Abstract
Members of the M13 family of zinc metalloendopeptidases have been shown to play critical roles in the metabolism of various neuropeptides and peptide hormones, and they have been identified as important therapeutic targets. Recently, a mouse NL1 protein, a novel member of the family, was identified and shown to be expressed mainly in the testis as a secreted protein. To define its physiological role(s), we used a gene targeting strategy to disrupt the endogenous murine Nl1 gene by homologous recombination and generate Nl1 mutant mice. The Nl1−/− mice were viable and developed normally, suggesting that zygotic expression of Nl1 is not required for development. However, Nl1−/− males produced smaller litters than their wild-type siblings, indicating specific male fertility problems. Reduced fertility may be explained by two impaired processes, decreased egg fertilization and perturbed early development of fertilized eggs. These two phenotypes did not result from gross anatomical modifications of the testis or from impaired spermatogenesis. Basic sperm parameters were also normal. Thus, our findings suggest that one of the roles of NL1 in mice is related to sperm function and that NL1 modulates the processes of fertilization and early embryonic development in vivo.
A wide variety of biologically active peptide hormones, neuropeptides, and regulatory peptides are proteolytically activated or inactivated by zinc metalloproteases (58, 59). The M13 family of zinc metalloproteases attracts much interest because its members are involved in the metabolism of several regulatory peptides of the mammalian nervous, cardiovascular, inflammatory, and immune systems as well as in the mineralization process (8, 56, 57, 59). For these reasons, several members of this family of peptidases have been selected as targets for therapeutic intervention by inhibitors or pharmacological agents.
NL1 (also called SEP or NEP2) is the newest member of the M13 family of zinc metallopeptidases (19, 25, 42), which also includes neprilysin (neutral endopeptidase 24.11; also called NEP and CALLA) (reviewed in reference 11), the endothelin-converting enzymes 1 and 2 (ECE-1 and ECE-2) (reviewed in reference 55), PHEX (formerly PEX; a phosphate regulating gene with homologies to endopeptidases on the X chromosome) (reviewed in reference 54), the Kell blood group protein (reviewed in reference 35), and ECEL1/DINE (endothelin-converting enzyme-like 1/damage-induced neuronal endopeptidase) (29, 60, 61). Nl1 cDNA was simultaneously cloned by reverse transcription-PCR technology with degenerate oligonucleotide primers based on conserved sequence similarity between members of the family and mRNA isolated from mouse and rat testis or from Ece-1−/− embryos (19, 25, 42). Finally, a clone coding for a putative human homologue has been obtained from a central nervous system cDNA library (4).
As observed for other members of the M13 family of peptidases, the Nl1 cDNA encodes a type II transmembrane glycoprotein encompassing a short N-terminal cytoplasmic tail, a single transmembrane domain, and a large C-terminal extracellular region that contains the zinc binding motif HEXXH, where X is any amino acid. However, in contrast to the other members, NL1 can be expressed as both a membrane-bound and a soluble protein (25). Differential splicing introduces an additional exon that causes the insertion of a furin-like cleavage site at the C-terminal end of the transmembrane domain and the release of the protein into the extracellular space. Data from reverse transcription-PCR experiments suggest that the secreted form of NL1 is mainly found in the testis, whereas the membrane-bound form is expressed in the remaining expressing sites (25).
NL1 degrades a broad variety of small peptides. Oligopeptides shorter than 3 kDa and containing neutral bulky aliphatic or aromatic amino acid residues are preferred substrates. These characteristics are close to those described for neprilysin. In fact, all peptide substrates identified so far for NL1 are also metabolized by neprilysin (48). Furthermore, for many peptides, both enzymes cleave at the same amide bond. As expected, prototypical transition state inhibitors of neprilysin, such as phosphoramidon and thiorphan, and dual neprilysin-angiotensin-converting enzyme inhibitors such as omapatrilat also inhibit NL1 enzymatic activity (19, 48). NL1 is more closely related to neprilysin, with 55% amino acid identity, than to any other members of the family.
Northern blot analyses with total RNA isolated from a variety of mouse tissues reveal strong expression of the Nl1 gene in the testis, while other tissues are negative (19). By in situ hybridization, Nl1 mRNA is detected in round and elongated spermatids in seminiferous tubules of the mature testis at all spermiogenesis maturational stages, suggesting that the predominant role of NL1 in the mouse may be related to sperm functions (19). Nevertheless, with the more sensitive reverse transcription-PCR technology, weak expression of Nl1 mRNA is also detected in several other tissues, such as heart, spleen, lungs, liver, kidney, intestine, and brain (19, 25). In situ hybridization experiments on mouse sections reveal the presence of Nl1 mRNA in neurons of the ventroposterior regions of the brain as well as in the anterior and intermediate lobes of the pituitary gland (16).
Although NL1 enzymatic activity and tissue expression has been studied extensively, no physiological function has yet been identified for this protein. To gain more insight into its role(s) in mammals, we disrupted the Nl1 gene by homologous recombination and produced Nl1-deficient mice. Here, we report the reduced fertility of Nl1−/− mutant male mice.
MATERIALS AND METHODS
Gene targeting.
The genomic organization of the Nl1 gene was characterized by PCR amplification as done previously (6). Briefly, several pairs of overlapping primers (Table 1) spanning the entire Nl1 cDNA sequence were used to amplify mouse 129/SvEv genomic DNA. The resulting PCR products were cloned, and the intron-exon junctions were sequenced with the Sanger dideoxynucleotide chain termination method (50). To construct the targeting vector, two genomic fragments were PCR amplified. The 5.6-kb 5′-end fragment and the 4.1-kb 3′-end fragment were amplified with the primer pairs 5′-AGTGACATCTGTCGACCACCACCCCAAGCTG-3′ (sense; SalI restriction site in italics) and 5′-ATGCGTCTCCAGTTCCAGCACCTC-3′ (antisense) and 5′-CCAGGAGAAAAGGCTCGAGATGATGTCACTGC-3′ (sense) and 5′-CCAAAATGTAGTCTCGAGAGGGTAGCCAATC-3′ (antisense; XhoI restriction site in italics), respectively. The 5′-end fragment was digested with SalI and HindIII (which removes a 729-bp fragment at the 3′ end of the amplification product), whereas the 3′-end fragment was digested with XhoI. The genomic fragments were cloned on each side of the neomycin selection cassette in the PGKneo vector. A counterselection herpes simplex virus thymidine kinase cassette was inserted at the 3′ end of the homology region of the targeting vector (2, 34).
TABLE 1.
Oligonucleotide sequences used for PCR amplification of genomic DNA
| Primer pair | Sense primer | Reverse primer |
|---|---|---|
| 1 | 5′-CAGCACCGTACCTCACACCC-3′ | 5′-CTGGTCTCTGCTCTCAATGCTG-3′ |
| 2 | 5′-TCTTAACTAGCCTGCTACACTTCTCC-3′ | 5′-TGCTATCACACAGCTTGGGGTG-3′ |
| 3 | 5′-TGCACCACCCCAAGCTGTGTGATAGC-3′ | 5′-TGGGATCACGTGGTGCCTCAGCC-3′ |
| 4 | 5′-TCGAGGAACCCCTGTGAAAACTTCTACC-3′ | 5′-TCGATGAGGACCCGCCTGTTGAACTG-3′ |
| 5 | 5′-AGTTCACCAGGCGGGTCCTCATCG-3′ | 5′-AGATGCGACTCCAGTTCCAGCACCTCC-3′ |
| 6 | 5′-ACTATGCTTAGGAAAGACCAGAACC-3′ | 5′-TCATCTGGCAACAGCTCGACTTCC-3′ |
| 7 | 5′-TTGTCTTCTGTGGAAGTCGAGCTGTTCC-3′ | 5′-TTGACATAGCTGACACACTCTCGCC-3′ |
| 8 | 5′-TGGCGAGAGTGTGTCAGCTATGTCAAC-3′ | 5′-TCTTCCAAAATGTAGTCAGGGTAGC-3′ |
| 9 | 5′-TGGCTACCCTGACTACATTTTGGAAG-3′ | 5′-AGGCTCCTCTGGGCATTGTTCTTG-3′ |
| 10 | 5′-ACTTCAGAACCTCAAGAACAATGC-3′ | 5′-TAGCCCCCAAAATTCAAGGAC-3′ |
| 11 | 5′-ATTCTCCAGCCGCCCTTCTTCAGC-3′ | 5′-ACCTCCGTTGTCGGCAATGTTCTCC-3′ |
| 12 | 5′-AGAACATTGCCGACAACGGAGG-3′ | 5′-TCCGTCTTGATGGACTGGACGG-3′ |
| 13 | 5′-AGTTCGCCGTCCAGTCCATCAAGACG-3′ | 5′-ACCTGCACATCTCTGCCAGCTAC-3′ |
WW6 embryonic stem (ES) cells were electroporated with 25 μg of SalI-linearized targeting vector DNA as previously described and plated on neomycin-resistant feeder layers (2). After 24 h, selection for G418 (400 μg/ml; Invitrogen, Burlington, Ontario, Canada) and ganciclovir (2 μM; Syntex, Palo Alto, Calif.) was applied. ES cell clones recovered from drug selection were screened by Southern analysis. Positive clones were injected into MF1 blastocysts and transferred into pseudopregnant foster mothers to generate chimeras that were tested for germ line transmission as previously described (2, 9, 20, 34).
Genotyping of ES cells and mice.
Genomic DNA was isolated from ES cells or mouse tail biopsies. Extracts were treated with proteinase K, followed by phenol-chloroform extraction and ethanol precipitation (24). Purified DNA was digested with restriction enzymes as indicated in Fig. 1, fractioned by electrophoresis through 0.8% agarose gels, blotted onto a Hybond-NX membrane (Amersham Biosciences, Baie d'Urfé, Québec, Canada), and hybridized by following the procedures recommended by the supplier for either the 5′ Nl1 probe (the first 733 bp of the Nl1 cDNA sequence, nucleotides 332 to 1064 of GenBank accession number AF176569) or the 3′ Nl1 probe (the last 1,000 bp of the Nl1 cDNA, nucleotides 1925 to 2925 of GenBank accession number AF176569).
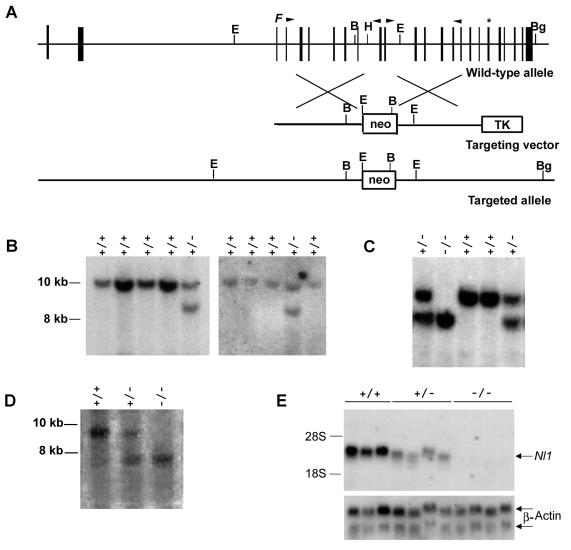
FIG. 1.
Disruption of the Nl1 gene. (A) Schematic representation of the wild-type mouse Nl1 gene, the targeting vector, and the targeted Nl1 allele. Vertical black boxes represent exons. The positions of the neomycin resistance gene (neo) and the herpes simplex virus thymidine kinase gene (TK) are shown. The neo cassette is flanked at its 5′ and 3′ ends by 4.9 and 4.1 kb of Nl1 genomic sequence, respectively. F indicates the position of the convertase cleavage site, and the asterisk shows the position of the NL1 active site. B, BamHI; Bg, BglII; H, HindIII; E, EcoRI. Arrowheads indicate the positions of oligonucleotide primers used to PCR amplify genomic DNA to generate the vector. (B and C) Southern blot analyses of DNA isolated from ES cell clones (B) and mouse tails (C). Genomic DNA was digested with BglII and BamHI and transferred to nylon membranes, and the blots were hybridized with an Nl1 probe located 3′ of the targeting vector sequence (described in Materials and Methods). Fragments from the wild-type (+/+; 9.6 kb) and mutated (−/−; 8.1 kb) alleles were detected. (D) Southern blot analysis of genomic DNA digested with EcoRI, transferred to nylon membranes, and hybridized with an Nl1 probe located 5′ of the targeting vector sequence (described in Materials and Methods). Fragments from the wild-type (+/+; 8.9 kb) and mutated (−/−; 7.3 kb) alleles were detected. (E) Northern blot analysis of total RNA (12.5 μg) isolated from the testes of three wild-type, four Nl1+/−, and four Nl1−/− mice. RNA was hybridized with either the 3′-derived Nl1 or actin probe.
Northern blot analysis.
Total RNA from testes was isolated with the RNeasy column kit (QIAGEN, Chatsworth, Calif.). Equal amounts (12.5 μg) of total RNA isolated from NlI+/+, Nl1+/−, and Nl1−/− specimens were electrophoresed on a 1.5% formaldehyde agarose gel, transferred onto a Hybond N+ blotting membrane, and hybridized with the 32P-labeled 3′ cDNA probe described above. The 645-bp β-actin fragment corresponding to nucleotides 333 to 978 of the coding sequence (GenBank accession number NM_007393) was used as a quantitative control. Samples from three Nl1+/+, four Nl1+/−, and four Nl1−/− adult mice aged between 3 and 6 months were analyzed.
Histology.
Testes were fixed in 4% paraformaldehyde for 16 h at 4°C and embedded in paraffin wax as previously described (27). Specimens were sectioned at 5 μm and stained with hematoxylin and eosin for general histological examination. Sections from four wild-type and five Nl1−/− males aged from 3 to 7 months were analyzed.
Sperm motility analysis.
Spermatozoa were collected from the caudal epididymides of six wild-type and six Nl1−/− males in HMB fertilization medium (62) and incubated for 30 min in 5% CO2-90% O2-air. Motility parameters were measured by computer-assisted analysis with a Ceros Analyzer (software version 12.0F; Hamilton-Thorn Research, Beverly, Mass.). The following parameters were measured: (i) average path velocity (velocity along the average path of the spermatozoon), (ii) straight-line velocity (calculated from the straight-line distance between the start and the end of the observed track), (iii) curvilinear velocity (the total distance divided by time), (iv) lateral head displacement (the deviation of the head from the mean head trajectory), (v) beat cross frequency (beats per second, the number of times the spermatozoon head crosses the head trajectory per seconds), (vi) straightness (calculated as straight-line velocity/average path velocity × 100), (vii) linearity (calculated as straight-line velocity/curved-line velocity × 100). The settings for the Ceros analyzer were 30 frames captured for analysis at 60 Hz (frame rate, 0.50), minimum contrast 30, and minimum cell size 4 pixels; progressive cells were defined as having an average path velocity of >50 μm/s and straightness of >50%, and slow cells were defined as having an average path velocity cutoff of 10 μm/s.
Blastocyst analysis.
F1 Nl1+/− females (aged 2 to 5 months) were mated with wild-type or mutant males (aged 2 to 5 months). Three days following the detection of a copulatory plug, females were sacrificed and embryos were recovered from uterine horns in phosphate-buffered saline. Embryos were counted and photographed for microscopic analysis. A total of five wild-type and five Nl1−/− males were mated with nine and 11 females, respectively.
In vivo fertilization analysis.
Thirty-two C57BL/6 × CBA F1 females aged between 24 and 28 days were superovulated by intraperitoneal injection of 5 U of pregnant mare’s serum (Intervet, Whitby, Ontario, Canada) at 1:00 p.m., followed by 10 U of human chorionic gonadotropin (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) 46 h later. The evening after the second injection, superovulated females were housed with single Nl1+/+ or Nl1−/− males. The next morning, the males were removed and mating was confirmed by detection of a copulatory plug. At 0.75 day postcoitus, the females were killed and the embryos were collected from the ampulla of the oviduct in M2 medium (24) and digested with 300 μg of hyaluronidase (embryo tested; Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) per ml. Embryos were first washed twice in M2 medium and then twice in M16 medium before incubation in M16 medium at 37°C in 5% CO2 for 4 days. Embryo development was monitored daily by microscopic inspection. Litters from the mating of 18 females with five different wild-type males and from 14 females with five different Nl1−/− mutant males were recovered. Males of both genotypes were age matched.
RESULTS
Generation of Nl1 mutant allele in ES cells.
As a first step toward the generation of a targeting vector for Nl1 genetic elimination, the structure of the murine Nl1 gene was defined. Based on the high conservation of the genomic organization of the members of the neprilysin family, genomic DNA was PCR amplified with several primer pairs derived from the Nl1 cDNA sequence. The resulting PCR products were sequenced, and the intron-exon junctions were determined (Table 2). The mouse Nl1 gene is divided into 24 small exons distributed over at least 27 kb of genomic DNA (Fig. 1A). The size of exons varied from 60 to 391 bp, whereas the size of introns ranged from 75 bp to over 8 kb. The ATG initiation start site was located in the second exon and the protein convertase cleavage site was in the third exon, whereas the HEITH motif, the signature of zinc metallopeptidases, was found in exon 19. The third exon is alternatively spliced in the membrane-bound form of NL1 (46). The positions of the intron-exon junctions were mostly conserved between the Nl1 and Neprilysin genes (12).
TABLE 2.
Exon-intron boundaries of the murine Nl1 gene
| Exon no. | Intron 3′ acceptor site | Exon
|
Intron 5′ donor site | Intron size (kb) | ||
|---|---|---|---|---|---|---|
| 5′ end | Size (kb) | 3′ end | ||||
| 1 | Undetermined | Undetermined | >0.141 | GCACCAG | GTAGGATT. . . | 1.7 |
| 2 | …CCCTCCTACACAG | GGTCCCC | 0.315 | AGCATAG | GTGAGCAAGGA. . . | >8 |
| 3a | …TTTCTCCCCAG | GGAAGCA | 0.069 | AAACGAG | GTAGCCTTTACC. . . | 0.55 |
| 4 | …CTGTTCCTCCAG | CCCTCAG | 0.060 | ATAGCAG | GTGAGCTTCTCC. . . | 0.84 |
| 5 | …CCTTAATACAAAG | CTGCCAG | 0.161 | CTCAAAG | GTGAAGAGTGG. . . | 0.28 |
| 6 | …CTGCCCACAG | GGGTGCT | 0.081 | AACCAAA | GTGAGTAGAGCC. . . | 1.24 |
| 7 | …CCCCTTCCCAG | GTGTGAT | 0.096 | ACCATGG | GTGAGTATCGG. . . | 0.46 |
| 8 | …AACTCCTCAG | GCCTCAA | 0.119 | TCATCTAC | GTATGCAATGCCCT. . . | 0.52 |
| 9 | …GTGTGTTTCCAG | ATAGACC | 0.065 | CCACAAG | GTAAGCAGGG. . . | 1.18 |
| 10 | …CCTTTGGCCCAG | GTACGGA | 0.075 | GGCCAAC | GTGAGGCGGGG. . . | 0.07 |
| 11 | …CCCCACACAG | GCCACAG | 0.090 | TCTGAAG | GTGAGACCCAGGCA. . . | 1.66 |
| 12 | …ACCCCTCTACAG | GGGTTTA | 0.137 | CAGCACG | GTGAGCCCAGGG. . . | 0.28 |
| 13 | …GCTACCTCTAG | GACCATG | 0.094 | CCGCAAG | GTGAGCGCAT. . . | 0.81 |
| 14 | …AACACACCAG | GCGCTGT | 0.129 | GAGCACG | GTGCGTATCCTTGA. . . | 0.51 |
| 15 | …GCATTCCCCCAG | GTCAGAG | 0.099 | GGAAAAG | GTGGGCAAGCC. . . | 0.36 |
| 16 | …CCTCCACTCAG | GCCATGA | 0.084 | CTCCAGT | GTGCGTACCTTCC. . . | 0.39 |
| 17 | …CTTCTCATAG | TTGACTT | 0.104 | AGAATCT | GTGAGTAGGATGG. . . | 0.30 |
| 18 | …ATTTACCACAG | CTGGATC | 0.059 | CAGATCG | GTAAGGTCCACC. . . | 0.34 |
| 19 | …GTGGGGGACAG | TCTTTCC | 0.120 | GATAATG | GTAAAGCAGCA. . . | 0.48 |
| 20 | …TCCTGCACAG | GTGAACG | 0.134 | CCAGAAT | GTGAGCATGTC. . . | 0.11 |
| 21 | …TCTTTTTTATGCAG | GTGAACG | 0.066 | ATACAAG | GTGGGCCCTA. . . | 0.46 |
| 22 | …TGCTCGCAGAG | GCTTACC | 0.096 | TGCCCAG | GTAATCCCTAG. . . | 0.27 |
| 23 | …ATGCATCTCCAG | GTGTGGT | 0.077 | AGTACAG | GCAAGTGTGC. . . | 0.09 |
| 24 | …TGGTCTACAG | GGTGCTG | ≥0.391 | Undetermined | ||
| Consensus | (y)12NCAGb | N | AG | GT | ||
Exon 3 is differentially spliced to generate the membrane-bound and secreted forms of NL1.
by, pyrimidine nucleotide; N, any nucleotide.
To construct the targeting vector designed to eliminate the mouse Nl1 gene in ES cells by homologous recombination, two noncontiguous DNA fragments from the Nl1 gene were PCR amplified and cloned on each side of the PGKneo cassette. A cDNA cassette coding for herpes simplex virus thymidine kinase was included at the 3′ end of the targeting vector for negative selection against random integration of the vector. The Nl1 5′-end (4.9-kb) and 3′-end (4.1-kb) fragments encompass regions that include exons 4 to 9 and exons 11 to 16, respectively. An intervening region of 823 bp was omitted between the 5′ and 3′ fragments. Therefore, integration of the vector into the Nl1 gene locus by homologous recombination deletes exon 10 and part of exon 11, modifying the open reading frame of the gene.
The vector was electroporated into ES cells, and positive and negative selections were applied to obtain clones that were resistant to G418 and insensitive to ganciclovir. Homologous recombination events were detected by Southern blot analyses. Genomic DNA from all recovered clones was digested with BglII and BamHI restriction enzymes, and membranes were hybridized with a probe generated from the Nl1 cDNA sequence flanking the 3′-end fragment of the targeting vector (Fig. 1B). Digestion of wild-type and Nl1 mutant alleles generated fragments of 9.6 and 8.1 kb, respectively. A targeting event was detected in 2 of 480 ES cell clones analyzed.
Generation of Nl1 mutant mice and Mendelian distribution of offspring.
Targeting ES cell clones were injected into mouse MF1 blastocysts, and chimeric transmitters were obtained. Chimeric males were bred either with MF1 females or with 129/SvEv females to establish the mutation in an inbred background. All subsequent phenotypic analyses were done with animals from the outbred background. Heterozygous mice developed normally and showed no obvious abnormalities. To determine the effect of the Nl1 mutation in homozygous animals, heterozygous intercrosses were performed and offspring were analyzed. Progeny from heterozygous intercrosses were genotyped, and Nl1−/− mice were recovered (Fig. 1C). Table 3 shows that the incidence of Nl1+/+, Nl1+/−, and Nl1−/− genotypes corresponds to a Mendelian distribution, as expected for random allele segregation and inheritance.
TABLE 3.
Nl1 genotypes of the F2 progeny from F1 heterozygote intercrosses
| Genotype | No. (%) of F2 mice per genotype | Expected Mendelian values (%) |
|---|---|---|
| +/+ | 20 (20.6) | 25 |
| +/− | 55 (56.7) | 50 |
| −/− | 22 (22.7) | 25 |
Mice were further genotyped to test whether integration at the 5′ end of the neo cassette also occurred by homologous recombination (Fig. 1D). Digestion of genomic DNA with EcoRI enzyme and hybridization of the membrane with a 5′ probe generated fragments of 8.9 kb for the wild-type allele and 7.3 kb for the mutated allele in Nl1+/− mice but only the 7.3-kb fragment in Nl1−/− mice. Altogether, these results indicate that the vector was integrated by homologous recombination and exclude any gross rearrangement at either end of the targeting site.
As a means to monitor Nl1 expression in mutant mice, Northern blot analyses were performed with RNA isolated from the testis. As expected, no Nl1 mRNA was detected in Nl1−/− specimens (Fig. 1E). The levels of Nl1 mRNA were reduced by about half in heterozygous mice compared to Nl1 expression in wild-type mice (Fig. 1E). These results clearly demonstrated that no Nl1 expression occurs in Nl1−/− mice, indicating that the mutation generated is a null allele.
Nl1−/− male mice show reduced fertility.
Mutant Nl1−/− mice did not exhibit obvious physical or behavioral abnormalities. Therefore, since NL1 is predominantly expressed in the round and elongated spermatids, we first tested whether disruption of the Nl1 gene impairs male fertility. The fertility of Nl1+/+, Nl1+/−, and Nl1−/− male and female mice was compared with different combinations of intercrosses (Table 4). First, heterozygous Nl1+/− females were crossed with wild-type, heterozygous, or homozygous mutant males. Wild-type males produced an average of 8.9 pups per litter, while Nl1−/− males generated only 4.7 pups per litter. Intercrosses with Nl1+/− males produced an intermediate average of 7.4 pups per litter. The differences observed in litter sizes produced by Nl1−/− males compared to those produced by wild-type and Nl1+/− males were statistically significant (Table 4). About 20% of the Nl1−/− males did not produce any progeny when mated with Nl1+/− females, even if they were housed together for several months. These males were not included in the calculation of the average litter size in Table 4. These results suggest that the loss of Nl1 function results in male hypofertility.
TABLE 4.
Reduced fertility of male Nl1−/− mice
| Parents (female × male) | No. of productive matings | Avg litter size (no. of pups) |
|---|---|---|
| +/− × +/+ | 18 | 8.9 |
| +/− × +/− | 20 | 7.85 |
| +/− × −/− | 21 | 4.7a |
| +/+ × +/− | 9 | 7.44 |
| −/− × +/− | 11 | 7.45 |
| +/+ × +/+ | 7 | 10.1 |
P = 0.0004 for variance between −/− and +/+ males and P = 0.0393 for variance between −/− and +/− males (one-way analysis of variance).
We also tested female fertility. Wild-type, heterozygous, and homozygous mutant females were bred with heterozygous Nl1+/− males. The average litter size obtained for matings between Nl1+/− males and Nl1−/− or Nl1+/+ females was 7.4 and 7.5, respectively (Table 4), which was equivalent to the values for heterozygous male and female intercrosses, indicating that fertility is not affected in female mice. The small difference in the litter size between wild-type breedings (10.1) and crosses between wild-type males and Nl1+/− females (8.9) was not statistically significant (P = 0.9348).
Testis development and spermatozoa are normal.
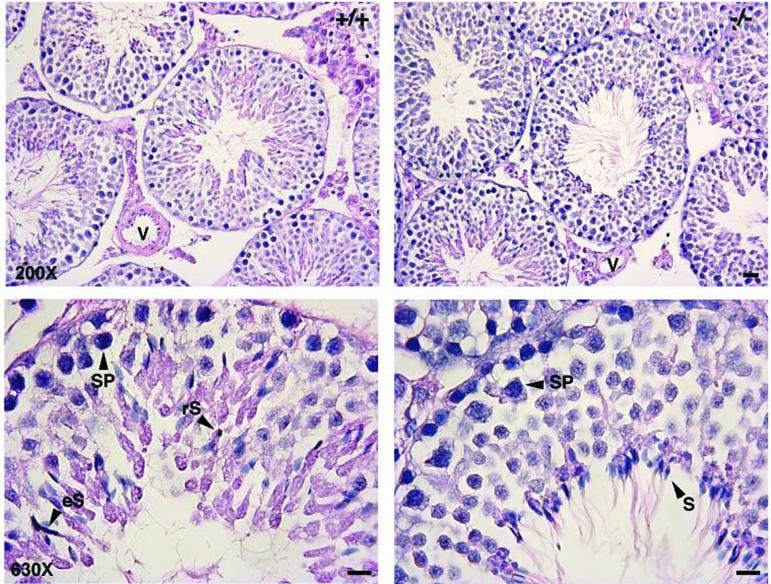
To determine whether the reduced fertility of Nl1−/− mice resulted from a spermatogenic defect, a microscopic analysis of the seminiferous tubules was conducted. Testis sections from four wild-type and five Nl1−/− males showing various stages of the spermatogenic cycle were stained with hematoxylin and eosin. No major difference was observed, and the morphology of the tubules as well as the relative numbers of spermatogonia, spermatocytes, and round and elongated spermatids were similar between control and Nl1−/− mice (Fig. 2). The histological characteristics of testes from mutant males that were determined to be infertile from the absence of progeny were also normal. Overall, the testis morphology of Nl1−/− mice was similar to that of wild-type littermates.
FIG. 2.
Histological analysis of testes from Nl1−/− and Nl1+/+ mice. Hematoxylin and eosin staining of thin sections from control (+/+) and Nl1 mutant (−/−) mice was performed. Arrowheads indicate round spermatid (rS), elongated spematids (eS), pachytene spermatocytes (SP), and spermatozoa (S). V, blood vessel. Bar, 10 μm.
We also compared the motility parameters of spermatozoa collected from wild-type and Nl1−/− males. Although most of the motility parameters of Nl1−/− spermatozoa were slightly lower than those of Nl1+/+ spermatozoa, the difference was not statistically significant (Table 5).
TABLE 5.
Comparison of sperm from wild-type and Nl1−/− micea
| Parameter | Mean value ±SD for mice with genotype:
|
P | |
|---|---|---|---|
| +/+ | −/− | ||
| Motile sperm (%) | 87.2 ± 6.66 | 78.3 ± 10.01 | 0.1184 |
| Progressively motile (%) | 40.5 ± 3.2 | 33.2 ± 3.2 | 0.1368 |
| Avg path velocity (μm/s) | 100.1 ± 4.6 | 86.8 ± 7.6 | 0.1642 |
| Straight-line velocity (μm/s) | 64.2 ± 3.6 | 55.5 ± 4.3 | 0.155 |
| Curved-line velocity (μm/s) | 186.9 ± 7.2 | 161.3 ± 12.9 | 0.1147 |
| Lateral head displacement (μm) | 9.8 ± 0.09 | 9.4 ± 0.2 | 0.0903 |
| Beat cross frequency (beat/s) | 24.3 ± 0.9 | 25.1 ± 1.2 | 0.6081 |
| Straightness | 58.8 ± 0.7 | 60.2 ± 0.8 | 0.2368 |
| Linearity | 32.7 ± 0.8 | 33.0 ± 0.4 | 0.7342 |
| Total count | 1109 ± 198 | 828 ± 181 | 0.3203 |
Seven mice of each genotype were studied; six fields per mouse were analyzed. The parameters examined are defined in Materials and Methods.
Nl1 mutation reduces both fertilizing capacity and early embryonic development.
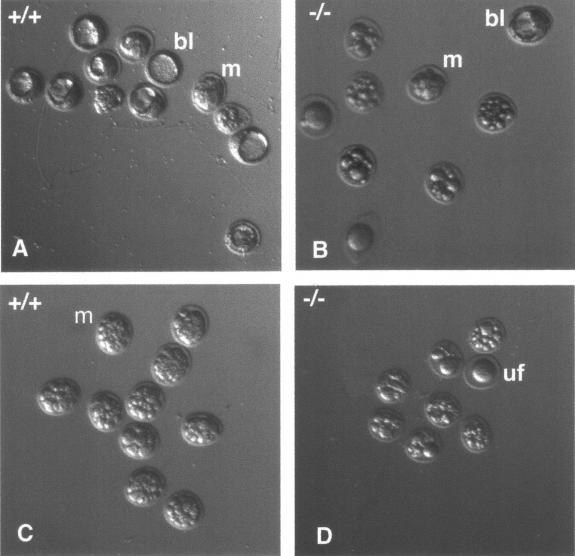
No morphological anomaly appears to be responsible for the reduced litter size phenotype observed with Nl1−/− males. To determine if defects in sperm fertilization ability and/or early embryonic development underlie the phenotype, outbred F1 Nl1+/− females were mated with either wild-type or Nl1−/− males, and embryos were collected at 3.5 days postcoitum. Microscopic examinations revealed that 74% of the embryos issued from crosses with Nl1+/+ males were in the late morula or blastocyst stage, compared to 33% of those fertilized by Nl1−/− males (Table 6; Fig. 3). This significant difference (chi square test, P < 0.0001) was very close to that observed in the litter size, suggesting that early development is perturbed when the sperm originates from Nl1−/− mice.
TABLE 6.
Fertilizing capacity of Nl1−/− males and early embryo development
| Genotype | No.a | No. of eggs harvested | No. (%) of embryos at 3.5 days after mating at development stageb:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | M | B | Dead | |||
| +/+ | 9 | 88 | 1 (1%) | 2 (2%) | 1 (1%) | 4 (5%) | 22 (25%) | 43 (49%) | 15 (17%) |
| −/− | 11 | 80 | 3 (4%) | 3 (4%) | 3 (5%) | 0 | 14 (17%) | 13 (16%) | 44 (54%) |
Total number of mated Nl1+/− females.
Each column represents the number of embryos for each developing stage: 1, 2, 3, 4 cells, morulae (M), and blastocyst (B) collected at 3.5 days postconception. Chi square test, P < 0.0001.
FIG. 3.
Embryo recovery at day 3.5 of gestation. Four representative litters from Nl1+/− females fertilized by Nl1+/+ (A and C) or Nl1−/− (B and D) males are shown. Morulae (m), blastocyst (bl), and unfertilized (uf) eggs are indicated. A larger proportion of embryos were well developed when fertilized by Nl1+/+ males compared to those fertilized by Nl1−/− males. A total of five wild-type or mutant males were mated with 9 and 11 females, respectively. Embryos were collected at 3.5 days postconception. Magnification, 50×.
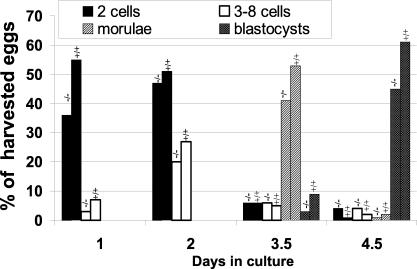
To determine whether fertilization was normal and define at which embryonic stage development was impaired, a time course experiment was performed. Superovulated C57BL/6 × CBA F1 virgin females were mated with either wild-type or Nl1−/− males. Embryos were collected around 0.75 day postcoitum and their development was pursued in culture for 4 days. Embryo development was monitored by daily microscopic observations. Around 1.0 day postcoitum, 62% of the embryos collected from matings with wild-type males were fertilized and had divided. In contrast, only 38% of the embryos obtained from matings with Nl1−/− males had reached cell division (Fig. 4). These results indicated that the fertilization rate was reduced by 39% when the sperm originated from Nl1−/− males. However, 2 days after fertilization, the Nl1−/− sperm fertilization ability was only decreased by 15% compared to wild-type sperm, suggesting that, in addition to fertilization defects, the absence of NL1 causes a delay in the first cell division(s). At 3.5 and 4.5 days postcoitum, the number of morulae and blastocysts generated following fertilization with Nl1−/− sperm was 30% lower than that obtained with wild-type sperm. These results suggest that the Nl1 mutation reduces the ability of fertilized eggs to develop normally. Differences in the distribution of embryos resulting from Nl1−/− and NL1+/+ males mated with Nl1+/− females were statistically significant at each day of development (Fig. 4). We cannot rule out the additional possibility that early processes of preimplantation development may also be perturbed by the mutation. Altogether, these anomalies explain the reduced litter size observed in matings with Nl1−/− males, since the differences closely reflect the variation detected in the litter size.
FIG. 4.
In vivo fertilization analysis. C57BL/6 × CBA F1 females were superovulated and mated with Nl1+/+ or Nl1−/− males. Embryos were collected at day 1 postconception and cultured in vitro for 4 days. Embryo development was monitored by daily microscopic analysis. Bars labeled −/− indicate eggs fertilized by Nl1−/− males, while bars labeled +/+ correspond to eggs fertilized by wild-type males. Chi square (χ2) and Jonkheere-Terpstra (JT) analyses revealed that the distribution of developing embryos was statistically different at each day (day 1: χ2, P < 0.0001, and JT, P < 0.0001; day 2, χ2, P < 0.0024, and JT, P < 0.0003; day 3.5, χ2, P < 0.0001, and JT, P < 0.0001; day 4.5, χ2, P < 0.0001, and JT, P < 0.0001).
DISCUSSION
In this study, we disrupted the Nl1 gene by homologous recombination and generated Nl1−/− mice. The mutation prevents the synthesis of both the transmembrane and secreted forms of NL1 protein. Although our anti-NL1 antibodies did not allow us to directly test whether the protein was present or not in the mutant mice, we demonstrated that Nl1 mRNA was not expressed in testes from Nl1−/− mice, supporting the notion that the protein is not produced. This result is in concordance with the fact that the designed targeting vector adds a new polyadenylation site after exon 9, interrupting transcription of the Nl1 gene. Matings of heterozygous mice produced the expected Mendelian ratio of wild-type, heterozygous, and homozygous mutant mice, and Nl1−/− mice were viable and anatomically normal. Thus, zygotic expression of Nl1 does not seem to be required for mouse development. The data are consistent with our recent in situ hybridization studies showing that Nl1 mRNA is not expressed in embryonic day 12 to 17 mouse embryos (7). Targeted disruption of other members of the M13 family has been reported. While elimination of the Neprilysin gene results in only a mild phenotype, disruption of the ECE-1, ECEL1, or Phex gene causes severe developmental defects (33, 49, 51, 63, 64).
We showed that NL1 plays a significant role in male fertility but is not essential for the fertility of females. This result is consistent with the expression profile of Nl1 mRNA in mouse tissues, which is strong in germ cells of the mature testis but weak in ovaries (25). The expression of the fertility impairment among Nl1−/− males was variable. For instance, approximately 20% of the Nl1−/− outbred males did not produce offspring. In contrast, about 10% of the Nl1−/− outbred males had litters approximating wild-type numbers. The variable phenotype in Nl1−/− males is not unprecedented and most probably reflects the segregation of genetic modifiers on the mosaic genetic background, as was shown for at least two other mutations in sperm (39, 43). For instance, male mice bearing mutations in the sprm-1 transcription factor gene or in the smcp mitochondrial protein gene are sterile only if these mutations are maintained in the 129/Sv genetic background (15, 43). Moreover, males deficient in the sperm protein SED1 show a similar variation in litter size (ranging from no pups to litter sizes approaching normal) in an outbred background (15). Testing this hypothesis, however, must await the characterization of the Nl1−/− male fertility phenotype in different inbred genetic backgrounds.
NL1 modulates two different processes related to fertility.
Our results suggest a role for NL1 in two developmental processes, egg fertilization and early embryo development. First, spermatozoa from Nl1−/− mice appeared to be less efficient than those produced by Nl1+/+ mice for egg fertilization despite normal sperm motility parameters and no major morphological defect of the testis. The fact that NL1 is mostly expressed as a secreted protein in spermatids is consistent with a role in a step that occurs outside of the testis. Accordingly, a neprilysin-like activity has been detected in the sperm of several species (3, 13, 23, 52), although such an activity has not been confirmed by others (10, 17). Generation of specific inhibitors and/or antibodies will be required to determine whether this activity is due to the presence of NL1. It is thus possible that the Nl1 phenotype is linked to defects in sperm maturation or capacitation or sperm-related processes during fertilization.
Several proteases are known to modulate the different steps of sperm maturation that lead to fertilization (31). Some of these are released during the acrosome reaction and play important roles in the fertilization process itself (31, 53). Consistently, several bioactive peptides are either coexpressed with NL1 in testicular germ cells, packaged in the sperm acrosome, and/or released during the acrosome reaction (1, 5, 22, 28). Among them, the enkephalins, which have been shown to be stored in the acrosome of human, hamster, and sheep spermatozoa and released following the acrosome reaction (28), decrease mouse fertility when overexpressed in the testis of transgenic mice (41). These results support the notion that the levels of neuropeptides are critical for proper fertilization processes. Since some of these bioactive peptides are known substrates of NL1 in vitro, the Nl1 phenotype could be partly explained by a lack of their regulation in vivo.
Second, a large proportion of eggs did not develop normally when fertilized with the Nl1−/− mutant spermatozoa. We must emphasize that defects in embryo development are not likely to be due to the lack of zygotic Nl1 expression, since we did not observe selective elimination of Nl1−/− offspring. The phenotype appears to be associated with the absence of NL1 in sperm. A growing body of evidences supports an important role(s) for spermatozoa and/or its components in early embryonic development (44, 45, 65). Therefore, through its action on active peptides, it is possible that NL1 may regulate sperm-dependent spatial patterning of the early mouse embryo, a sperm-activated block to polyspermy, and/or proteolytic activation of morphogens.
NL1 is downstream of the protein convertase PC4.
Cleavage and release into the extracellular fluids of the secreted testicular form of NL1 is accomplished by a member(s) of the prohormone convertase family (19). In the testis, the nature of the prohormone convertase that cleaves NL1 remains to be defined. However, a good candidate is the prohormone convertase PC4 because it is abundantly and specifically expressed in the spermatocytes and round spermatids of rodents (38). Moreover, PC4 homozygous mutant males show reduced fertility in the absence of any evidence of spermatogenic abnormality (36), similar to what we observed in Nl1-deficient mice. Interestingly, the reduced fertility of Pc4−/− males is due to a combination of both reduced efficiency in egg fertilization and abnormal embryo development after fertilization. Therefore, it is possible that some of the phenotypes attributed to Pc4 gene elimination are directly related to the failure of Pc4-deficient mice to properly process NL1, the lack of secreted NL1 protein in Pc4−/− mice causing the phenotypes. Analysis of NL1 maturation in Pc4−/− mice would help in answering this question.
The Pc4 phenotype is broader than that observed in Nl1−/− mice, suggesting that NL1 acts downstream of PC4. For instance, the hypofertility observed in males is more important in Pc4 mutants than in Nl1 mutant mice. Second, Pc4−/− females are less fertile, while the fertility of Nl1−/− females appears normal. Finally, in intercrosses of heterozygous mutant mice, the recovery of Pc4−/− mutant animals was reduced, conversely to what was observed for transmission of the Nl1 mutation. These observations indicate that NL1 is not the only substrate for PC4 and that the absence of maturation of another bioactive polypeptide(s) may be responsible for the additional phenotypes detected in Pc4 mutants. Alternatively, the differences can be explained by variations in the genetic background in which the two studies were conducted (C57BL/6J versus outbred).
Putative functional compensation by other peptidases.
The relatively mild phenotype observed after genetic elimination of the Nl1 gene suggests that other peptidases may compensate for the loss of NL1. A strong candidate is neprilysin, since NL1 and neprilysin have overlapping sites of expression and similar enzymatic properties and degrade the same set of peptides (32). It may not be surprising then that mutant mice lacking Neprilysin function are viable and appear developmentally normal except for some minor effects on lymphoid development. Therefore, it is possible that the NL1 and neprilysin enzymes have redundant functions and that the expression of one enzyme can compensate for the absence of the other in mutant animals. The production of compound Nl1 Neprilysin mutants should test this hypothesis.
Although neprilysin has been shown to be expressed in the testis, Neprilysin−/− mice have no reported fertility problems. Since neprilysin is membrane bound, it might compensate for the lack of some of the membrane-bound NL1-related functions in the testis of Nl1−/− mice, but it might not rescue the phenotypes associated with loss of the secreted form of NL1 protein. In other tissues, NL1 and neprilysin are both believed to be membrane-bound proteins. In the brain, their sites of expression are rather complementary, although many regions expressed both enzymes (7, 16). It will be interesting to study the effect of NL1 deficiency on brain function. For example, injection of thiorphan in the rat brain region that controls nociception causes analgesia resulting from enhanced extracellular concentration of enkephalins (47). This effect was believed to be due to the pharmacological inhibition of neprilysin activity. However, Neprilysin mutant animals were unexpectedly reported to exhibit hyperalgesia in the hot plate and tail withdrawal tests (18). This suggests that neprilysin may not be involved in this function and/or that other enzymes closely related to neprilysin, such as NL1, may be involved in the analgesic effect of thiorphan.
Other zinc proteases are known to be involved in processes related to fertility (10, 13, 14, 40). Any of these peptidases could play a role in the partial rescue of the NL1 phenotype. For example, the testicular form of the angiotensin-converting enzyme could be a good candidate because it is specifically expressed in developing spermatids, and angiotensin-converting enzyme mutant mice show reduced fertility (21). Furthermore, NL1 and angiotensin-converting enzyme have overlapping substrate specificities and hydrolyze several common peptides in vitro. Other peptidases of the M13 family of metallopeptidases such as Phex, ECE-1, ECE-2, and ECEL1 have also been detected in the testis (26, 30, 37, 60), but their role in male fertility has not yet been established.
The production of Nl1 null mice constitutes an important step in the understanding of the physiological importance of NL1 in mammals. NL1 deficiency has been proven to impair normal fertilization processes. Further investigation will be necessary to precisely determine the molecular basis of the fertility defect, at the level both of the fertilization processes and of the early steps in embryonic development. Moreover, these basic investigations could be clinically relevant because the human homolog of NL1, MMEL2, has recently been identified (4). Therefore, mutations in this gene may be responsible for some of the fertility problems found in humans. Future studies evaluating the importance of MMEL2 in human reproduction processes will contribute to our understanding of the multiple factors involved in male fertility.
Acknowledgments
We thank Marcelle Carter and Margot Lemieux for technical support and François Harel for performing the statistical analyses.
This work was supported by grants MT-13052 (to L.D.G. and G.B.) and MOP-42523 (to J.C.) from the Canadian Institutes for Health Research of Canada (CIHR). M.C. was supported by studentships from the National Sciences and Engineering Research Council of Canada (NSERC) and the Fonds pour la Formation des Chercheurs et l'Aide à la Recherche (FCAR). L.J. holds a Chercheur National award from the Fonds de la Recherche en Santé du Québec (FRSQ), and J.C. is a Senior Scholar from the FRSQ.
REFERENCES
- 1.Ayer-Lelievre, C., L. Olson, T. Ebendal, F. Hallbook, and H. Persson. 1988. Nerve growth factor mRNA and protein in the testis and epididymis of mouse and rat. Proc. Natl. Acad. Sci. USA 85:2628-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bélanger, L. F., S. Roy, M. Tremblay, B. Brott, A. M. Steff, W. Mourad, P. Hugo, R. Erikson, and J. Charron. 2003. Mek2 is dispensable for mouse growth and development. Mol. Cell. Biol. 23:4778-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boettger, A., U. Kertscher, C. Steinmann, U. Baeger, W. E. Siems, and G. Heder. 1993. Degradation of bradykinin in semen of ram and boar. Biochem. Pharmacol. 45:1983-1988. [DOI] [PubMed] [Google Scholar]
- 4.Bonvouloir, N., N. Lemieux, P. Crine, G. Boileau, and L. DesGroseillers. 2001. Molecular cloning, tissue distribution, and chromosomal localization of MMEL2, a gene coding for a novel human member of the neutral endopeptidase-24.11 family. DNA Cell Biol. 20:493-498. [DOI] [PubMed] [Google Scholar]
- 5.Breyer, P. R., J. K. Rothrock, N. Beaudry, and O. H. Pescovitz. 1996. A novel peptide from the growth hormone releasing hormone gene stimulates Sertoli cell activity. Endocrinology 137:2159-2162. [DOI] [PubMed] [Google Scholar]
- 6.Brizard, F., M. Luo, and L. DesGroseillers. 2000. Genomic organization of the human and mouse stau genes. DNA Cell Biol. 19:331-339. [DOI] [PubMed] [Google Scholar]
- 7.Carpentier, M., M. Marcinkiewicz, G. Boileau, and L. DesGroseillers. 2003. The neuropeptide-degrading enzyme NL1 is expressed in specific neurons of mouse brain. Peptides 24:1083-1091. [DOI] [PubMed] [Google Scholar]
- 8.Carson, J. A., and A. J. Turner. 2002. Beta-amyloid catabolism: roles for neprilysin (NEP) and other metallopeptidases? J. Neurochem. 81:1-8. [DOI] [PubMed] [Google Scholar]
- 9.Charron, J., B. A. Malynn, P. Fisher, V. Stewart, L. Jeannotte, S. P. Goff, E. J. Robertson, and F. W. Alt. 1992. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 6:2248-2257. [DOI] [PubMed] [Google Scholar]
- 10.Correa, L. M., C. Cho, D. G. Myles, and P. Primakoff. 2000. A role for a TIMP-3-sensitive, Zn2+-dependent metalloprotease in mammalian gamete membrane fusion. Dev. Biol. 225:124-134. [DOI] [PubMed] [Google Scholar]
- 11.Crine, P., N. Dion, and G. Boileau. 1997. Endopeptidase 24:11, p. 79-98. In A. J. Kenny and C. M. Boustead (ed.), Cell-surface peptidases in health and disease. M. BIOS Scientific Publisher Ltd., Oxford, United Kingdom.
- 12.D'Adamio, L., M. A. Shipp, E. L. Masteller, and E. L. Reinherz. 1989. Organization of the gene encoding common acute lymphoblastic leukemia antigen (neutral endopeptidase 24.11): multiple miniexons and separate 5′ untranslated regions. Proc. Natl. Acad. Sci. USA 86:7103-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz-Perez, E., and S. Meizel. 1992. Importance of mammalian sperm metalloendoprotease activity during the acrosome reaction to subsequent sperm-egg fusion: inhibitor studies with human sperm and zona-free hamster eggs. Mol. Reprod. Dev. 31:122-130. [DOI] [PubMed] [Google Scholar]
- 14.Diaz-Perez, E., P. Thomas, and S. Meizel. 1988. Evidence suggesting a role for sperm metalloendoprotease activity in penetration of zona-free hamster eggs by human sperm. J. Exp. Zool. 248:213-221. [DOI] [PubMed] [Google Scholar]
- 15.Ensslin, M. A., and B. D. Shur. 2003. Identification of mouse sperm SED1, a bimotif EGF repeat and discoidin-domain protein involved in sperm-egg binding. Cell 114:405-417. [DOI] [PubMed] [Google Scholar]
- 16.Facchinetti, P., C. Rose, J. C. Schwartz, and T. Ouimet. 2003. Ontogeny, regional and cellular distribution of the novel metalloprotease neprilysin 2 in the rat: a comparison with neprilysin and endothelin-converting enzyme-1. Neuroscience. 118:627-639. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez, D., A. Valdivia, J. Irazusta, C. Ochoa, and L. Casis. 2002. Peptidase activities in human semen. Peptides 23:461-468. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, H. S., G. Zernig, K. F. Hauser, C. Gerard, L. B. Hersh, and A. Saria. 2002. Neutral endopeptidase knockout induces hyperalgesia in a model of visceral pain, an effect related to bradykinin and nitric oxide. J. Mol. Neurosci. 18:129-134. [DOI] [PubMed] [Google Scholar]
- 19.Ghaddar, G., A. F. Ruchon, M. Carpentier, M. Marcinkiewicz, N. G. Seidah, P. Crine, L. DesGroseillers, and G. Boileau. 2000. Molecular cloning and biochemical characterization of a new mouse testis soluble-zinc-metallopeptidase of the neprilysin family. Biochem. J. 347:419-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giroux, S., M. Tremblay, D. Bernard, J. F. Cardin-Girard, S. Aubry, L. Larouche, S. Rousseau, J. Huot, J. Landry, L. Jeannotte, and J. Charron. 1999. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr. Biol. 9:369-372. [DOI] [PubMed] [Google Scholar]
- 21.Hagaman, J. R., J. S. Moyer, E. S. Bachman, M. Sibony, P. L. Magyar, J. E. Welch, O. Smithies, J. H. Krege, and D. A. O'Brien. 1998. Angiotensin-converting enzyme and male fertility. Proc. Natl. Acad. Sci. USA 95:2552-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannibal, J., and J. Fahrenkrug. 1995. Expression of pituitary adenylate cyclase activating polypeptide (PACAP) gene by rat spermatogenic cells. Regul. Pept. 55:111-115. [DOI] [PubMed] [Google Scholar]
- 23.Heder, G., A. Bottger, W. E. Siems, M. Rottmann, and U. Kertscher. 1994. The enzymatic degradation of bradykinin in semen of various species. Andrologia 26:295-301. [DOI] [PubMed] [Google Scholar]
- 24.Hogan, B., F. Constantini, and E. Lacy. 1986. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Ikeda, K., N. Emoto, S. B. Raharjo, Y. Nurhantari, K. Saiki, M. Yokoyama, and M. Matsuo. 1999. Molecular identification and characterization of novel membrane-bound metalloprotease, the soluble secreted form of which hydrolyzes a variety of vasoactive peptides. J. Biol. Chem. 274:32469-32477. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda, S., N. Emoto, H. Alimsardjono, M. Yokoyama, and M. Matsuo. 2002. Molecular isolation and characterization of novel four subisoforms of ECE-2. Biochem. Biophys. Res. Commun. 293:421-426. [DOI] [PubMed] [Google Scholar]
- 27.Jaffe, L., L. Jeannotte, E. K. Bikoff, and E. J. Robertson. 1990. Analysis of beta 2-microglobulin gene expression in the developing mouse embryo and placenta. J. Immunol. 145:3474-3482. [PubMed] [Google Scholar]
- 28.Kew, D., K. E. Muffly, and D. L. Kilpatrick. 1990. Proenkephalin products are stored in the sperm acrosome and may function in fertilization. Proc. Natl. Acad. Sci. USA 87:9143-9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiryu-Seo, S., M. Sasaki, H. Yokohama, S. Nakagomi, T. Hirayama, S. Aoki, K. Wada, and H. Kiyama. 2000. Damage-induced neuronal endopeptidase (DINE) is a unique metallopeptidase expressed in response to neuronal damage and activates superoxide scavengers. Proc. Natl. Acad. Sci. USA 97:4345-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korth, P., R. M. Bohle, P. Corvol, and F. Pinet. 1999. Cellular distribution of endothelin-converting enzyme-1 in human tissues. J. Histochem. Cytochem. 47:447-462. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S. L., and Y. H. Wei. 1994. The involvement of extracellular proteinases and proteinase inhibitors in mammalian fertilization. Biotechnol. Appl. Biochem. 19:31-40. [PubMed] [Google Scholar]
- 32.Li, C., R. M. Booze, and L. B. Hersh. 1995. Tissue-specific expression of rat neutral endopeptidase (neprilysin) mRNAs. J. Biol. Chem. 270:5723-5728. [DOI] [PubMed] [Google Scholar]
- 33.Lu, B., N. P. Gerard, L. F. Kolakowski, O. Finco, M. C. Carroll, and C. Gerard. 1996. Neutral endopeptidase modulates septic shock. Ann. N. Y. Acad. Sci. 780:156-163. [DOI] [PubMed] [Google Scholar]
- 34.Ma, S., J. Charron, and R. L. Erikson. 2003. Role of Plk2 (Snk) in mouse development and cell proliferation. Mol. Cell. Biol. 23:6936-6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsh, W. L. 1992. Molecular biology of blood groups: cloning the Kell gene. Transfusion 32:98-101. [DOI] [PubMed] [Google Scholar]
- 36.Mbikay, M., H. Tadros, N. Ishida, C. P. Lerner, E. De Lamirande, A. Chen, M. El-Alfy, Y. Clermont, N. G. Seidah, M. Chrétien, C. Gagnon, and E. M. Simpson. 1997. Impaired fertility in mice deficient for the testicular germ-cell protease PC4. Proc. Natl. Acad. Sci. USA 94:6842-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer, M. H., and R. A. Meyer, Jr. 2000. mRNA expression of Phex in mice and rats: the effect of low phosphate diet. Endocrine 13:81-87. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama, K., W. S. Kim, S. Torii, M. Hosaka, T. Nakagawa, J. Ikemizu, T. Baba, and K. Murakami. 1992. Identification of the fourth member of the mammalian endoprotease family homologous to the yeast Kex2 protease. Its testis-specific expression. J. Biol. Chem. 267:5897-5900. [PubMed] [Google Scholar]
- 39.Nayernia, K., I. M. Adham, E. Burkhardt-Gottges, J. Neesen, M. Rieche, S. Wolf, U. Sancken, K. Kleene, and W. Engel. 2002. Asthenozoospermia in mice with targeted deletion of the sperm mitochondrion-associated cysteine-rich protein (Smcp) gene. Mol. Cell. Biol. 22:3046-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimura, H., C. Cho, D. R. Branciforte, D. G. Myles, and P. Primakoff. 2001. Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin beta. Dev. Biol. 233:204-213. [DOI] [PubMed] [Google Scholar]
- 41.O'Hara, B. F., D. M. Donovan, I. Lindberg, M. T. Brannock, D. D. Ricker, C. A. Moffatt, B. A. Klaunberg, C. Schindler, T. S. Chang, R. J. Nelson, and G. R. Uhl. 1994. Proenkephalin transgenic mice: a short promoter confers high testis expression and reduced fertility. Mol. Reprod. Dev. 38:275-284. [DOI] [PubMed] [Google Scholar]
- 42.Ouimet, T., P. Facchinetti, C. Rose, M. C. Bonhomme, C. Gros, and J. C. Schwartz. 2000. Neprilysin II: a putative novel metalloprotease and its isoforms in CNS and testis. Biochem. Biophys. Res. Commun. 271:565-570. [DOI] [PubMed] [Google Scholar]
- 43.Pearse, R. V., D. W. Drolet, K. A. Kalla, F. Hooshmand, J. R. Bermingham, Jr., and M. G. Rosenfeld. 1997. Reduced fertility in mice deficient for the POU protein sperm-1. Proc. Natl. Acad. Sci. USA 94:7555-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piotrowska, K., and M. Zernicka-Goetz. 2001. Role for sperm in spatial patterning of the early mouse embryo. Nature 409:517-521. [DOI] [PubMed] [Google Scholar]
- 45.Piotrowska, K., and M. Zernicka-Goetz. 2002. Early patterning of the mouse embryo—contributions of sperm and egg. Development 129:5803-5813. [DOI] [PubMed] [Google Scholar]
- 46.Raharjo, S. B., N. Emoto, K. Ikeda, R. Sato, M. Yokoyama, and M. Matsuo. 2001. Alternative splicing regulates the endoplasmic reticulum localization or secretion of soluble secreted endopeptidase. J. Biol. Chem. 276:25612-25620. [DOI] [PubMed] [Google Scholar]
- 47.Roques, B. P., M. C. Fournie-Zaluski, E. Soroca, J. M. Lecomte, B. Malfroy, C. Llorens, and J. C. Schwartz. 1980. The enkephalinase inhibitor thiorphan shows antinociceptive activity in mice. Nature 288:286-288. [DOI] [PubMed] [Google Scholar]
- 48.Rose, C., S. Voisin, C. Gros, J. C. Schwartz, and T. Ouimet. 2002. Cell-specific activity of neprilysin 2 isoforms and enzymic specificity compared with neprilysin. Biochem. J. 363:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowe, P. S. 1998. The role of the PHEX gene (PEX) in families with X-linked hypophosphataemic rickets. Curr. Opin. Nephrol. Hypertens. 7:367-376. [DOI] [PubMed] [Google Scholar]
- 50.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schweizer, A., O. Valdenaire, A. Koster, Y. Lang, G. Schmitt, B. Lenz, H. Bluethmann, and J. Rohrer. 1999. Neonatal lethality in mice deficient in XCE, a novel member of the endothelin-converting enzyme and neutral endopeptidase family. J. Biol. Chem. 274:20450-20456. [DOI] [PubMed] [Google Scholar]
- 52.Siems, W. E., B. Maul, B. Wiesner, M. Becker, T. Walther, L. Rothe, and A. Winkler. 2003. Effects of kinins on mammalian spermatozoa and the impact of peptidolytic enzymes. Andrologia 35:44-54. [DOI] [PubMed] [Google Scholar]
- 53.Snell, W. J., and J. M. White. 1996. The molecules of mammalian fertilization. Cell 85:629-637. [DOI] [PubMed] [Google Scholar]
- 54.Tenenhouse, H. S. 1999. X-linked hypophosphataemia: a homologous disorder in humans and mice. Nephrol. Dial. Transplant. 14:333-341. [DOI] [PubMed] [Google Scholar]
- 55.Turner, A. J. 1997. Endothelin converting enzymes, p. 137-153. In A. J. Kenny and C. M. Boustead (ed.), Cell-surface peptidases in health and disease. BIOS Scientific Publishers Ltd., Oxford, United Kingdom.
- 56.Turner, A. J., C. D. Brown, J. A. Carson, and K. Barnes. 2000. The neprilysin family in health and disease. Adv. Exp. Med. Biol. 477:229-240. [DOI] [PubMed] [Google Scholar]
- 57.Turner, A. J., R. E. Isaac, and D. Coates. 2001. The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays 23:261-269. [DOI] [PubMed] [Google Scholar]
- 58.Turner, A. J., R. Matsas, and A. J. Kenny. 1985. Are there neuropeptide-specific peptidases? Biochem. Pharmacol. 34:1347-1356. [DOI] [PubMed] [Google Scholar]
- 59.Turner, A. J., and K. Tanzawa. 1997. Mammalian membrane metallopeptidases: NEP, ECE, KELL, and PEX. FASEB J. 11:355-364. [DOI] [PubMed] [Google Scholar]
- 60.Valdenaire, O., J. G. Richards, R. L. Faull, and A. Schweizer. 1999. XCE, a new member of the endothelin-converting enzyme and neutral endopeptidase family, is preferentially expressed in the CNS. Brain Res. Mol. Brain Res. 64:211-221. [DOI] [PubMed] [Google Scholar]
- 61.Valdenaire, O., and A. Schweizer. 2000. Endothelin-converting enzyme-like 1 (ECEL1; ′XCE'): a putative metallopeptidase crucially involved in the nervous control of respiration. Biochem. Soc. Trans. 28:426-430. [PubMed] [Google Scholar]
- 62.Visconti, P. E., G. D. Moore, J. L. Bailey, P. Leclerc, S. A. Connors, D. Pan, P. Olds-Clarke, and G. S. Kopf. 1995. Capacitation of mouse spermatozoa. II. Protein tyrosine phosphorylation and capacitation are regulated by a cAMP-dependent pathway. Development 121:1139-1150. [DOI] [PubMed] [Google Scholar]
- 63.Yanagisawa, H., R. E. Hammer, J. A. Richardson, N. Emoto, S. C. Williams, S. Takeda, D. E. Clouthier, and M. Yanagisawa. 2000. Disruption of ECE-1 and ECE-2 reveals a role for endothelin-converting enzyme-2 in murine cardiac development. J. Clin. Investig. 105:1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yanagisawa, H., M. Yanagisawa, R. P. Kapur, J. A. Richardson, S. C. Williams, D. E. Clouthier, D. de Wit, N. Emoto, and R. E. Hammer. 1998. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development 125:825-836. [DOI] [PubMed] [Google Scholar]
- 65.Zernicka-Goetz, M. 2002. Patterning of the embryo: the first spatial decisions in the life of a mouse. Development 129:815-829. [DOI] [PubMed] [Google Scholar]