotide sequence of HSUR 5 RNA from Herpesvirus saimiri. Nucleic Acids Res. 17:1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wassarman, K. M., and J. A. Steitz. 1992. The low-abundance U11 and U12 small nuclear ribonucleoproteins (snRNPs) interact to form a two-snRNP complex. Mol. Cell. Biol. 12:1276-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weiss, A., J. Imboden, D. Shoback, and J. Stobo. 1984. Role of T3 surface molecules in human T-cell activation: T3-dependent activation results in an increase in cytoplasmic free calcium. Proc. Natl. Acad. Sci. USA 81:4169-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Werner, F. J., G. W. Bornkamm, and B. Fleckenstein. 1977. Episomal viral DNA in a Herpesvirus saimiri-transformed lymphoid cell line. J. Virol. 22:794-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Will, C. L., and R. Luhrmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 82.Wilson, G. M., J. Lu, K. Sutphen, Y. Sun, Y. Huynh, and G. Brewer. 2003. Regulation of A + U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 278:33029-33038. [DOI] [PubMed] [Google Scholar]
- 83.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu, N., C. Y. Chen, and A. B. Shyu. 2001. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 21:6960-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang, M., and K. Tracey. 1998. Tumor necrosis factor, p. 517-548. In A. Thomson (ed.), The cytokine handbook. Academic Press, San Diego, Calif.