Abstract
In resting cells, the NFAT1 transcription factor is kept inactive in the cytoplasm by phosphorylation on multiple serine residues. These phosphorylated residues are primarily contained within two types of serine-rich motifs, the SRR-1 and SP motifs, which are conserved within the NFAT family. Several different kinases have been proposed to regulate NFAT, but no single candidate displays the specificity required to fully phosphorylate both types of motifs; thus, the identity of the kinase that regulates NFAT activity remains unclear. Here we show that the NFAT1 serine motifs are regulated by distinct kinases that must coordinate to control NFAT1 activation. CK1 phosphorylates only the SRR-1 motif, the primary region required for NFAT1 nuclear import. CK1 exists with NFAT1 in a high-molecular-weight complex in resting T cells but dissociates upon activation. GSK3 does not phosphorylate the SRR-1 region but can target the NFAT1 SP-2 motif, and it synergizes with CK1 to regulate NFAT1 nuclear export. We identify a conserved docking site for CK1 in NFAT proteins and show that mutation of this site disrupts NFAT1-CK1 interaction and causes constitutive nuclear localization of NFAT1. The CK1 docking motif is present in proteins of the Wnt, Hedgehog, and circadian-rhythm pathways, which also integrate the activities of CK1 and GSK3.
The activity of transcription factors is tightly regulated by a variety of mechanisms. One class of transcription factors achieves strict regulation through control of subcellular localization, existing in an inactive or unstable state in the cytoplasm until they are activated in response to signals transduced from cell surface receptors (reviewed in reference 7). Upon activation, the transcription factors translocate from the cytoplasm to the nucleus, where they induce gene expression. This strategy is employed by diverse families of transcription factors, including those in the Wnt, Notch, Hedgehog, STAT, SMAD, NF-κB, NFAT, and Pho4 signaling pathways (reviewed in references 7, 27, and 31). In each of these cases, phosphorylation is utilized as a means of regulating nuclear translocation of the latent cytoplasmic transcription factor, although the precise mechanisms and signaling pathways involved differ widely.
NFAT is an example of a latent transcription factor whose subcellular localization, DNA binding, and transcriptional activity are all dictated by its phosphorylation state. The NFAT family is composed of four calcium-regulated members, NFAT1 (also known as NFATc2 or NFATp), NFAT2 (NFATc1, NFATc), NFAT3 (NFATc4), and NFAT4 (NFATc3, NFATx), that play an essential role in immune function as well as in development of the cardiac, vascular, and immune systems (reviewed in references 12 and 18). NFAT proteins are located in the cytoplasm of resting cells, where they exist in a heavily phosphorylated inactive state. In response to calcium-mobilizing stimuli they are partially dephosphorylated by calcineurin, a calcium-dependent phosphatase, and translocate into the nucleus (reviewed in references 11, 23, 29, and 45). Treatment of stimulated cells with calcium chelators or the calcineurin inhibitor cyclosporin A (CsA) results in the rapid rephosphorylation of NFAT and its export out of the nucleus.
The responsiveness of NFAT proteins to calcium signaling is mediated through their regulatory domains, which contain both the calcineurin binding site and the majority of the serine residues that are phosphorylated in resting cells. Mass spectrometric analysis has demonstrated that the regulatory domain of NFAT1 is constitutively phosphorylated at 18 serine residues in resting T cells (41). Twelve of these phosphorylated residues are contained within two distinct types of serine-rich sequence motifs, the SRR-1 and SPXX repeat motifs (referred to below as SP motifs), which are recognizably conserved throughout the NFAT family (see Fig. 1A). The SRR-1 region is the primary region involved in NFAT nuclear import, whereas the SP motifs have been reported to control DNA-binding affinity and nuclear export (5, 40, 41). Several kinases, including glycogen synthase kinase 3 (GSK3), casein kinase 1 (CK1), p38, and c-Jun N-terminal kinase 1 (JNK1), have been suggested to regulate NFAT function (6, 10, 17, 42, 56, 58; reviewed in references 23 and 29). However, no single proposed kinase displays the specificity required for full phosphorylation of both the SRR-1 and SP motifs.
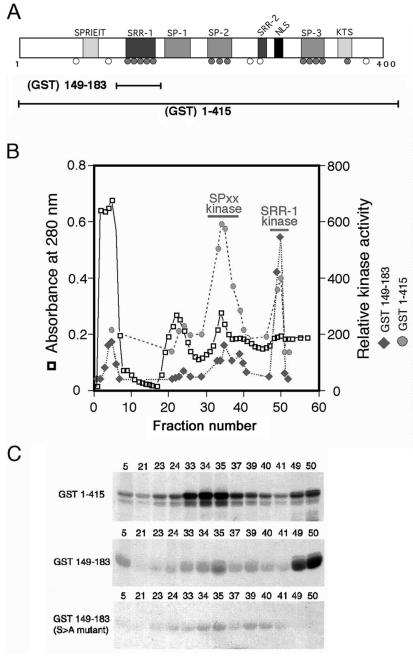
FIG. 1.
NFAT1 is phosphorylated by two distinct kinases. (A) N-terminal regulatory domain of NFAT1, showing the conserved serine motifs and relative positions of phosphorylated serine residues identified in NFAT1 from resting T cells. Shaded circles, conserved phosphoserines that become dephosphorylated upon stimulation; open circles, nonconserved or constitutively phosphorylated serines. GST(149-183) includes only the region containing the NFAT1 SRR-1 motif, while GST(1-415) encompasses the entire NFAT1 regulatory domain. (B) Fractionation of T-cell lysates over a cation-exchange column reveals two separate peaks of kinase activity when GST(1-415) is used as a substrate (shaded circles). Assay of the same fractions using GST(149-183) as a substrate shows that only one of these peaks contains an activity that is able to phosphorylate the SRR-1 motif (solid diamonds). (C) Autoradiograph of kinase assays from the fractionation shown in panel B. NFAT1 kinase activity was assessed by incubating an aliquot from each fraction with either GST(1-415) or GST(149-183) in the presence of [γ-32P]ATP. For GST(1-415), there are two peaks of activity: fractions 33 to 37 and fractions 49 and 50 (top panel). For GST(149-183), only the activity present in fractions 49 and 50 is capable of efficiently phosphorylating the fusion protein (center panel). Mutation of the conserved serines in the SRR-1 motif to alanine completely abolishes phosphorylation of GST(149-183) by the kinase activity present in fractions 49 and 50 (bottom panel).
Here we demonstrate that NFAT1 regulation involves the concerted effort of distinct kinases to phosphorylate each of the two types of serine motifs. CK1 specifically phosphorylates only the NFAT1 SRR-1 motif by docking at a site that is conserved in the N-terminal regions of all four NFAT proteins, and disruption of CK1 docking is sufficient to cause aberrant nuclear translocation of NFAT1. CK1 and NFAT1 are present in a high-molecular-weight complex in resting T cells that dissociates upon activation. GSK3 is not present in this complex but can phosphorylate NFAT SP motifs if suitably primed and can synergize with CK1 to promote NFAT1 nuclear export. The NFAT pathway resembles the Wnt, Hedgehog, and circadian-rhythm pathways in being regulated by the combined activity of CK1 and GSK3, and we demonstrate that the circadian rhythm protein mPER2 also contains a functional CK1 docking motif.
MATERIALS AND METHODS
Expression plasmids and GST fusion constructs.
The expression plasmid pEFTAGmNFAT1c, encoding full-length murine NFAT1 with three copies of the hemagglutinin (HA) epitope at the N terminus, and the glutathione S-transferase (GST) fusion proteins GST-NFAT1(1-415) and GST-NFAT2(1-418) have been described previously (36, 37). GST-NFAT4(1-371) was generated by subcloning a PCR product encoding the first 371 amino acids of human NFAT4 into the pGEX6P-2 vector (Pharmacia). Plasmids pV367, encoding full-length HA-tagged CK1α, pV371, encoding full-length CK1ɛ with an HA tag, and pV570, encoding HA-tagged CK1ɛ truncated at amino acid 319, have been described elsewhere (46). The NFAT1 fusion constructs GST-NFAT1(149-183) and GST-NFAT1(1-100) were created by subcloning PCR products generated from murine NFAT1 into pGEX6P-2. The NFAT1(LDFS), NFAT1(DE), NFAT1(F30F34), and NFAT1(F34) point mutations were created by synthesizing oligonucleotides containing the appropriate substitutions, which were then used as PCR primers. The resulting PCR products were used to replace the corresponding sequences encoded in the GST-NFAT1(1-100) and NFAT1(1-460)-GFP (4) expression vectors.
Protein purification and kinase assays.
Whole-cell extracts were prepared by Dounce homogenization of Jurkat cells in hypotonic lysis buffer (50 mM Tris [pH 7.9], 10 mM NaCl, 10% glycerol, 0.05% Triton X-100, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol [DTT], 20 mM β-glycerol-phosphate, 10 mM sodium pyrophosphate, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of aprotinin/ml, 10 μg of leupeptin/ml). Lysates were spun for 10 min at 20,000 × g, followed by a high-speed spin for 1 h at 100,000 × g in the ultracentrifuge. The supernatant was loaded onto a POROS HQ column (Applied Biosystems) and eluted with a 0 to 400 mM NaCl gradient. In vitro kinase assays were performed on GST-NFAT1 fusion proteins by incubating column fractions and kinase buffer (20 mM HEPES, 20 mM MgCl2, 1 mM DTT, 10 mM β-glycerol-phosphate) in a final volume of 30 μl in the presence of 20 μM ATP and 2 μCi of [γ-32P]ATP for 20 min at 30°C. Fractions enriched for NFAT1 kinase activity were pooled and dialyzed against buffer B (50 mM HEPES [pH 7.2], 10% glycerol, 1 mM DTT, 1 mM PMSF) before fractionation over a POROS HS column (Applied Biosystems). Proteins were eluted by using a NaCl step gradient, and the bulk of SRR-1 kinase activity was observed between 400 and 500 mM NaCl. After concentration on a Centriplus 30 spin column (Amersham), the sample was loaded onto a Superdex 200 gel filtration column (Pharmacia). Fractions were analyzed for NFAT1 SRR-1 kinase activity and were subjected to silver staining to identify candidate bands that coincided with the fractionation of this activity. Six bands that cofractionated with SRR-1 kinase activity were excised and analyzed by mass spectrometry. A prominent band between 35 and 40 kDa whose fractionation coincided most closely with kinase activity was identified as CK1α. Two other candidate bands of slightly lower molecular weight (presumably degradation products) also contained multiple peptides corresponding to CK1. No other kinases were identified in the mass spectrometry analysis.
The ability of GSK3 or CK1 to phosphorylate NFAT1 was examined by first prephosphorylating GST-NFAT fusion proteins with 100 U of recombinant protein kinase A (PKA) (New England Biolabs [NEB]) in the presence of 1 mM cold ATP for 4 h at 30°C. After being primed with PKA, fusion proteins were washed repeatedly to remove the kinase and ATP. The prephosphorylated fusion proteins were then incubated with 100 U of GSK3 (NEB) or 50 U of CK1 (NEB) in kinase buffer along with 3 μCi of [γ-32P]ATP for 20 min at 30°C.
Coprecipitation and Western blotting.
Jurkat T cells were used for immunoprecipitation of endogenous NFAT1 and CK1. Either resting cells or cells that had been stimulated for 15 min with 1 μM ionomycin in the presence of 2 mM CaCl2 were lysed by resuspension in Triton X-100 lysis buffer (50 mM Tris [pH 7.5], 140 mM NaCl, 10% glycerol, 0.05% Triton X-100, 1 mM EDTA, 1 mM EGTA, 10 mM β-glycerol-phosphate, 1 mM PMSF, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml). Lysates were spun for 10 min at 14,000 rpm in an Eppendorf 5417R microcentrifuge before spinning for 30 min at 100,000 × g in an ultracentrifuge. Supernatants were then precleared for 30 min with protein A-Sepharose before immunoprecipitation for 3 h with an NFAT1 antibody. For GST pulldown experiments, HEK 293 cells were transiently transfected with expression vectors containing CK1α (pV367) or CK1ɛ (pV371). Cells were lysed in a buffer containing Triton X-100, and lysates were clarified by spinning at 20,000 × g for 15 min at 4°C. Cell lysates were incubated with GST or a GST-NFAT fusion protein bound to glutathione beads for 2 h at 4°C. Fusion proteins were washed with Triton X-100 lysis buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blotted with antibodies against CK1α (Santa Cruz Biotechnology) and CK1ɛ (Transduction Laboratories) to detect bound proteins.
Immunocytochemistry.
For the nuclear export studies, the murine T-cell clone D5 (44) was incubated with 150 to 300 μM CKI-7 inhibitor (Seikagaku) and/or 10 mM LiCl (or an equivalent amount of dimethyl sulfoxide [DMSO] and 10 mM NaCl) for 1 h before being stimulated with 1 μM ionomycin in the presence of 2 mM CaCl2. After 10 min, 1 μM CsA was added to inhibit calcineurin activity and allow the rephosphorylation of NFAT1. Aliquots of cells were taken at the indicated intervals and fixed on coverslips, and localization of the endogenous NFAT1 was detected with a polyclonal anti-NFAT1 antibody (22) followed by Cy3-conjugated anti-rabbit immunoglobulin G (Jackson Immunoresearch Laboratories). For experiments examining the effect of CKI-7 or LiCl on resting NFAT1, we utilized a HeLa cell line stably expressing the regulatory domain of NFAT1 fused to green fluorescent protein (GFP) [NFAT(1-460)-GFP] (4). Cells were incubated for 2 h with the export inhibitor leptomycin B (LMB; Sigma) at 20 nM in addition to either DMSO, 10 mM LiCl, or 300 μM CKI-7. NFAT1 was detected by GFP fluorescence. To examine the effects of CKI-7 on CK1 and GSK3 activities, GST(1-415) was first primed with PKA as described above and then added to purified GSK3 (NEB) or CK1 (NEB) that had been preincubated with increasing concentrations of CKI-7 or 10 mM LiCl, along with [γ-32P]ATP.
RESULTS
Identification of the SRR-1 kinase as CK1.
To identify potential NFAT1 regulatory kinases, we fractionated T-cell lysates over a sequence of ion-exchange columns. The activity of kinases capable of phosphorylating the NFAT1 regulatory domain was monitored by using a GST fusion protein encompassing this region [GST(1-415)] (Fig. 1A). These experiments revealed the presence of two distinct peaks of kinase activity capable of phosphorylating the NFAT1 regulatory domain (Fig. 1B and 1C, top panel; fractions 30 to 40 and fractions 49 and 50). To further investigate the target of these kinases, we constructed a shorter fusion protein [GST(149-183)] that encompasses the SRR-1 motif but does not include any of the SP motifs (Fig. 1A). Only one of the peaks (fractions 49 and 50) contained a potential SRR-1 kinase activity, which phosphorylated the GST(149-183) fusion protein but not a mutant derivative with serine-to-alanine changes of five of the conserved serines (Fig. 1C). The two peaks of kinase activity showed distinct elution patterns upon subsequent gel filtration, indicating that they corresponded to different NFAT1 kinases.
To identify the SRR-1 kinase, we fractionated whole-cell lysates from Jurkat cells over a series of ion-exchange and chromatography columns (see Materials and Methods). Fractions containing the largest amounts of SRR-1 kinase activity were pooled and loaded onto a gel filtration column (Fig. 2A). Silver-stained SDS-PAGE gels of the eluted fractions consistently showed two to three bands between 35 and 40 kDa that correlated strongly with the peak of SRR-1 kinase activity (Fig. 2A, fractions 87 to 90). Mass spectrometry demonstrated that these bands contained CK1α (see Materials and Methods). Western blot analysis of gel filtration fractions confirmed that peak SRR-1 kinase activity coincided precisely with the fractions containing the highest levels of CK1α (Fig. 2B, fractions 61 to 66). These fractions did not contain GSK3, which has been suggested to be the main regulatory kinase for two NFAT family members, NFAT2 and NFAT3 (6, 19) (see below).
FIG. 2.
Identification of CK1 as an NFAT1 SRR-1 kinase. (A) (Top) Silver stain gel showing kinase-active fractions from a size exclusion column. Arrow indicates the position of a band that cofractionates with SRR-1 kinase activity. (Bottom) The GST(149-183) fusion protein was incubated with an aliquot from each column fraction in the presence of [γ-32P]ATP to assay for the presence of NFAT1 SRR-1 kinase activity. (B) Western blot analysis confirms that the distribution of CK1α coincides precisely with fractions containing NFAT1 SRR-1 kinase activity after fractionation of T-cell lysates through several columns. (C) Western blot analysis of CK1ɛ and CK1α from an early anion-exchange column shows that CK1α and CK1ɛ cofractionate with SRR-1 kinase activity, whereas both isoforms of GSK3 elute earlier and are distinct from the SRR-1 kinase activity. (D) The NFAT1 SRR-1 motif is phosphorylated by purified CK1 but not by GSK3. For priming with PKA, GST fusion proteins were prephosphorylated by PKA in the presence of cold ATP, washed thoroughly to remove PKA, and then used as a substrate for in vitro kinase assays with GSK3 and [γ-32P]ATP. Prephosphorylation of the fusion protein by PKA enables GSK3 to target GST-NFAT1(1-415) (lanes 1 and 2). Within this domain, GSK3 is able to target the SP-2 motif when it is primed by PKA (lanes 3 and 4) but is not able to phosphorylate the SRR-1 region (lanes 5 and 6). In contrast, purified recombinant CK1 phosphorylates GST-NFAT1(1-415) (lane 7) and the SRR-1 region (lane 8) without the need for prior phosphorylation.
The CK1 family is composed of several different gene products, including CK1α, CK1δ, and CK1ɛ (reviewed in reference 21). Unlike CK1α, which has a very short C terminus, CK1ɛ and CK1δ possess a longer C-terminal region that becomes autophosphorylated in the presence of ATP, resulting in the rapid downregulation of kinase activity (8, 20, 46). Consequently, CK1ɛ and CK1δ are inefficiently detected by in vitro kinase assays (8), leading us to ask whether these kinases may have been overlooked during the multistep chromatography protocol used to purify CK1α as an NFAT kinase. Western blot analysis of fractions from each of the chromatography columns revealed that CK1ɛ and CK1δ cofractionated with CK1α and SRR-1 kinase activity through all but the final gel filtration step (Fig. 2C and data not shown), while GSK3 fractionated away from SRR-1 kinase activity at the very first chromatography step (Fig. 2C). These results strongly implicate CK1 proteins as SRR-1 kinases for NFAT1.
We tested whether GSK3 could also serve as an SRR-1 kinase for NFAT1 (Fig. 2D). Purified recombinant GSK3 was able to phosphorylate a fusion protein that encompassed the entire regulatory domain of NFAT1, but only if the substrate was previously phosphorylated in a “priming” step by PKA (Fig. 2D; compare lanes 1 and 2), similar to what has been reported for NFAT2 (6). Under identical conditions, GSK3 was also able to phosphorylate a shorter fusion protein containing the isolated NFAT1 SP-2 motif (Fig. 2D, lanes 3 and 4). However, GSK3 was incapable of phosphorylating the fragment that contained only the NFAT1 SRR-1 region, even after preincubation with PKA (Fig. 2D, lanes 5 and 6). In contrast, a recombinant CK1δ kinase (lacking its autoinhibitory domain) phosphorylated the SRR-1 region robustly (lane 8). Similarly, a truncated version of CK1ɛ lacking its autoinhibitory region also had strong SRR-1 kinase activity (data not shown). Taken together, these results demonstrate that CK1 is an NFAT1 SRR-1 kinase, while GSK3 phosphorylates residues in the NFAT1 SP-2 motif in conjunction with a priming kinase.
A high-molecular-weight complex containing NFAT1 and CK1.
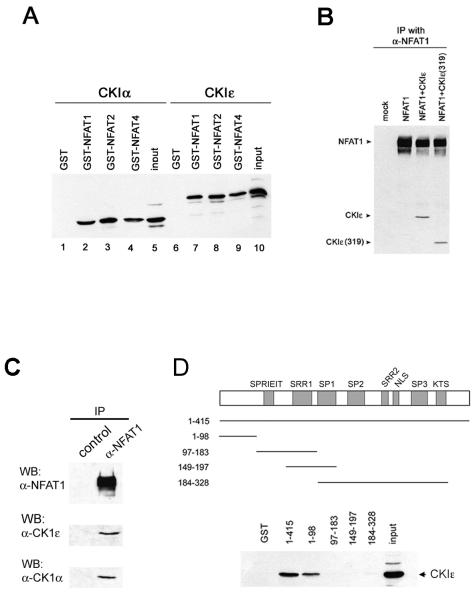
Since enzymes in a signal transduction pathway often bind to their substrates, we asked whether CK1 proteins associated with NFAT (Fig. 3). We generated GST fusion proteins encompassing the N-terminal regulatory domains of NFAT family members NFAT1, NFAT2, and NFAT4. Stable interactions were readily detectable between the GST-NFAT fusion proteins and CK1α, CK1ɛ (Fig. 3A), and CK1δ (data not shown) from HEK 293 lysates. No interaction was detected between any CK1 kinase and GST alone (Fig. 3A). The interactions of CK1ɛ and CK1α with NFAT1 and NFAT3 were also established by coimmunoprecipitating full-length proteins expressed in HEK 293 cells (Fig. 3B and data not shown) as well as endogenous proteins present in Jurkat T cells (Fig. 3C). The interaction mapped to the first 319 amino acids of CK1ɛ (Fig. 3B, last lane), which contain its kinase domain; this domain is virtually identical to the kinase domain of CK1δ and is closely related to that of CK1α (21). On NFAT1, the region of interaction mapped to the beginning of the N terminus, which contains its transactivation domain (amino acids 1 to 98) (Fig. 3D). These results show that there are stable interactions between CK1 kinases and each NFAT family member.
FIG. 3.
Interaction of NFAT family members with CK1. (A) GST fusion proteins containing the regulatory domains of NFAT proteins bind CK1 isoforms. GST or GST fusion proteins encompassing the regulatory domains of NFAT1, NFAT2, or NFAT4 were incubated with HEK 293 cell lysates that were either mock transfected or transfected with CK1 isoform α or ɛ. Bound proteins were visualized by Western blotting and sequential probing with antibodies against CK1α and CK1ɛ. (B) NFAT1 interacts with the kinase domain of CK1. Full-length HA-tagged NFAT1 was expressed in HEK 293 cells either alone, in combination with HA-tagged full-length CK1ɛ, or in combination with C-terminally truncated CK1ɛ [CK1ɛ(319)]. NFAT1 was immunoprecipitated with an anti-NFAT1 antibody, and precipitated proteins were visualized by Western blotting with an antibody against the HA tag. (C) Endogenous NFAT1 and CK1 stably associate. NFAT1 was immunoprecipitated from Jurkat T cells with either an anti-NFAT1 antibody or preimmune serum. The presence of associated CK1 in the immunoprecipitates was detected by Western blotting (WB) with antibodies against CK1α or CK1ɛ. (D) The region of NFAT1 that mediates CK1 binding maps to the N-terminal transactivation domain. GST fusion proteins encompassing various regions of the NFAT1 regulatory domain were incubated with cell lysates transfected with CK1ɛ. Fusion proteins able to associate with CK1 were detected by Western blot analysis using anti-CK1ɛ.
Based on these interaction experiments, we asked whether NFAT1 might exist in a stable protein complex with CK1 (Fig. 4). T-cell extracts containing endogenous NFAT1 and CK1 were fractionated through a size exclusion column. Although the fully phosphorylated form of NFAT1 migrates with an apparent molecular mass of about 130 to 140 kDa on denaturing SDS gels, NFAT1 in T-cell extracts eluted in gel filtration fractions corresponding to an apparent molecular mass between 350 and 400 kDa (Fig. 4A, top panel). A significant portion of CK1α and CK1ɛ kinases coeluted with NFAT1 (Fig. 4A, second panel; also data not shown). CK1α, which has a molecular mass of about 37 kDa, migrated through the column with two distinct peaks of elution: a fraction that eluted at the expected molecular weight for a CK1α monomer (fractions 88 to 90) and a second, high-molecular-weight fraction that coeluted with NFAT1 at an apparent molecular mass of ∼350 kDa (fractions 62 to 64). CK1ɛ also demonstrated altered mobility on the size exclusion column, eluting mainly in two peaks: one that coincided with NFAT1 elution and a second at its expected molecular mass (∼47 kDa) (data not shown). This is consistent with our findings that endogenous CK1α and CK1ɛ can be coimmunoprecipitated with NFAT1 in T cells (Fig. 3C and 4C). In contrast, both isoforms of GSK3 eluted at their expected molecular masses, well separated from the fractions containing NFAT1 and CK1 (Fig. 4A, third panel). Calcineurin, the NFAT phosphatase, eluted at a molecular mass higher than that expected for the holoenzyme containing the catalytic A subunit and the regulatory B subunit but did not cofractionate with the NFAT1 high-molecular-weight complex.
FIG. 4.
CK1 is complexed with NFAT1 in resting cells but dissociates upon stimulation. (A) Analysis of resting T-cell lysates fractionated over a size exclusion column shows that NFAT1 exists in a high-molecular-weight complex that coelutes with a subpopulation of CK1. GSK3 and calcineurin are not part of this complex. Aliquots from each fraction were separated by SDS-PAGE, and the elution profile of each protein was visualized by Western blotting. (B) Stimulation of T cells with the calcium ionophore ionomycin before fractionation of lysates over a size exclusion column results in dissociation of CK1 from the NFAT1 complex. CK1 no longer elutes in two peaks but elutes primarily in fractions corresponding to the expected molecular weight for a monomer of the kinase. (C) Jurkat cells that were either resting (R) or stimulated with 1 μM ionomycin (iono) in the presence of 2 mM CaCl2 were lysed and subjected to immunoprecipitation with an antibody against the C terminus of NFAT1. Endogenous CK1α can be coimmunoprecipitated with NFAT1 from resting cells but not from cells that have been stimulated. WB, Western blot.
The NFAT1-CK1 association is dynamic and sensitive to the stimulation state of the cell. Gel filtration analysis of lysates from ionomycin-stimulated T cells showed that the NFAT1-CK1 interaction became weaker after stimulation with the calcium ionophore ionomycin (Fig. 4B). After stimulation, CK1 showed a pronounced change in its elution profile, with only a minor fraction coeluting with NFAT1 while the majority of CK1 shifted into fractions corresponding to the expected size of the kinase alone (Fig. 4B, center panel). This effect was prominent for both CK1α and CK1ɛ (Fig. 4 and data not shown). In contrast, there was only a minor change in the elution profile of NFAT1, suggesting the presence of other proteins in the NFAT1 complex. The elution profiles of GSK3 and calcineurin showed no change (Fig. 4B and data not shown). These results mirror those from immunoprecipitation experiments showing that NFAT1 from Jurkat cells associates with endogenous CK1α but that this interaction is almost undetectable in cells that have been stimulated with ionomycin (Fig. 4C). Thus, cell stimulation concomitantly activates the calcineurin phosphatase and leads to dissociation of the SRR-1 kinase.
CK1 and GSK3 synergize to effect NFAT1 rephosphorylation and nuclear export.
Termination of calcium and/or calcineurin signaling results in rephosphorylation of NFAT1 and its export from the nucleus. We used untransformed T cells in which we could monitor the behavior of endogenous NFAT1 to investigate whether CK1 or GSK3 plays a role in this process (Fig. 5). Since several different CK1 family members are able to bind and phosphorylate NFAT1 in vitro, we used CKI-7, a cell-permeant inhibitor that affects all known family members of CK1 (9). LiCl was used to inhibit GSK3 (30). To minimize nonspecific effects, we exposed cells to the inhibitors for 2 h or less.
FIG. 5.
CK1 and GSK3 coordinate to effect rephosphorylation and nuclear export of NFAT1. (A) D5 T cells were preincubated for 1 h with CKI-7, LiCl, or a combination of both inhibitors. Control cells were incubated with a corresponding amount of DMSO and/or NaCl. Cells were stimulated with ionomycin to induce dephosphorylation and nuclear translocation of NFAT1. Calcineurin activity was subsequently inhibited by addition of CsA to the cells, which were attached to coverslips and fixed in paraformaldehyde at the indicated time points. The subcellular localization of endogenous NFAT1 was visualized by indirect immunofluorescence. (B) D5 T cells were preincubated with inhibitors and thenstimulated with ionomycin (iono) as for panel A. CsA was added to cells, and the phosphorylation state of NFAT1 at various time points after CsA addition was determined by quickly lysing cells in boiling SDS sample buffer. Western blotting was performed with an anti-NFAT1 antibody. (C) CKI-7 does not inhibit GSK3 kinase activity. GST(1-415) was prephosphorylated by use of PKA with cold ATP before being washed extensively and incubated with increasing concentrations of CKI-7 in the presence of [γ-32P]ATP and either GSK3 or CK1. LiCl and NaCl (each at 10 mM) were used as positive and negative controls, respectively, for inhibition of GSK3 activity. (D) Quantitation of the ability of CKI-7 to inhibit GSK3 and CK1. GST(1-415) was incubated with either GSK3 or CK1 in the presence of CKI-7 as described for panel C. Radiolabeled GST(1-415) was then separated by SDS-PAGE, and incorporation of 32P into the fusion protein was quantified by phosphorimager analysis. (E) Inhibition of CK1 or GSK3 mimics the effect of mutating the SRR-1 or SP motifs. Resting HeLa cells stably expressing the NFAT1(1-460)-GFP fusion protein were incubated for 2 h in the presence of LMB in addition to the inhibitor DMSO, LiCl, or CKI-7 before being fixed and visualized by GFP fluorescence. Inhibition of CK1 results in partial NFAT1 nuclear localization in resting cells, reminiscent of NFAT1 localization after SRR-1 mutation. LiCl has no effect on endogenous NFAT1 localization in resting cells, in agreement with a role for GSK3 in phosphorylation of the SP motifs but not the NFAT1 SRR-1 motif.
Resting T cells were first incubated with LiCl, CKI-7, or a combination of both inhibitors and then stimulated with ionomycin to induce dephosphorylation and nuclear import of endogenous NFAT1. The calcineurin inhibitor CsA was then added directly to the cells to synchronously initiate NFAT1 rephosphorylation and nuclear export. Immunofluorescence revealed a striking difference in the kinetics of NFAT1 nuclear export between control T cells and those treated with the inhibitors (Fig. 5A). After 30 min of CsA treatment (Fig. 5A), nuclear export of NFAT1 was essentially complete in control cells, well under way in LiCl-treated cells, and barely initiated in CKI-7-treated cells. One hour after CsA addition, NFAT1 nuclear export was complete in control and LiCl-treated cells, in which the protein was predominantly cytoplasmic, but still incomplete in CKI-7-treated cells, where NFAT1 was equally distributed between the nucleus and cytoplasm. The combination of CKI-7 and LiCl was significantly more effective at blocking NFAT1 nuclear export than either inhibitor alone; in cells treated with both kinase inhibitors, NFAT1 remained almost completely nuclear even 1 h after CsA addition (Fig. 5A).
Consistent with the delay in nuclear export, Western blotting demonstrated that the combination of CK1 and GSK3 kinase inhibitors resulted in a significant delay in the rephosphorylation of NFAT1 in T cells after CsA addition (Fig. 5B). The finding that the inhibitors delay NFAT1 rephosphorylation and nuclear export rather than blocking it completely most likely reflects the fact that even when tested in vitro, high concentrations of the inhibitors do not completely abolish the ability of recombinant CK1 or GSK3 to phosphorylate NFAT1 (Fig. 5C and D). Approximately 10 to 20% of kinase activity remains in vitro, and this amount of kinase activity is likely sufficient for slow rephosphorylation of NFAT1 in vivo. To ensure that CKI-7 and LiCl were acting specifically on CK1 and GSK3, respectively, we asked whether CKI-7 inhibited the kinase activity of purified GSK3 (Fig. 5C). Using GST(1-415) that had been prephosphorylated by PKA as a substrate, we showed that concentrations of CKI-7 that efficiently inhibited CK1 kinase activity did not alter the ability of GSK3 to phosphorylate the NFAT1 regulatory domain (Fig. 5C and D). Conversely, LiCl impaired GSK3 kinase activity by >70%, as expected, but had only a minor effect on CK1 kinase activity (Fig. 5C).
CK1 controls NFAT1 nuclear import.
The greater effectiveness of CKI-7 than LiCl in prolonging the nuclear residence of NFAT1 (Fig. 5A) is consistent with the proposed role for CK1 in phosphorylating the SRR-1 motif, the primary region controlling nuclear localization signal exposure. It has been shown previously that phosphorylation of conserved serines within the SRR-1 region is critical for establishing and maintaining the cytoplasmic localization of NFAT1 (41). Serine-to-alanine mutation of conserved serines within the SRR-1 motif is sufficient to cause partial constitutive nuclear localization of NFAT1 in resting cells, even when the SP motifs remain fully phosphorylated. In contrast, mutation of the SP motifs alone has no effect on NFAT1 localization in resting cells (41). If CK1 is the major constitutive kinase for the NFAT1 SRR-1 region, inhibition of CK1 activity or disruption of CK1-NFAT1 association would be expected to gradually decrease phosphorylation of the SRR-1 region and promote partial nuclear localization of NFAT1 in resting cells, whereas inhibition of GSK3 or other SP motif kinases should have little or no effect.
HeLa cells stably expressing a GFP-NFAT1 fusion protein (Fig. 5E) were incubated with CKI-7 or LiCl in the presence of LMB (54), an inhibitor of NFAT nuclear export which facilitates detection of nuclear translocation by preventing any nucleocytoplasmic shuttling of NFAT (28, 41, 57). In control HeLa cells treated for 2 h with LMB but no kinase inhibitors, the bulk of NFAT1 is excluded from the nucleus (Fig. 5E, left panel). Cells treated with LMB and LiCl did not differ detectably from control cells (center panel); in contrast, cells treated with LMB and CKI-7 showed a noticeable increase in nuclear NFAT1 staining relative to that in cells treated with LMB alone, with similar intensities of NFAT1 staining in the nucleus and the cytoplasm (right panel). Identical results were observed for endogenous NFAT1 in T cells (data not shown). These results are completely consistent with our findings that CK1 is responsible for SRR-1 phosphorylation and that this region is, in turn, primarily responsible for NFAT1 nuclear import. Similarly, the fact that the GSK3 inhibitor LiCl did not promote the nuclear import of NFAT1 is consistent with our findings that GSK3 is an SP motif kinase but not an SRR-1 kinase and that mutations which mimic dephosphorylation of the SP motifs do not alter the subcellular distribution of NFAT1 in resting cells.
Mutation of the CK1 docking site deregulates NFAT1 localization.
Although use of the CKI-7 inhibitor altered NFAT1 localization in cells, general concerns about inhibitor specificity led us to seek an additional method to validate the role of CK1 in NFAT1 regulation. Since NFAT1 and CK1 are stably associated in resting cells (Fig. 4), we asked whether disruption of the NFAT1-CK1 association would affect NFAT1 localization. We had mapped CK1 binding to the first 98 amino acids of NFAT1 (Fig. 3D). To identify potential docking motifs present in this region, we aligned the N-terminal sequences of the four NFAT proteins with the PER circadian-rhythm proteins (see Fig. 7A), which have also been shown to associate directly with CK1. Remarkably, this comparison identified a common F-X-X-X-F sequence, present both in the N-terminal region of NFAT proteins and in the region of PER proteins shown to bind CK1 (51, 53) (Fig. 7A). The CK1-binding region of NFAT1 also contains an LDFS motif (Fig. 7A), previously identified in a subset of helix-loop-helix transcription factors (38); however, this motif is only loosely conserved among members of the NFAT family and does not appear to be conserved in other CK1 targets.
FIG. 7.
The F-X-X-X-F motif also serves as a CK1 docking site in the PER circadian-rhythm proteins. (A) Sequences of F-X-X-X-F motifs in NFAT and PER proteins thought to interact with CK1. (B) Mutation of the F-X-X-X-F motif in mPER2 abolishes binding to CK1ɛ. Mutations targeting the phenylalanines shown in panel A were introduced into GST fusion proteins containing mPER2 residues 583 to 765, which encompass most of the CK1 binding domain. Fusion proteins were incubated with HEK cell lysates and analyzed for the ability to interact with endogenous CK1 by Western blotting (WB) using an antibody against CK1ɛ. GST fusion proteins containing the wild-type CK1 binding domain of mPER2 (amino acids 583 to 765) or NFAT1 (amino acids 1 to 100) were able to associate with CK1ɛ, while GST alone or fusion proteins carrying mutations in the F-X-X-X-F motif were unable to interact with the kinase. (C) Processive phosphorylation by CK1. See Discussion for details.
We tested whether mutating the F-X-X-X-F and LDFS motifs affected NFAT1-CK1 association or NFAT1 nuclear localization. Mutation of the entire LDFS motif or the phenylalanine residues within the overlapping F-X-X-X-F motif drastically reduced the ability of GST-NFAT1 fusion proteins to bind CK1 (Fig. 6A), whereas mutation of other, nearby residues did not alter NFAT1-CK1 interaction (“DE” in Fig. 6A; data not shown for other mutations). In cells, NFAT1-GFP fusion proteins with alterations in either the LDFS or the F-X-X-X-F motif were constitutively localized to the nucleus, in this respect resembling proteins with mutations of the SRR-1 serines themselves (Fig. 6B). In contrast, mutations of adjacent residues that did not affect CK1 binding did not alter the cytoplasmic localization of NFAT1 (DE in Fig. 6B; data not shown for other mutations). Remarkably, a single point mutation (F34A) which disrupted NFAT1-CK1 binding (Fig. 6A) sufficed to upset proper localization of NFAT1 in resting cells (Fig. 6B), presumably by interfering with phosphorylation of the SRR-1 motif by its regulatory kinase. These data provide strong genetic evidence that the F-X-X-X-F motif controls NFAT1 phosphorylation at the SRR-1 motif by serving as a docking site for CK1.
FIG. 6.
Docking-site mutations that disrupt NFAT1-CK1 association also result in aberrant nuclear localization of NFAT1. (A) Mutation of residues within the F-X-X-X-F motif disrupts association of NFAT1 and CK1. (Left) The mutations shown were introduced into GST proteins containing the first 100 amino acids of NFAT1. (Right) The GST proteins were incubated with cell lysates expressing CK1ɛ, and the ability of the fusion proteins to interact stably with CK1 was detected by Western blotting. Mutations that altered either phenylalanine within the docking motif of NFAT1 virtually abolished NFAT1-CK1 interactions, but substitutions in nearby amino acids not affecting these residues did not diminish the association. (B) Mutation of the CK1 docking site interferes with proper localization of NFAT1. (Top) GFP-NFAT1 proteins containing mutations in the CK1 binding motif were transfected into HeLa cells, and their localization was visualized by fluorescence. Mutations that affected the ability of CK1 and NFAT1 to associate stably, as seen in panel A, also resulted in improper localization of NFAT1. The constitutive nuclear localization of CK1 docking-site mutants is similar to that observed upon mutation of the target serine residues in the SRR-1 motif (bottom panel). Mutations that did not affect CK1-NFAT1 associations, as shown in panel A, also did not affect NFAT1 localization. (Bottom) Two representative fields from an NFAT mutant with substitutions of the target serine residues in the SRR-1 motif, showing different degrees of nuclear localization.
Since the F-X-X-X-F motif is also present in PER proteins (Fig. 7A), we tested whether mutating this motif would affect the binding of CK1 to PER. Mutation of this motif in the context of a fusion protein containing the CK1 binding domain of mPER2 abolished the ability of this known CK1ɛ target to bind to its regulatory kinase (Fig. 7B). Thus, the F-X-X-X-F motif may prove to be a conserved CK1 docking site in diverse classes of proteins.
DISCUSSION
A modular mechanism for NFAT phosphorylation and function.
Although a number of candidate kinases have been proposed for each NFAT protein, it seems unlikely, in light of the characteristic sequences of the conserved motifs and the distinct sequence preferences of the proposed kinases, that a single kinase phosphorylates the almost 20 different phosphorylation sites that exist in the regulatory domains of NFAT proteins. Here we provide evidence that the two major classes of conserved serine motifs in the NFAT1 regulatory domain are phosphorylated by different kinases. Specifically, CK1 selectively targets the SRR-1 motif, and disruption of CK1-NFAT1 association is sufficient to cause aberrant nuclear localization of NFAT1 in resting cells. CK1 shows a preference for phosphorylating targets three residues downstream of an acidic or prephosphorylated amino acid [(pS/pT/E/D)-X-X-(S/T), where pS/pT is a phosphoserine or phosphothreonine and S/T is the target serine or threonine residue] (16, 39). Thus, phosphorylation of the first serine in a repeating sequence, S-X-X-S, would create a CK1 recognition site for the next serine, enabling processive phosphorylation of five of the seven serines in the SRR-1 motif (Fig. 7C), as observed in cells (41). Such a pattern of processive phosphorylation by CK1 has also been suggested for the PER proteins (51) (Fig. 7C).
We have also shown that GSK3 does not phosphorylate the SRR-1 region but can phosphorylate the SP motifs after a priming phosphorylation. Notably, previous mass spectrometric analysis of cytoplasmic NFAT1 from resting T cells did not yield any evidence of stable, stoichiometric phosphorylation at the postulated priming sites (41) and suggested that GSK3 functions as an NFAT1 export kinase rather than as a constitutive kinase that maintains SP motif phosphorylation in resting cells. Consistent with this, the GSK3 inhibitor LiCl did not cause nuclear localization of NFAT1 even in LMB-treated cells, but it did delay NFAT1 export from the nucleus (Fig. 5). In contrast, CK1 inhibitors caused NFAT1 nuclear import in LMB-treated cells and also delayed NFAT1 export from the nucleus (Fig. 5); these results suggest that CK1 both maintains SRR-1 phosphorylation under resting conditions and plays a role in its rephosphorylation in the nucleus.
It is clear from our own previous work and that of others that the conserved SRR and SP motifs behave as individual modules that regulate NFAT function. Serine-to-alanine mutations which mimic dephosphorylation of the SRR-1 motif result in constitutive nuclear localization of NFAT proteins but do not affect the DNA binding of NFAT, indicating that the phosphorylation state of the SRR-1 motif primarily regulates exposure of the nuclear localization sequence in the regulatory domain (5, 41, 42). In contrast, the phosphorylation state of the SP motifs appears to be more important for controlling DNA binding affinity: mutations that mimic dephosphorylation of the SP motifs increase NFAT DNA binding (40) without affecting NFAT1 localization in resting cells (41). As a result, NFAT1 SRR-1 mutants are constitutively nuclear but exhibit only partial transcriptional activity under conditions where the SP motifs remain phosphorylated (J. Aramburu, H. Okamura, and A. Rao, unpublished data). Our present work demonstrating that these motifs are targeted by two different kinases sheds light on why a single protein requires so many different phosphorylation sites. Regulation of each modular cluster of phosphorylation sites by a separate kinase allows for finely tuned control over each aspect of NFAT activation, from subcellular localization to DNA binding to transcription.
Implications for differential regulation of individual NFAT family members.
The level of NFAT activity at any given time is determined by the balance of calcineurin-dependent activation and kinase-mediated deactivation (23, 47, 52). Regardless of which NFAT family member is being considered, a single phosphatase, calcineurin, recognizes and dephosphorylates all phosphoserine residues relevant for NFAT activation. Yet there is growing evidence that even in a single cell, where all NFAT proteins are subject to the same parameters of Ca2+-calcineurin signaling, individual NFAT proteins may show very different patterns of subcellular localization (reviewed in reference 23). For instance, NFAT1, NFAT2, and NFAT4 are present in skeletal muscle cells at all stages of myogenesis but exhibit distinct patterns of nuclear translocation depending on the specific stage of muscle differentiation (1). Similarly, only NFAT2 translocates to the nucleus in human pulmonary valve endothelial cells treated with vascular endothelial growth factor, even though these cells also contain NFAT1 and NFAT4 (26). Likewise, NFAT2 is dephosphorylated and nuclear in T cells stimulated for 5 to 6 h, at the same time that NFAT1 is being rephosphorylated and exported to the cytoplasm (35; H. Okamura and A. Rao, unpublished data). In yet another example, NFAT3 is completely localized to the nucleus in resting astrocytes, while NFAT1 is predominantly cytoplasmic in the same cells (A. Benedito, A. Rao, and A. Bonni, unpublished data). Finally, short calcium pulses lead to sustained localization of NFAT3 in the nuclei of hippocampal neurons (19), whereas a similar short calcium pulse in T cells leads only to transient, low-level activation of NFAT1 (14, 15).
If all NFAT proteins are dephosphorylated by calcineurin, what is the basis for the differences in regulation among the NFAT family members? The concept that NFAT is regulated by multiple kinases provides a new insight into the mechanisms of NFAT activation, allowing for individual control of each family member even in the presence of an active phosphatase that is common to the regulation of all. NFAT deactivation by CK1 and GSK3 is likely to be sensitive to changes in a variety of other intracellular signaling pathways. For instance, the activity of GSK3 is suppressed by Akt, a kinase activated in response to diverse signaling pathways in different cell types (13). Additionally, GSK3 prefers targets that have been prephosphorylated by a priming kinase. Thus, after exposure to stimuli that activate calcineurin-mediated dephosphorylation, direct modification of NFAT by an inducible kinase could provide a priming phosphorylation site that would preferentially increase its sensitivity to GSK3 and facilitate rephosphorylation. The rate of NFAT deactivation would therefore be influenced by which inducible kinases were active under the particular stimulation conditions imposed and whether the NFAT proteins available to the kinases possessed the appropriate sequences for docking and target recognition.
An interesting illustration of such a mechanism is provided by mitogen-activated protein (MAP) kinases, which have been reported by several groups to oppose the nuclear accumulation of NFAT (10, 17, 42, 56). MAP kinases can selectively phosphorylate NFAT proteins at the Ser-Pro sequence at the beginning of their SRR-1 regions (see Fig. 7C); JNK1 phosphorylates NFAT2 and NFAT4 (10, 42), while p38 may selectively target NFAT1 and NFAT3 (17, 56). As inducible kinases that phosphorylate only a limited subset of residues in the NFAT regulatory domain, MAP kinases are unlikely to be responsible for maintaining NFAT proteins in a phosphorylated and inactive state in resting T cells. However, phosphorylation of the initial Ser-Pro sequence in the SRR-1 region could promote processive phosphorylation of the entire SRR-1 region by CK1. Although CK1 can clearly phosphorylate NFAT1 without any requirement for priming activity (Fig. 2D), as predicted for a constitutive kinase that maintains NFAT phosphorylation in resting cells, the MAP kinase-mediated mechanism would selectively enhance nuclear export in stimulated cells of those NFAT family members that were specific targets for the MAP kinase in question.
Parallels with other signaling pathways regulated by CK1.
In both the Wnt and Hedgehog signaling pathways, CK1 coordinates with GSK3 to phosphorylate and negatively regulate a transcription factor anchored in the cytoplasm. This echoes the theme of NFAT regulation documented in this study, although in each of these cases the kinases coordinate their activities in a slightly different way. In the case of Wnt signaling, CK1 acts as a priming kinase for GSK3 on the β-catenin transcriptional activator (3, 34). In the Hedgehog pathway, PKA serves as a priming kinase for adjacent GSK3 and CK1 sites on the transcription factor Ci (25, 32, 43). In the Drosophila circadian clock, the PER proteins are phosphorylated by the CK1ɛ homolog Double-time, while the heterodimeric partner of PER, Timeless, is phosphorylated by the GSK3 homolog Shaggy, which in turn may be primed by CK2 (2, 33). In contrast, for NFAT regulation, CK1 acts independently of GSK3 to phosphorylate the conserved SRR-1 motif, while the combination of a priming kinase and GSK3 could phosphorylate the conserved SP motifs to facilitate NFAT nuclear export. Notably, PKA and CK2 could conceivably both serve as priming kinases for GSK3, since they would target the same residues in the SP motifs (42, 50).
Interestingly, the F-X-X-X-F motif, which we have identified as an NFAT docking site for CK1, is also present in proteins regulated by CK1 in the Wnt and Hedgehog pathways (e.g., β-catenin, APC), but whether the motif serves as a docking site for CK1 in these cases has not been determined. The situation is clearest for the PER proteins, which serve as both substrates and direct binding partners for CK1ɛ, and in which the F-X-X-X-F motif is necessary for mediating CK1 association (Fig. 7B). While the presence or absence of an accessible F-X-X-X-F motif may have predictive value in determining whether or not a given protein binds directly to CK1, it is likely that other CK1 binding motifs exist, since proteins such as axin and diversin, which are thought to bind CK1, do not contain a detectable F-X-X-X-F motif.
What coordinates the activities of multiple kinases that function concurrently to control the activity of a single transcription factor sequestered in the cytoplasm? In the canonical Wnt/β-catenin pathway, negative control of the transcriptional activator β-catenin is exerted in the context of a cytoplasmic regulatory complex that includes the scaffolding protein axin, the tumor suppressor APC, the protein phosphatase PP2A, and the CK1 and GSK3 kinases (3, 24, 34, 48, 49, 55). In an analogous manner, it is possible that scaffolding proteins serve to assemble NFAT1 and its regulatory kinases into a multicomponent complex. The high-molecular-weight complex that contains NFAT1 and CK1 in resting T cells does not appear to contain GSK3 or calcineurin (Fig. 4), although we cannot rule out the possibility that these proteins do not remain stably associated under the conditions used. Since the apparent molecular weight of this complex is higher than that expected for NFAT1 and CK1 alone, an intriguing possibility is that adaptor proteins and additional kinase activities are present in this complex. This would be an efficient and sensitive method of finely modulating the phosphorylation status of NFAT in response to different intracellular signals, thereby regulating the subcellular localization, DNA binding, and transcriptional activity of this important transcription factor.
Acknowledgments
This work was supported by NIH grants AI 40127 (A.R.) and GM 60387 (D.M.V.) and by American Cancer Society grant IRG199A, a grant from the L. E. Gordy Cancer Research Fund (J.Q.), and a grant from the Sandler Program for Asthma Research (A.R.). H.O. is a recipient of a Cancer Research Institute Postdoctoral Fellowship and an NIH Program in Cancer Biology training grant (T32 CA72320). C.G.-R. was a recipient of a Lady Tata Memorial Trust Fellowship.
REFERENCES
- 1.Abbott, K. L., B. B. Friday, D. Thaloor, T. J. Murphy, and G. K. Pavlath. 1998. Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol. Biol. Cell 9:2905-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akten, B., E. Jauch, G. K. Genova, E. Y. Kim, I. Edery, T. Raabe, and F. R. Jackson. 2003. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 6:251-257. [DOI] [PubMed] [Google Scholar]
- 3.Amit, S., A. Hatzubai, Y. Birman, J. S. Andersen, E. Ben-Shushan, M. Mann, Y. Ben-Neriah, and I. Alkalay. 2002. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 16:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aramburu, J., F. García-Cózar, A. Raghavan, H. Okamura, A. Rao, and P. G. Hogan. 1998. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell 1:627-637. [DOI] [PubMed] [Google Scholar]
- 5.Beals, C. R., N. A. Clipstone, S. N. Ho, and G. R. Crabtree. 1997. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 11:824-834. [DOI] [PubMed] [Google Scholar]
- 6.Beals, C. R., C. M. Sheridan, C. W. Turck, P. Gardner, and G. R. Crabtree. 1997. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science 275:1930-1933. [DOI] [PubMed] [Google Scholar]
- 7.Brivanlou, A. H., and J. E. Darnell, Jr. 2002. Signal transduction and the control of gene expression. Science 295:813-818. [DOI] [PubMed] [Google Scholar]
- 8.Cegielska, A., K. F. Gietzen, A. Rivers, and D. M. Virshup. 1998. Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J. Biol. Chem. 273:1357-1364. [DOI] [PubMed] [Google Scholar]
- 9.Chijiwa, T., M. Hagiwara, and H. Hidaka. 1989. A newly synthesized selective casein kinase I inhibitor, N-(2-aminoethyl)-5-chloroisoquinoline-8-sulfonamide, and affinity purification of casein kinase I from bovine testis. J. Biol. Chem. 264:4924-4927. [PubMed] [Google Scholar]
- 10.Chow, C. W., M. Rincon, J. Cavanagh, M. Dickens, and R. J. Davis. 1997. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science 278:1638-1641. [DOI] [PubMed] [Google Scholar]
- 11.Crabtree, G. R. 1999. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell 96:611-614. [DOI] [PubMed] [Google Scholar]
- 12.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl.):S67-S79. [DOI] [PubMed] [Google Scholar]
- 13.Cross, D. A., D. R. Alessi, P. Cohen, M. Andjelkovich, and B. A. Hemmings. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378:785-789. [DOI] [PubMed] [Google Scholar]
- 14.Dolmetsch, R. E., K. Xu, and R. S. Lewis. 1998. Calcium oscillations increase the efficiency and specificity of gene expression. Nature 392:933-936. [DOI] [PubMed] [Google Scholar]
- 15.Feske, S., R. Draeger, H. H. Peter, K. Eichmann, and A. Rao. 2000. The duration of nuclear residence of NFAT determines the pattern of cytokine expression in human SCID T cells. J. Immunol. 165:297-305. [DOI] [PubMed] [Google Scholar]
- 16.Flotow, H., P. R. Graves, A. Q. Wang, C. J. Fiol, R. W. Roeske, and P. J. Roach. 1990. Phosphate groups as substrate determinants for casein kinase I action. J. Biol. Chem. 265:14264-14269. [PubMed] [Google Scholar]
- 17.Gomez del Arco, P., S. Martinez-Martinez, J. L. Maldonado, I. Ortega-Perez, and J. M. Redondo. 2000. A role for the p38 MAP kinase pathway in the nuclear shuttling of NFATp. J. Biol. Chem. 275:13872-13878. [DOI] [PubMed] [Google Scholar]
- 18.Graef, I. A., F. Chen, and G. R. Crabtree. 2001. NFAT signaling in vertebrate development. Curr. Opin. Genet. Dev. 11:505-512. [DOI] [PubMed] [Google Scholar]
- 19.Graef, I. A., P. G. Mermelstein, K. Stankunas, J. R. Neilson, K. Deisseroth, R. W. Tsien, and G. R. Crabtree. 1999. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature 401:703-708. [DOI] [PubMed] [Google Scholar]
- 20.Graves, P. R., and P. J. Roach. 1995. Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J. Biol. Chem. 270:21689-21694. [DOI] [PubMed] [Google Scholar]
- 21.Gross, S. D., and R. A. Anderson. 1998. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 10:699-711. [DOI] [PubMed] [Google Scholar]
- 22.Ho, A. M., J. Jain, A. Rao, and P. G. Hogan. 1994. Expression of the transciption factor NFATp in a neuronal cell line and in the murine nervous system. J. Biol. Chem. 269:181-186. [PubMed] [Google Scholar]
- 23.Hogan, P. G., L. Chen, J. Nardone, and A. Rao. 2003. Transcriptional regulation by calcium, calcineurin and NFAT. Genes Dev. 17:2205-2232. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda, S., S. Kishida, H. Yamamoto, H. Murai, S. Koyama, and A. Kikuchi. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and beta-catenin and promotes GSK-3β-dependent phosphorylation of beta-catenin. EMBO J. 17:1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia, J., K. Amanai, G. Wang, J. Tang, B. Wang, and J. Jiang. 2002. Shaggy/GSK3 antagonizes Hedgehog signaling by regulating Cubitus interruptus. Nature 416:548-552. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, E. N., Y. M. Lee, T. L. Sander, E. Rabkin, F. J. Schoen, S. Kaushal, and J. Bischoff. 2003. NFATc1 mediates vascular endothelial growth factor-induced proliferation of human pulmonary valve endothelial cells. J. Biol. Chem. 278:1686-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalderon, D. 2002. Similarities between the Hedgehog and Wnt signaling pathways. Trends Cell Biol. 12:523-531. [DOI] [PubMed] [Google Scholar]
- 28.Kehlenbach, R. H., A. Dickmanns, and L. Gerace. 1998. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J. Cell Biol. 141:868-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiani, A., A. Rao, and J. Aramburu. 2000. Manipulating immune responses with immunosuppressive agents that target NFAT. Immunity 12:359-372. [DOI] [PubMed] [Google Scholar]
- 30.Klein, P. S., and D. A. Melton. 1996. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA 93:8455-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komeili, A., and E. K. O'Shea. 2001. New perspectives on nuclear transport. Annu. Rev. Genet. 35:341-364. [DOI] [PubMed] [Google Scholar]
- 32.Lefers, M. A., and R. Holmgren. 2002. Ci proteolysis: regulation by a constellation of phosphorylation sites. Curr. Biol. 12:R422-R423. [DOI] [PubMed] [Google Scholar]
- 33.Lin, J. M., V. L. Kilman, K. Keegan, B. Paddock, M. Emery-Le, M. Rosbash, and R. Allada. 2002. A role for casein kinase 2 alpha in the Drosophila circadian clock. Nature 420:816-820. [DOI] [PubMed] [Google Scholar]
- 34.Liu, C., Y. Li, M. Semenov, C. Han, G. H. Baeg, Y. Tan, Z. Zhang, X. Lin, and X. He. 2002. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108:837-847. [DOI] [PubMed] [Google Scholar]
- 35.Loh, C., J. A. Carew, J. Kim, P. G. Hogan, and A. Rao. 1996. T-cell receptor stimulation elicits an early phase of activation and a later phase of deactivation of the transcription factor NFAT1. Mol. Cell. Biol. 16:3945-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo, C., E. Burgeon, J. A. Carew, P. G. McCaffrey, T. M. Badalian, W. S. Lane, P. G. Hogan, and A. Rao. 1996. Recombinant NFAT1 (NFATp) is regulated by calcineurin in T cells and mediates transcription of several cytokine genes. Mol. Cell. Biol. 16:3955-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, C., K. T. Shaw, A. Raghavan, J. Aramburu, F. Garcia-Cozar, B. A. Perrino, P. G. Hogan, and A. Rao. 1996. Interaction of calcineurin with a domain of the transcription factor NFAT1 that controls nuclear import. Proc. Natl. Acad. Sci. USA 93:8907-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massari, M. E., P. A. Grant, M. G. Pray-Grant, S. L. Berger, J. L. Workman, and C. Murre. 1999. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell 4:63-73. [DOI] [PubMed] [Google Scholar]
- 39.Meggio, F., J. W. Perich, E. C. Reynolds, and L. A. Pinna. 1991. A synthetic beta-casein phosphopeptide and analogues as model substrates for casein kinase-1, a ubiquitous, phosphate directed protein kinase. FEBS Lett. 283:303-306. [DOI] [PubMed] [Google Scholar]
- 40.Neal, J. W., and N. A. Clipstone. 2001. Glycogen synthase kinase-3 inhibits the DNA binding activity of NFATc. J. Biol. Chem. 276:3666-3673. [DOI] [PubMed] [Google Scholar]
- 41.Okamura, H., J. Aramburu, C. Garcia-Rodriguez, J. P. Viola, A. Raghavan, M. Tahiliani, X. Zhang, J. Qin, P. G. Hogan, and A. Rao. 2000. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell 6:539-550. [DOI] [PubMed] [Google Scholar]
- 42.Porter, C. M., M. A. Havens, and N. A. Clipstone. 2000. Identification of amino acid residues and protein kinases involved in the regulation of NFATc subcellular localization. J. Biol. Chem. 275:3543-3551. [DOI] [PubMed] [Google Scholar]
- 43.Price, M. A., and D. Kalderon. 2002. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by glycogen synthase kinase 3 and casein kinase 1. Cell 108:823-835. [DOI] [PubMed] [Google Scholar]
- 44.Rao, A., S. J. Faas, and H. Cantor. 1984. Activation specificity of arsonate-reactive T cell clones: structural requirements for hapten recognition and comparison with monoclonal antibodies. J. Exp. Med. 159:479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao, A., C. Luo, and P. G. Hogan. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707-747. [DOI] [PubMed] [Google Scholar]
- 46.Rivers, A., K. F. Gietzen, E. Vielhaber, and D. M. Virshup. 1998. Regulation of casein kinase I epsilon and casein kinase I delta by an in vivo futile phosphorylation cycle. J. Biol. Chem. 273:15980-15984. [DOI] [PubMed] [Google Scholar]
- 47.Salazar, C., and T. Höfer. 2003. Allosteric regulation of the transcription factor NFAT1 by multiple phosphorylation sites: a mathematical analysis. J. Mol. Biol. 327:31-45. [DOI] [PubMed] [Google Scholar]
- 48.Schwarz-Romond, T., C. Asbrand, J. Bakkers, M. Kuhl, H. J. Schaeffer, J. Huelsken, J. Behrens, M. Hammerschmidt, and W. Birchmeier. 2002. The ankyrin repeat protein diversin recruits casein kinase Iɛ to the beta-catenin degradation complex and acts in both canonical Wnt and Wnt/JNK signaling. Genes Dev. 16:2073-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seeling, J. M., J. R. Miller, R. Gil, R. T. Moon, R. White, and D. M. Virshup. 1999. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science 283:2089-2091. [DOI] [PubMed] [Google Scholar]
- 50.Sheridan, C. M., E. K. Heist, C. R. Beals, G. R. Crabtree, and P. Gardner. 2002. Protein kinase A negatively modulates the nuclear accumulation of NF-ATc1 by priming for subsequent phosphorylation by glycogen synthase kinase-3. J. Biol. Chem. 277:48664-48676. [DOI] [PubMed] [Google Scholar]
- 51.Toh, K. L., C. R. Jones, Y. He, E. J. Eide, W. A. Hinz, D. M. Virshup, L. J. Ptacek, and Y. H. Fu. 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291:1040-1043. [DOI] [PubMed] [Google Scholar]
- 52.Tomida, T., K. Hirose, A. Takizawa, F. Shibasaki, and M. Iino. 2003. NFAT functions as a working memory of Ca2+ signals in decoding Ca2+ oscillation. EMBO J. 22:3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vielhaber, E., E. Eide, A. Rivers, Z. H. Gao, and D. M. Virshup. 2000. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol. Cell. Biol. 20:4888-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 55.Yanagawa, S., Y. Matsuda, J. S. Lee, H. Matsubayashi, S. Sese, T. Kadowaki, and A. Ishimoto. 2002. Casein kinase I phosphorylates the Armadillo protein and induces its degradation in Drosophila. EMBO J. 21:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, T. T., Q. Xiong, H. Enslen, R. J. Davis, and C. W. Chow. 2002. Phosphorylation of NFATc4 by p38 mitogen-activated protein kinases. Mol. Cell. Biol. 22:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, J., and F. McKeon. 1999. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature 398:256-260. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, J., F. Shibasaki, R. Price, J.-C. Guillemot, T. Yano, V. Dotsch, G. Wagner, P. Ferrara, and F. McKeon. 1998. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell 93:851-861. [DOI] [PubMed] [Google Scholar]