Abstract
This review provides an overview of diterpene synthases which were initially identified via genetic and/or biochemical means, traversing all organisms researched to date.
Keywords: Biosynthesis, cDNA, Cyclase, Diterpenoid, Eriocalyxin B
Introduction
Naturally occurring cyclic diterpenoids are the foundation for a number of biologically important compounds, which confer anti-microbial and anti-tumor action i.e. tanshinones, Taxol, and platensimycin. They are generally derived from the linear primary metabolite (E,E,E)-geranylgeranyl diphosphate (GGPP) by diterpene synthases (diTPS). These enzymes undergo complex electrophilic cyclizations and/or rearrangements leading to diverse backbone structures. According to the established general patterns of diterpene biosynthesis, diTPS can be divided into two mechanistically distinct categories: class I and class II enzymes. Structural and mechanistic studies have demonstrated that class I diTPS contain a characteristic aspartate (D)-rich DDXXD motif that binds divalent metal ions required for the catalysis of diphosphate ionization. On the other hand, class II diTPS exhibit a typical DXDD motif which is abundant in aspartate, that confers a role in initiating cyclization by protonating the terminal C=C double bond of GGPP [1, 2]. Usually, the class II diTPS yield terpene diphosphates that can serve as substrates of class I diTPS. Highly reactive carbocation intermediates arise from the cleavage of diphosphate group on the substrate by class I diTPS that end the cyclization cascade, by quenching the carbocation via deprotonation or electrophilic attack by water. A significant amount of cyclic diterpenoids are known to originate biosynthetically from dual cyclization and/or rearrangement reactions, which proceed via a bicyclic diphosphate intermediate [2]. In the past decades, there has been some remarkable progress in the research of biosynthetic genes and enzymes responsible for scaffold cyclization to diterpenoids in plants, fungi, bacteria, and even animals. This review attempts to describe diTPS cloned as cDNAs and functionally characterized in all organisms researched to date, and further focusing on those first identified genetically and/or biochemically. Information regarding chemical structures of enzyme products and their relevant diterpenoid natural products will be listed in tables according to hydrocarbon backbones of the natural diterpenoids, in parallel with organisms and enzyme accession numbers released on GenBank. The same enzyme relevant to a different diterpenoid product is specifically included here.
Enzymes for Bicyclic Hydrocarbon Skeletons (Table 1)
Table 1.
Enzymes for bicyclic hydrocarbon skeletons
| Genus species (organisms), enzymes/GenBank accession numbers | Enzyme products | Natural diterpenoids |
|---|---|---|
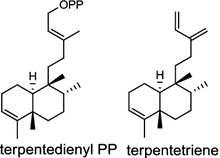
| Kitasatospora griseola (bacterium) [3, 4]; terpentedienyl diphosphate synthase/BAB39206; terpentetriene synthase/BAB39207 |
|
|
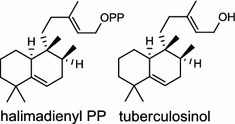
| Mycobacterium tuberculosis (bacterium) [5–7]; halimadienyl (tuberculosinyl) diphosphate synthase/CCP46198 (Rv3377c); tuberculosinol and isotuberculosinol synthase/CCP46199 (Rv3378c) |
|
|
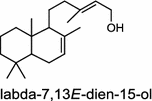
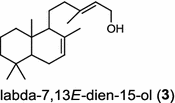
| Selaginella moellendorffii (lycophyte) [8]; labda-7,13E-dien-15-ol synthase (bifunctional)/AEK75338 |
|
|
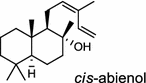
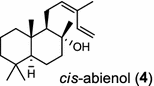
| Abies balsamea (gymnosperm) [9]; cis-abienol synthase (bifunctional)/AEL99953 |
|
|
| Nicotiana tabacum (dicot) [10]; labda-13E-8-ol diphosphate synthase/CCD33018; cis-abienol synthase/CCD33019 |
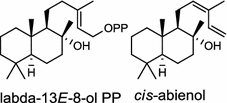
|
|
| Cistus creticus subsp. creticus (dicot) [11]; labda-13E-8-ol diphosphate synthase/ADJ93862 | labda-13E-8-ol PP |
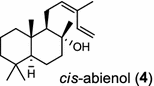
|
| Salvia sclarea (dicot) [12]; labda-13E-8-ol diphosphate synthase/AFU61897; sclareol and manool synthase/AFU61898 |
labda-13E-8-ol PP sclareol |
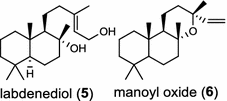
|
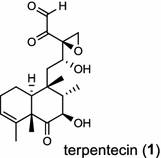
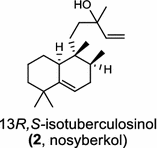
The first diTPS of prokaryotic origin was reported in Streptomyces griseolosporeus MF730-N6 (currently referred to as Kitasatospora griseola), a diterpenoid anti-biotic terpentecin (1) producer. Using the general diterpenoid precursor GGPP as its substrate, the class II terpentedienyl diphosphate synthase produces a bicyclic terpentedienyl diphosphate (PP) which is then transformed to terpentetriene by the class I terpentetriene synthase. These two enzymes are involved in terpentecin (1) biosynthesis [3, 4]. In Mycobacterium tuberculosis H37, a bacterium causing fatal infections of tuberculosis, via the bicyclization of GGPP by the class II Rv3377c, produces halimadienyl (tuberculosinyl) PP that undergoes subsequent ionization-dependent re-arrangement catalyzed by the class I Rv3378c to give tuberculosinol {5(6),13-halimadiene-15-ol} and isotuberculosinol (2, also known as nosyberkol) [5–7]. Interestingly, the two diTPS genes are specifically found in the virulent species, but not in the avirulent species [7].
The basal vascular land plant lycophyte Selaginella moellendorffii contains a class I/II bi-functional enzyme labda-7,13E-dien-15-ol synthase that directly generates the endocyclic double bond and the hydroxyl group to form the bicyclic product labda-7,13E-dien-15-ol (3) [8]. The labdanoid diterpene alcohol cis-abienol (4) is a major component of the aromatic oleoresin of the gymnosperm balsam fir (Abies balsamea) and serves as a valuable bioproduct material for the fragrance industry. A bi-functional class I/II cis-abienol synthase (AbCAS) was discovered using high-throughput 454 transcriptome sequencing and metabolite profiling of balsam fir bark tissue [9]. AbCAS-catalyzed formation of cis-abienol (4) proceeds via cyclization and hydroxylation at carbon C-8 of a postulated carbocation intermediate in the class II active site, followed by cleavage of the diphosphate group and termination of the reaction sequence without further cyclization in the class I active site [9]. In the leaves of the angiosperm tobacco (Nicotiana tabacum) there are two consecutive reactions catalyzed by separate diTPS which lead to the production of cis-abienol (4). Accordingly, a labda-13E-8-ol PP synthase (NtCPS2) encodes a class II diterpene synthase that produces labda-13E-8-ol PP from GGPP; NtABS encodes a class I diterpene synthase that uses labda-13E-8-ol PP to form cis-abienol (4) [10].
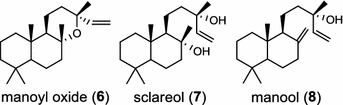
Another labda-13E-8-ol PP synthase from Cistus creticus subsp. creticus was found to catalyze cyclization of GGPP to labda-13E-8-ol (copal-8-ol) PP, a likely intermediate in the biosynthesis of the oxygen-containing labdane-type diterpenes labdenediol (5) and manoyl oxide (6) abundant in the resin of this plant [11]. The bicyclic sclareol (7) extracted from cultivated clary sage (Salvia sclarea) is a diterpene natural product of high value for the fragrance industry and a valued starting material for semi-synthesis of numerous commercial substances, including production of Ambrox®. Similar to the labdanoid alcohols in two dicot plants mentioned above, sclareol (7) and manool (8) were produced in a two-step process mediated sequentially by labda-13E-8-ol PP synthase and sclareol/manool synthase [12]. Only a few diTPS generate the hydroxylated diterpenoids without requirement for additional oxidation by enzymes like cytochrome P450 monooxygenases.
Enzymes for Tricyclic Hydrocarbon Skeletons (Table 2)
Table 2.
Enzymes for tricyclic hydrocarbon skeletons. (Color table online)
| Genus species (organisms), enzymes/GenBank accession numbers | Enzyme products | Natural diterpenoids |
|---|---|---|
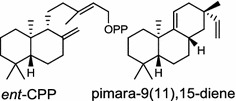
| Streptomyces sp. (bacterium) [13, 14]; ent-CPP synthase/BAD86797; pimara-9(11),15-diene synthase/BAD86798 |
|
|
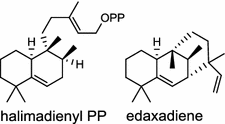
| Mycobacterium tuberculosis (bacterium) [15]; halimadienyl diphosphate synthase/CCP46198 (Rv3377c); edaxadiene synthase/CCP46199 (Rv3378c) |
|
|
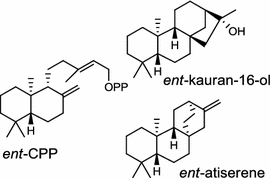
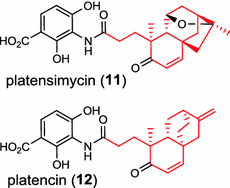
| Streptomyces platensis (bacterium) [16, 17]; ent-CPP synthases/ACO31276 (PtmT2), ADD83015 (PtnT2); ent-kaurene synthase/ACO31279 (PtmT3); ent-atiserene synthase/ACO31274 (PtmT1), ADD83014 (PtnT1); Diterpene moieties are highlighted in red |
|
|
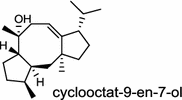
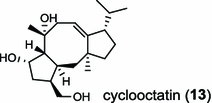
| Streptomyces melanosporofaciens (bacterium) [18]; cyclooctat-9-en-7-ol synthase/BAI44338 (CotB2) |
|
|
| Phomopsis amygdali (fungus) [19]; fusicoccadiene synthase/BAF45924 |
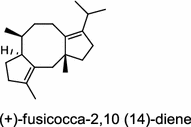
|
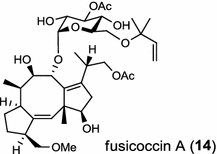
|
| Alternaria brassicicola (fungus) [20]; fusicoccadiene synthase/BAI44849 |
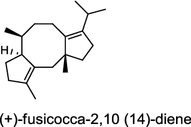
|
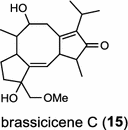
|
| Abies grandis (gymnosperm) [21]; abietadiene synthase/AAB05407 |
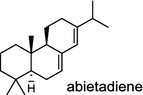
|
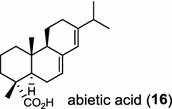
|
| Picea abies (gymnosperm) [22, 23]; isopimara-7,15-diene synthase/AAS47690; levopimaradiene/abietadiene synthase/AAS47691 |
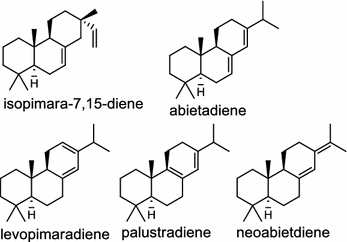
|
diterpene resin acids (rosin) |
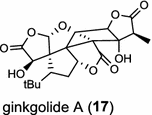
| Ginkgo biloba (gymnosperm) [24]; levopimaradiene synthase/AAL09965 |
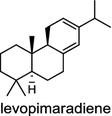
|
|
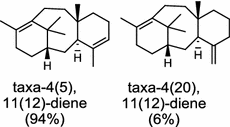
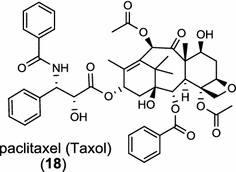
| Taxus brevifolia (gymnosperm) [25, 26]; taxadiene synthase/AAC49310 |
|
|
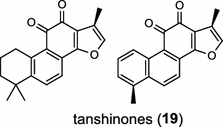
| Salvia miltiorrhiza Bunge (dicot) [27]; CPP synthase/ABV57835; miltiradiene synthase/ABV08817 |

|
|
| Oryza sativa (monocot) [28–30]; ent-CPP synthase/AAT11021 (OsCPS2), BAD42452 (OsCyc2); syn-CPP synthase/BAD42451 (OsCyc1), AAS98158 (OsCPS4) |
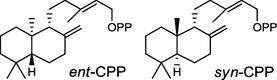
|
phytoalexins |
|
Oryza sativa (monocot) [31, 32] ent-cassa-12,15-diene synthase/BAC56714 (OsDTC1), ABH10734 (OsKSL7); A: R1=R2=H, R3=OH, R4=R5=O B: R1=OH, R2=H, R3=OH, R4=H, R5=OH C: R1=OH, R2=R3=R4=H, R5=OH D: R1=H, R2=R3=O, R4=R5=H E: R1=OH, R2=R3=H, R4=R5=O |
(ent-CPP) 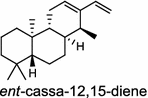
|
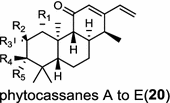
|
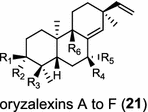
| Oryza sativa (monocot) [32, 33]; ent-sandaracopimaradiene synthase/ABH10735 (OsKSL10), BAD54752 (OsKS10) A: R1=H, R2=OH, R3=CH3, R4=R5=O, R6=H B: R1=R2=O, R3=CH3, R4=OH, R5=R6=H C: R1=R2=O, R3=CH3, R4=R5=O, R6=H D: R1=H, R2=OH, R3=CH3, R4=R5=H, R6=OH E: R1=H, R2=OH, R3=CH3, R4=R5=H, R6=OH F: R1=H, R2=OH, R3=CH2OH, R4=R5=R6=H | (ent-CPP) 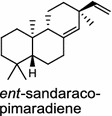
|
|
| Oryza sativa (monocot) [32, 34, 35]; ent-pimara-8(14),15-diene synthase/BAE72100 (OsKS5, OsKSL5j) | (ent-CPP) 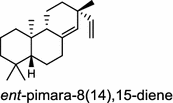
|
currently unknown |
| Oryza sativa (monocot) [33, 36]; syn-pimara-7,15-diene synthase/BAD54751(OsKS4), AAU05906 (OsKSL4) | (syn-CPP) 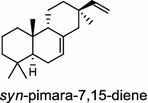
|
|
| Pseudopterogorgia elisabethae (gorgonian, marine invertebrate) [37, 38]; natively purified enzyme |
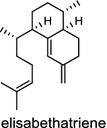
|
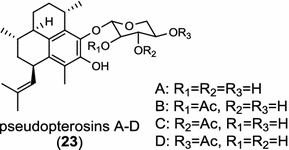
|
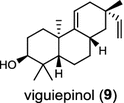
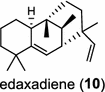
The ent-copalyl diphosphate (CPP) synthase and pimara-9(11),15-diene synthase from Streptomyces sp. strain KO-3988, is responsible for ent-CPP and pimara-9(11),15-diene formation, respectively, and were confirmed to participate in viguiepinol (9) {3-hydroxypimara-9(11),15-diene} biosynthesis by a heterologous expression experiment [13, 14]. Notably, Rv3378c from Mycobacterium tuberculosis H37 was found to encode an unusual halimadienyl PP specific class I diterpene synthase that is primarily involved in the production of the tricyclic edaxadiene (10) [15]. Platensimycin (PTM, 11) and platencin (PTN, 12) harbor diterpene moieties were isolated from Streptomyces platensis strains. They exhibit inhibitory activity toward bacterial and mammalian fatty acid synthases and have great potential in anti-bacterial and anti-diabetic drug development [16, 17]. Their biosyntheses are controlled by dedicated ent-kaurene synthase (PtmT3) and ent-atiserene synthases (PtmT1 and PtnT1) that channel the common ent-CPP precursor formed by the ent-CPP synthases (PtmT2 and PtnT2) to the characteristic PTM and PTN scaffolds, respectively. Therefore, the class I diterpene synthases PtmT1 and PtnT1 represent the first ent-atiserene synthases discovered, unveiling a previously undescribed pathway for the biosynthesis of diterpenoid natural products from the common precursor ent-CPP [16, 17]. The lysophospholipase inhibitor cyclooctatin (13) from Streptomyces melanosporofaciens MI614-43F2 is characterized by a 5-8-5 fused ring system. Sequence analysis in combination with sub-cloning and gene deletion revealed that the CotB2 gene encodes an enzyme that acts as a class I diTPS to produce cyclooctat-9-en-7-ol from GGPP [18]. The reaction is most likely to be initiated by ionization of the diphosphate of GGPP and followed by proton-induced cyclization, employing a similar mechanism underlying the taxadiene formation in taxol biosynthesis [18]. Harboring the same 5-8-5 fused ring scaffold, fusicoccins (14) are the major metabolites produced by the plant-pathogenic fungus Phomopsis (originally known as Fusicoccum) amygdali (Del.). They function as potent activators of plasma membrane H+-ATPase in plants and also exhibit unique biological activity in animal cells. The tricyclic hydrocarbon skeleton of fusicoccin A (14) is biosynthesized by an unusual multifunctional enzyme, P. amygdali fusicoccadiene synthase (PaFS) showing both prenyltransferase and terpene synthase activities [19]. Equipped with a terpene synthase domain at the N terminus and a GGPP synthase domain at the C terminus, PaFS is able to form GGPP and the resulting (+)-fusicocca-2,10(14)-diene using C5 isoprene units as precursors [19]. Another fusicoccadiene synthase was identified in the fungus Alternaria brassicicola strain ATCC 96836 for the brassicicene C (15) biosynthesis in a similar manner to that of PaFS [20].
Diterpene resin acids (16, rosin) secreted by the gymnosperm conifer species are important defense compounds against herbivores and pathogens. The abietadiene synthase from grand fir (Abies grandis; AgAS) is the first diterpene synthase in conifers to be cloned and functionally characterized, and also a bifunctional diTPS catalyzing both protonation-initiated and ionization-initiated cyclization steps for rosin biosynthesis [21]. The AgAS’s homologues from Norway spruce (Picea abies), isopimara-7,15-diene synthase and levopimaradiene/abietadiene synthase (PaLAS), produce isopimara-7,15-diene and a mixture of levopimaradiene, abietadiene, neoabietadiene, and palustradiene, respectively, when incubating with GGPP as substrate [22]. The initial product of PaLAS, however, may be epimers of the thermally unstable allylic tertiary alcohol 13-hydroxy-8(14)-abietene that is likely by dehydration to yield a mixture of the above four diterpene hydrocarbons [23]. Another homologue from Ginkgo biloba encodes a bifunctional levopimaradiene synthase catalyzing the initial cyclization step in ginkgolide (17) pathway [24]. Biosynthesis of the anticancer drug Taxol (18) in Taxus (yew) species involves 19 steps from GGPP that first cyclizes to the taxane skeleton by taxadiene synthase [25, 26].
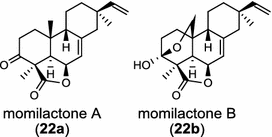
Tanshinones (19) are abietane-type norditerpenoid quinone natural products in the Chinese medicinal herb Dānshēn (Salvia miltiorrhiza Bunge). The CPP synthase and miltiradiene synthase cDNAs have been cloned from the induced root tissue and demonstrated to be involved in the biosynthesis of tanshinones [27]. In the monocot plant rice (Oryza sativa, O. glaberrima), the diterpenoid phytoalexins have been identified to derive via the common biosynthetic origins in the initiating dual cyclization and/or rearrangement of GGPP precursor. The ent-CPP synthase (OsCPS2/OsCyc2) paired with ent-cassa-12,15-diene and ent-sandaracopimaradiene synthases, respectively, give rise to phytocassanes A–E (20) and oryzalexins A–F (21) [28–33]. An ent-CPP specific ent-pimara-8(14),15-diene synthase was discovered in spite of its currently unknown products [32, 34, 35]. In addition, the dual phytoalexin/allelochemical momilactones (22a and 22b) are formed by tandem catalysis of syn-CPP synthase (OsCPS4/OsCyc1) and syn-pimara-7,15-diene synthase [33, 36]. The only animal diTPS which has been reported is a natively purified enzyme from a marine invertebrate gorgonian (Pseudopterogorgia elisabethae), which shows elisabethatriene synthase activity presumably linked to the formation of pseudopterosins (23) [37, 38].
Enzymes for Tetracyclic Hydrocarbon Skeletons (Table 3)
Table 3.
Enzymes for tetracyclic hydrocarbon skeletons
| Genus species (organisms), enzymes/GenBank accession numbers | Enzyme products | Natural diterpenoids | |
|---|---|---|---|
| Bradyrhizobium japonicum (bacterium) [39]; CPP synthase/BAC47414; kaurene synthase/BAC47415 |
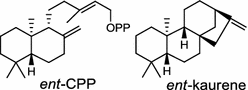
|
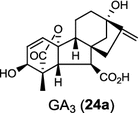
|
|
| Phaeosphaeria sp. (fungus) [40]; ent-kaurene synthase/BAA22426 |
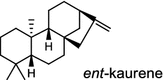
|
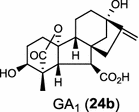
|
|
| Phoma betae PS-13 (fungus) [41]; aphidicolan-16β-ol synthase/BAB62102 |
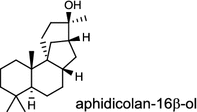
|

|
|
| Phomopsis amygdali (fungus) [42]; CPP synthase/BAG30962; phyllocladan-16α-ol synthase/BAG30961 |
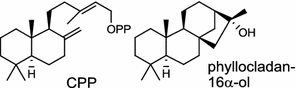
|
|
|
| Phomopsis amygdali (fungus) [43]; phomopsene synthase/AB252833 (DDBJ: mRNA) |
|
|
|
| Physcomitrella patens (bryophyte) [44–46]; ent-CPS/KS/BAF61135 |
ent-kaurene; 16α-hydroxy-ent-kaurane |
|
|
|
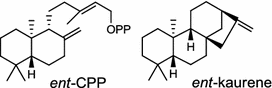
Arabidopsis thaliana (dicot) [47]; ent-CPP synthase/AAA53632 Cucurbita maxima (dicot) [48]; ent-kaurene synthase/AAB39482 |
|
|
|
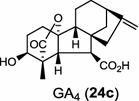
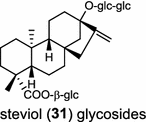
| Stevia rebaudiana (dicot) [49]; ent-CPP synthase/AAB87091; ent-kaurene synthase/AAD34294 |
ent-CPP ent-kaurene |
|
|
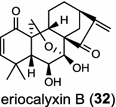
| Isodoneriocalyx (dicot) [50]; ent-CPP synthase/AEP03175 | ent-CPP |
 Isodonent-kauranoids Isodonent-kauranoids |
|
| Zea mays (monocot) [51, 52]; ent-CPP synthase (AN2)/AAT70083, AAT70084 | ent-CPP |
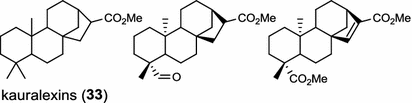
|
|
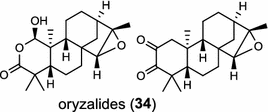
| Oryza sativa (monocot) [28–30] ent-CPP synthase/AAT11021 (OsCPS2), BAD42452 (OsCyc2) syn-CPP synthase/BAD42451 (OsCyc1), AAS98158 (OsCPS4) |
|
phytoalexins | |
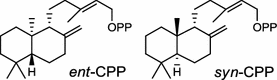
| Oryza sativa (monocot) [32, 34, 35] ent-isokaurene synthase: OsKS6/BAE72101, OsKSL6/ABH10733, OsKSL5i/ABH10732 | (ent-CPP) 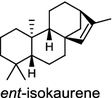
|
|
|
| Oryza sativa (monocot) [53]; syn-stemar-13-ene synthase/BAD34478 (OsKSL8) | (syn-CPP) 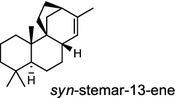
|
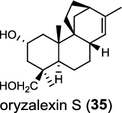
|
|
| Oryza sativa (monocot) [54]; syn-stemodene synthase/AAZ76733 (OsKSL11) | (syn-CPP) 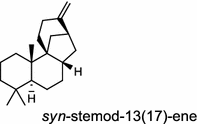
|
currently unknown | |
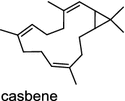
| Sapium sebiferum (dicot) [55]; casbene synthase/ADB90272 |
|
|
|
| Euphorbia esula (dicot) [55]; casbene synthase/ADB90273 | Casbene |
|
|
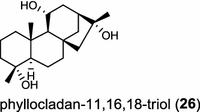
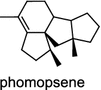
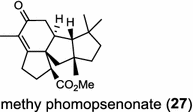
The ubiquitous gibberellin phytohormones (24a-c) are tetracyclic diterpenoids produced via distinct biosynthetic pathways by plants, fungi, and bacteria. Like in plants, the bacterium Bradyrhizobium japonicum was demonstrated to encodes for separate ent-CPP and ent-kaurene synthases for gibberellic acid GA3 production (24a) [39]; while fungi use only a single bi-functional ent-kaurene synthase for GA1 (24b) [40]. Aphidicolin (25) isolated from the fungus Phoma betae PS-13 has a unique hydrocarbon skeleton and remarkable bio-activity such as anti-tumor and specific inhibition of DNA polymerase α. Aphidicolan-16β-ol synthase was identified as a bifunctional diTPS involved in the biosynthesis of aphidicolin (25) from GGPP via syn-CPP [41]. However, the metabolite phyllocladan-11α,16α,18-triol (26) is likely to result from a two-step cyclization via CPP synthase coupled with phyllocladan-16α-ol synthase in the fusicoccin (14)-producing fungus Phomopsis amygdali [19, 42]. Using homology based PCR of GGPP synthase genes and the subsequent genome walking in the fungus Phomopsis amygdali N2, phomopsene synthase was identified and led to discovery of a new diterpene, methyl phomopsenonate (27) [43].
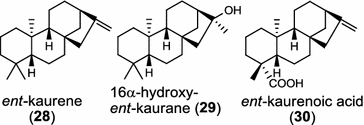
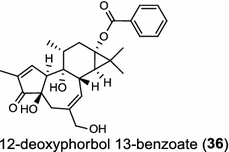
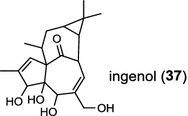
In the basal vascular land plant moss Physcomitrella patens, a bifunctional ent-CPS/KS directly generates the ent-kaurene skeleton from GGPP to give ent-kaurene (28), 16α-hydroxy-ent-kaurane (29) and ent-kaurenoic acid (30) [44–46]. Instead, most angiosperms use normal, ent, or syn-CPP synthase and kaurene synthase or kaurene synthase-like (KSL) for the two sequential cyclization reactions that initiate biosyntheses of gibberellins such as GA4 (24c) and labdane-related diterpenoids. The plant gibberellin diTPS were the first identified in Arabidopsis thaliana and Cucurbita maxima, respectively [47, 48]. Interestingly, the intensely sweet steviol (31) producing plant Stevia rebaudiana was found to share the same ent-CPP synthase and ent-kaurene synthase for both gibberellin and steviol (31) biosyntheses [49]. In contrast, Isodon plants are most likely to recruit novel a diTPS that functions as an ent-CPP synthase specific for the production of ent-kaurane diterpenoids (also ent-kauranoids) like eriocalyxin B (32) [50]. The induced accumulation of six ent-kaurane–related diterpenoid phytoalexins, collectively termed kauralexins (33) was observed in maize in response to stem attack by the European corn borer (Ostrinia nubilalis) and fungi. The maize An2 encoding an ent-CPP synthase, is probably relevant to kauralexin (33) production [51, 52]. Besides tri-cyclic diterpenoid phytoalexins as described in Table 2, rice produces tetracyclic phytoalexins such as oryzalides (34) and oryzalexin S (35). The ent-CPP synthase (OsCPS2/OsCyc2) and ent-isokaurene synthase in tandem catalyze the formation of hydrocarbon skeletons of oryzalides (34) [32; 34, 35]; wherase the syn-CPP synthase (OsCPS4/OsCyc1) coupled with syn-stemar-13-ene synthase generate those of oryzalexin S (35) [53]. Despite the demonstration of syn-CPP specific syn-stemodene synthase activity in rice, its relevant natural product has remained elusive [54]. The latent HIV-1 activator prostratin (and related 12-deoxyphorbol esters), the analgesic resiniferatoxin, and the anti-cancer drug candidate ingenol 3-angelate are Euphorbiaceae diterpenoids of medical interest and likely derived from casbene. Casbene synthase genes were considered for carbon skeletons formation of 12-deoxyphorbol 13-benzoate (36) and ingenol (37), respectively in Sapium sebiferum and Euphorbia esula [55].
Enzymes for Macrocyclic Hydrocarbon Skeletons (Table 4)
Table 4.
Enzymes for macrocyclic hydrocarbon skeletons
| Genus species (organisms), enzymes/GenBank accession numbers | Enzyme products | Natural diterpenoids |
|---|---|---|
| Streptomyces sp. (bacterium) [56]; DtcycA/BAM78697; DtcycB/BAM78698 |
|
currently unknown |
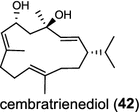
| Nicotiana tabacum (dicot) [57]; cembratrieneol synthase/AAL25826 |
|
|
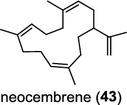
| Ricinus communis (dicot) [55]; neocembrene synthase (CAS2)/XP_002513334 | Neocembrene |
|
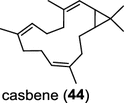
| Ricinus communis (dicot) [55, 59]; casbene synthase/EEF48743 (CAS1), EEF48746 (CAS3) | Casbene |
|
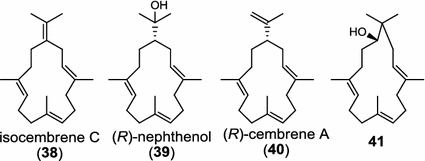
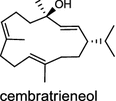
Two novel class I diTPS DtcycA and DtcycB, derived from Streptomyces sp. SANK 60404, are capable of forming multiple diterpene products with macrocyclic skeletons. The reaction products catalyzed by DtcycA were determined to be the isopropylidene isomer of cembrene C (38) and (R)-nephthenol (39); while the outcome of DtcycB included (R)-nephthenol (39), (R)-cembrene A (40), and a novel diterpene {(4E,8E,12E)-2,2,5,9,13-pentamethylcyclopentadeca-4,8,12-trien-1-ol} (41) [56]. However, none of these diterpene products were detected in the culture broths under any of the culture conditions used in this study, as revealed by GC–MS analysis. Furthermore, DtcycA and DtcycB show no sequence similarity to cembratrieneol synthase for cembratrienediol (42) in Nicotiana tabacum [57] or to neocembrene (43) (also known as cembrene A) synthase (CAS2) from castor bean (Ricinus communis) [55]. Casbene (44) is a macrocyclic diterpene hydrocarbon that serves as a phytoalexin in castor bean [58]. The casbene synthase from castor bean is the first diTPS cloned as cDNA as early as in 1994 [59].
In summary, angiosperm plants and bacteria generally share the common two-step process that shapes the hydrocarbon cyclic skeletons of diterpenoids by two mono-functional diTPS in tandem catalysis. Such a correlation seems plausible considering that the plastidial MEP (2-C-methyl-D-erythritol phosphate) pathway dominates the C5 isoprene precursor supplies and plastids are thought to have originated from endosymbiotic cyanobacteria. Conversely, bi-functional class I/II diTPS are more likely found in gymnosperms, basal vascular land plants, and fungi. Currently, only class I diTPS can produce macrocyclic skeletons of natural diterpenoids. In the near future, ‘Omics’ approaches and metabolite profiling can be extensively exploited in an effort to reveal genes and enzymes responsible for cyclization and subsequent modification reactions that take place in diterpenoid biosyntheses of plants, fungi, and bacteria.
Acknowledgments
This study was supported by grants from the National Key Laboratory of Plant Molecular Genetics, and from the State Key Laboratory of Phytochemistry and Plant Resources in West China.
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Christianson DW. Chem. Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 2.Peters RJ. Nat. Prod. Rep. 2010;27:1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dairi T, Hamano Y, Kuzuyama T, Itoh N, Furihata K, Seto H. J. Bacteriol. 2001;183:6085–6094. doi: 10.1128/JB.183.20.6085-6094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamano Y, Kuzuyama T, Itoh N, Furihata K, Seto H, Dairi T. J. Biol. Chem. 2002;277:37098–37104. doi: 10.1074/jbc.M206382200. [DOI] [PubMed] [Google Scholar]
- 5.Nakano C, Okamura T, Sato T, Dairi T, Hoshino T. Chem. Commun. 2005;8:1016–1018. doi: 10.1039/b415346d. [DOI] [PubMed] [Google Scholar]
- 6.Nakano C, Hoshino T. ChemBioChem. 2009;10:2060–2071. doi: 10.1002/cbic.200900248. [DOI] [PubMed] [Google Scholar]
- 7.Nakano C, Ootsuka T, Takayama K, Mitsui T, Sato T, Hoshino T. Biosci. Biotechnol. Biochem. 2011;75:75–81. doi: 10.1271/bbb.100570. [DOI] [PubMed] [Google Scholar]
- 8.Mafu S, Hillwig ML, Peters RJ. ChemBioChem. 2011;12:1984–1987. doi: 10.1002/cbic.201100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zerbe P, Chiang A, Yuen M, Hamberger B, Hamberger B, Draper JA, Britton R, Bohlmann J. J. Biol. Chem. 2012;287:12121–12131. doi: 10.1074/jbc.M111.317669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sallaud C, Giacalone C, Töpfer R, Goepfert S, Bakaher N, Rösti S, Tissier A. Plant J. 2012;72:1–17. doi: 10.1111/j.1365-313X.2012.05068.x. [DOI] [PubMed] [Google Scholar]
- 11.Falara V, Pichersky E, Kanellis AK. Plant Physiol. 2010;154:301–310. doi: 10.1104/pp.110.159566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A. Caniard, P. Zerbe, S. Legran, A. Cohade, N. Valot, J.L. Magnard, J. Bohlmann, L. Legendre, BMC Plant Biol. 12, 119 (2012) [DOI] [PMC free article] [PubMed]
- 13.Kawasaki T, Kuzuyama T, Kuwamori Y, Matsuura N, Itoh N, Furihata K, Seto H, Dairi T. J. Antibiot. 2004;57:739–747. doi: 10.7164/antibiotics.57.739. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda C, Hayashi Y, Itoh N, Seto H, Dairi T. J. Biochem. 2007;141:37–45. doi: 10.1093/jb/mvm004. [DOI] [PubMed] [Google Scholar]
- 15.Mann FM, Xu M, Chen X, Fulton DB, Russell DG, Peters RJ. J. Am. Chem. Soc. 2009;131:17526–17527. doi: 10.1021/ja9019287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smanski MJ, Yu Z, Casper J, Lin S, Peterson RM, Chen Y, Wendt-Pienkowski E, Rajski SR, Shen B. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13498–13503. doi: 10.1073/pnas.1106919108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smanski MJ, Peterson RM, Huang S-X, Shen B. Curr. Opin. Chem. Biol. 2012;16:132–141. doi: 10.1016/j.cbpa.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SY, Zhao P, Igarashi M, Sawa R, Tomita T, Nishiyama M, Kuzuyama T. Chem. Biol. 2009;16:736–743. doi: 10.1016/j.chembiol.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Toyomasu T, Tsukahara M, Kaneko A, Niida R, Mitsuhashi W, Dairi T, Kato N, Sassa T. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3084–3088. doi: 10.1073/pnas.0608426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minami A, Tajima N, Higuchi Y, Toyomasu T, Sassa T, Kato N, Dairi T. Bioorg. Med. Chem. Lett. 2009;19:870–874. doi: 10.1016/j.bmcl.2008.11.108. [DOI] [PubMed] [Google Scholar]
- 21.Vogel BS, Wildung MR, Vogel G, Croteau R. J. Biol. Chem. 1996;271:23262–23268. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- 22.Martin DM, Fäldt J, Bohlmann J. Plant Physiol. 2004;135:1908–1927. doi: 10.1104/pp.104.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeling CI, Madilao LL, Zerbe P, Dullat HK, Bohlmann J. J. Biol. Chem. 2011;286:21145–21153. doi: 10.1074/jbc.M111.245951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schepmann HG, Pang J, Matsuda SP. Arch. Biochem. Biophys. 2001;392:263–269. doi: 10.1006/abbi.2001.2438. [DOI] [PubMed] [Google Scholar]
- 25.Wildung MR, Croteau R. J. Biol. Chem. 1996;271:9201–9204. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- 26.Croteau R, Ketchum REB, Long RM, Kaspera R, Wildung MR. Phytochem. Rev. 2006;5:75–97. doi: 10.1007/s11101-005-3748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ. Org. Lett. 2009;11:5170–5173. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prisic S, Xu M, Wilderman PR, Peters R. J. Plant Physiol. 2004;136:4228–4236. doi: 10.1104/pp.104.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otomo K, Kenmoku H, Oikawa H, König WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T. Plant J. 2004;39:886–893. doi: 10.1111/j.1365-313X.2004.02175.x. [DOI] [PubMed] [Google Scholar]
- 30.Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ. Plant J. 2004;39:309–318. doi: 10.1111/j.1365-313X.2004.02137.x. [DOI] [PubMed] [Google Scholar]
- 31.Cho EM, Okada A, Kenmoku H, Otomo K, Toyomasu T, Mitsuhashi W, Sassa T, Yajima A, Yabuta G, Mori K, Oikawa H, Toshima H, Shibuya N, Nojiri H, Omori T, Nishiyama M, Yamane H. Plant J. 2004;37:1–8. doi: 10.1046/j.1365-313X.2003.01926.x. [DOI] [PubMed] [Google Scholar]
- 32.Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya NM, Coates RM, Peters RJ. Phytochemistry. 2007;68:312–326. doi: 10.1016/j.phytochem.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 33.Otomo K, Kanno Y, Motegi A, Kenmoku H, Yamane H, Mitsuhashi W, Oikawa H, Toshima H, Itoh H, Matsuoka M, Sassa T, Toyomasu T. Biosci. Biotechnol. Biochem. 2004;68:2001–2006. doi: 10.1271/bbb.68.2001. [DOI] [PubMed] [Google Scholar]
- 34.Kanno Y, Otomo K, Kenmoku H, Mitsuhashi W, Yamane H, Oikawa H, Toshima H, Matsuoka M, Sassa T, Toyomasu T. Biosci. Biotechnol. Biochem. 2006;70:1702–1710. doi: 10.1271/bbb.60044. [DOI] [PubMed] [Google Scholar]
- 35.Xu M, Wilderman PR, Peters RJ. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7397–7401. doi: 10.1073/pnas.0611454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilderman PR, Xu M, Jin Y, Coates RM, Peters R. J. Plant Physiol. 2004;135:2098–2105. doi: 10.1104/pp.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohl AC, Kerr RG. Arch. Biochem. Biophys. 2004;424:97–104. doi: 10.1016/j.abb.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Brück TB, Kerr RG. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2006;143:269–278. doi: 10.1016/j.cbpb.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 39.Morrone D, Chambers J, Lowry L, Kimc G, Anterola A, Bender K, Peters RJ. FEBS Lett. 2009;583:475–480. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- 40.Kawaide H, Imai R, Sassa T, Kamiya Y. J. Biol. Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- 41.Oikawa H, Toyomasu T, Toshima H, Ohashi S, Kawaide H, Kamiya Y, Ohtsuka M, Shinoda S, Mitsuhashi W, Sassa T. J. Am. Chem. Soc. 2001;123:5154–5155. doi: 10.1021/ja015747j. [DOI] [PubMed] [Google Scholar]
- 42.Toyomasu T, Niida R, Kenmoku H, Kanno Y, Miura S, Nakano C, Shiono Y, Mitsuhashi W, Toshima H, Oikawa H, Hoshino T, Dairi T, Kato N, Sassa T. Biosci. Biotechnol. Biochem. 2008;72:1038–1047. doi: 10.1271/bbb.70790. [DOI] [PubMed] [Google Scholar]
- 43.Toyomasu T, Kaneko A, Tokiwano T, Kanno Y, Kanno Y, Niida R, Miura S, Nishioka T, Ikeda C, Mitsuhashi W, Dairi T, Kawano T, Oikawa H, Kato N, Sassa T. J. Org. Chem. 2009;74:1541–1548. doi: 10.1021/jo802319e. [DOI] [PubMed] [Google Scholar]
- 44.Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H. FEBS Lett. 2006;580:6175–6181. doi: 10.1016/j.febslet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 45.Hayashi K, Horie K, Hiwatashi Y, Kawaide H, Yamaguchi S, Hanada A, Nakashima T, Nakajima M, Mander LN, Yamane H, Hasebe M, Nozaki H. Plant Physiol. 2010;153:1085–1097. doi: 10.1104/pp.110.157909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawaide H, Hayashi K, Kawanabe R, Sakigi Y, Matsuo A, Natsume M, Nozaki H. FEBS J. 2011;278:123–133. doi: 10.1111/j.1742-4658.2010.07938.x. [DOI] [PubMed] [Google Scholar]
- 47.Sun TP, Kamiya Y. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaguchi S, Saito T, Abe H, Yamane H, Murofushi N, Kamiya Y. Plant J. 1996;10:203–213. doi: 10.1046/j.1365-313X.1996.10020203.x. [DOI] [PubMed] [Google Scholar]
- 49.Richman AS, Gijzen M, Starratt AN, Yang ZY, Brandle JE. Plant J. 1999;19:411–421. doi: 10.1046/j.1365-313X.1999.00531.x. [DOI] [PubMed] [Google Scholar]
- 50.Li JL, Chen QQ, Jin QP, Gao J, Zhao PJ, Lu S, Zeng Y. Phytochemistry. 2012;76:32–39. doi: 10.1016/j.phytochem.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 51.Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ. Plant Mol. Biol. 2005;59:881–894. doi: 10.1007/s11103-005-1674-8. [DOI] [PubMed] [Google Scholar]
- 52.Schmelz EA, Kaplan F, Huffaker A, Dafoe NJ, Vaughan MM, Ni X, Rocca JR, Alborn HT, Teal PE. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5455–5460. doi: 10.1073/pnas.1014714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nemoto T, Cho EM, Okada A, Okada K, Otomo K, Kanno Y, Toyomasu T, Mitsuhashi W, Sassa T, Minami E, Shibuya N, Nishiyama M, Nojiri H, Yamane H. FEBS Lett. 2004;571:182–186. doi: 10.1016/j.febslet.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Morrone D, Jin Y, Xu M, Choi SY, Coates RM, Peters RJ. Arch. Biochem. Biophys. 2006;448:133–140. doi: 10.1016/j.abb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Kirby J, Nishimoto M, Park JG, Withers ST, Nowroozi F, Behrendt D, Rutledge EJG, Fortman JL, Johnson HE, Anderson JV, Keasling JD. Phytochemistry. 2010;71:1466–1473. doi: 10.1016/j.phytochem.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Meguro A, Tomita T, Nishiyama M, Kuzuyama T. ChemBioChem. 2013;14:316–321. doi: 10.1002/cbic.201200651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang E, Wagner GJ. Planta. 2003;216:686–691. doi: 10.1007/s00425-002-0904-4. [DOI] [PubMed] [Google Scholar]
- 58.Sitton D, West C. A. Phytochemistry. 1975;14:1921–1925. doi: 10.1016/0031-9422(75)83098-6. [DOI] [Google Scholar]
- 59.Mau CJ, West CA. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8497–8501. doi: 10.1073/pnas.91.18.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]


